Abstract
Human neural progenitors from a variety of sources present new opportunities to model aspects of human neuropsychiatric disease in vitro. Such in vitro models provide the advantages of a human genetic background, combined with rapid and easy manipulation, making them highly useful adjuncts to animal models. Here, we examined whether a human neuronal culture system could be utilized to assess the transcriptional program involved in human neural differentiation and in modeling some of the molecular features of a neurodevelopmental disorder, such as autism. Primary normal human neuronal progenitors (NHNPs) were differentiated into a post-mitotic neuronal state through addition of specific growth factors and whole-genome gene expression was examined throughout a time course of neuronal differentiation. After four weeks of differentiation, a significant number of genes associated with autism spectrum disorders (ASD) are either induced or repressed. This includes the ASD susceptibility gene neurexin 1, which showed a distinct pattern from neurexin 3 in vitro, and which we validated in vivo in fetal human brain. Using weighted gene co-expression network analysis (WGCNA), we visualized the network structure of transcriptional regulation, demonstrating via this unbiased analysis that a significant number of ASD candidate genes are coordinately regulated during the differentiation process. Since NHNPs are genetically tractable and manipulable, they can be used to study both the effects of mutations in multiple ASD candidate genes on neuronal differentiation and gene expression in combination with the effects of potential therapeutic molecules. These data also provide a step towards better understanding of the signaling pathways disrupted in ASD.
Keywords: Model system, neuropsychiatric disease, pharmacogenomics, high-throughput drug screen, neurodevelopment
Introduction
Genetic advances in human neuropsychiatric conditions have provided us with a new window through which to begin to study and understand these complex diseases. Perhaps this is most evident in autism spectrum disorders (ASD) where recent studies have identified many relatively rare monogenic causes of ASD, as well as some common variants associated with the disease (1–7). These studies have revealed that autism, like other neurodevelopmental disorders is genetically very heterogeneous, involving many potential mutational mechanisms and potentially distinct molecular pathways (2, 4, 8).
While this complexity may at first appear daunting, the study of monogenic ASD risk genes has proven to be instrumental for understanding the basic biology of these genes during brain development. Thus, even though many of the more Mendelian forms of ASDs are rare, they provide an important opportunity to begin to understand the normal function of these autism risk genes and loci through their manipulation in transgenic mice (9–11). Mouse models serve an important function in understanding the role of these risk genes in brain development and conserved behavioral traits. While extremely useful for understanding monogenic disorders, one drawback to the use of mouse as a model is the challenge of perturbing multiple genes to mimic the genetics of disease. In addition, lengthy breeding and gestational times can also make experimental design prolonged and less amenable to high-throughput screening. On the other hand, models such as zebrafish (Danio rerio) do allow for rapid and high-throughput in vivo screening of genes and small molecules (12).
So, while animal models are necessary for examining circuits and behavior correlates, it is likely that no single model of a complex neurodevelopmental disease will completely recapitulate the human disorder. In fact, the study of gene expression pathways provides mounting evidence that human and mouse have significant areas of divergence. Comparison of global differences in gene co-expression between human and mouse revealed many instances of human-specific patterns of expression; several of these modules are associated with human neurodegenerative disorders that are challenging to model in mice (13). Moreover, a specific comparison of human and chimpanzee signaling pathways downstream of FOXP2, a transcription factor involved in speech and language, again identified differential gene expression and co-expression networks that could be characterized in vitro (14). Therefore, even the most closely related species to humans, the chimpanzee, has evolved different signaling networks in some cases. Thus, the use of human-derived model systems should be considered an important adjunct for a complete understanding of neuropsychiatric disorders involving human higher cognition and behavior. In addition, both of the studies mentioned above (13, 14), and many others (15–20) highlight how the identification of gene co-expression networks can uncover new functional insights—including genes involved in neurodegeneration or genes involved in human language and cognition. Therefore, the employment of a network approach to any gene expression data set generated from a human model system is a significant step towards a complete understanding of how gene interactions promote a particular function.
One new approach for modeling disease-related phenotypes in human tissue is the use of induced pluripotent stem cells (iPSCs) (21). Using this technique, fibroblasts from patients are converted to the cells of interest and the resultant phenotypes analyzed. At least one study has used iPSCs to study Rett syndrome, a disorder within the autism spectrum (22). Such a process is a step in the right direction as the cells are human and originate from specific patient populations. However, working with these cells can be challenging because they need to be manipulated genetically to reprogram into a pluripotent state and the conversion to neurons can be lengthy and low in efficiency (23). There is little doubt though that a thorough comparative study of the gene expression profile of neurons from iPSCs to neural tissue from the same individuals will be very useful.
Another potential model system that also has not been well tested, are human cells that are derived from brain tissue and grown in vitro. For the study of neurodevelopmental disorders, the use of embryonic brain tissue from early brain development provides the advantage of capturing the correct time points in development when genetic and environmental insults are thought to be causative (24–26). We therefore developed a human neural progenitor system to assess the functional genomics of both normal human neuronal development and autism using human fetal brain tissue. These normal human neural progenitors, or NHNPs, are derived from early human embryos at approximately 8–19 gestational weeks. These cells have previously been utilized to examine neuronal phenotypes during Wnt signaling (27), as well as targets of the transcription factor FOXP2, recapitulating patterns observed in vivo (14). Here, we find that during the differentiation process of NHNPs, many genes already previously linked to ASD are highly co-expressed, as assessed using whole genome microarrays. Moreover, a number of ASD genes demonstrate conserved patterns of gene co-expression during the time course of differentiation. Since little is known about the transcriptional program accompanying human neuronal differentiation, the comprehensive analysis of gene expression reported here provides a foundational tool for assessing the interaction of numerous signaling cascades in the pathophysiology of ASD and other neurodevelopmental disorders. The ability to easily manipulate gene expression in these cells (14) also makes NHNPs attractive for studying the functional consequences of altered signaling pathways involved in ASD in human neurons.
Methods and Materials
Cell Culture
NHNP cells were obtained from Lonza or generated as previously described (28, 29). See Supplementary Table 1 for detailed cell line characteristics. Cells were used at a low passage (30), typically around passage 20. Proliferating cells were grown on tissue culture plates coated with 5μg/mL of fibronectin and 50μg/mL of polyornithine. Growth media of proliferating cells was Neurobasal A media (Invitrogen) containing 2.5μM heparin, 10% BIT (Stem-Cell Tech), 1% Glutamax (Invitrogen) and 2% penicillin and streptomycin. Growth media was supplemented every other day with 10 ng/mL of epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF). For differentiation followed by RNA extraction, 100,000 cells were plated per well of a 6-well tissue culture dish coated with poly-ornithine and laminin. For differentiation followed by immunocytochemistry, 5,000 cells were plated per well of a 24-well tissue culture dish containing glass coverslips coated with poly-ornithine and laminin (5μg/mL). Two days after plating, samples were collected for the undifferentiated, or time zero, time point. At this time, we initiated the differentiation process and fed cells with a 50% media change containing retinoic acid (10 ng/mL), neurotrophin-3 (NT-3) (10ng/mL), brain-derived neurotrophic factor (BDNF) (10ng/mL), and potassium chloride (KCl) (10mM). We continued to provide a 50% media change containing these growth factors every other day until the samples were collected. Samples were harvested at three time points after initiating the differentiation process: 2 weeks, 4 weeks, and 8 weeks.
Genetic Association Data
All human transcripts from build hg18 that map to a unique location in the genome were utilized. SNPs from the Illumina 550k platform were analyzed from 20k upstream of the transcript start site to 10k downstream of the transcript end. P values for association with ASD were drawn from the discovery phase of (6), using 780 multiplex families from the Autism Genetics Research Exchange (AGRE). Data are included in Supplementary Table 6.
Genotyping
Genomic DNA was extracted from four independently generated cell lines using Qiagen’s DNeasy kit according to the manufacturer’s instructions. Approximately 1.5ug of gDNA from each line was hybridized to Illumina’s Human CytoSnp-12 genotyping arrays. SNP genotypes were filtered for quality and analyzed for gender in PLINK (31). Race determinations were made in PLINK using multi-dimensional scaling. Existing genotype data (6) and self-reported race information from the AGRE cohort were used to establish racial groups. CNVs were called using Penn CNV (32)with wave adjustment and subjected to the following quality filters: log R ratio SD <0.28, B allele frequency drift <0.002, waviness factor <0.05.
Immunocytochemistry
Coverslips were fixed in 2% PFA in PBS at RT for 10mins and permeabilized in 0.5% Triton-X at RT for 10mins. Next, non-specific staining was blocked using TBS containing 10% normal donkey serum and 0.1% Triton-X at RT for 1hr. Incubations with primary antibodies diluted in blocking solution were done at 4C overnight. Incubations with secondary antibodies diluted in blocking solution were done at RT for 1hr. Cell nuclei were stained using DAPI. Images were captured using a Zeiss Axio Imager. D1. Antibodies utilized were: rabbit anti-nestin (1:200, Millipore); mouse anti-Tuj1 (1:2000, Covance); Alexa Fluor 488 donkey anti-rabbit IgG and Alexa Fluor 555 donkey anti-mouse IgG (1:1000, Invitrogen).
In situ Hybridization
In situ hybridization was performed as previously described (33)using human fetal brain as template. Primers are included in Supplementary Table 8. Sense control probes did not show any signal (data not shown). Fresh frozen human fetal brains were obtained from the University of Maryland Brain and Tissue Bank. Demographics on the fetuses from which the brains were derived are as follows: Figure 2B and 2F: #1137, 18GW, Female, African American, 2hr PMI; Figure 2C and 2G: #1110, 19GW, Female, African American, 2hr PMI; Figure 2D and 2H: #1010, 20GW, Female, Caucasian, 3hr PMI; Figure 2E and 2I: #1057, 19GW, Female, African American, 1hr PMI. All fetuses were considered “normal” except for #1010, which had polyhydramnios, twin-to-twin transfusion syndrome.
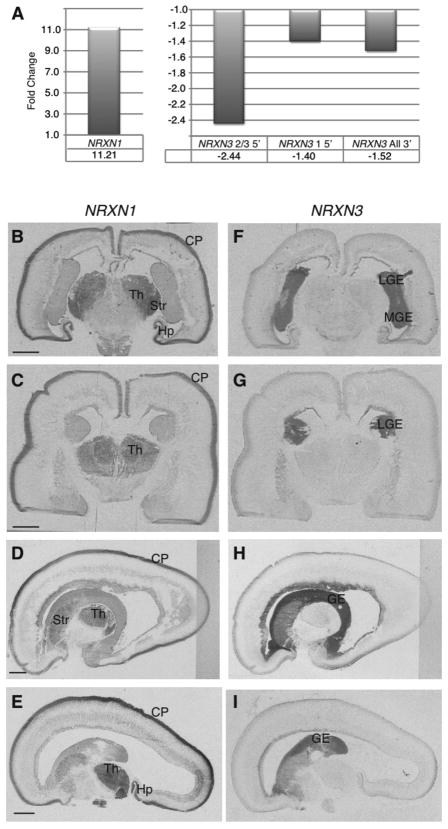
Figure 2. Inverse expression patterns of NRXN1 and NRXN3.
A) qRT-PCR of NRXN1 and NRXN3 shows an increase in NRXN1 and a decrease in NRXN3 with differentiation at four weeks in NHNPs. Three different primer pairs amplifying either the 5′ end of isoforms 2 and 3 or isoform 1, and the 3′ end of all isoforms of NRXN3 were used. In situ hybridization of NRXN1 (B–E) and NRXN3 (F–I) in human fetal brain demonstrates an enrichment of NRXN1 in areas of post-mitotic neurons whereas NRXN3 is expressed in proliferative areas. Gestational week age of tissue is 18 weeks (B and F), 19 weeks (C, E, G, and I), and 20 weeks (D and H). Both coronal (B, C, F, and G) and sagittal (D, E, H, and I) were used. CP=cortical plate, GE=ganglionic eminence (L=lateral, M=medial), Hp=hippocampus, Str=striatum, Th=thalamus. Scale bars=5mM.
Microarray Analysis
Total RNA was extracted using Qiagen’s RNeasy kit, according to the manufacturer’s instructions. Samples were hybridized to Illumina Human Ref8-v3 microarrays. Quantile normalization was conducted as previously described (14). Heatmaps and dendrograms were generated using the limma package in R. Gene ontology (GO) and disease association analysis was done as previously described (14)using DAVID (http://david.abcc.ncifcrf.gov) and all of the probes on the microarrays as background. Gene expression data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible using GEO accession number GSE28046.
qRT-PCR
cDNA was generated from total RNA using random hexamers and Superscript III (Invitrogen). Real-time PCR was conducted using Sybr-Rox SuperMix (BioRad) and an HT-7900 (Applied Biosystems). Intron-spanning primers are detailed in Supplementary Table 8.
WGCNA
Weighted gene co-expression analysis was conducted as previously described (14, 15). See supplemental methods for R code used to generate and analyze the network. Genes denoted as being implicated in ASD were based on the SFARI gene database (http://gene.sfari.org), and are demarcated or used for analysis in Figure 3, Supplemental Figure 2, and Supplementary Tables 6 and 7.
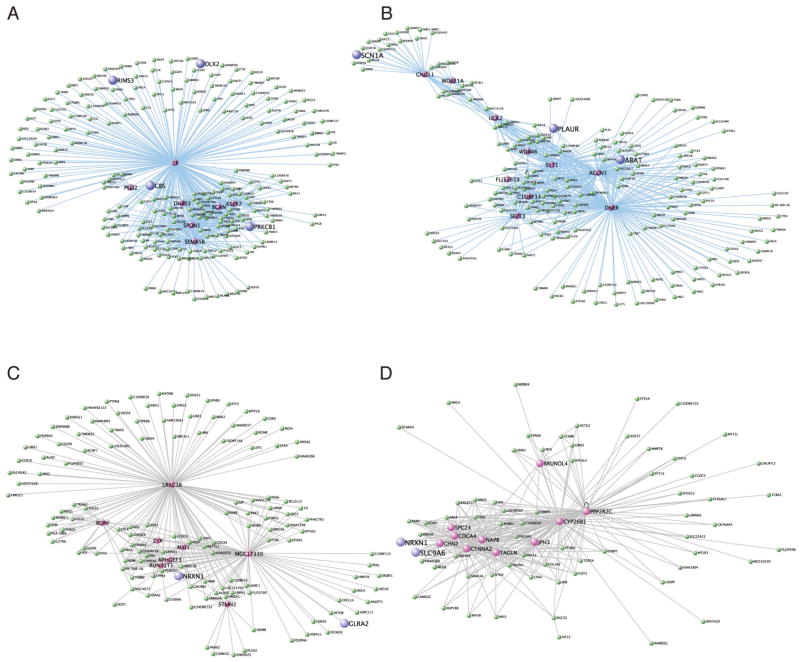
Figure 3. Network visualization of modules containing ASD genes.
A) Royal blue module. B) Pink module. C) Black module. D) Turquoise module. Color names are randomly assigned to each module for descriptive purposes. ASD genes are in blue and hub genes are depicted in purple. Lines represent connecting edges in the network based on topological overlap. The top 250 connections are depicted and line length and thickness are arbitrary.
Results
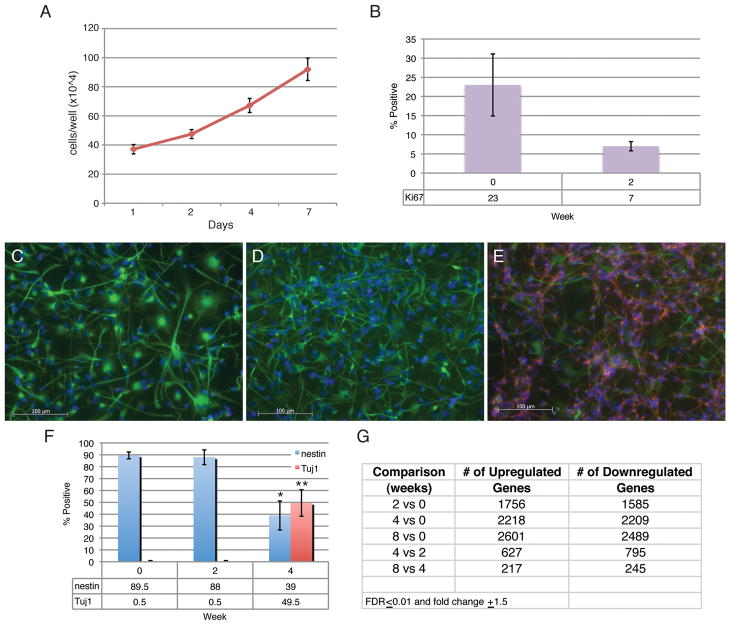
We first assessed the feasibility of using NHNPs as a model system for studying neuronal differentiation. Low passage NHNPs were utilized for all experiments, and when the cells were provided with growth-inducing signals in the form of EGF and bFGF they maintained a proliferative state (Figure 1A; see Materials and Methods). While the doubling time of these human cells is prolonged compared to embryonic rodent neural progenitors, the NHNPs can be kept in culture in a proliferative state for at least a year, if not longer (data not shown). We then tested the characteristics of the NHNPs after receiving differentiation cues, which was achieved by replacing EGF and bFGF with retinoic acid to promote cell cycle exit (see Materials and Methods). In addition, we also supplemented the media with BDNF, NT-3, and KCl to provide supportive growth factors and appropriate conductive ions for neurons. Under these conditions, we found a three-fold decrease in proliferating, Ki67+ cells after two weeks (Figure 1B).
Figure 1. Characterization of NHNPs.
A) NHNPs are mitotically active in the absence of differentiation growth factors. Growth curve of proliferating cells. Error bars are ± s.e.m., n=5. B) NHNPs can be induced to a post-mitotic state using the right combination of differentiation cues. The number of Ki67+ cells decreases at least three fold within two weeks. Error bars are ±s.e.m., n=3. C–E) NHNPs display markers and morphological characteristics of neurons with differentiation from zero weeks (C) to two (D) and four weeks (E). Green is nestin-positive staining denoting uncommitted progenitors. Red is Tuj1-positive staining marking neurons, and blue is DAPI. F) The percentage of nestin-positive cells significantly decreases with four weeks differentiation whereas the percentage of Tuj1-positive cells increases. Asterisk indicates P≤0.005, double asterisk denotes P≤0.001, and error bars are ±s.e.m. (ANOVA, n=4). G) Table showing the number of genes changing at each time point comparison using microarrays with thresholds of an FDR ≤0.01 and fold change ±1.5.
We next assessed whether we could generate neurons under these differentiation conditions. We conducted immunostaining for nestin, a marker of proliferating progenitors (34); Tuj1, a marker of immature neurons (35); MAP2, a marker of mature neurons (36); and GFAP, a marker of glia (37). We found a significant decrease in nestin-positive cells and a significant increase in both Tuj1 and MAP2-positive cells with four weeks of differentiation (Figure 1C–F and data not shown). Tuj1-positive cells represent about 50% of the cells in culture at four weeks (Figure 1F), whereas the number of GFAP-positive cells stayed consistent throughout differentiation (data not shown). Thus, using our differentiation conditions, we can efficiently differentiate human neural progenitors into post-mitotic neurons. Moreover, based on gene expression profiling (see below), the NHNPs may differentiate into a number of neuronal types including both excitatory and inhibitory neurons. This is evidenced by an increase in expression of genes such as DRD3, DRD4, GABBR1, GABRB3, GAD1, GAD2, GRIK1, GRIN1, GRM2, GRM3, SLC23A1, and SLC6A1 for example (Supplementary Table 2).
Next, we characterized this process of differentiation by conducting whole-genome microarray analyses on a time course of differentiating NHNPs. We selected four time points to assess: time 0 or undifferentiated cells, two weeks, four weeks, and eight weeks of differentiation (T0, D2, D4, and D8). These time points were chosen for the following reasons: D2 because we observed a significant decrease in proliferation at this time (Figure 1B), D4 because there was a significant increase in Tuj1 at this time (Figure 1F), and D8 because it represented a period of time twice as long as D4 for direct comparison with the D2 to D4 transition during neuronal maturation. Four biological replicates from each time point were included for analysis. Hierarchical clustering demonstrates that the biological replicates clustered with one another and that the time point most different from the other three when considering expression of all genes was the one containing the undifferentiated cells (Supplemental Figure 1A). When only the top 500 most variable genes were analyzed, T0 and D2 clustered separately from D4 and D8 (Supplementary Figure 1B), suggesting that the genes changing the most during the time course represent differential patterns of expression between the early and late phase of differentiation. This pattern also suggested that neural specification may be entrained at time point D4.
To investigate the possibility that D4 was a critical time point in NHNP differentiation, we examined the genes changing at D4 compared to T0. Using criteria of a false discovery rate (FDR) ≤0.01 together with a fold change of ±1.5, 2218 genes increased in abundance and 2209 genes decreased at D4 compared to T0 (Figure 1G). Known key markers of neuronal differentiation increased at this time point including MAP1B (38), PAX6 (39), SNAP25 (40), ephrins (EFNB3, EFNB1) (41), and semaphorins (SEMA5A, SEMA5B, SEMA6C) (42). In addition, the expression pattern of genes in the NHNPs is consistent with the cells being derived from forebrain progenitors. For example, SP8 and SHH are expressed in the proliferating cells (43), whereas RELN, SOX5, TBR1, BCL11B (CTIP2), CALB2, and NGFR are induced with differentiation (44). Supplementary Table 2 lists all of the genes changing at the D4 time point. Gene Ontology (GO) analysis revealed that the most significantly enriched categories for upregulated genes were nervous system development, neurogenesis, neuron differentiation, and synaptogenesis while those for downregulated genes were cell cycle and mitosis (Table 1 and Supplementary Table 3), consistent with cessation of the proliferative state. Disease association data were also derived from the DAVID database based on Gene Ontology classifications (see Materials and Methods). All of these data were derived from gene expression profiling in one cell line. However, we found significant overlap (P<3.73E-85 – P=0.0) using similar differentiation parameters with two different lines of NHNPs (line 4, Supplemental Table 1 and an additional third line, data not shown).
Table 1.
Most significantly enriched gene ontology (biological function) and disease categories of differentially expressed genes at D4 compared to T0
| Upregulated genes at D4 vs T0 | ||
|---|---|---|
| Gene Ontology Category | Count | PValue |
| nervous system development | 195 | 2.79E-17 |
| neurogenesis | 111 | 1.06E-10 |
| neuron differentiation | 84 | 5.52E-09 |
| synaptogenesis | 16 | 1.21E-07 |
| neuron development | 66 | 1.77E-07 |
| cell development | 104 | 5.78E-07 |
| cell morphogenesis involved in differentiation | 51 | 6.90E-07 |
| negative regulation of cellular process | 221 | 1.65E-06 |
| cell morphogenesis | 64 | 4.78E-06 |
| neuron projection morphogenesis | 44 | 5.94E-06 |
| Disease Association | Count | PValue |
| schizophrenia | 56 | 3.40E-04 |
| Parkinson’s disease | 27 | 5.76E-03 |
| deafness | 4 | 1.60E-02 |
| measles | 5 | 1.82E-02 |
| frontotemporal lobar degeneration | 3 | 4.30E-02 |
| epilepsy, temporal lobe | 4 | 4.63E-02 |
| ALS/amyotrophic lateral sclerosis | 7 | 4.79E-02 |
| Downregulated genes at D4 vs T0 | ||
| Gene Ontology Category | Count | PValue |
| cell cycle phase | 115 | 4.93E-26 |
| M phase | 96 | 1.67E-23 |
| DNA metabolic process | 117 | 3.17E-20 |
| nuclear division | 71 | 6.11E-20 |
| mitosis | 71 | 6.11E-20 |
| M phase of mitotic cell cycle | 71 | 1.09E-19 |
| sterol biosynthetic process | 19 | 4.31E-10 |
| regulation of cell cycle | 67 | 2.20E-08 |
| interphase | 31 | 5.89E-08 |
| interphase of mitotic cell cycle | 30 | 1.09E-07 |
| Disease Association | Count | PValue |
| colorectal cancer | 39 | 8.51E-05 |
| breast cancer | 48 | 4.99E-04 |
| methotrexate toxicity | 7 | 2.74E-03 |
| drug hypersensitivity | 7 | 4.01E-03 |
| neural tube defects | 11 | 4.40E-03 |
| lymphoma, non-Hodgkin | 9 | 1.45E-02 |
| lung cancer | 26 | 2.92E-02 |
| nemaline myopathy | 3 | 3.04E-02 |
| folate, erythrocyte homocysteine thromboembolism | 3 | 3.04E-02 |
| methotrexate efficacy | 3 | 3.04E-02 |
| stomach cancer | 17 | 3.49E-02 |
| leukemia | 15 | 3.85E-02 |
| bladder cancer | 17 | 4.49E-02 |
We next compared the genes changing at D4 to those changing at least 1.3-fold in human brain in vivo between 18 and 23 gestational weeks (45)to determine to what extent the in vitro changes recapitulated those observed in vivo. Out of a potential 575 genes that are available for overlap comparison using our criteria for fold change, 167 genes are increased (P=7.56E-89) and 202 are decreased (P=5.64E-114) under both maturation conditions (Supplementary Table 4), an overlap of about two-thirds. These data are even more remarkable considering that the microarrays utilized are different, only a relatively short period of in vivo development is represented and the in vivo data for comparison consists of pooled data that includes subcortical areas. Thus, these data support the hypothesis that four weeks of differentiation of NHNPs recapitulates the appropriate timing for robust neuronal differentiation in human cells and tissue and reflects molecular processes relevant to in vivo development.
To assess whether this model system could be appropriate for understanding ASD, we cross-referenced two lists of genes with strong evidence for association with ASD (8, 46) with the genes changing during D4 (FDR≤0.01 and fold change ±1.3). Out of a total of 28 genes implicated in ASD, we found a significant overlap of genes going both up (7 genes; P=2.6E-02) and down (7 genes; P=2.4E-02) at D4 (Table 2). Next, we examined whether we could independently confirm the genes changing on the microarrays using real-time RT-PCR (qRT-PCR). Using a threshold of 1.5-fold change, we confirmed approximately 80% of the genes tested (Table 2). Therefore, the microarrays indicate that a significant number of ASD candidate genes are regulated during human neuronal differentiation in vitro.
Table 2.
Expression and confirmation of ASD-candidate genes during differentiation
| Fold Change D4 vs T0 Microarrays |
Pvalue FDR | Fold Change D4 vs T0 qRT-PCR |
|
|---|---|---|---|
| A2BP1 | 1.78 | 7.79E-05 | |
| AHI1 | −2.14 | 8.23E-08 | 1.06 |
| AUTS2* | 1.95 | 1.67E-09 | 3.64 |
| CACNA1C | 1.44 | 9.45E-03 | 21.82 |
| CADPS2 | −1.06 | 6.28E-01 | |
| CNTNAP2 | −1.19 | 2.97E-01 | |
| CYFIP1 | −1.18 | 6.21E-02 | |
| DHCR7 | −3.75 | 5.41E-11 | −3.66 |
| EN2 | 1.18 | 1.34E-01 | |
| FMR1 | −1.06 | 6.06E-01 | |
| FZD9* | −2.28 | 2.28E-10 | −2.44 |
| GABRB3 | 1.86 | 8.93E-07 | 5.60 |
| GRIK2 | −1.18 | 7.43E-02 | |
| LIMK1* | −1.77 | 9.70E-06 | −1.58 |
| MECP2 | 1.72 | 4.77E-03 | 3.28 |
| MET | −1.45 | 3.06E-03 | |
| NLGN3 | −1.33 | 4.90E-04 | 1.18 |
| NLGN4X | 1.16 | 6.27E-02 | |
| NRXN1 | 4.33 | 4.48E-10 | 46.40 |
| OXTR | 1.06 | 7.60E-01 | |
| PLAUR | −7.08 | 4.19E-13 | |
| PRKCB1 | −6.28 | 4.37E-13 | |
| PTEN | 1.53 | 2.16E-06 | 3.37 |
| RELN | 1.22 | 1.38E-02 | |
| SERPINE1 | −10.19 | 9.07E-13 | |
| SHANK3 | 1.32 | 1.66E-03 | 2.36 |
| SLC25A12 | 1.09 | 3.20E-01 | |
| SLC6A4 | −1.01 | 9.63E-01 | |
| TSC1 | 1.66 | 1.50E-04 | 2.05 |
| TSC2 | 1.27 | 1.29E-02 | |
| UBE3A | −1.32 | 5.58E-05 | 2.04 |
Genes not included in refs 8 and 46, but identified through the SFARI database and confirmed by RT-PCR (http://gene.sfari.org)
One interesting observation was that neurexin 1 (NRXN1), an autism and schizophrenia susceptibility gene, and one probe for neurexin 3 (NRXN3) change in opposite directions on the microarrays. We subsequently confirmed by qRT-PCR that NRXN1 was upregulated during NHNP differentiation (Figure 2A) consistent with its known role in synaptic function (47, 48). However, using three different primer pairs to different NRXN3 isoforms, we found NRXN3 was downregulated (Figure 2A). This was somewhat surprising, since there was no previous suggestion that these two highly homologous genes had different functions or expression patterns in human brain. To see if this corresponded to the in vivo situation, we performed in situ hybridization in human fetal brain, which remarkably confirmed these results. NRXN3 is expressed in areas of progenitor cells in human fetal brain, while NRXN1 is expressed in areas of post mitotic neurons (Figures 2B–I). Inspection of the mouse in situ hybridization data in Allen Brain Atlas also shows a similar pattern using different probes (http://www.brain-map.org/). This proof of principle demonstrates how a simple in vitro human system can be used to guide in vivo discovery. These particular data suggest that NRXN3 has a previously unknown role in neural progenitor biology, distinct from canonical neurexin function at the synapse, which is likely conserved from mice to humans.
While distinct up or down changes in gene expression are important parameters of any cellular process, we have begun to appreciate that gene expression datasets contain other inherent patterns of gene co-expression that can be mined, so as to reach new functional insights (49). Specifically, we have used weighted gene expression co-expression network analysis (WGCNA; (50)) as previously described (14, 15)to provide a systems-level view of the organization of the neural transcriptome. Genes that are highly co-expressed are grouped into “modules.” Modules’ correspondence to different aspects of function, such as neuronal phenotypes, sample characteristics (disease vs. control), or cellular organelles is determined by analysis of the module eigen gene or first principle component (15, 50). In addition, central or “hub” genes can be identified within modules (15, 20).
WGCNA of the NHNP differentiation time course identified a total of 22 modules, 14 of which could be easily characterized (Supplementary Table 5). Four modules were clearly associated with differentiation over time throughout the entire time course from T0 through D8. Four other modules correlated best with the undifferentiated cells (T0), while one module eigengene was highly correlated with D2 and another with D8. The module whose eigengene is most correlated with D2 (the green module) is of interest since these genes are likely those involved in defining the onset of neural specification and the cessation of proliferation and cell division. The D8, or sea green, module may contain genes involved in neuronal maturation and phenotype maintenance. Finally, four other modules depicted the transition from D2 to D4. We hypothesized that these modules likely contain genes that are critical for the differentiation process, as D4 was the time point when a significant increase in Tuj1 positive cells was observed (Figure 1) and the GO of genes changing at D4 was enriched for CNS differentiation.
We next sought to determine whether there may be shared biological processes or pathways between distinct ASD risk genes in an unbiased manner using WGCNA. To broaden the number of genes available for this analysis, we queried the SFARI gene database for the most inclusive list of genes with a relationship to ASD (http://gene.sfari.org). While many of these genes may currently have a weak association to ASD, inclusion of as many genes as possible in this analysis should yield networks of genes with the greatest potential for ASD-related gene co-expression and provides an unbiased view. Using the SFARI database, we demarcated 224 genes with a potential role in ASD. Ninety of these genes overlapped with the top 6000 most-connected genes in the current gene expression dataset and were included in the network analysis. Although these 90 genes were distributed across 17 modules (Supplementary Table 6), there were four modules with a particular concentration of ASD genes: the royal blue module (with 18 ASD genes), the pink module (with 17 ASD genes), the turquoise module (with 8 ASD genes), and the black module (with 7 ASD genes) (Figure 3). This non-random concentration of genes within this cohort of modules suggested some shared pathways and previously un-recognized connections between ASD susceptibility genes.
The module eigengene of the royal blue module is related to T0; thus these genes are most related to the proliferative state of the neural progenitors. Not surprisingly, therefore, the top enriched GO categories for the royal blue module include cell adhesion, nervous system development, and neurogenesis (Supplementary Table 3). While none of the ASD genes are “hub” (or the most connected) genes in the royal blue module, at least four of the candidate ASD genes, CBS, DLX2, RIMS3 and PRKCB1, are included in the top 250 connections within this module (Supplemental Figure 2A).
The pink module contains genes related to differentiation across the entire time course. Therefore, these genes are most correlated with the process of differentiation. The pink module contains genes that are enriched in cell cycle GO categories (P=2.29E-05) and axon guidance (P=1.74E-02) (Supplementary Table 3). Visualization of the top connected genes in the pink module reveals ABAT, PLAUR, and SCN1A, all ASD candidate genes, as being among the most connected genes within the module (Figure 5B). Interestingly, MET, another ASD gene that likely coordinates with PLAUR to transmit hepatocyte growth factor signaling (8, 51), is also contained within the pink module, but does not make the cutoff for visualization.
The black and turquoise modules contain genes involved in the D2 to D4 transition; therefore, these genes are likely to be important for early neuronal differentiation. The black module is enriched for genes involved in signal transduction (P=4.53E-07). Two out of the seven ASD candidate genes, NRXN1 and GLRA2, can be visualized among the most connected genes in the black module (Figure 5C). The most significant GO categories for the turquoise module are neuron projection (P=1.27E-06) and nervous system development (P=5.41E-06). Both NRXN1 and SLC9A6 are highly connected within the turquoise module (Figure 5D). Strikingly, of the almost 600 gene connections to ASD genes within the four ASD modules, only seven, or 1%, were previously known or could be surmised based on the literature, while the remaining 590 are novel, demonstrating the power of this analysis.
Another manner with which to identify potentially novel ASD signaling pathways is to focus on the interconnectedness of only the known ASD genes. For example, upon filtering for only known ASD-gene related connections in the turquoise module, not only are the ASD genes always connected by one node or fewer, but now other genes are prioritized, such as the cell adhesion genes CTNNA2 and TAGLN (Supplemental Figure 2A). Using an identical approach with the royal blue module shows inclusion of 17 out of the 18 ASD genes in the module (Supplemental Figure 2B). In addition, well-characterized genes such as SEMA5B and GFAP are easily identifiable as connected among these ASD genes. This level of connectivity is non-random, as the mean overall network connectivity degree is 17 times less than the ASD genes alone, and permutation analysis (See Supplemental Methods) shows that the ASD gene connectivity is much more significant compared with a randomized list of genes of the same size (P=0). This demonstrates that within the complex and enormous process of neuronal differentiation, specific aspects and pathways are more clearly associated with ASD.
To validate whether there were any genetic associations increased in these four modules, we first examined the literature using GO. The royal blue module is enriched for genes with a genetic association with schizophrenia (although this P value did not reach our threshold for significance; P=5.86E-02). These data make sense in light of some of the overlapin the genetic origins of schizophrenia and ASD (52–54). In contrast, other neurological disease genes were not significantly enriched, e.g. dementia (P=1.1E-01), Parkinson’s disease (P=3.9E-01), or bipolar disorder (P=4.9E-01). The pink module also contains genes that are enriched in schizophrenia (P=2.12E-02). The black module is enriched for genes implicated in autism (P=2.45E-02), bipolar disorder (P=2.50E-04), and Alzheimer’s disease (P=4.05E-02). Interestingly, the GO analysis reports four additional genes in the autism category that were not included in the SFARI.gene database as potential ASD candidate genes (BDNF, TH, HLA-DRB4, and NRG1). This highlights that the criteria for inclusion as an “autism gene” can vary among databases. Finally, the turquoise module is enriched for genes involved in panic disorder (P=1.28E-02) and attention deficit disorder (P=1.56E-02). Thus, these data illustrate that there are likely overlapping signal networks during brain development that are disrupted in various permutations to lead to a number of neuropsychiatric disorders.
Recent studies have shown that although genome-wide association studies (GWAS) may not reveal strongly associated genes with large effect sizes in neuropsychiatric disease, overall there may be a weaker signal spread across many genes, as has been demonstrated in schizophrenia and bipolar disorder (6, 7, 55). Thus, it has been suggested that identification of potentially causal pathways could be a means with which to increase power to detect genetic association. To further assess whether there is independent genetic evidence for pathway enrichment, we tested for enhancement of genetic association signals for ASD within the four modules defined above. When the genes used for network analysis were analyzed for genetic association to ASD over 50% of genes with a P Value < 1.0E-02 fell into one of these four modules (Supplementary Table 6). While this percentage of genes is not statistically enriched, it provides another level of information for prioritizing unannotated genes that are highly connected to known ASD genes in these modules.
Discussion
The use of human neural progenitors is an important asset for uncovering the genomic patterns of expression in normal human neuronal differentiation and for the study of neuropsychiatric diseases. These cells recapitulate the morphology and markers of differentiation observed in other model systems. However, the use of a few cell markers is limiting, and screening the cells one gene at a time for possible usefulness in the study of a particular disease is tedious. Whole-genome expression profiling is a powerful tool for viewing the entire transcriptional program of a model system under various conditions. Transcriptional profiling has proven extremely informative in the study of many diseases of the CNS as well as in understanding the development and evolution of the brain (49, 56). We therefore employed a model system of human neuronal cells in combination with whole genome profiling and observed an enrichment of the expression of the genes and signaling pathways involved in ASD, and to a lesser extent other psychiatric disorders with a neurodevelopmental origin, such as schizophrenia. The expression of many of these genes is modulated during neuronal differentiation, lending credence to the neurodevelopmental origin of ASD (57). Moreover, ASD genes cluster non-randomly in modules of gene co-expression leading to the potential identification of additional genes with links to ASD. The ease with which gene expression can be manipulated in these cells (14)makes them an attractive tool for studying signaling pathways involved in ASD and other neurodevelopmental disorders.
As mentioned above, the expression pattern of genes such as SHH and RELN in the NHNPs suggests that the cells are derived from forebrain progenitors. Since one of the underlying pathologies of ASD is thought to be a developmental disconnection syndrome involving frontal –cortical and frontal -subcortical pathways (25, 58, 59), the use of cells derived from human fetal cortex provides a potentially powerful and efficient method of examining signaling pathways disrupted in the disease. Moreover, it has also been postulated that ASD is a disorder of axon outgrowth and/or neuronal migration due to the number of cell adhesion molecules that have been implicated in the disorder (60, 61). In line with this, we have found a significant enrichment of genes involved in cell adhesion that are changing with differentiation (Supplementary Table 3) such as CDH1, FBLN2, NRG1, several laminins and numerous protocadherins. In addition, cell adhesion genes are also highly enriched within all four ASD modules identified, and cell adhesion is the most significantly enriched biological process category in the royal blue module (Supplementary Table 7), which is also enriched in ASD candidate susceptibility genes. Parsing the gene expression data to identify groups of genes involved in a particular cellular function should prove useful as the genetic causes of many of the subtypes of ASD are uncovered. For example, these data can be connected to future genes associated with immune dysfunction or head circumference in ASD. In addition, perturbation of ASD genes can reveal potential shared common pathways within the rubric of the normal network topology.
Here, we study ASD as a prototypical neurodevelopmental disorder for the examination of signaling pathways. Many of the genes associated with ASD have known roles in normal brain development and have been associated with other neuropsychiatric diseases such as schizophrenia (54, 62, 63). Thus, this model system should also serve the study of other neurodevelopmental disorders such as epilepsy, or intellectual disability (ID) well. To this end, we have uncovered at least one module of gene co-expression containing an enrichment of epilepsy genes (64), the pink module, which is also one of the ASD-associated modules identified (Supplemental Table 6). This is not surprising given the high co-morbidity of these neurodevelopmental disorders, with between 10–25% of ASD patients experiencing seizures or epilepsy (65). In addition, genetic data support common causal etiologies for these disorders in several cases (66–70). Moreover, GO analyses highlights an enrichment of genes with known associations to schizophrenia or other neuropsychiatric disorders.
The application of network analysis to these data can yield important biological insights. Not only does WGCNA highlight the interconnectedness of known ASD genes in this model system (Supplementary Figure 2), it also serves to uncover potentially novel genes that may be affected in the disorder. By conducting our own analysis of publicly available ASD association data, we show that the majority of the most significant SNPs are associated with genes in our four ASD modules (Supplementary Table 6; See Materials and Methods). These data illustrate that the combination of genetic and genomic data is a powerful tool for identifying new genes in a disorder, particularly for those that might not make a statistical threshold using only one of the methods. For example, while CTNNA2 has been previously associated with schizophrenia (71) and attention deficit hyperactivity disorder (72), it has no known association with ASD. However, we find it co-expressed with five ASD genes in the turquoise module (CACNA1C, FRMPD4, HS3ST5, NRXN1, and SLC9A6) and exhibiting a suggestive association signal with ASD (4.77E-04). Thus, our analysis would suggest that CTNNA2 has a high probability of playing a role in ASD and is a good candidate for re-sequencing in affected patients. In addition to using genetic association data for prioritizing candidate genes, future studies can employ the wealth of gene dosage information in ASD becoming available (e.g. (1, 2)) to further rank genes in terms of potential pathogenicity.
In addition, we have previously shown that co-expression can be used to annotate gene function (15, 17). For example, highly co-expressed genes in the human brain transcriptome are also physically co-expressed at the protein level in an adult human neural stem cell niche (15). Through the principle of guilt by association we can identify and annotate new gene functions. This is based on the hypothesis that similar expression levels among the most highly co-expressed genes in a module are the result of shared regulatory mechanisms. Thus, it is likely that some of the genes co-expressed with the ASD genes in the NHNPs are also key physical partners of the ASD genes in performing certain intrinsic neurobiological features in vivo. This fits with both the synapse and developmental dysconnection models of ASD. Genes correlated with known ASD genes found at the synapse such as NRXN1 (73)and RIMS3 (74)could themselves bekey players in synapse biology. For example, both NRXN1 and SLC9A6 are co-expressed with FLJ30596 (or c5orf33). Currently, there is nothing known about the biology of FLJ30596, however, the expression of SLC9A6 in endosomal vesicles (75)and NRXN1 at the synapse suggests that FLJ30596 may also have some role in synaptic function. Remarkably, we also find that NRXN3 shows an inverse expression pattern to NRXN1, suggesting a role in progenitor biology, which is validated in vivo in human fetal brain. In a similar vein, genes correlated with known ASD genes that are integral in axon path finding and hence cortico-cortico connections such as EPHA6 (76), NRP2 (77), and SEMA5A (78) (Supplementary Table 6) may themselves be involved in such processes. Thus, the network approach to gene expression analysis may not only identify novel disease-related genes but also may point to the functional role of previously uncharacterized genes.
One area of neuropsychiatric research in need of increased innovation is the discovery of novel therapeutics. Understanding the genes involved in a disorder is a necessary first step; however, having an appropriate model within which to test new drugs is the next critical step. The use of the NHNP system fulfills both criteria. Here, we have demonstrated that there are robust gene expression changes during the differentiation process that follow what is known in vivo in other model organisms as well as in developing human fetal brain. While we have found significant overlap with these model systems and in vivo expression, the expression of some genes that do not agree (e.g. genes going in the opposite direction in Supplementary Table 4) may provide insight into the limitations of this model system and should also be considered in future functional studies. In particular, the effects of processes such as imprinting on cultured cells versus intact or dissociated tissue is an important caveat of this study that may be able to be teased apart using this information. Nonetheless, many of the genes in agreement are not only important for normal differentiation, but are also disrupted or are co-expressed with genes that are disrupted in ASD. Combining these data with previously published (6) and ongoing GWAS in ASD and other neuropsychiatric diseases (79) will solidify the role of particular genes and pathways in the disease. Moreover, as the cells are genetically tractable through modification by lentiviral over-expression or knockdown, multiple genes can be modified simultaneously to mimic the genetic heterogeneity of diseases such as ASD. Then together with high-throughput chemical library screenings (80), therapeutics for particular genetic signatures can be rapidly tested. This combinatorial approach therefore is particularly advantageous over animal models where only one or a few genes can be modified concurrently, or high-throughput chemical screening is time intensive.
The combination of these human tools with non-human models clearly provides a synergistic approach for elucidating the signaling cascades important in both normal human brain development as well as in neuropsychiatric disorders. Datasets such as the one presented here will identify new disease-related genes and present opportunities for the testing and development of personalized therapeutics.
Supplementary Material
Acknowledgments
This work is supported by grants from the NIMH (R37MH060233 and R01MH081754) to DHG and the Shappel-Guerin Foundation. GK is supported by an A.P. Giannini Foundation Medical Research Fellowship, a NARSAD Young Investigator Award, and the NIMH (K99MH090238). EW is supported by the NIMH (K08MH074362). Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland (NICHD Contract numbers N01-HD-4–3368 and N01-HD-4–3383). The role of the NICHD Brain and Tissue Bank is to distribute tissue, and therefore cannot endorse the studies performed or the interpretation of results.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009 Jun;5(6):e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009 May 28;459(7246):569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009 May;73(Pt 3):263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 Jul 15;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007 Apr 20;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009 May 28;459(7246):528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009 Oct 8;461(7265):802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bill BR, Geschwind DH. Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev. 2009 Jun;19(3):271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010 Jul;11(7):490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009 Jun 26;137(7):1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flint J, Shifman S. Animal models of psychiatric disease. Curr Opin Genet Dev. 2008 Jun;18(3):235–240. doi: 10.1016/j.gde.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Kabashi E, Champagne N, Brustein E, Drapeau P. In the swim of things: recent insights to neurogenetic disorders from zebrafish. Trends Genet. 2010 Aug;26( 8):373–381. doi: 10.1016/j.tig.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009 Nov 12;462(7270):213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008 Nov;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaisier CL, Horvath S, Huertas-Vazquez A, Cruz-Bautista I, Herrera MF, Tusie-Luna T, et al. A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 2009 Sep;5(9):e1000642. doi: 10.1371/journal.pgen.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4(8):e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008 Mar 27;452(7186):429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006 Nov 14;103(46):17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010 Oct 15;24(20):2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010 Nov 12;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010 Mar 2;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005 Apr-May;23(2–3):183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007 Feb;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathol. 2007 Oct;17( 4):434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wexler EM, Paucer A, Kornblum HI, Palmer TD, Geschwind DH. Endogenous Wnt signaling maintains neural progenitor cell potency. Stem Cells. 2009 May;27( 5):1130–1141. doi: 10.1002/stem.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, et al. A new method for the rapid and long term growth of human neural precursor cells. J Neurosci Methods. 1998 Dec 1;85(2):141–152. doi: 10.1016/s0165-0270(98)00126-5. [DOI] [PubMed] [Google Scholar]
- 29.Palmer TD, Schwartz PH, Taupin P, Kaspar B, Stein SA, Gage FH. Cell culture. Progenitor cells from human brain after death. Nature. 2001 May 3;411(6833):42–43. doi: 10.1038/35075141. [DOI] [PubMed] [Google Scholar]
- 30.Esquenet M, Swinnen JV, Heyns W, Verhoeven G. LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids. J Steroid Biochem Mol Biol. 1997 Aug;62(5–6):391–399. doi: 10.1016/s0960-0760(97)00054-x. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007 Nov;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abu-Khalil A, Fu L, Grove EA, Zecevic N, Geschwind DH. Wnt genes define distinct boundaries in the developing human brain: implications for human forebrain patterning. J Comp Neurol. 2004 Jun 21;474(2):276–288. doi: 10.1002/cne.20112. [DOI] [PubMed] [Google Scholar]
- 34.Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res Dev Brain Res. 1995 Jan 14;84( 1):109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- 35.Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17(2):118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 36.Izant JG, McIntosh JR. Microtubule-associated proteins: a monoclonal antibody to MAP2 binds to differentiated neurons. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4741–4745. doi: 10.1073/pnas.77.8.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eng LF. Glial fibrillary acidic protein (GFAP): the major protein of glial intermediate filaments in differentiated astrocytes. J Neuroimmunol. 1985 Jun;8(4–6):203–214. doi: 10.1016/s0165-5728(85)80063-1. [DOI] [PubMed] [Google Scholar]
- 38.Scales TM, Lin S, Kraus M, Goold RG, Gordon-Weeks PR. Nonprimed and DYRK1A-primed GSK3 beta-phosphorylation sites on MAP1B regulate microtubule dynamics in growing axons. J Cell Sci. 2009 Jul 15;122(Pt 14):2424–2435. doi: 10.1242/jcs.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002 Apr;5(4):308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 40.Bailey JA, Lahiri DK. Neuronal differentiation is accompanied by increased levels of SNAP-25 protein in fetal rat primary cortical neurons: implications in neuronal plasticity and Alzheimer’s disease. Ann N Y Acad Sci. 2006 Nov;1086:54–65. doi: 10.1196/annals.1377.001. [DOI] [PubMed] [Google Scholar]
- 41.Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 42.Pasterkamp RJ, Kolodkin AL. Semaphorin junction: making tracks toward neural connectivity. Curr Opin Neurobiol. 2003 Feb;13(1):79–89. doi: 10.1016/s0959-4388(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 43.O’Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008 Feb;18(1):90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaspard N, Gaillard A, Vanderhaeghen P. Making cortex in a dish: in vitro corticopoiesis from embryonic stem cells. Cell Cycle. 2009 Aug 15;8(16):2491–2496. doi: 10.4161/cc.8.16.9276. [DOI] [PubMed] [Google Scholar]
- 45.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009 May 28;62(4):494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat RevGenet. 2008 May;9(5):341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010 Jun 11;141(6):1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 48.Fairless R, Masius H, Rohlmann A, Heupel K, Ahmad M, Reissner C, et al. Polarized targeting of neurexins to synapses is regulated by their C-terminal sequences. J Neurosci. 2008 Nov 26;28(48):12969–12981. doi: 10.1523/JNEUROSCI.5294-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009 Oct 15;461(7266):908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 51.Mars WM, Zarnegar R, Michalopoulos GK. Activation of hepatocyte growth factor by the plasminogen activators uPA and tPA. Am J Pathol. 1993 Sep;143(3):949–958. [PMC free article] [PubMed] [Google Scholar]
- 52.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008 Oct 16;455(7215):919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 53.Crespi B, Badcock C. Psychosis and autism as diametrical disorders of the social brain. Behav Brain Sci. 2008 Jun;31(3):241–261. doi: 10.1017/S0140525X08004214. discussion 261–320. [DOI] [PubMed] [Google Scholar]
- 54.Crespi B, Stead P, Elliot M. Evolution in health and medicine Sackler colloquium: Comparative genomics of autism and schizophrenia. Proc Natl Acad Sci U S A. 2009 Jan 26;107(Suppl 1):1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009 Aug 6;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konopka G. Functional genomics of the brain: uncovering networks in the CNS using a systems approach. Wiley Interdiscip Rev Syst Biol Med. 2010 Dec 31; doi: 10.1002/wsbm.139. [DOI] [PubMed] [Google Scholar]
- 57.Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, Fauchereau F, et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010 Aug;26(8):363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010 Nov 3;2(56):56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melillo R, Leisman G. Autistic spectrum disorders as functional disconnection syndrome. Rev Neurosci. 2009;20(2):111–131. doi: 10.1515/revneuro.2009.20.2.111. [DOI] [PubMed] [Google Scholar]
- 60.Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009 Jul;32(7):402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008 Oct 16;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cantor RM, Geschwind DH. Schizophrenia: genome, interrupted. Neuron. 2008 Apr 24;58(2):165–167. doi: 10.1016/j.neuron.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008 Apr 25;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 64.Noebels JL. The biology of epilepsy genes. Annu Rev Neurosci. 2003;26:599–625. doi: 10.1146/annurev.neuro.26.010302.081210. [DOI] [PubMed] [Google Scholar]
- 65.Spence SJ, Schneider MT. The role of epilepsy and epileptiform EEGs in autism spectrum disorders. Pediatr Res. 2009 Jun;65(6):599–606. doi: 10.1203/01.pdr.0000352115.41382.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Bon BWM, Mefford HC, de Vries BBA. 15q13.3 Microdeletion. 1993. [Google Scholar]
- 67.Hamdan FF, Daoud H, Piton A, Gauthier J, Dobrzeniecka S, Krebs MO, et al. De Novo SYNGAP1 Mutations in Nonsyndromic Intellectual Disability and Autism. Biol Psychiatry. 2011 Jan 14; doi: 10.1016/j.biopsych.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Walsh CA, Engle EC. Allelic diversity in human developmental neurogenetics: insights into biology and disease. Neuron. 2010 Oct 21;68(2):245–253. doi: 10.1016/j.neuron.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brooks-Kayal A. Epilepsy and autism spectrum disorders: are there common developmental mechanisms? Brain Dev. 2010 Oct;32(9):731–738. doi: 10.1016/j.braindev.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 70.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006 Mar 30;354(13):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 71.Chu TT, Liu Y. An integrated genomic analysis of gene-function correlation on schizophrenia susceptibility genes. J Hum Genet. 2010 May;55(5):285–292. doi: 10.1038/jhg.2010.24. [DOI] [PubMed] [Google Scholar]
- 72.Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008 Nov;115(11):1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 73.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008 Jan;82(1):199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar RA, Sudi J, Babatz TD, Brune CW, Oswald D, Yen M, et al. A de novo 1p34.2 microdeletion identifies the synaptic vesicle gene RIMS3 as a novel candidate for autism. J Med Genet. 2010 Feb;47(2):81–90. doi: 10.1136/jmg.2008.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garbern JY, Neumann M, Trojanowski JQ, Lee VM, Feldman G, Norris JW, et al. A mutation affecting the sodium/proton exchanger, SLC9A6, causes mental retardation with tau deposition. Brain. 2010 May;133(Pt 5):1391–1402. doi: 10.1093/brain/awq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007 May;17(5):230–238. doi: 10.1016/j.tcb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 77.Roffers-Agarwal J, Gammill LS. Neuropilin receptors guide distinct phases of sensory and motor neuronal segmentation. Development. 2009 Jun;136(11):1879–1888. doi: 10.1242/dev.032920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hilario JD, Rodino-Klapac LR, Wang C, Beattie CE. Semaphorin 5A is a bifunctional axon guidance cue for axial motoneurons in vivo. Dev Biol. 2009 Feb 1;326(1):190–200. doi: 10.1016/j.ydbio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 79.Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010 Oct 21;68(2):182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barbaric I, Gokhale PJ, Andrews PW. High-content screening of small compounds on human embryonic stem cells. Biochem Soc Trans. 2010 Aug;38( 4):1046–1050. doi: 10.1042/BST0381046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.