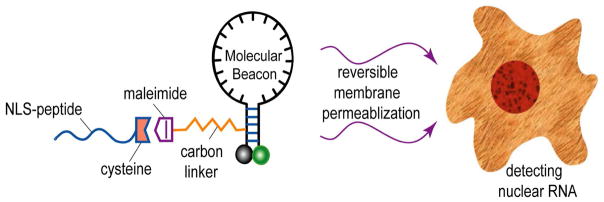
Abstract
Imaging the expression and localization of RNAs in live-cell nucleus can provide important information on RNA synthesis, processing and transport. Here we report the development of a bi-functional molecular beacon (NLS-MB) composed of a single nuclear localization sequence (NLS) peptide conjugated to a molecular beacon for efficient delivery and imaging of endogenous RNAs in the nuclei of living cells. We characterized the NLS-MBs by comparing their signal to noise ratios with unmodified molecular beacons and determined their efficiency of nuclear import. We demonstrated the specificity and sensitivity of the method by observing in living cells the localization and co-localization of small nuclear RNAs (snRNA) U1 and U2 at discrete foci in the nucleoplasm, and the localization of small nucleolar RNA U3 in the nucleolus. These snRNAs were chosen because of their essential roles in RNA biogenesis. The results were validated using in situ hybridization as positive control and random beacons as negative control. This novel approach may be applied to imaging other nuclear RNAs and pre-mRNAs in living cells.
INTRODUCTION
Imaging RNA molecules in the nuclei of living cells can provide important spatial and temporal information on the dynamics of RNA synthesis, processing and transport. Specifically, in eukaryotic cells, RNA molecules especially messenger RNAs (mRNA) are processed post-transcriptionally in the nucleus, including splicing, capping, polyadenylation, and methylation. These processing steps are carried out by multiple protein nanomachines and ribonucleoprotein (RNP) complexes (1, 2). The mature RNA molecules (including mRNA and ribosomal RNA) are then exported to the cytoplasm. Further, many viral genes are processed in the cell nucleus, including transcription and replication of viral RNAs. Therefore, the ability to detect and localize RNA molecules (including small nuclear RNAs) in live-cell nuclei may provide a powerful tool not only for studies of basic biology but also disease detection and diagnosis. This ability will also complement the approaches of imaging RNA in the cytoplasm of living cells (3, 4) so that an integrated picture of RNA transport, distribution and localization in the cell cytoplasm and nucleus can emerge.
Extensive efforts are being made to develop new approaches to image RNA in living cells. One novel approach uses a fusion protein, GFP-MS2, to track the localization and dynamics of RNA in living cells with single molecule sensitivity (5). Although a very powerful technique in tracking RNA dynamics, this method relies on transfecting cells to express GFP-MS2 as a reporter, and add to the target RNA multiple (20–25) MS2 binding sequences. Therefore this technique lacks the ability to image endogenous RNAs, which is important for diagnostic applications especially in-vivo, and may not be ideal for imaging small RNAs since binding multiple MS2-GFPs may significantly perturb their structure and dynamics. Further, imaging assays have been performed by introducing fluorescently labeled linear oligonucleotide (ODN) probes into living cells for RNA expression, tracking and localization studies (6–8) . This approach, however, lacks the ability to distinguish background from true signal. In addition, although 2’-O-methyl backbone chemistry has been used to improve the affinity of linear ODN probes for binding to the target sequence (7, 8), in many cases the (9) increased affinity is associated with decreased specificity of the probe (9). To enhance detection specificity and increase signal-to-background ratio, two linear probes with a fluorescence resonance energy transfer (FRET) pair of (donor and acceptor) fluorophores have been used (6). This approach, however, may still have a high background signal due to direct excitation of the acceptor and emission detection of the donor fluorescence. Further, this dual-probe approach may be difficult to be used in targeting small nuclear RNAs.
Molecular beacon based approaches have been developed to image endogenous RNA in live cells. Molecular beacons are dual-labeled antisense oligonucleotide (ODN) probes with a fluorophore at one end and a quencher at the other end. In contrast to fluorescently-labeled linear ODN probes, molecular beacons are designed to form a stem-loop (hairpin) structure in the absence of a complementary target so that fluorescence of the fluorophore is quenched (10). Hybridization with target RNA opens the hairpin and physically separates the reporter from the quencher, allowing a fluorescence signal to be emitted upon excitation. Molecular beacons have been used to image endogenous RNA localization in the cytoplasm of living cells as well as in live embryos during development (3, 4, 7, 10–14). However, most of the studies on live-cell RNA detection used microinjection to introduce linear or hairpin probes in to cells. This delivery approach is labor-intensive when used for studying a large number of cells. Further, probes delivered using microinjection tend to non-specifically accumulate in the nuclei of cells and thus have a high background signal. In some cases no localized signal was observed (12, 14).
Here we report the development of a bi-functional probe (NLS-MB) composed of a single nuclear localization sequence (NLS) peptide (15–18) conjugated to a molecular beacon (MB) for efficient delivery and imaging of endogenous snRNA and snoRNA in living cells. Using a combination of reversible membrane permeabilization and peptide based delivery, we demonstrated the high specificity and sensitivity of the method by observing in living cells the localization and co-localization of snRNA U1 and U2 at discrete foci in the nucleoplasm, and the localization of snoRNA U3 in the nucleolus. These small RNAs were chosen because U1 and U2 snRNAs are the key members of the spliceosome responsible for splicing of nuclear pre-mRNA (15, 19–21), and U3 snoRNA is one of the best characterized small nucleolar RNAs involved in the endonucleolytic cleavage of various pre-rRNAs (22). Our results clearly demonstrated the ability of the NLS-MBs in detecting snRNAs and snoRNAs in cell nucleus, which can be used to image other nuclear RNAs and pre-mRNAs in studying transcriptional and post-transcriptional processing of RNAs in living cells.
EXPERIMENTAL PROCEDURES
Molecular Beacon Design and Synthesis
Probes (as shown in Table 1) with modification in the stem region (dT-NH2 on the quencher side) were synthesized by MWG Biotech, NC and Biosearch Corp, CA. The molecular beacons were designed with regular DNA chemistry (deoxyribonucleotide backbone). The probes were purified using double HPLC. Probes for U1 and U2 snRNPs and U3 snoRNPs were designed with the Cy3 (fluorophore) and BHQ-II (quencher) pair. U2 snRNP probes were also designed to have Cy5 fluorophore and BHQ-III quencher. In all cases the quencher was conjugated to the 3’ end, while the fluorophore was on the 5- end (Table 1).
Table 1.
Design of NLS-peptide linked molecular beacons
| NLS Peptide |
| (N terminus) C(Ahx)GGGPKKKRKVED (C terminus) |
| U1 snRNA Molecular Beacon |
| 5’-Cy3-GCAGCCTGCCAGGTAAGTATGC(dT-C6-NH2)GC-BHQ2-3' |
| U2 snRNA Molecular Beacon |
| 5’-Cy3-CGAGCACAGATACTACACTTGGC(dT-C6-NH2)CG-BHQ2-3' |
| U3 snoRNA Molecular Beacon |
| 5’-Cy3-CGACCGGCTTCACGCTCAGGGG(dT-C6-NH2)CG -BHQ2-3' |
| Negative Control Molecular Beacon |
| 5’-Cy3-CGACGCGACAAGCGCACCGATACG(dT-C6-NH2)CG-BHQ2-3' |
Peptide Conjugation and Synthesis
In the conjugation scheme as shown in Figure 1, a nucleotide on the quencher-arm of a molecular beacon was modified with the dT-amine group and a 6-carbon spacer. To generate a stable linkage between a peptide and the modified molecular beacon, the NLS peptide was modified with a cysteine group at its N terminus. Amine modified molecular beacons were reacted with a heterofunctional crosslinker sulfo-SMCC (Pierce Biotech) for 3 hours at room temperature with intermediate shaking. The reaction was carried out with 1× (2 μm) molecular beacons and 20× (40 μm) sulfo-SMCC in PBS buffer. After 3 hours, the excess of sulfo-SMCC was removed from the reaction mixture using centrifugal filter (YM-10) with a cut off of 10 kDa. The resultant solution was brought to the same initial volume by adding PBS and reacted with 2× (4 μm) concentration of Cys modified NLS peptide for overnight at room temperature. Following this reaction, the peptide linked molecular beacon complex was dialyzed overnight using Slide-A-Lyzer Dialysis Unit, 10K MWCO (Pierce Biotech Inc., Rockford, IL) to remove unconjugated peptide. The NLS peptide (99% purity) as shown in Table 1 was synthesized at SynPep Inc. The NLS peptide sequence was modified at its N terminus to introduce a cysteine residue. The aminohexanoic acid spacer (Ahx) equivalent to a length of ~2 amino acids was introduced between the N terminus residue of NLS sequence and the cysteine residue to improve conjugation efficiency. Reaction between the thiol group (Cys) on an NLS peptide with the maleimide group introduced on a molecular beacon resulted in the formation of a thiol-maleimide bond which is stable under physiological conditions, including the reducing intracellular environment.
Figure 1.
A schematic illustration of the design of a peptide-linked molecular beacon and its delivery into cell nucleus. The NLS peptide is covalently linked to the molecular beacon using a modified nucleotide (dT-C6-NH2) in its quencher arm. The NLS linked molecular beacons are delivered into the cytoplasm first using a reversible membrane permeabilization agent Streptolysin O (SLO), and the NLS peptide actively transports the probes into the nucleus of a living cell.
Cell Lines
All experiments in this study were carried out using HeLa cells. This cell line has been used in several nuclear transport studiesand also in study of various snRNPs in fixed cells HeLa cells were cultured using Dulbecco’s modified Eagle's medium + fetal calf serum (10%) + horse serum (2.5 %). HeLa cells were plated in T-25 (cell culture flask) using 1:10 dilution after each splitting cycle. A batch of HeLa cultured only for about 10 splitting cycles. For imaging experiments, cells were plated on 4 well or 8 well Nalgene-Nunc chamber cover slides 24 hours prior to the experiment.
Solution Assay for S/N of Modified and Unmodified Molecular Beacons
Measurement of signal-to-background ratio of peptide linked molecular beacons and unmodified beacons were carried out using a SAFIRE microplate monochromator reader (TECAN, Austria). To determine the signal-to-background ratios, 200 nM conventional and peptide-linked molecular beacons were mixed with 200 nM of complementary target respectively in the microplate reader, and the fluorescence intensity at equilibrium (10 minute incubation time) was recorded. The fluorescence signal of each molecular beacon type (conventional and peptide-linked) in the absence of target was recorded as background signal.
Cellular Delivery and Confocal Imaging
For cellular delivery of peptide-linked molecular beacons, experiments were carried out using HeLa cells (ATCC, VA). For imaging experiments, cells were cultured in a 4 or 8-well Nalge Nunc culture plate with a glass coverslip bottom in their respective cell culture media (as described above) for 24 hours before the experiment. For delivery, peptide-linked molecular beacons were mixed with activated streptolysin O (SLO) in serum free media and incubated with the specific cells at 37°C for 10 min. After incubation, the medium was changed to regular serum rich media and the cells were incubated for 1 hr after changing the medium. We also conducted a control study in which NLS-conjugated, unquenched beacon probes were incubated with cells without streptolysin-O treatment. Our results indicate that there was essentially no uptake of unquenched beacons after 1 hour of incubation without SLO (data not shown). Cells were then imaged alive using a confocal microscope (Axiovert LSM-100, Zeiss). For detection and imaging of localization of U1, U2 snRNPs and U3 snoRNP, we used laser excitation at 543 nm with emission detection using a band pass filter from 560–610 nm. For imaging co-localization of U1 and U2 snRNPs we used both 543 nm and 640 nm laser excitation. The signal was detected using band pass filter from 560–610 nm for Cy3 signal detection and long pass filter from 660 nm for Cy5 signal detection.
RESULTS
Design of Peptide Linked Molecular Beacons
In this study we have designed and synthesized the peptide-linked molecular beacons targeting U1, U2 snRNAs and U3 snoRNAs, as well as molecular beacons with a ‘random’ probe sequence. The specific oligonucleotide sequences of these MBs and the sequence of the NLS peptide are shown in Table 1, with the stem domain of a molecular beacon underlined. The design of these probes to target specific segments of small RNAs in the nuclei of living cells was based on previous fluorescence in-situ hybridization (FISH) studies in fixed cells with linear oligonucleotide probes, which were shown to be able to access the target (15, 16, 23–27). The ‘random’ beacon was designed as a negative control, with a 17-base probe sequence that does not have any exact match in the mammalian genome (3, 4). The probes used in this study are with deoxyribonucleotide backbone; the dye and quencher pairs (Cy3-BHQ-II and Cy5-BHQ-III) were selected to ensure effective quenching of the probes in their unhybridized state (stem-loop hairpin).
Signal-to-Noise Ratio of Peptide-linked Molecular Beacons
To determine the possible effect of peptide conjugation on the molecular beacon function, in-solution hybridization assays were carried out to compare the signal-to-noise (S/N) ratios of the peptide-linked molecular beacons and unmodified beacons. When the positively-charged NLS peptide is conjugated to a molecular beacon, the electrostatic interactions of the peptide with the negatively-charged oligonucleotide hairpin may interfere with the probe-target (RNA) binding. As shown by the results in Figure 2, there were no significant changes in the S/N for each of the molecular beacons after modification with NLS peptide. This result is in agreement with our earlier results with Tat peptide-linked MBs for cytoplasmic delivery and mRNA detection (4).
Figure 2.
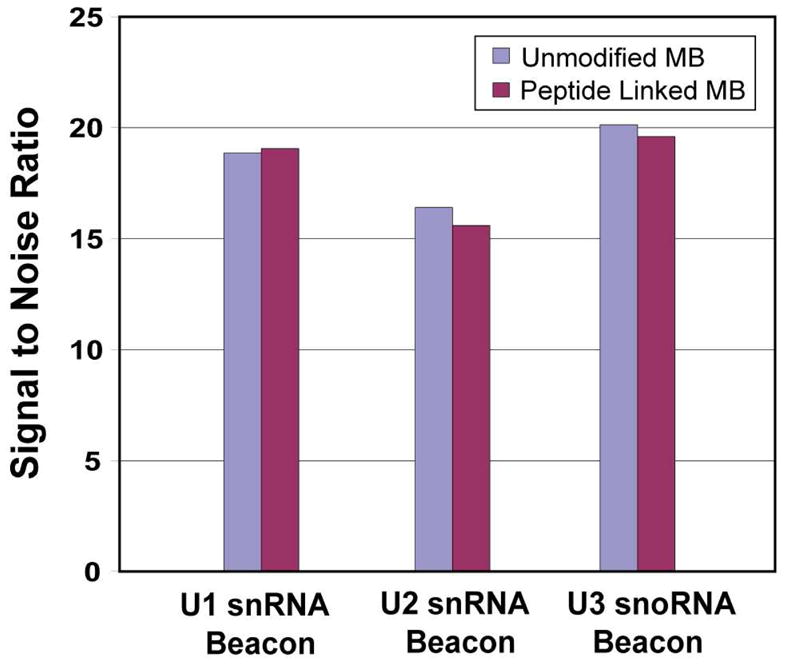
Signal-to-noise ratios of the NLS-linked and unmodified molecular beacons. For molecular beacons designed to target U1, U2 snRNAs and U3 snoRNA, with and without the NLS peptide there is no significant difference in the signal-to-noise ratios.
Efficiency of Nuclear Delivery Using Peptide Linked Molecular Beacons
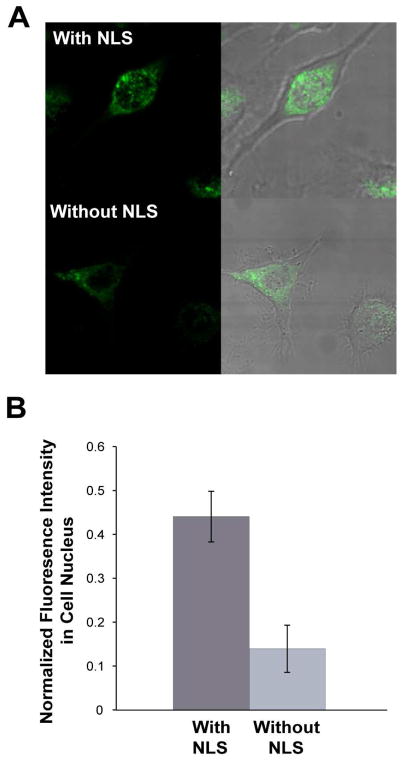
To provide an estimate for the total concentration of NLS-MBs delivered into nucleus, we synthesized unquenched, U3-targeting hairpin probes with and without NLS peptide respectively and delivered them into the cytoplasm of living cells using SLO. Since U3 snoRNA does not have any target in the cytoplasm, there should be no specific signal in the cytoplasm; thus this approach gives a good measure of probe distribution in the cytoplasm due to delivery but not probe-target hybridization. The total fluorescence signal was quantified in the cytoplasm and nucleus of approximately 40 cells. The average ratio of fluorescence intensity in the nucleus to total fluorescence in each cell was determined. A representative image of nuclear and cytoplasmic distribution of probes with and without NLS is shown in Figure 3A. There was a significant increase (~3.3 fold increase) in the nuclear localization of NLS-conjugated probes compared to probes without the NLS peptides, as demonstrated in Figure 3B. Specifically, about 40% of the NLS-conjugated probes were delivered into the cell nucleus as compared to only about ~10% of the probes without NLS peptide. This result is in agreement with a recent study using NLS linked PNA probes (28).
Figure 3.
Efficiency of nuclear delivery of NLS linked MB probes in living cells. (a) Fluorescence images of unquenched probes delivered to the cytoplasm of living cells using SLO. The NLS-conjugated probes show a much higher signal level in cell nucleus as compared with probes without the NLS peptide. (b) Quantification of nuclear delivery efficiency of probes with and without NLS peptide. The average fluorescence signal from NLS-conjugated probes in cell nucleus has a ~3.3 fold increase compared with probes without NLS peptide.
Imaging of U3 snoRNP, U1 and U2 snRNPs in the Nucleus of Living cells
To demonstrate the delivery of NLS-MBs into cell nuclei, and specific detection and imaging of an endogenous small RNA in the nuclei of living cells, we have performed detection assays to target U3 snoRNA and U1 and U2 snRNAs in the nucleus. The NLS-linked MBs targeting the respective small RNAs were introduced into cell cytoplasm using a reversible cell membrane permeabilization method with Streptolysin O (SLO). The results for the detection of U3 snoRNP in the nucleus of a living cell are shown in Figure 4a. We observed very discrete localization of signal in the nucleolus and discrete peri-nucleolar foci in living cells with no detectable signal in the cytoplasm, consistent with the results obtained in previous studies (24, 26, 27, 29), indicating that U3 snoRNA is localized in the nucleolus and in certain cases associated with the cajal bodies. It is known that U3 snoRNA is processed in the cajal bodies prior to its transport and localization to the nucleolus. Furthermore, we observed heterogeneous localization (punctate pattern) of the U3 snoRNA in some nucleoli and a uniform fluorescence pattern in others, also consistent with previous observations (24).
Figure 4.
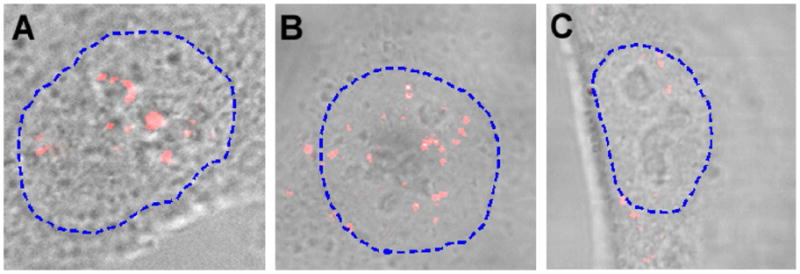
Imaging of U3 snoRNA, U1 and U2 snRNAs in the nucleus of living cells using NLS-linked molecular beacons. (a) Fluorescence signal of molecular beacons targeting U3 snoRNA in the nucleus of a living cell. Note that most of the localized signal was in the nucleolars, with no signal in the cytoplasm. (b) Imaging of U1 snRNA showing localized fluorescence signal in the nucleoplasm of a living cell excluding the nucleolus. Note that the discrete foci of U1 snRNPs are widely distributed in the nucleoplasm, with a few foci in the peri-nucleolar region. Note also the localized signal of U1 snRNPs in the peri-nuclear region of the cytoplasm, indicating the population of the U1 snRNAs exported to the cytoplasm for processing of its 5’ cap. (c) The fluorescence signal suggests that the localization of U2 snRNAs in the nucleus is similar to that of U1 snRNAs but with a much lower abundance.
Shown in Figure 4b are the results of detection and imaging of U1 snRNPs using NLS peptide-linked MBs, clearly indicating the discrete localization of the U1 snRNPs in the nucleoplasm of HeLa cells but not in the nucleolus. The localized foci of U1 snRNPs are widely distributed in the nucleoplasm, with a few foci at discrete peri-nucleolar regions (marked with an arrow). This observation of widespread U1 snRNP localization in the nucleoplasm is in agreement with the results of the previous studies (16, 23, 25, 30). These studies have shown in fixed cells that U1 snRNPs are associated with both cajal bodies (processing of U1 snRNP after its import from cytoplasm) and speckles (site of assembly of splicing complex). In addition to widespread nucleoplasmic localization, we also observed some localized signal at discrete spots in the cytoplasmic, peri-nuclear region. This signal indicates a fraction of the U1 snRNPs exported to the cytoplasm. It has been suggested that U1 and some other snRNPs (excluding U6) are exported from the nucleus to the cytoplasm for assembly of the snRNP complexes and the processing of their cap (31–33). They are then imported back into the nucleus and further processed in the cajal bodies before transported to the speckle sites. This export process has been directly observed in xenopus oocytes using labeled RNAs as well as EM based high resolution imaging (17, 32). These studies have also reported a rapid import of the snRNPs from the cytoplasm to nucleus after processing (32). The detection of both the cytoplasmic and nuclear portions of U1 snRNPs in live cells demonstrated clearly the high sensitivity of the molecular beacon based method.
Molecular beacon based imaging assays were performed to detect U2 snRNPs in live cells and the results, displayed in Figure 4c, showed clear U2 localization at discrete foci in the nucleoplasm, excluding the nucleoli. In contrast to widespread localization of the U1 snRNPs in the nucleoplasm, the fluorescence signal of U2 snRNPs appeared at only a few discrete foci. This observation is in agreement with the results of the localization pattern obtained in previous studies (16, 23). We also observed discrete signal of U2 snRNAs in the peri-nuclear region in the cytoplasm of living cells, confirming that U2 snRNPs, similar to U1 snRNPs, are exported to the cytoplasm for processing.
Taken together, the results in Figure 4 demonstrated that single NLS-peptide linked molecular beacons could be transported from the cytoplasm to the nucleus. To highlight the high specificity of the multifunctional probes in the nucleus, we have imaged RNA at discrete localizations (nucleoplasm vs nucleolus) with high signal to background ratio in the nuclei of living cells. However, it is unclear whether U1 and U2 snRNPs are restricted to the cajal bodies, or the pattern observed in this study represents both the cajal bodies and the speckle sites. Answering this question through live-cell imaging studies would require the simultaneous labeling of some of the proteins in the speckle domains such as SF2/ASF (34, 35) as well as snRNAs. The combination of fusion proteins such as GFP with an exogenous probe for snRNA can provide valuable insight into the dynamics and function of RNAs in living cells. This will be pursued in a subsequent study.
Co-localization of U1 and U2 snRNAs in Living Cells
U1, U2 and other members of the snRNP family, together with some non-snRNP proteins form a splicing complex responsible for the trans-splicing of pre-mRNA transcripts. The assembly of this complex requires various snRNPs to come together to form a define structure. Although in vitro characterization of the complex has been performed, there are still many unknowns concerning the assembly and inner-workings of the spliceosome complex. For example, it is important to know the dynamics of U1 and U2 snRNPs as they form a complex. As an initial effort, we have studied the co-localization of U1–U2 snRNAs in cell nucleus to demonstrate the ability to simultaneously detect multiple RNAs in the nuclei of living cells. As shown in Figures 5a–5c, some U1 and U2 snRNAs are co-localized at discrete foci in the nucleoplasm, with a more widespread distribution of U1 snRNPs, consistent with the results obtained using FISH with fixed cells (15, 16). This U1–U2 co-localization may be the first step towards the formation of the splicesosome. To our knowledge this is the first direct observation of the co-localization of U1–U2 snRNAs in living cells.
Figure 5.
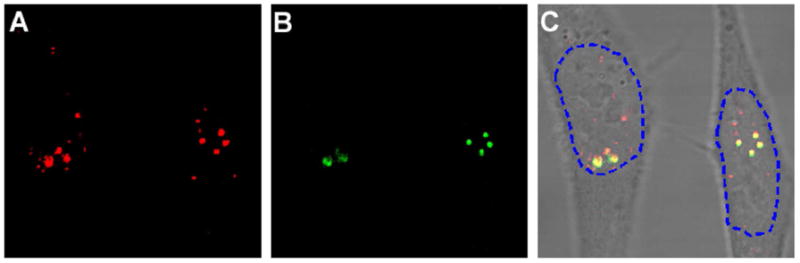
Co-localization of U1 and U2 snRNAs in the nucleoplasm of living cells imaged using peptide-linked molecular beacons. Shown in (a) & (b) are respectively images of fluorescence signal from U1 and U2 snRNAs in the same living cell. (c) The overlay of the images in (a) and (b) indicating the co-localization of U1 and U2 snRNAs at discrete foci in nucleoplasm. In addition to the U1–U2 co-localized foci, U1 snRNAs can be observed at other foci in the nucleoplasm.
In-situ Hybridization Studies with Fixed Cells
To confirm the observations in live cell studies, as a positive control we carried out fluorescence in-situ hybridization (FISH) studies in fixed cells. The cells were fixed and permeablized according to the protocol described in the material and methods section. The result of detecting U3 snoRNAs in fixed cells, shown in Figure 6a, clearly demonstrated the localization of U3 snoRNAs in the nucleoli, similar to what was observed in live cells. To validate our observations of the localization and co-localization of U1 and U2 snRNAs in living cells, we performed fluorescence in situ hybridization in fixed cells using probes targeted to U1 and U2 snRNAs. The results of the FISH studies are displayed in Figures 6b – 6d, which clearly show the discrete foci of co-localized U1 and U2 snRNAs. We also observed more widespread localization of the U1 snRNAs in the nucleoplasm of fixed cells. These observations are in good agreement with the results obtained from the live cell studies. In addition, we also observed few discrete foci of U1 and U2 snRNAs in the peri-nuclear region of cytoplasm in agreement with our live cell assay.
Figure 6.
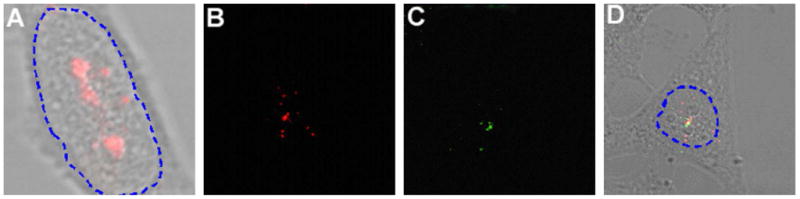
Fluorescence In-situ hybridization (FISH) images of the U3 localization and U1–U2 co-localization in the nuclei of fixed cells. (a) FISH image of the U3 snoRNA in a fixed cell indicating that most of the signal was in the nucleolus, consistent with the live-cell results. Shown in (b) & (c) are respectively FISH images of the U1 and U2 snRNAs localized at discrete foci in the nucleoplasm of the same fixed cell, in good agreement with that observed in live cells. (d) Overlay of the images in (b) and (c) shows the co-localization of U1 and U2 snRNAs in the nucleus of a fixed cell, confirming that observed in live cells.
Negative Control Studies
In order to determine the level of non-specific interaction of NLS-MB in living cells, we carried out control studies with ‘random’ molecular beacons, which have a specific sequence generated using ‘random walk’, with no exact match in the mammalian genome. Figure 7a shows the signal from the NLS peptide-linked ‘random’ beacons. Evidently there was very low signal under the same imaging conditions as compared to the results shown in Figure 4. This clearly indicates that peptide-linked molecular beacons delivered to cells maintained their functionality during the course (~ 1–1.5 hr) of the study.
Figure 7.
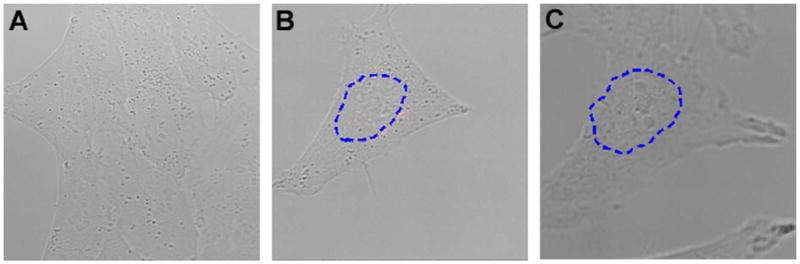
The results of negative control studies. (a) The low background signal from ‘random’ beacons in the nuclei of living cells. These negative-control molecular beacons have no target in the mammalian genome thus indicating non-specific opening of probes during delivery to and diffusion in the nucleus. (b) Fluorescence signal obtained by delivering U1-targeting molecular beacons without the NLS peptide. Only a few discrete foci in the peri-nuclear region in the cytoplasm could be observed but no significant signal in the nucleus of the living cell. (c) Fluorescence signal as a result of delivering unmodified molecular beacons targeting U3 snoRNA, indicating that without the NLS peptide attached to the U3-targeting beacons no significant signal could be detected in the nucleus or cytoplasm of a living cell.
To further demonstrate that active transport of probes from the cytoplasm to nucleus is necessary for detecting nuclear RNAs (or RNAs in cell nuclei), we did a control study in which the molecular beacons designed to target U1 snRNAs and U3 snoRNA without any NLS peptide conjugated to them were delivered into the cytoplasm using SLO. Shown in Figures 7b and 7c are respectively the resulting images of U1- and U3-targeting molecular beacons. The image in Fig. 7b shows discrete foci of U1 snRNA in the peri-nuclear region of the cytoplasm without any detectable signal in the nucleus, while no signal could be detected in the cytoplasm or in the nucleus from U3-targeting beacons (Figure 7c). This clearly indicates that without conjugation of NLS peptides, molecular beacons cannot be actively transported to the nuclei of living cells. Since U1 snRNAs are exported from nucleus for their processing before being re-imported back to the nucleus, their detection in cytoplasm is in good agreement with our earlier results. Since U3 snoRNAs are being processed in the nucleus only, not exported to the cytoplasm, they should not be detected in the cytoplasm.
DISCUSSION
To detect specific RNAs in the nuclei of living cells, we have developed peptide-linked molecular beacon, a bi-functional oligonucleotide probe that has both RNA targeting and nuclear-delivery functions. Specifically, a single peptide with an NLS sequence was conjugated to a molecular beacon that targets a snRNA or snoRNA. To internalize the peptide-linked molecular beacons into the cytoplasm of living cells, we adopted a reversible permeabilization approach using streptolysin O (SLO). In some previous studies (6, 8, 11, 12, 14), probes (including molecular beacons) were introduced into the cytoplasm of living cells by microinjection; however upon microinjection most probes were non-specifically sequestered in nucleus, and molecular beacons had non-specific opening in both cytoplasm and nucleus (7, 12).
Unlike previous molecular beacon based studies in which high background signal was present in detecting RNAs in the nuclei of cells, the results presented in this study show highly specific targeting of nuclear RNAs with very low background levels (based on ‘random’ beacon images and other negative controls). The significant difference between our results and that reported previously is believed to be a consequence of the delivery approach in introducing molecular beacons into living cells. The approach developed in this study allows for delivery of probes into the nucleus with high efficiency while maintaining the normal function of molecular beacons. Our results of detecting U1, U2 snRNAs and U3 snoRNA, together with the control studies provide strong evidence that the peptide-linked molecular beacons can be used not only to study fundamental RNA biology but also as a diagnostic imaging tool (36, 37). In combination with our previous approaches (4), this method has the potential to image transcriptional and post-transcriptional processing of RNAs in living cells, and provide an integrated picture of RNA distribution in cell nucleus and cytoplasm. Although the delivery method presented in this study was used in cell culture assays, the NLS peptide can be combined with Tat and other cell penetrating peptides (CPPs) for in vivo delivery of probes. With molecular beacons designed to have near infrared (NIR) reporter dyes, this approach can be further used to image RNAs in cell nucleus of tissue samples.
In summary, in this study we have developed a novel method that allows for direct imaging of endogenous RNAs in the nuclei of living cells. This approach combines the high specificity and signal-to-noise ratio of molecular beacons with the ability of NLS peptide to delivery effectively oligonucleotide probes into the cell nucleus. This approach can be further combined with various protein tags to develop a comprehensive understanding of RNA biogenesis, transport, translation and metabolism. This understanding will potentially lead to novel targets for therapeutic interventions in various diseases as well as significantly enhance our understanding of cell biology.
Acknowledgments
This work was supported by the NIH Roadmap Initiative in Nanomedicine through a Nanomedicine Development Center award PN2EY018244 (GB). We thank Phillip J. Santangelo for helpful discussions.
References
- 1.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–42. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 3.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei-Iida Y, Kondo S, Ishibashi K. Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys J. 2000;78:3260–74. doi: 10.1016/S0006-3495(00)76862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molenaar C, Marras SA, Slats JC, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2' O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:E89–9. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demidov VV, Frank-Kamenetskii MD. Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem Sci. 2004;29:62–71. doi: 10.1016/j.tibs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–8. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys J. 2004;87:4153–62. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlette J, Tan W. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal Chem. 2001;73:5544–50. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 13.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc Natl Acad Sci U S A. 2003;100:13308–13. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA.RNA hybridization in living cells. Proc Natl Acad Sci U S A. 1998;95:11538–43. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol1. 1992;17:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol1. 1993;21:715–27. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldherr CM, Akin D. EM visualization of nucleocytoplasmic transport processes. Electron Microsc Rev. 1990;3:73–86. doi: 10.1016/0892-0354(90)90014-j. [DOI] [PubMed] [Google Scholar]
- 18.Feldherr CM, Lanford RE, Akin D. Signal-mediated nuclear transport in simian virus 40-transformed cells is regulated by large tumor antigen. Proc Natl Acad Sci U S A. 1992;89:11002–5. doi: 10.1073/pnas.89.22.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins CA, Guthrie C. The question remains: is the spliceosome a ribozyme? Nat Struct Biol. 2000;7:850–4. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T, Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987;325:673–8. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- 21.Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980;283:220–4. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- 22.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–70. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. Embo J. 1991;10:1863–73. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matera AG, Tycowski KT, Steitz JA, Ward DC. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–99. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S, Spector DL. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci U S A. 1992;89:305–8. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange TS, Ezrokhi M, Borovjagin AV, Rivera-Leon R, North MT, Gerbi SA. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol Biol Cell. 1998;9:2973–85. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan A, Lukowiak A, Jady BE, Dragon F, Kiss T, Terns RM, Terns MP. Nucleolar localization signals of box H/ACA small nucleolar RNAs. Embo J. 1999;18:5120–30. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cogoi S, Codognotto A, Rapozzi V, Meeuwenoord N, van der Marel G, Xodo LE. Transcription Inhibition of Oncogenic KRAS by a Mutation-Selective Peptide Nucleic Acid Conjugated to the PKKKRKV Nuclear Localization Signal Peptide. Biochemistry. 2005;44:10510–10519. doi: 10.1021/bi0505215. [DOI] [PubMed] [Google Scholar]
- 29.Narayanan A, Eifert J, Marfatia KA, Macara IG, Corbett AH, Terns RM, Terns MP. Nuclear RanGTP is not required for targeting small nucleolar RNAs to the nucleolus. J Cell Sci1. 2003;16:177–86. doi: 10.1242/jcs.00176. [DOI] [PubMed] [Google Scholar]
- 30.Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell. 1992;3:555–69. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–12. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 32.Pante N, Jarmolowski A, Izaurralde E, Sauder U, Baschong W, Mattaj IW. Visualizing nuclear export of different classes of RNA by electron microscopy. Rna. 1997;3:498–513. [PMC free article] [PubMed] [Google Scholar]
- 33.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–90. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 34.Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci1. 2000;13 (Pt 11):1841–9. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- 35.Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–7. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence JB, Marselle LM, Byron KS, Johnson CV, Sullivan JL, Singer RH. Subcellular localization of low-abundance human immunodeficiency virus nucleic acid sequences visualized by fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 1990;87:5420–4. doi: 10.1073/pnas.87.14.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ambinder RF, Mann RB. Epstein-Barr-encoded RNA in situ hybridization: diagnostic applications. Hum Pathol. 1994;25:602–5. doi: 10.1016/0046-8177(94)90227-5. [DOI] [PubMed] [Google Scholar]