Abstract
Signaling between neurons requires highly specialized subcellular structures, including dendrites and axons. Dendrites exhibit diverse morphologies yet little is known about the mechanisms controlling dendrite formation in vivo. We have developed methods to visualize the stereotyped dendritic morphogenesis in living Drosophila embryos. Dendrite development is altered in prospero mutants and in transgenic embryos expressing a constitutively active form of the small GTPase cdc42. From a genetic screen, we have identified several genes that control different aspects of dendrite development including dendritic outgrowth, branching, and routing. These genes include kakapo, a large cytoskeletal protein related to plectin and dystrophin; flamingo, a seven-transmembrane protein containing cadherin-like repeats; enabled, a substrate of the tyrosine kinase Abl; and nine potentially novel loci. These findings begin to reveal the molecular mechanisms controlling dendritic morphogenesis.
Keywords: Drosophila, dendrite, genetic screen, morphogenesis, neuronal branching
The nervous system is composed of a vast number of neurons that vary in shape, size, position, and connection with other neurons. The richness in neuronal morphology is thought to reflect the richness in the functional complexity of the nervous system (Ramón y. Cajal 1911). Neurons are highly polarized cells with distinct subcellular compartments, including one or multiple dendritic processes arising from the cell body, and a single, extended axon. Dendrites can be highly branched and may account for >90% of the postsynaptic surface of some neurons (Sholl 1956). To wire a nervous system, dendrites need to have correct branching patterns. Not only are dendrites used by neurons to receive signals, but they also actively participate in the computation and storage of information (Schuman 1997; Sejnowski 1997).
Substantial progress has been made in uncovering mechanisms underlying axon guidance and target recognition (Goodman and Tessier-Lavigne 1997). In contrast, the molecular mechanisms regulating dendrite morphogenesis in vivo are not well understood. Previous studies have implicated both extrinsic and intrinsic mechanisms in this developmental process. In vertebrates, neurotrophin 3 (NT-3), brain-derived neurotrophic factor (BDNF), and nerve growth factor (NGF) can act as extrinsic factors to influence the dendritic morphology of cortical neurons (McAllister et al. 1995). Furthermore, the same neurotrophic factors can either inhibit or promote dendritic outgrowth in cortical slices, depending on the neuronal subtype (McAllister et al. 1997). Dendritic arborization of cultured neurons can also be modulated by bone morphogenetic proteins (BMPs) (Lein et al. 1995). CPG15, a putative glycosylphosphatidyl-inositol (GPI)-linked molecule, promotes dendritic growth of projection neurons in virally infected tadpoles of Xenopus laevis (Nedivi et al. 1998). In Drosophila, bipolar dendrite neurons fail to extend dendrites in glial cells missing mutants, suggesting that glial cells play a role in dendrite development (Hosoya et al. 1995; Jones et al. 1995). In addition to these extrinsic influences, intrinsic factors have been found to regulate dendritic morphology. Rho family GTPases affect external sensory neuron dendrites in fly (Luo et al. 1994), Purkinje cell spine density in mice (Luo et al. 1996), and dendrite development in cultured cortical neurons (Threadgill et al. 1997). The mitotic motor protein CHO1/MKLP1 is required for dendritic differentiation of culture neurons (Sharp et al. 1997). Endogenous CaMKII activity limits dendritic growth and stabilizes dendritic arbor structure in Xenopus (Wu and Cline 1998); the Gα protein ODR-3 regulates the morphogenesis of the olfactory cilia in Caenorhabditis elegans (Roayaie et al. 1998).
Neuronal activity can also have profound effects on dendritic morphogenesis. It has been shown that dendritic branching and the number of dendritic spines are greater in cortical neurons of rodents reared in a stimulation-enriched environment than in a deprived environment (Holloway 1966; Valverde 1967; Volkmar and Greenough 1972). A number of recent studies strengthens this notion. For example, glutamatergic activities mediated by AMPA and NMDA receptors differentially influence dendritic growth at different developmental stages in Xenopus tadpoles (Rajan and Cline 1998). Local synaptic stimulation results in the enhanced growth of filopodia-like protrusions (Maletic-Savatic et al. 1999) and increased number of spines (Engert and Bonhoeffer 1999) on dendrites of hippocampal CA1 neurons.
Systematic genetic analysis offers a powerful approach to unravel complex biological processes (Nüsslein-Volhard and Wieschaus 1980). Useful insights in understanding axon guidance and target recognition in Drosophila and C. elegans have been obtained through screens for anatomical or behavioral mutants (Hedgecock et al. 1990; Seeger et al. 1993;Van Vactor et al. 1993; Salzberg et al. 1994; Kolodziej et al. 1995; Martin et al. 1995; Zallen et al. 1998; for review, see Tear 1999). However, to date no such screen has been reported for dendrite development, due to the lack of a suitable assay system. We have chosen the embryonic peripheral nervous system (PNS) of Drosophila to study the molecular mechanisms that control dendrite morphogenesis. Neurogenesis and cell fate determination in the Drosophila PNS have been studied extensively (Jan and Jan 1993). The location, cell lineage, and dendritic morphology of these neurons have been described in some detail (Bodmer and Jan 1987; Bodmer et al. 1989; Brewster and Bodmer 1995). Analysis of dendrite morphogenesis in wild-type and mutant embryos is facilitated by the availability of neuronal specific markers (Jan and Jan 1982; Goodman et al. 1984; Zipursky et al. 1984) and a technique that allows the expression of markers in a subset of neurons (Brand and Perrimon 1993). These resources make it possible to conduct genetic screens to identify the genes involved in dendritic morphogenesis.
Here, we describe the establishment of an assay on the basis of the GAL4–UAS system (Brand and Perrimon 1993) and green fluorescence protein (GFP) to visualize the development of the dendrites of the dorsal sensory neurons in Drosophila. Dendritic outgrowth and branching are dynamic during development. Nonetheless, the complex dendritic pattern in late embryos is sufficiently invariant to suit the purpose of genetic analysis. We demonstrate that the transcription factor Prospero and the small GTPase Dcdc42 regulate dendritic morphogenesis. Furthermore, we have conducted a genetic screen and identified mutants that affect various aspects of dendrite development, including dendritic outgrowth, branching, and routing. Several of these dendritic mutants correspond to genes identified previously for other functions: kakapo (also known as short stop) encodes a large cytoskeletal protein involved in axonal outgrowth and muscle attachment (Van Vactor et al. 1993; Kolodziej et al. 1995; Gregory and Brown 1998; Strumpf and Volk 1998; Lee et al. 1999); flamingo encodes a seven-transmenbrane protein with cadherin-like repeats and is involved in planar polarity (Usui et al. 1999); and enabled encodes a substrate of the tyrosine kinase Abl, which is involved in actin polymerization (Gertler et al. 1996). Other mutants may lead to the identification of potentially novel genes important for dendrite development.
Results
An assay system to study dendrite development
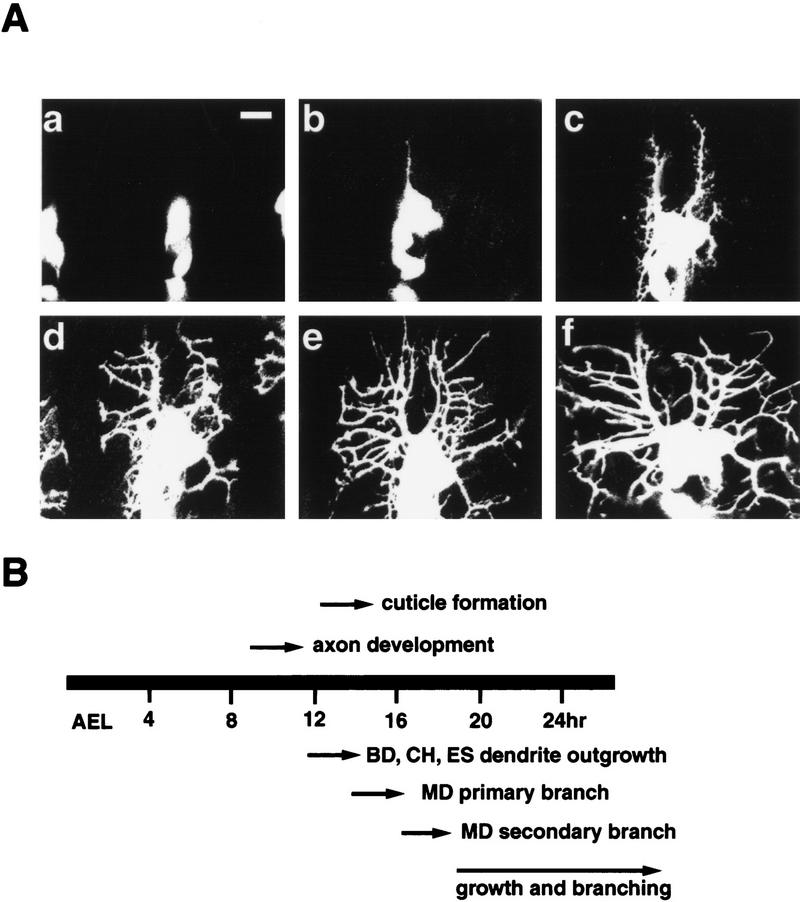
The peripheral neurons in each hemisegment of the Drosophila embryo are grouped into dorsal, lateral, and ventral clusters (Campos-Ortega and Hartenstein 1985; Ghysen et al. 1986) (Fig. 1A). The neurons within each cluster can be further classified on the basis of their dendritic morphology, external sensory (es) neurons and chordotonal (ch) neurons, each containing a single dendrite; bipolar dendrite (bd) neurons, each with two simple unbranched dendritic projections; and multiple dendrite (md) neurons with extensive dendritic arborizations. The md neurons are thought to function as touch receptors or proprioceptors to sense body surface tension or deformation (Bodmer and Jan 1987). The dendritic branching of md neurons does not begin until 16 hr after egg laying (AEL) and continues until and beyond hatching. Because impermeable cuticle already forms at 16 hr AEL, md neuron dendrites can not be visualized by standard antibody staining of whole mount embryos. It is possible to manually dissect individual embryos to allow antibody access (Bodmer and Jan 1987); however, this technique is too laborious to be useful for a large-scale mutant screen. To circumvent these technical problems, we developed an assay system on the basis of expression of GFP in living embryos. First, we screened a panel of Gal4 enhancer trap lines (E. Giniger and Y.N. Jan, unpubl.) to identify those that allow high levels of UAS-driven GFP expression in a subset of PNS neurons at the appropriate developmental stages. Of these, the Gal4 line 109(2)80 was chosen (Fig. 1A). Recombination was performed to create a second chromosome harboring both the Gal4 109(2) 80 transgene and a UAS–GFP transgene but no background lethal mutations. A fly line homozygous for the Gal4 109(2) 80/GFP chromosome (denoted as Gal4 80/GFP) was then used in this study. In the dorsal clusters of abdominal segments A1–A7, GFP expression labels both axons and dendrites of all six md neurons, one bd neuron, and one tracheal innervating neuron, but not the es neurons (Fig. 1B). In addition, high levels of GFP expression are detected in the lateral and ventral clusters, and in the antennomaxillary complex (Fig. 1A). Low levels of GFP fluorescence are also observed in a small subset of neurons in the central nervous system (CNS) (not shown). The dendrites of dorsal cluster md neurons elaborate just underneath the epidermal layer. In larvae, these dendrites as revealed by Gal4 80/GFP are in tight association with a layer of epidermal cells labeled by Krüppel–Gal4/GFP. It is thus possible to visualize the md neuron dendrites in the dorsal cluster in living animals. In this report, we focus on the development of these md neuron dendrites in wild-type as well as mutant embryos. To simplify our description, we define two types of easily detectable dendrites, dorsal branches grow toward the dorsal midline, and lateral branches grow approximately along the anterior–posterior axis toward segment boundaries (Fig. 1C).
Figure 1.
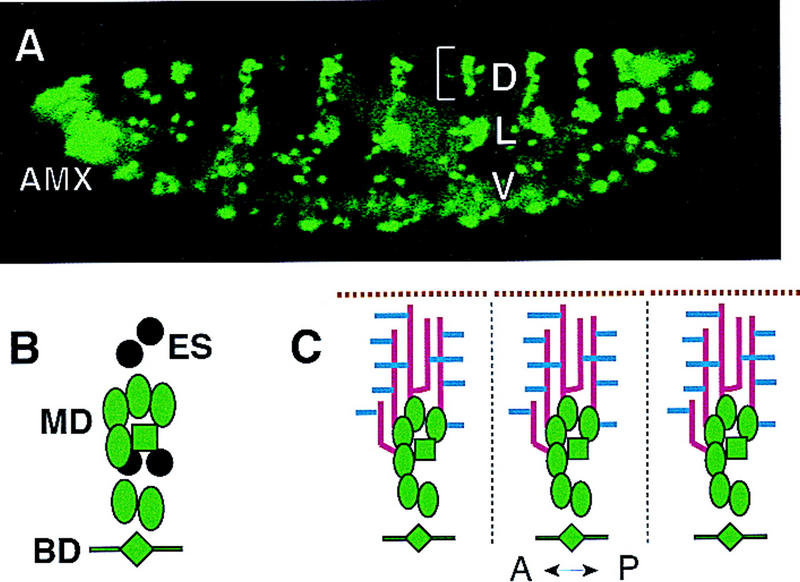
Drosophila PNS neurons in wild-type embryos labeled by GFP. (A) A lateral view of PNS neurons in a stage 17 wild-type embryo. In all figures, anterior is toward the left and dorsal is up. (AMX) Antennomaxillary complex; (D) dorsal cluster; (L) lateral cluster; (V) ventral cluster. (B) Schematic representation of all the dorsal cluster neurons bracketed in A. There are six md neurons (ovals), four es neurons (circles), one bd neuron (diamond), and one tracheal innervating neuron (square). Gal4 109(2) 80 drives UAS–GFP expression (green) in all but the four es neurons (black) in the dorsal cluster. (C) Schematic representation of the general dendritic pattern in the dorsal cluster. The lines represent: segment boundaries (broken black line), dorsal midline (broken brown line), dorsal dendrite branches (red), and lateral dendrite branches (blue).
Dorsal md neuron dendrite development
The projection pattern of md neuron dendrites in a specific hemisegment is largely invariant from embryo to embryo, on the basis of our observations on over thousands of embryos. The major characteristics of dendritic morphogenesis are summarized here. By 12–13 hr AEL, ch and es neurons have already sent out their initial dendritic projections. At this stage, bd neurons have also extended their dendrites. The primary dendrites of md neurons emerge at 13–14 hr AEL (Fig. 2Aa,b), 2 hr after the axons of PNS neurons have reached the CNS. The location of initial dendritic outgrowth and the orientation of this outgrowth are fairly invariant for md neurons (Bodmer and Jan 1987; this work). At 13 hr AEL, a dorsal dendrite first emerges from one md neuron in the anterior of the dorsal cluster, shortly after, a second dorsal dendrite emerges from a posterior md neuron of the same cluster. Both dorsal dendrites extend perpendicular to the anterior–posterior axis towards the dorsal midline. Each md neuron in the dorsal cluster only sends out one dorsally oriented primary dendrite; however, some md neurons have additional primary lateral dendrites. The dorsal extension essentially stops between 15 and 16 hr AEL, before the lateral branches start to develop. Between 15 and 17 hr AEL, numerous transient lateral branches extend and retract (Figs. 2Ab,c, and 3). These branches undergo constant remodeling (see below). Only a subset is eventually stabilized between 18 and 20 hr AEL to become the final lateral branches (Fig. 2Ad,e). At this stage, dorsal and lateral branches are clearly distinguishable. The number of lateral branches in a particular segment is similar from embryo to embryo. In addition, the anterior and posterior dorsal branches within a hemisegment are clearly separated by an area devoid of dendrites. Before hatching (23–24 hr AEL), most lateral branches further elaborate tertiary branches before and after they reach the segment boundary, but only a small number of branches cross over into neighboring segments (Bodmer and Jan 1987; this work). At hatching, the dorsal branches have not yet reached the dorsal midline so there is a clear dendrite-free zone near the dorsal midline. After hatching, the dorsal branches resume elongation and reach the dorsal midline by the second instar stage. The length and the thickness of dendritic processes continue to increase with increasing larval body size.
Figure 2.
Dorsal md neuron dendrites develop late in embryogenesis. (A) Pictures were taken from homozygous Gal4 109(2) 80-UAS–GFP embryos at different stages. (a) Dendrite budding begins at 12–13 hr AEL; (b) dorsal dendrites extend significantly by 14–15 hr AEL; (c) dorsal dendrite extension is almost complete at 16–17 hr AEL; (d) lateral dendrites continue extending and retracting at 18–19 hr AEL; (e) branching pattern is fixed by 20–21 hr AEL; (f) lateral branches cover the hemisegment before hatching (22–23 hr AEL). (B) The time line of dorsal cluster dendrite development. The tail of each arrow indicates the approximate time point at which a specific developmental process begins. Impermeable cuticle forms before dorsal branches fully extend. Bar, 10 μm.
Dendritic remodeling during development
Before hatching, the lateral branches are regularly spaced and project toward the segment boundaries. This pattern is relatively invariant from embryo to embryo for a specific hemisegment. To investigate how the dendritic patterning develops, we monitored dendrite formation in living embryos from 15 to 16 hr AEL and carried out time-lapse analysis (Fig. 3). Numerous lateral growth buds emerge anteriorly or posteriorly from the dorsal branches and then retract. Only a subset of these lateral branches elongates toward the segment boundaries and becomes stabilized. During this process, the length and orientation of dorsal branches remains largely unchanged. Numerous thin processes at the tips of the lateral branches undergo rapid extension and retraction (Fig. 3, arrow). These thin processes were not labeled by a Tau–GFP fusion protein (not shown), indicating that they might not contain microtubules. This analysis reveals that dendritic development is a dynamic process.
Figure 3.
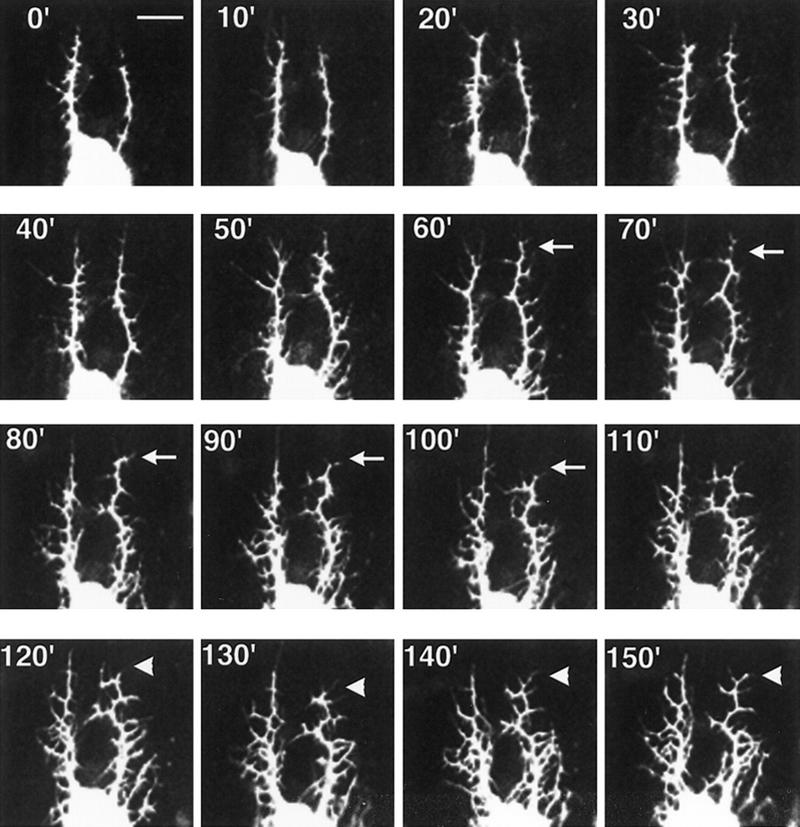
Dynamic lateral branching revealed by time lapse analysis. Dendrite development beginning at 15 hr AEL was recorded at room temperature by confocal microscopy. Pictures were taken at 10-min intervals. (Arrows) One of many areas in which dendritic retraction is observed; (arrowheads) one of many areas in which dendritic extension occurs. Bar, 10 μm.
prospero mutations cause misrouting of dorsal branches
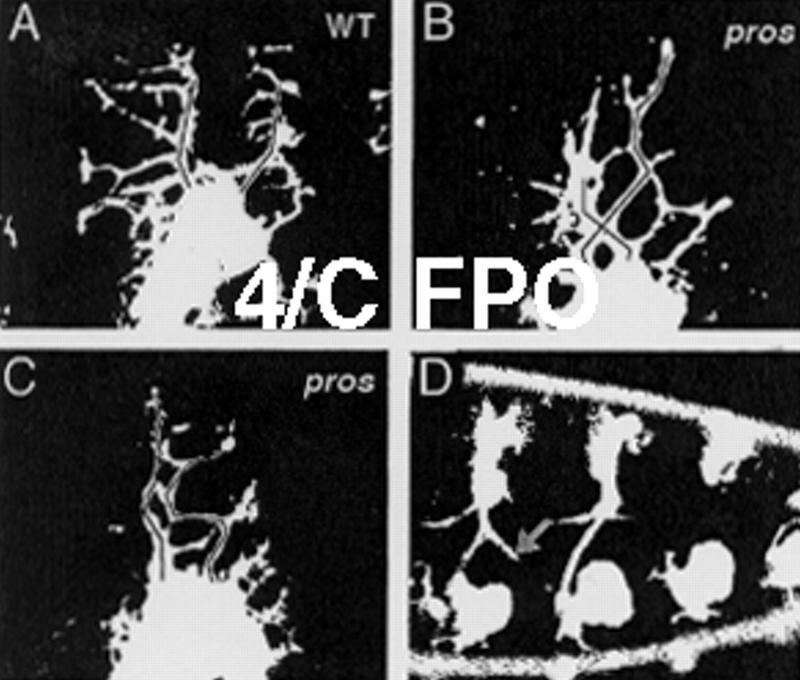
We used two approaches to identify genes involved in dendritic morphogenesis: (1) to investigate the effects of previously isolated mutations; and (2) to carry out a systematic mutant screen. We reasoned that dendrite development might share some common molecular mechanisms with axon and tracheal development, because all of these processes exhibit subcellular outgrowth and branching. Among the mutants we examined (Table 1), prospero mutants and embryos expressing a constitutively active form of Dcdc42 showed detectable dendrite phenotypes. prospero encodes a nuclear protein with multiple homopolymeric amino acid stretches and is expressed in neuronal precursor cells (Doe et al. 1991; Vaessin et al.1991). It has been suggested that prospero controls the expression of neuronal precursor genes and is required for proper neuronal differentiation (Vaessin et al. 1991). Two different alleles of prospero were used in our study, pros17 (Broadus et al. 1998) and prosjo13 (Vaessin et al. 1991). The prospero mutant embryos did not show any obvious cell fate change in the embryo PNS on the basis of available cell type-specific markers, instead they exhibited abnormal outgrowth and misrouting of axons from dorsal clusters of sensory neurons (Fig. 4D), as reported previously (Vaessin et al. 1991). In addition, we observed abnormal dendritic patterning in these mutant embryos. The anterior and posterior dorsal branches in wild-type embryos are roughly parallel to each other (Figs. 2,4A). However, in ∼70% of pros17 embryos, these branches make dramatic turns and occasionally criss-cross each other (Fig. 4B,C). A similar phenotype was observed in ∼10% of prosjo13 embryos. These studies indicate that proper development of both dendrites and axons requires the function of prospero.
Table 1.
The effects of previously isolated mutations on dendritic morphogenesis
| Mutant
|
Functions ina
|
Gross dendritic defects
|
|---|---|---|
| roundabout | axon guidance (1) | no |
| semaphorin I | axon guidance (2) | no |
| slit | axon guidance/ branching (3, 4) | no |
| branchless | tracheal development (5) | no |
| sprouty | tracheal development (6) | no |
| Dcdc42 | axon outgrowth (7) | outgrowth and branching |
| prospero | axon outgrowth/ cell fate (8, 9) | misrouting |
References: (1) Seeger et al. (1993); (2) Kolodkin et al. (1993); (3) Rothberg et al. (1990); (4) Wang et al. (1999); (5) Sutherland et al. (1996); (6) Hacohen et al. (1998); (7) Luo et al. (1994); (8) Vaessin et al. (1991); (9) Doe et al. (1991).
Figure 4.
Dendritic routing defects in prospero mutant embryos. (A) Anterior (red) and posterior (blue) dorsal branches stay clear of each other in wild-type embryos. (B,C) In prospero17 mutant embryos, dorsal branches may cross each other or veer sharply toward the other dorsal branch. (D) Outgrowth (red arrow) and routing (yellow arrow) defects of dorsal cluster axons in prospero mutant embryos.
Dcdc42 mutants exhibit supernumerary primary branches, but fewer secondary branches
Small GTPases of the Rac/Rho/Cdc42 subfamily have been implicated in interactions between the membrane and the actin cytoskeleton (Hall 1998). In yeast, the Cdc42 protein is essential for bud site assembly (Johnson and Pringle 1990). In Drosophila, expression of mutant forms of Dcdc42 can disturb axonal outgrowth and es dendrite outgrowth (Luo et al. 1994). We reasoned that this family of proteins may also function in md neuron dendritic morphogenesis. The constitutively active form of Dcdc42 was expressed in postmitotic neurons driven by Gal4 60 (Luo et al. 1994), and the dendrites were visualized with Tau–GFP (Brand 1995). The Tau–GFP signal driven by Gal4 60 is stronger than the expression of GFP driven by a single copy of Gal4 109(2) 80. As shown in Figure 5B, the dendritic pattern in the dorsal cluster was dramatically altered. Detailed examination revealed that the number of secondary branches was greatly reduced, whereas the primary branches often failed to fully extend and appear abnormally thick. Normally, the md neuron indicated by the arrow in Figure 5C only extends one dorsal primary dendrite. Expression of the constitutively active form of Dcdc42 induces more than one dorsal primary branch (Fig. 5D). The constitutively active form of Drac1 gave rise to a similar dendritic phenotype as Dcdc42, but with much lower penetrance (data not shown). Aberrant axonal outgrowth and CNS defects were also detected for both constitutively active Dcdc42 and Drac1, as reported previously (Luo et al. 1994). This study indicates that perturbation of Cdc42 and Rac GTPase activity can affect the outgrowth of primary dendrites and secondary branching.
Figure 5.
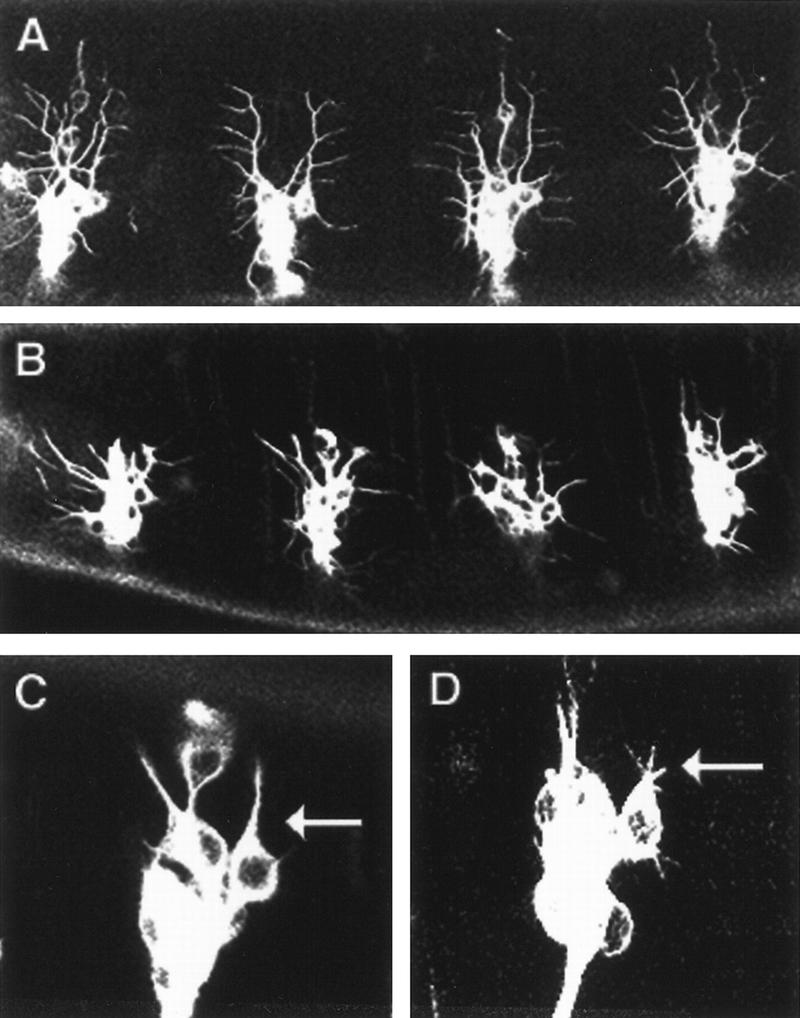
The constitutively active form of Dcdc42 promotes primary dendrite outgrowth and inhibits lateral branching. (A) Wild-type 16–18 hr AEL embryo. The neurons were labeled by Tau–GFP, which binds to microtubules. Tau–GFP expression was driven by Gal4 60. (B) 16–18 hr AEL embryo with the constitutively active form of Dcdc42 expressed in PNS neurons driven by Gal4 60. Dorsal branches fail to fully extend and the number of lateral branches is reduced. (C) One dorsal primary dendrite extends from the neuron (arrow) in wild-type 14–15 hr AEL embryo. (D) Multiple dorsal primary dendrites (arrow) extend in14–15 hr AEL embryo expressing the constitutively active form of Dcdc42.
Identification of mutants with dendritic patterning defects
The dendrites of dorsal sensory neurons labeled by GFP offer an excellent model system for identifying mutants with abnormal dendrites. To screen for mutations that alter dendritic patterning, we used EMS to mutagenize flies homozygous for the Gal4 80/GFP chromosome. We established mutant stocks using a GFP-tagged balancer in which Krüppel–Gal4 and UAS–GFP transgenes together generate a specific GFP expression pattern (D. Casso, F. Ramirez-Weber, and T.B. Kornberg, in prep.). We collected embryos from each line and examined the dendritic pattern in homozygous embryos, which could be unambiguously identified because they did not exhibit the balancer GFP expression pattern. Of 3157 lines containing lethal mutations induced by EMS on the second chromosome, 25 lines showed clearly abnormal dendritic patterns. These lines fall into 12 complementation groups and exhibit defects in several aspects of dendritic development, including dendritic outgrowth, branching, and routing. Some of these mutants exhibit defects in multiple developmental aspects. The major phenotype and the cytological location for each mutant are given in Table 2. We chose the following mutants for further study: sequoia, tumbleweed, shrub, shrinking violet, flamingo, kakapo, enabled, because they appear to have interesting dendritic phenotypes and their phenotypes could be verified when trans-heterozygous with a deficiency (Table 2). Cuticle preparations of these seven mutants did not reveal gross defects in anterior–posterior or dorsal–ventral patterning (data not shown).
Table 2.
Mutations on the second chromosome affecting dendrite development
| Mutants
|
Major dendritic phenotypes
|
Alleles
|
cytological interval
|
|---|---|---|---|
| sequoia | excessive dorsal branch outgrowth | 1 | 49C–50D |
| tumbleweed | excessive branching | 2 | 44F–45E |
| shrub | weak elaboration | 1 | 49C–50D |
| shrinking violet | reduced lateral branches | 1 | 37F–51F |
| flamingo | excessive dorsal branch outgrowth and misrouting | 9 | 47A–47B |
| kakapo | reduced lateral branches | 4 | 50A–50C |
| enabled | misrouting | 1 | 55F–56C |
| yew | misrouting | 2 | 44D |
| limber | abnormal branching pattern | 1 | 50A–51B |
| ponderosa | reduced lateral branches | 1 | N.D. |
| cypress | misrouting | 1 | N.D. |
| saguaro | reduced lateral branches | 1 | N.D. |
Cytological interval indicates the extent of the chromosomal deficiency that uncovers the mutation causing the dendritic phenotype. For some mutants [(N.D.) not determined], we did not find a deficiency in the Bloomington deficiency kit (DK-2) that uncovers the mutation.
Genes affecting dorsal branch outgrowth
In late first instar larvae, the entire dorsal surface is covered with dendrites, although dendrites from opposing hemisegments rarely intermingle with each other (Fig. 6A). By the end of embryogenesis, however, dendrites from each dorsal cluster have not reached the dorsal midline and are thus separated by a large area devoid of any dendrites (Fig. 6B). We have discovered 10 mutations in 2 complementation groups that affect the length of dorsal branches. In sequoia (seq22) mutant embryos, dorsal branches in most hemisegments fail to stop at the appropriate time, and instead continue to extend toward the dorsal midline, eventually crossing the midline and mixing with dendrites of the dorsal cluster in the other hemisegment by stage 17 (Fig. 6C). The dendritic phenotype in seq22 mutant embryos is highly penetrant and seen in every embryo examined. The dendritic phenotype in mutant embryos in which seq22 is trans-heterozygous with the deficiency covering the region is indistinguishable from that in seq22 mutant embryos, suggesting that seq22 may be a null or a strong hypomorphic allele. Similar phenotype is observed for md neurons in the lateral cluster, that is, the dendrites extend over abnormally long distances (data not shown). The failure of dorsal branches to stop before reaching the dorsal midline does not appear to reflect a general defect in dorsal positional information, because seq22 mutants exhibit no gross defects in muscle patterning or dorsal vessel formation, as indicated by anti-MEF-2 antibody staining (Fig. 6D); in midline epidermal cells, as indicated by anti-Numb antibody staining (Fig. 6E); or in anterior–posterior and dorsal–ventral patterning, as revealed by cuticle preparations (not shown). seq22 mutant embryos also exhibit axonal outgrowth defects; the axons from dorsal cluster neurons or from chordotonal neurons fail to extend toward the CNS (Fig. 6F). The axonal breaks are also seen in the CNS as revealed by anti-HRP antibody staining (data not shown). These results suggest that sequoia causes specific defects in both axons and dendrites. Even though the number of md neurons in some segments in seq22 mutant embryos is reduced, almost all of the segments, including ones with a normal number of md neurons, exhibit dendritic outgrowth phenotype, suggesting that the defects in dendritic growth is not due to the change in md neuron fates.
Figure 6.
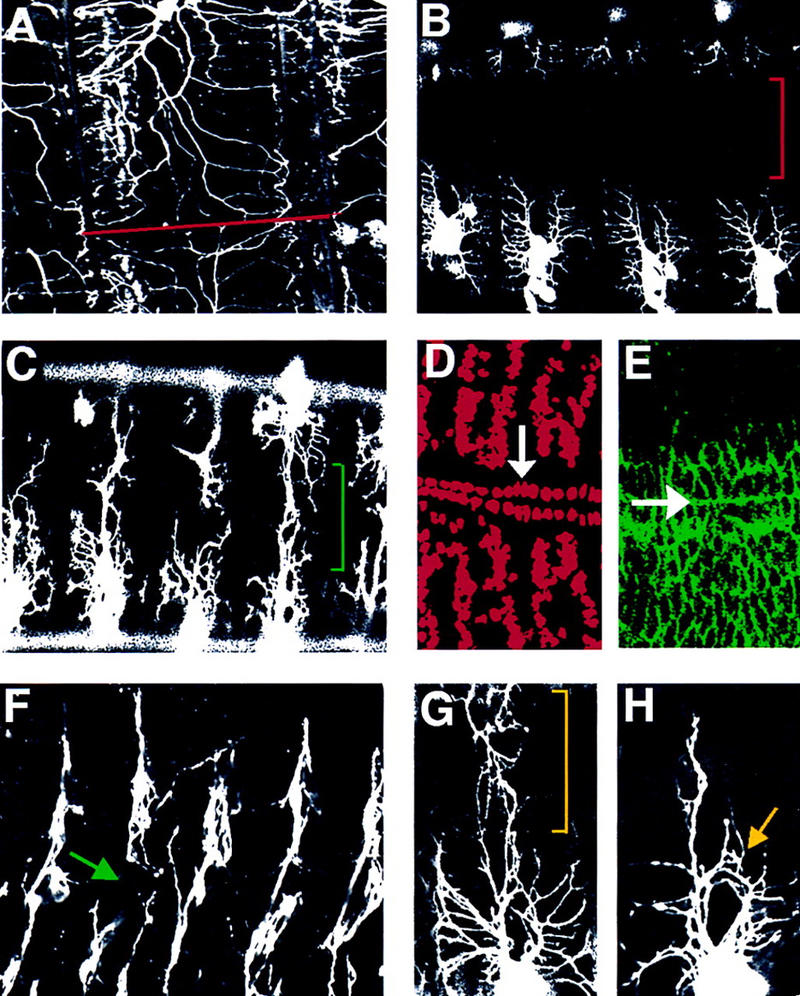
Mutations causing excessive dorsal branch outgrowth. (A) A dorsal view of a late first instar larva shows dendrites from dorsal clusters of the two hemisegments meet at the dorsal midline (red line), but do not mix extensively. (B) A dorsal view of a 20–21 hr AEL wild-type embryo shows the large area near the dorsal midline not covered by dendrites (red bracket). (C) A dorsal view of a 20–21 hr AEL sequoia (seq22) embryo reveals abnormal dendritic outgrowth (green bracket). (D) Normal muscle differentiation in seq22 embryos illustrated by anti-MEF-2 antibody staining (Red). The white arrow indicates cardioblasts of the dorsal vessel that underlie the dorsal midline. (E) Normal epidermal cell layer organization in seq22 embryos stained with anti-Numb antibody (green). The white arrow points to the dorsal midline. (F) seq22 embryos exhibit axonal outgrowth defects. The green arrow indicates the breakpoint of the axon. Dendrites in fmi72 mutant embryos exhibit aberrant dendritic outgrowth phenotype (yellow bracket) in G or misrouting phenotype (yellow arrow in H).
We identified nine mutants in another complementation group that exhibit dendritic outgrowth phenotypes similar to that of sequoia mutants. Unlike sequoia mutants that exhibit a reduced number of md neurons in some segments, this complementation group does not affect the neuron number as shown by 22C10 antibody and anti-Elav antibody staining, nor does it affect the number of md neurons. We mapped the lethal mutation to cytological region 47A–47B and found that the lethality could not be complemented by alleles of flamingo, fmiE45 and fmiE59, which have been mapped to 47B1-2 (Usui et al. 1999). Flamingo is a seven transmembrane protein containing cadherin repeats and is involved in planar polarity (Usui et al. 1999). To confirm that the dendritic phenotypes are caused by mutations in flamingo, we verified the presence of similar dendritic phenotypes in mutant embryos in which either fmiE45 or fmiE59 is trans-heterozygous with an allele in this complementation group, fmi72 (data not shown). In 70% of fmi72 mutant embryos, the dorsal branches overextend beyond the dorsal midline, intermingling with dorsal branches from the corresponding cluster in the other hemisegment (Fig. 6G). In addition, within the same embryos, dorsal dendrites exhibit misrouting defects and extend branches into the normally vacant space between the anterior and posterior dorsal branches (Fig. 6H). There is no gross defect in muscle patterning or in midline epidermal cells, as indicated by anti-MEF2 antibody and anti-Numb antibody staining . These studies demonstrate a function for flamingo in dendrite development.
Genes regulating lateral branching
The formation of lateral branches proceed in three steps, budding from the dorsal branches, extension of numerous lateral branches, and stabilization of a subset of these branches (Fig. 2). We have identified eight mutations in four complementation groups that result in reduced numbers of lateral branches in the dorsal cluster of sensory neurons. In one mutant the number of anterior lateral branches is reduced from ∼10 per hemisegment to ∼4, whereas more branches extend dorsally and appear to contact each other more extensively (Fig. 7B,C). The dendrite phenotype is seen in >90% of the mutant embryos. The individual dorsal branches may appear to have fused. This is an illusion due to the strong GFP signals. Overall, this mutation appears to primarily affect the extension of lateral branches. We mapped the lethal mutation to the cytological region between 50A3 and 50C6. The lethality of this mutant line was not complemented by l(2)k03010, l(2)k04204, l(2)k15606, l(2)k05821, l(2)k05434, l(2)k10821, and two alleles of short stop, shot1, and shot2. The shot gene has been implicated in axonal outgrowth (Van Vactor et al. 1993; Kolodziej et al. 1995; Lee and Luo 1999) and has been shown to be allelic to kakapo (Lee et al. 1999). kakapo encodes a protein of 5385 amino acids with an α-actinin-type actin-binding domain and dystrophin-like repeats (Gregory and Brown 1998; Strumpf and Volk 1998). In late embryos, kakapo is expressed predominantly at muscle attachment sites, with low levels of expression in the epidermal cell layer and in neurons (Gregory and Brown 1998; Prokop et al. 1998; Strumpf and Volk 1998). It was shown previously that the line l(2)k03010 carries a P-element insertion in an intron of kakapo, disrupting the normal kakapo expression (Gregory and Brown 1998). The other five lethal P lines that fail to complement our mutant map to the same region as kakapo. Thus, it appears that the lethal mutation we have isolated is an allele of kakapo; we named it kak30. Indeed, kak30/kakk03010embryos exhibited the same dendritic phenotype as homozygous kak30 (data not shown). In addition, md neurons in kak30/shot2 embryos also exhibited fewer lateral branches. Like other kak mutants, kak30 homozygotes exhibit defects in axonal outgrowth (data not shown). It has been reported that kakapo mutations alter the dendritic sprouting of motoneurons (Prokop et al. 1998). Taken together, these studies indicate that the kakapo gene is required for the normal development of both dendrites and axons in the CNS and PNS of Drosophila.
Figure 7.
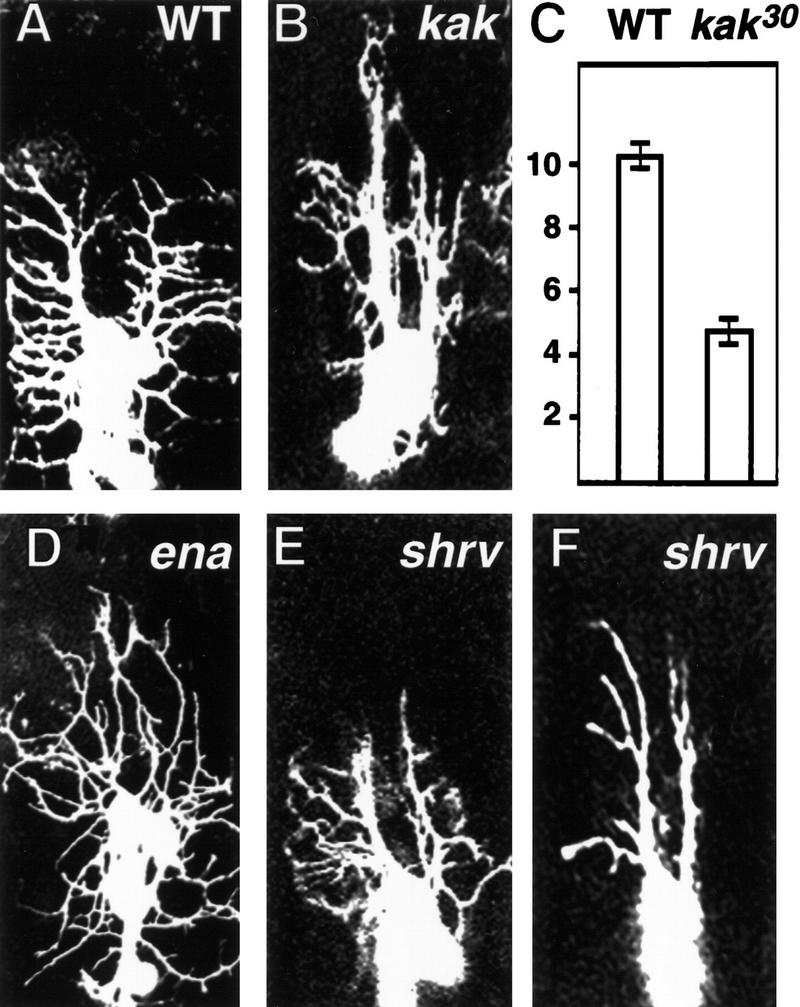
kakapo (kak30) and shrinking violet (shrv13) mutant embryos display reduced numbers of lateral branches in the dorsal cluster of the PNS. Wild-type embryos (A) have more anterior lateral branches than kak30 mutant embryos (B). (C) The numbers of anterior lateral branches for wild-type (10.2 ± 0.2) and kak30 (4.4 ± 0.2) embryos (mean±s.d. for 100 segments). (D) Some lateral branches in enabled (ena46) embryos fail to grow toward segment boundaries, but turn dorsally instead. shrv13 mutant embryos exhibit fairly normal lateral branch number at stage 16 (E) but do not maintain them, resulting in fewer lateral branches at stage 17 (E). (E,F) are representative images of dorsal clusters at different developmental stages.
Another mutant shows a dendritic phenotype that is somewhat analogous to that in kak30 mutant (Fig. 7D). The early budding and extension of lateral branches appear to be normal; however, at a late stage, some lateral branches turn dorsally instead of extending toward segment boundaries, resulting in an apparent reduction in the number of lateral branches. Approximately 60% of the mutant embryos exhibit the phenotype. The mutation that causes lethality and the dendritic phenotype is not complemented by a chromosomal deletion of the region 55F to 56C. Of the available lines carrying lethal P insertions in this region, l(2)02029 did not complement this mutant line. L(2)02029 carries a P-element insertion in the 5′ region of the gene enabled. Our mutant, which we named ena46, did not complement four different enabled alleles: ena210, enaGC5, enaGC8, and ena23 (Ahern-Djamali et al. 1998). enabled encodes an Abl tyrosine kinase substrate (Gertler et al. 1995). It has been shown that enabled cooperates with Dlar to control motor axon guidance (Wills et al. 1999). These results indicate that enabled affects dendrite as well as axon development.
In contrast to kak30, shrinking violet (shrv13) appears to affect the stabilization step of lateral branching. Between 15 and 17 hr AEL, the number of immature lateral branches and the direction of their extension seem to be roughly normal (Fig. 7E). However, between 19 and 21 hr AEL, the lateral dendrites start to retract, consequently there is a reduced number of lateral branches extending from dorsal branches, although their length and orientation stay roughly normal (Fig. 7F). This dendrite phenotype is observed in ∼85% of shrv13 embryos. The number of PNS neurons in shrv13 embryos is same as that in wild type (stained by anti-Elav antibody or by counting GFP positive cells). There is no gross defect in axonal outgrowth in both the PNS and the CNS of shrv13 embryos shown by 22C10 antibody, 49C4 antibody, and anti-HRP antibody staining. The development of epidermis and muscle patterning appears to be normal in kak30, ena46, and shrv13, judged by cuticle preparations and staining with anti-Numb and anti-MEF-2 antibodies (data not shown).
Genes affecting dendritic elaboration
Coordinated outgrowth of dorsal and lateral branches during development regulates the number and the length of these branches, so that the dorsal surface is covered with dendrites at a certain density. We have identified two genes that disrupt the coordination of dendritic patterning in apparently opposite manners. In shrub (shrb4) mutant embryos, the dorsal dendrites extend to only a fraction of the normal length and produce a few weakly elaborating lateral branches. The dendritic field in stage 17 shrb4 embryos is dramatically reduced (Fig. 8B). This strong phenotype is highly penetrant and seen in >95% of the shrb4 embryos. The epidermal layer (labeled by anti-Numb antibody), muscle patterning (shown by anti-MEF-2 antibody) appear to be normal. The number of PNS neurons in shrb4 mutant embryos is the same as that in wild-type embryos, suggesting that there is no neuron to non-neuronal cell fate change. es and ch neurons in these mutants retain their single dendrite morphology, suggesting that there is no fate change between different types of neurons. In addition, there is no detectable abnormality in both PNS and CNS axons (shown by 22C10 antibody, 49C4 antibody, and anti-HRP antibody staining). In ∼80% of tumbleweed (tum14) embryos, both dorsal and lateral dendrites branch excessively to produce numerous overlapping branches (Fig. 8C), unlike wild-type embryos, in which branching of lateral dendrites occurs only once or twice before the segment boundary is reached. tum14 embryos also exhibit axonal outgrowth and misrouting defects (Fig. 8D). Furthermore, the number of neurons (stained by anti-Elav antibody) is reduced by 25%-30% in each hemisegment in tum14 embryos. This reduction in cell number is not restricted to the PNS; both epidermal cells (stained by anti-Numb antibody) and the muscle cells (stained by anti-MEF-2 antibody) are also reduced in number. However, the sizes of these cell types are increased in every tum14 mutant embryo, whereas the size of the embryo appears to be normal (data not shown). Neuron size in CNS is also bigger than wild type, and the axon tracks are less tightly packed. Mutant embryos, in which tum14 is trans-heterozygous with the deficiency covering the region, exhibit similar phenotypes as tum14 homozygous mutant embryos.
Figure 8.
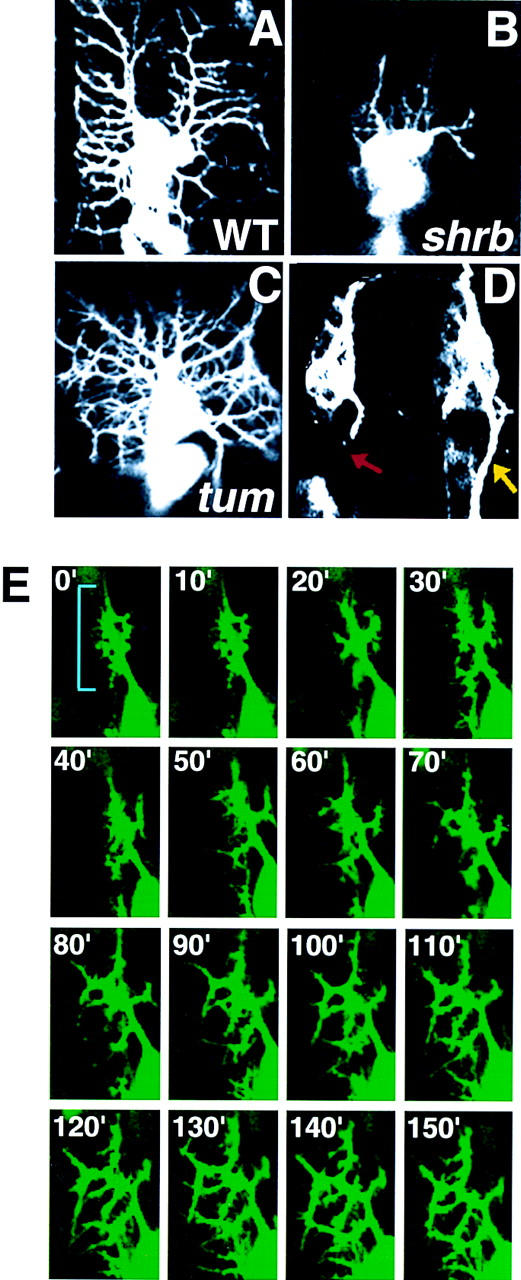
Mutations affecting dendritic branching lead to changes in density of dendrites. shrub (shrb4) mutant embryos (B) show decreased dorsal and lateral dendritic elaboration compared with wild-type (WT) embryos (A). tumbleweed (tum14) mutant embryos exhibit increased dendritic branching resulting in increased dendritic coverage per segment (C), as well as axonal outgrowth defects (D). Time-lapse analysis of dendrite development beginning at 15 hr AEL in a tum14 embryo reveals that dendrites undergo premature and excessive branching (blue bracket) prior to complete outgrowth of the dorsal primary dendrites (E). Pictures were taken at 10-min intervals and chronological sequence in E is from left to right and from top to bottom.
To further examine the dendritic branching process in tum14 mutant embryos, we carried out a time lapse analysis. Normally, numerous lateral buds emerge in a regular pattern from the dorsal branches (Fig. 3). Immature lateral branches extend and retract constantly, with only a subset of them eventually stabilized as permanent lateral branches (Fig. 2). However, in tum14 mutant embryos, many dendritic outgrowths emerge in many different directions from the primary dorsal dendrite (Fig. 8E). In addition, the branching occurs prematurely before the primary dorsal dendrite has elongated.
Discussion
Essential to proper neuronal signaling, the control of dendritic branching is a complex and important problem. Visualization of the dorsal md neurons in Drosophila embryos with GFP provides an assay system for the genetic dissection of the mechanisms controlling dendritic morphogenesis. This system has several desirable features. First, there is a wealth of knowledge about the genetic program that specifies the fate of these neurons (Jan and Jan 1993). Second, they develop elaborate dendritic branching patterns in a stereotyped manner. This branching process can be visualized and followed in living embryos in real time, thus allowing high resolution analysis in both wild-type and mutant embryos. Third, the branching process takes place in a relatively simple environment. The md neurons and their dendrites are sandwiched between the epithelial layer and the layer of body wall muscle. Thus, the dendritic branching of these neurons is essentially a two-dimensional process and is simpler to analyze than the 3-dimensional branching process often encountered in the vertebrate nervous system.
md neurons develop stereotyped patterns during embryogenesis
Dendrite development of the dorsal cluster md neurons has several invariant characteristics. At 12–13 hr AEL, dendrite morphogenesis begins. Each neuron sends out one dorsally oriented primary dendrite toward the dorsal midline. It is not known how the initial dendritic bud sites are selected in Drosophila. It is possible that the already projected axon specifies the opposite side as the site of dendrite outgrowth; alternatively, dendritic extensions may be guided by extrinsic signals from the dorsal midline in the same way that motor neuron axons are guided toward their targets (Goodman and Tessier-Lavigne 1997). These putative signaling molecules, if they exist, remain to be identified.
Between 12- and 14-hr AEL, dendrite growth is predominantly in the dorsal direction. By 16-hr AEL, dorsal dendrite extension is nearly completed. Interestingly, the dorsal dendrites stop short of reaching the dorsal midline, so that at the end of embryogenesis, there is a clear dendrite-free zone near the dorsal midline (Fig. 6B). This suggests that there is some sort of stop or repulsive signal(s). During larval stages, the dorsal dendrites resume their growth and eventually reach the dorsal midline.
Lateral branching takes place mostly after the extension of dorsal dendrites has stopped. Initial budding of lateral branches occurs from both sides of each dorsal branch. By 19–20 hr AEL, most lateral branches are oriented roughly perpendicular to the dorsal branches and project toward the closest segment boundary. Toward the end of embryogenesis, lateral branches of the anterior and posterior bundles are roughly evenly spaced and constant in number for a given hemisegment. However, earlier in development, there are numerous lateral growth buds that emerge and retract (Fig. 3). By unknown mechanisms, only a subset of the lateral growth buds that grow toward the closest segment boundary becomes stabilized to form mature lateral branches. In addition, numerous thin processes extend and retract from the tip of many immature branches. It was observed nearly a century ago by Ramón y Cajal (1911) that Purkinje cell dendrites undergo dynamic remodeling during development. The dynamic dendritic remodeling observed here in living Drosophila embryos bears some resemblance to what has been observed for Purkinje cells and may serve as a model system to dissect the underlying mechanisms.
Genetic analysis of dendritic outgrowth and branching
Our analysis of dendritic morphogenesis in wild-type Drosophila embryos raises a number of questions: (1) How are the initial budding sites for dendrite growth determined? (2) What limits each neuron to extend only one dorsally oriented primary dendrite? (3) What stops the dorsal dendrites extension prior to reaching the dorsal midline? (4) How are the spacing and the direction of the lateral branches controlled? (5) How are some of the lateral branches chosen to be stabilized? (6) Does neuronal activity have a role in modulating any of the dendritic branching processes? (7) Each md neuron has a defined receptive field. What controls the formation of the receptive field of each neuron?
To begin to address these questions, we have conducted a genetic screen searching for mutations on the second chromosome that affect dendritic outgrowth and branching. We have also examined a number of candidate genes for their possible roles in controlling dendritic morphogenesis. We estimate that we have examined >4000 of the estimated 15,000 genes in the Drosophila genome (Rubin 1998). Although the screen has not been conducted to the extent of saturating the genome, it has provided a significant sampling of genes involved in this biological process. We have found genes affecting several aspects of dendritic morphogenesis and have begun to gain some insight into the molecular mechanisms.
Unlike wild-type dorsal md neurons, which each extends only one dorsally oriented primary dendrite, expression of the constitutively active forms of Dcdc42 in neurons causes the initial outgrowth of multiple dorsal dendrites and reduces the branching of lateral dendrites. The dorsal dendrites extend dorsally after their initial outgrowth. Invariably, they stop short of the dorsal midline, suggesting the presence of certain stop signals. From our screen, we have identified two genes, flamingo and sequoia, that affect the extension and routing of dorsal dendrites. In mutants of either genes, the dorsal dendrites grow past the dorsal midline and give rise to excessively elongated dorsal branches. General dorsal–ventral patterning does not seem to be affected, as judged by the cuticle preparations (data not shown). Moreover, the patterning of the epithelial and mesodermal layers also appears to be normal (Fig. 6). These mutants may affect a putative stop signaling system. In this context, it is interesting to note that Flamingo may function in a signaling pathway because it encodes a seven-transmembrane spanning molecule with sequences indicative of G-protein coupled receptors. We found that the dendritic mutant phenotype can be partially rescued by expressing Flamingo in md neurons (unpublished data). Flamingo also has extracellular cadherin-like repeats and functions in planar polarity determination (Usui et al. 1999). The molecular nature of sequoia is presently unknown. Future cloning of genes like sequoia may lead to the identification of potential stop signals.
Dorsal dendrite growth is followed by extensive lateral branching. We have identified several genes important for different aspects of lateral dendrite development. The kak30 mutant exhibits fewer lateral branches. The CNS motoneurons in kakapo embryos also produce fewer dendritic branches (Prokop et al. 1998). kakapo encodes a large protein with an actinin-like actin-binding motif and dystrophin-like domains. It is expressed in neurons (Prokop et al. 1998) as well as at muscle attachment sites (Gregory and Brown 1998). It has been shown recently that Kakapo family proteins contain a conserved microtubule-binding domain, suggesting that these proteins may play an essential role in the integration of the microtubule network with actin cytoskeleton (Yang et al. 1999). Another mutant we found to affect lateral branching pattern is an allele of enabled. Enabled contains an SH-3 domain and is a substrate of the Abl tyrosine kinase (Gertler et al. 1995). It has been shown that Enabled family members bind to profilin and regulate actin filament dynamics (Reinhard et al. 1995; Gertler et al. 1996). The fact that proper lateral branching requires Dcdc42, enabled, and kakapo, likely regulators of the actin cytoskeleton, underscores the importance of actin in dendritic development. In contrast to kakapo mutants, md neurons in shrinking violet mutants seem to extend lateral branches normally but fail to maintain them. shrinking violet may therefore act at a later step in dendrite development.
In wild-type embryos, each md neuron has its own unique receptive field. The md neuron dendrites tend to respect the territory of neighboring neurons and avoid intermingling. For example, the anterior and posterior bundles of the dorsal dendrites do not intersect each other. Mutations in the gene that encodes the transcription factor Prospero cause misrouting of not only axons (Doe et al.1991; Vaessin et al. 1991) but also dorsal dendrites, including the crossing-over of the anterior and posterior bundles (Fig. 4; this work). Prospero may therefore function as a regulator of a machinery essential for correct routing of neuronal processes. The downstream targets of Prospero remain to be identified. The recent progress of microarray technology and Drosophila genome projects (Rubin 1998) should facilitate the identification of these target genes.
In addition to genes that affect certain specific aspects of dendrite development, mutations in several genes have rather general effects. In shrub mutants, the overall growth and branching of dendrites are much reduced. The dendritic phenotype of tumbleweed mutants is particularly intriguing. In wild-type embryos, the dendrites branch along more or less Cartesian coordinates (i.e., along dorsal–ventral and anterior–posterior axis). In contrast, the dendrites of tumbleweed mutants branch in a radial manner. It will be interesting to find out what tumbleweed encodes for and how alteration of tumbleweed function can cause such drastically different branching patterns.
Differences and similarities between dendritic and axonal development
Both dendrites and axons are elongated and sometimes branched structures. However, dendrites differ from axons in many aspects, including different cytoskeletal filament structures and the employment of different membrane protein sorting and targeting mechanisms (Craig and Banker 1994; Higgins et al. 1997; Stowell and Craig 1999). One might expect that dendritic branching and axon guidance share some but not all mechanisms. Indeed, it is the case. The transcription factor Prospero functions in both axonal and dendritic development. We also find that mutations of Dcdc42, kakapo, and enabled can alter md neuron dendritic morphology. The roles of these molecules in axonal outgrowth have been well documented (Van Vactor et al. 1993; Luo et al. 1994; Kolodziej et al. 1995; Wills et al. 1999). Several mutants identified in our screen, such as sequoia and tumbleweed, also result in abnormalities in both axons and dendrites.
We have identified several mutants that appear to affect only or predominantly dendrite development. There are no gross defects in epidermal cell and muscle cell differentiation in shrinking violet mutants, nor obvious axonal abnormalities of PNS neurons. It appears that shrinking violet specifically affects the lateral branching process. The shrub mutants exhibit dramatically reduced outgrowth of md neuron dendrites, but there is no gross defect in axonal development of the same neurons as visualized by GFP or antibody staining. However, we do not exclude the possibility that some axonal defects exist for certain neurons in these mutants. We found that a number of the genes involved in axon guidance do not appear to be involved in the formation of dendrites of the dorsal md neurons (Table 1). We note, however, that these observations do not preclude a role for these genes in the development of other dendrites.
Concluding remarks
Although some of the mutants identified from our screen correspond to known genes, the majority are probably mutations of novel genes that remain to be cloned and analyzed. Those mutants were identified on the basis of dendritic phenotypes of the dorsal md neurons, and we have not systematically studied whether these mutants exhibit abnormal dendrite formation in general, although several mutations have already been found to affect the dendrites of neurons in addition to md neurons. It is thus likely that many of the genes identified will have roles in dendritic morphogenesis of many neurons. One theme that has emerged from the recent rapid progress made in the axon guidance field is the remarkable conservation of the molecular mechanisms used to guide axons in worms, flies, and vertebrates (for review, see Goodman and Tessier-Lavigne 1997; Tear 1999). We are optimistic that the genetic analysis described here will help us understand how the formation of highly complex dendritic branching patterns, such as that of Purkinje cells in the vertebrate CNS, is controlled.
Materials and methods
Fly stocks
The following Gal4 lines were used in this study: Gal4 109(2) 80 (E. Giniger); Gal4 60 (G. Technau); and elav–Gal4 (Luo et al. 1996). The GFP-tagged Cyo balancer fly line is described (D. Casso, F. Ramirez-Weber, and T.B. Kornberg, in prep.). The fly line containing UAS–GFP on the second chromosome, deficiency lines, and lethal P insertion lines were obtained from the Bloomington Stock Center (IN), the Umea Stock Center (Sweden), or the Drosophila Genome Center (Berkeley, CA). The following fly lines were also used here: UAS–constitutively active form of Dcdc42 (Luo et al. 1994), UAS-Tau–GFP (Brand 1995), shot1 and shot2 (Kolodziej et al. 1995), pros17 (Broadus et al. 1998), ena alleles (Ahern-Djamali et al. 1998), bnl and sty (Hacohen et al. 1998), fmiE59 and fmiE45 (Usui et al. 1999).
Gal4–GFP screening and EMS mutagenesis
The Gal4 109(2)80 and UAS–GFP P elements were recombined onto the same chromosome. Mutations were induced by 20 mm EMS in males containing isogenized second chromosomes homozygous for Gal4 109(2)80, UAS–GFP (Lewis and Bacher 1968). Mutagenized males were mated en masse to yw/yw; Pin/Cyo (GFP-tagged) females and each F1 male was then backcrossed to yw/yw; Pin/Cyo (GFP-tagged) females. F2 siblings of the genotype Gal4 109(2)80 UAS–GFP/Cyo (GFP tagged) were crossed to each other to establish balanced stocks. Altogether, 3157 lethal lines (F3) were expanded. The lethal line percentage varied between 70% and 80%, indicating one to two lethal hits per chromosome if a Poisson distribution is assumed. Flies were kept in grape agar vials at 25°C overnight and embryos (F4) were collected, dechorionated with 2.5% sodium hypochlorite, then washed with 0.7% NaCl, 0.1% Triton X-100, and mounted in 90% glycerol in PBS (0.8% NaCl, 0.02% KCl, 10 mm phosphate buffer at pH 7.2). Confocal images of dendritic morphology were obtained with a Bio-Rad MRC600 confocal microscope. The fly lines exhibiting gross dendritic defects were kept for further analysis.
Mapping
We reasoned that in most cases lethality and dendrite phenotypes would be caused by the same mutation. As a first approach, we used the second chromosome deficiency kit (DK-2, Bloomington Stock Center) to identify deficiencies that uncover the EMS-induced lethal mutations. Once such a deficiency was identified, lethal P insertion lines and known mutations mapped to the region were tested for failure to complement lethality of the original EMS-induced mutations. In all cases, we confirmed the presence of a dendritic phenotype in embryos trans-heterozygous for EMS-induced mutations and P elements, deficiencies, or known mutant chromosomes; to do so, we introduced elav–Gal4 and UAS–GFP on the third chromosome to aid dendrite visualization.
Immunohistochemistry and cuticle preparations
Flies were kept overnight in yeast–grape agar vials and embryos were collected and dechorionated as described above. The embryos were fixed for 20 min at room temperature in a 1:1 mixture of heptane and 4% formaldehyde in PBS and incubated for 30 min in blocking solution containing 5% normal goat serum and 0.1% Triton in PBS. Antibodies used were monoclonal antibody mAb22C10 (provided by S. Benzer, 1:200 dilution), rabbit anti-MEF2 antibody (provided by B. Paterson, 1:500 dilution), rabbit anti-Numb antibody (1:1000 dilution) and anti-Elav antibody (1:5 dilution). Secondary antibodies used were Cy2 or Cy3-conjugated goat anti-mouse or anti-rabbit IgG (Jackson Laboratories, used at 1:200). The embryos were mounted in 90% glycerol in PBS and confocal images were obtained as described above.
Cuticle preparation was performed as descibed (Wieschaus and Nüsslein-Volhard 1986). In brief, the embryos were collected and dechorionated as described above and fixed in glycerol:acetic acid (1:1) at 60°C for 1 hour. The cuticles were visualized after incubation in Hoyers:lactate overnight at 60°C.
Time-lapse analysis
Staged embryos were collected and dechorionated as described above. The embryos were placed on top of a piece of air-permeable membrane and mounted in Halocarbon oil (River Edge, NJ). Confocal images of dendritic morphology were obtained with a BioRad MRC600 confocal microscope at 10-min intervals at room temperature.
Acknowledgments
We thank S. Benzer, A.H. Brand, D. Casso, C.Q. Doe, F.M. Hoffmann, P.A. Kolodziej, M.A. Krasnow, L. Luo, B, Paterson, T. Uemura, Bloomington Stock Center, Umea Stock Center, and Berkeley Drosophila Genome Center for antibodies and fly lines, and T. Uemura and P. Kolodziej for communication of unpublished results. We are grateful to L. Ackerman for microscopy, W. Walantus, L. Sharp, and S. Barbel for technical assistance, S. Younger and other laboratory members for suggestions. We also thank C. Bargmann and Y.-M. Chan for critical reading of the manuscript. F.-B.G. is supported by the Burroughs Wellcome Fund and the American Cancer Society, California Chapter. J.E.B. is supported by the Damon Runyon-Walter Winchell Foundation. L.Y.J. and Y.N.J. are Howard Hughes Investigators.
The publlication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked 'advertisement' in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ynjan@itsa.ucsf.edu; FAX (415) 476-5774.
References
- Ahern-Djamali SM, Comer AR, Bachmann C, Kastenmeier AS, Reddy SK, Beckerle MC, Walter U, Hoffmann FM. Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer R, Jan YN. Morphological differentiation of the embryonic peripheral neurons in Drosophila. Wilhelm Rous's Arch Dev Biol. 1987;196:69–77. doi: 10.1007/BF00402027. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Carretto R, Jan YN. Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron. 1989;3:21–32. doi: 10.1016/0896-6273(89)90112-8. [DOI] [PubMed] [Google Scholar]
- Brand AH. GFP in Drosophila. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brewster R, Bodmer R. Origin and specification of type II sensory neurons in Drosophila. Development. 1995;121:2923–2936. doi: 10.1242/dev.121.9.2923. [DOI] [PubMed] [Google Scholar]
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Berlin, Germany: Springer; 1985. [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes & Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;18:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudière C, Aceves E, Jan LY, Jan YN. Sensory neurons and peripheral pathways in Drosophila embryos. Wilhelm Roux's Arch Dev Biol. 1996;195:281–289. doi: 10.1007/BF00376060. [DOI] [PubMed] [Google Scholar]
- Goodman CS, Tessier-Lavigne M. Molecular mechanisms of axon guidance and target recognition. In: Cowan WM, Jessell TM, Zipursky L, editors. Molecular and cellular approaches to neural development. Oxford, UK: Oxford University Press; 1997. pp. 108–178. [Google Scholar]
- Goodman CS, Bastiani MJ, Doe CQ, du Lac S, Helfand SL, Kuwada JY, Thomas JB. Cell recognition during neuronal development. Science. 1984;225:1271–1279. doi: 10.1126/science.6474176. [DOI] [PubMed] [Google Scholar]
- Gregory SL, Brown NH. kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J Cell Biol. 1998;143:1271–1282. doi: 10.1083/jcb.143.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sporuty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Higgins D, Burack M, Lein P, Banker G. Mechanisms of neuronal polarity. Curr Opin Neurobiol. 1997;7:599–604. doi: 10.1016/s0959-4388(97)80078-5. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- Holloway RL. Dendritic branching: Some preliminary results of training and complexity in rat visual cortex. Brain Res. 1966;2:393–396. doi: 10.1016/0006-8993(66)90009-6. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: A binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. The peripheral nervous system. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. pp. 1207–1244. [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. Glial cells missing: A genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- Kolodziej PA, Jan LY, Jan YN. Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron. 1995;15:273–286. doi: 10.1016/0896-6273(95)90033-0. [DOI] [PubMed] [Google Scholar]
- Lee S, Harris K, Kolodziej P. 40th annual Drosophila research conference, Bellevue, WA. 1999. [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lein P, Johnson M, Guo X, Rueger D, Higgins D. Osteogenic protein-1 induces dendritic growth in rat sympathetic neurons. Neuron. 1995;15:597–605. doi: 10.1016/0896-6273(95)90148-5. [DOI] [PubMed] [Google Scholar]
- Lewis EB, Bacher F. Method of feeding ethyl methane sulfonate to Drosophila males. Dros Inf Serv. 1968;43:193. [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes & Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Martin KA, Poeck B, Roth H, Ebens AJ, Ballard LC, Zipursky SL. Mutations disrupting neuronal connectivity in the Drosophila visual system. Neuron. 1995;14:229–240. doi: 10.1016/0896-6273(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Prokop A, Uhler J, Roote J, Bate M. The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophila motorneurons. J Cell Biol. 1998;143:1283–1294. doi: 10.1083/jcb.143.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan I, Cline HT. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivio. J Neurosci. 1998;18:7836–7846. doi: 10.1523/JNEUROSCI.18-19-07836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón. Cajal S. Histology of the nervous system of man and vertebrates, 1995 translation. Oxford, UK: Oxford University Press; 1911. [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. Slit: An extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes & Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- Rubin GM. The Drosophila genome project: A progress report. Trends Genet. 1998;14:340–343. doi: 10.1016/s0168-9525(98)01568-6. [DOI] [PubMed] [Google Scholar]
- Salzberg A, D'Evelyn D, Schulze KL, Lee JK, Strumpf D, Tsai L, Bellen HJ. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- Schuman EM. Synapse specificity and long-term information storage. Neuron. 1997;18:339–342. doi: 10.1016/s0896-6273(00)81234-9. [DOI] [PubMed] [Google Scholar]
- Seeger M, Tear G, Ferres-Marco D, Goodman CS. Mutations affecting growth cone guidance in Drosophila: Genes necessary for guidance toward or away from the midline. Neuron. 1993;10:409–426. doi: 10.1016/0896-6273(93)90330-t. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ. The year of the Dendrite. Science. 1997;275:178–179. doi: 10.1126/science.275.5297.178. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Yu W, Ferhat L, Kuriyama R, Rueger DC, Baas PW. Identification of a microtubule-associated motor protein essential for dendritic differentiation. J Cell Biol. 1997;138:833–843. doi: 10.1083/jcb.138.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. The organization of the cerebral cortex. London, UK: Methuen; 1956. [Google Scholar]
- Stowell JN, Craig AM. Axon/dendrite targeting of metabotropic glutamate receptors by their cytoplasmic carboxy-terminal domains. Neuron. 1999;22:525–536. doi: 10.1016/s0896-6273(00)80707-2. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Volk T. Kakapo, a novel cytoskeletal-associated protein is essential for the restricted localization of the neuregulin-like factor, vein, at the muscle-tendon junction site. J Cell Biol. 1998;143:1259–1270. doi: 10.1083/jcb.143.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Tear G. Neuronal guidance. A genetic perspective. Trends Genet. 1999;15:113–118. doi: 10.1016/s0168-9525(98)01686-2. [DOI] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates epithelial planar polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- Valverde F. Apical dendritic spines of the visual cortex and light deprivation in the mouse. Exp Brain Res. 1967;3:337–352. doi: 10.1007/BF00237559. [DOI] [PubMed] [Google Scholar]
- Yan Vactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Greenouch WT. Rearing complexicity affects branching of dendrites in the visual cortex of the rat. Science. 1972;176:1445–1447. doi: 10.1126/science.176.4042.1445. [DOI] [PubMed] [Google Scholar]
- Wang KH, Brose K, Arnott D, Kidd T, Goodman CS, Henzel W, Tessier-Lavigne M. Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila: A practical approach. Oxford, UK: RL Press; 1986. pp. 199–286. [Google Scholar]
- Wills Z, Bateman J, Korey CA, Comer A, Van Vactor D. The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron. 1999;22:301–312. doi: 10.1016/s0896-6273(00)81091-0. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- Yang Y, Bauer C, Strasser G, Wollman R, Julien J-P, Fuchs E. Integrators of the cytoskeleton that stabilize microtubules. Cell. 1999;98:229–238. doi: 10.1016/s0092-8674(00)81017-x. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Yi BA, Bargmann CI. The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell. 1998;92:217–227. doi: 10.1016/s0092-8674(00)80916-2. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Venkatesh TR, Teplow DB, Benzer S. Neuronal development in the Drosophila retina: Monoclonal antibodies as molecular probes. Cell. 1984;26:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]