Summary
The botulinum neurotoxins (BoNTs) are the most potent protein toxins for humans. There are seven serotypes of BoNTs (A-G) based on a lack of cross anti-sera neutralization. BoNT/C and BoNT/D serotypes include mosaic toxins that are organized as D-C and C-D toxins. One BoNT D-C mosaic toxin, BoNT/D-South Africa (BoNT/D-SA), was not fully neutralized by immunization with a vaccine composed of either prototype BoNT/C-Stockholm or BoNT/D-1873. While several BoNT serotypes utilize dual receptors (gangliosides and proteins) to bind and enter neurons, the basis for BoNT/C and BoNT/D entry into neurons is less well understood. Recent studies solved the crystal structures of the receptor binding domains of BoNT/C, BoNT/D, and BoNT/D-SA. Comparative structural analysis showed that BoNT/C, BoNT/D, and BoNT/D-SA lacked components of the ganglioside binding pocket that exist within other BoNT serotypes. Utilizing structure based alignments, biochemical analyses, and cell binding approaches, BoNT/C and BoNT/D-SA have been shown to possess a unique ganglioside binding domain, the ganglioside binding loop. Defining how BoNTs enter host cells provides insight towards understanding the evolution and extending the potential therapeutic and immunologic values of the BoNT serotypes.
Introduction
The Botulinum neurotoxins (BoNTs) are the most potent protein toxins for humans and the etiological agent of botulism [1]. BoNTs are produced by Clostridium botulinum and several other species of clostridia [2]. The BoNTs are grouped into 7 serotypes (termed A-G) based upon anti-sera neutralization [3]. Serotypes A, B, E, and F are associated with natural human intoxication, while serotypes C and D are associated with natural intoxication of animals.
BoNTs are AB toxins composed of independent functional domains linked by disulphide bond. The N-terminal light chain (LC) is the enzymatic domain, while the heavy chain (HC) includes two independent domains, the translocation domain (HCT) and the receptor binding domain (HCR) (Figure 1). The crystal structure of BoNT/A revealed that the three functional domains were structurally distinct and arranged in a linear fashion [4]. The LC protease active site is composed of a zinc atom coordinated by a HExxH…E motif that can access SNARE proteins as substrates. The identifying features of the HCT include a pair of α-helices ~105Å long that twist around each other and a “belt” region within the N terminus of the HC that wraps around the LC partially occluding the active site [5]. The HCR consists of two sub-domains; the N-terminal subdomain is predominantly β-sheets arranged into a jelly roll motif and the C-terminal sub-domain folds into a β-trefoil. The structures of each BoNT serotype have similar three dimensional organization [6]. Using single particle electron microscopy, Fischer et al. visualized the holotoxin architecture, which revealed a heterogeneous unique globular organization for BoNT/E, in contrast to homogenous conformation for BoNT/A that reflects the crystal structure [4, 7]. A recent crystal structure showed that BoNT/E is composed of three independent structural domains like BoNT/A and /B, but unlike BoNT/A and BoNT/B where the LC and HCR are separated by the HCT, BoNT/E forms a compact, globular structure with the three domains in direct contact with each other [8].
Figure 1. Structure-Function Organization of the Botulinum Neurotoxins.
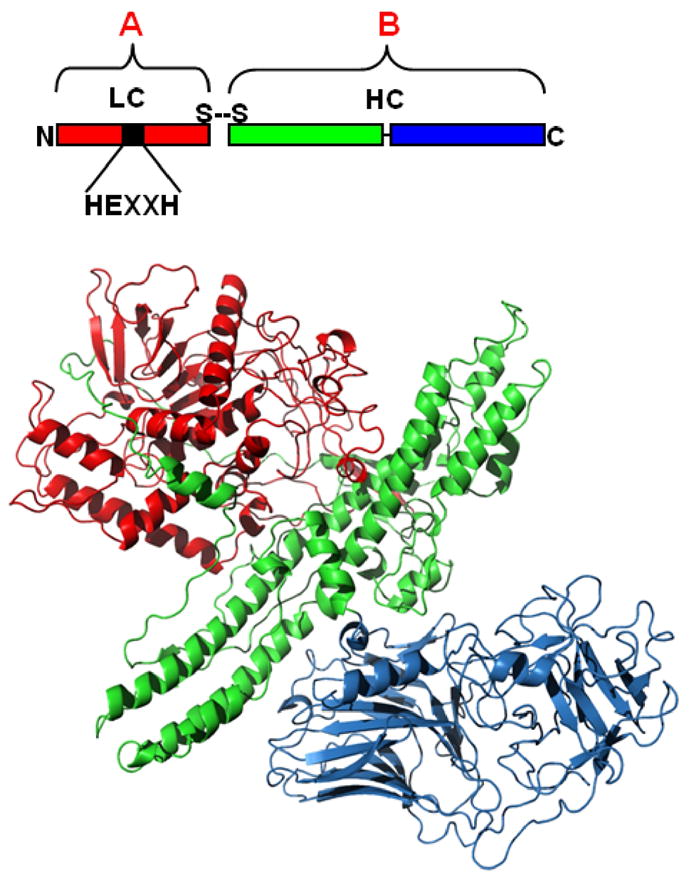
(Upper Panel) BoNTs are AB toxins composed of independent functional domains linked by a disulphide bond. The N-terminal light chain (LC, Red) encodes the enzymatic domain, while the heavy chain (HC) encodes two independent domains, the receptor binding domain (HCR, Blue) and the translocation domain (HCT, Green). (Lower Panel) Crystal structure of BoNT/A showed three functional domains: LC (Red), (HCR (Blue), and HCT (Green). PDB:3BTA; solved by Lacy and Stevens [4].
Retention of function by the three individual domains (LC, HCT, and HCR) has facilitated the structure-function characterization of BoNT and Tetanus neurotoxin (TeNT). The molecular identities of the LC substrate(s) as well as the structural intricacies of substrate recognition have been defined using recombinant LC domains [9, 10]. Using a catalytically inactive LC/A and a deletion peptide of SNAP-25, Breidenbach et al. generated a crystal structure of LC/A bound to SNAP-25 [5]. The structure revealed that LC-SNAP-25 interactions extended through multiple sites with SNAP-25 wrapping around a cleft that spanned the circumference of LC/A. The N terminus of SNAP-25 assumed a helical conformation when contacted to a hydrophobic region of LC/A termed the α-exosite, while the C terminus of SNAP-25 interacted with the β-exosite located on the opposite face of LC/A. The region of SNAP-25 between the α- and β-exosites of LC/A was positioned to align the catalytic active site with the scissile bond of SNAP-25. Unexpectedly, SNAP-25 wraps around LC/A in an orientation similar to the belt region of the translocation domain of the HC. This implicates the belt region as a safeguard against premature proteolysis until LC is delivered into the host cell [10]. Similarly, LC/F interacts with VAMP by binding with three exosites and wrapping the SNARE protein in the same direction as LC/A [11, 12]. Building on biochemical and structural data regarding LC-SNARE protein interactions [13], a LC/E was engineered to cleave the non-neuronal SNARE protein SNAP-23. Incorporation of a point mutation at Lys224 expanded LC/E substrate specificity, where LC/E(Lys224Asp) cleaved endogenous SNAP-23 in HeLa cells and effectively reduced TNF-α induced mucin and IL-8 secretion [14]. Studies addressing the role of the translocation domain in BoNT toxicity have also benefited from the use of recombinant BoNT domains [15-18]. Recombinant HCR domains have been utilized for structural and cellular studies and have been shown to represent minimal essential components required to host cell interactions [19, 20]. The C-terminal β-trefoil domain alone contains known receptor binding sites and has been shown to retain ganglioside interactions for TeNT and BoNT/A [21, 22], although binding SV2 and neuronal entry has not yet been demonstrated for BoNT/A. HCR domains bind and enter target neurons and antagonize the action of full-length BoNTs [23], while co-crystal structural studies have defined the HCR-receptor interactions with atomic resolution [24-30].
Entry of Botulinum Toxins into Neurons
When interactions with gangliosides (sialylated glycosphingolipids) were not sufficient to explain the affinity and specificity of BoNTs for nerve terminals, Montecucco proposed a dual receptor model for BoNTs, inferring the presence of a protein co-receptor to facilitate entry [31]. Nishiki and colleagues subsequently identified synaptotagmin II as a functional protein receptor for BoNT/B in complex with ganglioside GT1b [32-35], while Rummel et al found that synaptotagmins I and II facilitate BoNT/G entry [36]. Several groups showed that neuronal stimulation led to rapid BoNT toxicity, prior to the identification of functional BoNT protein receptors [37, 38], when Chapman and coworkers showed that BoNT/B entered neurons bound to synaptotagmin upon membrane depolarization [39]. Subsequent studies showed that BoNT/A, BoNT/E, and BoNT/F utilized SV2 as protein receptor and that BoNT/G utilized synaptotagmin I and II as co-receptor [36, 40-42]. Thus, the BoNT protein co-receptor comprises luminal domains of synaptic vesicle membrane proteins exposed through fusion of the synaptic vesicle to the plasma membrane [40-42]. The general entry mechanism of BoNT is shown in Figure 2. BoNT initially binds ganglioside on the plasma membrane of resting neurons. A depolarization event triggers an influx of extracellular calcium which is recognized by the cytoplasmic calcium-binding domains of synaptotagmin on synaptic vesicles. This initiates vesicle fusion to the plasma membrane where luminal domains of synaptic vesicle proteins are exposed and function as the protein co-receptors for BoNT. Recent studies have identified a synaptic vesicle protein complex as a high affinity receptor for BoNTs [43, 44]. Upon BoNT binding, plasma membrane bound synaptic vesicles are recycled by an endocytic mechanism [45] and the BoNT-receptor complex is sequestered into the lumen of the vesicle. Upon maturation, the lumen of the synaptic vesicle is acidified by the H+ v-ATPase. Acidification triggers insertion of the HCT domain of BoNTs into the synaptic vesicle membrane which facilitates translocation of LC into the cytosol.
Figure 2. Entry of BoNTs into Neurons.
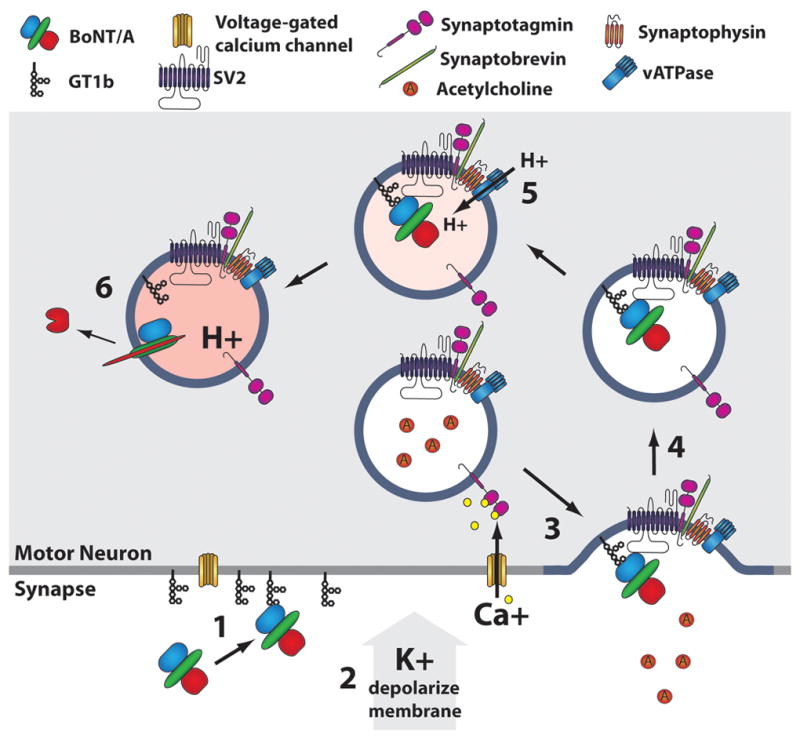
Several BoNT serotypes enter neurons upon membrane depolarization, using BoNT/A several steps that can be resolved include: Step 1 HCR of BoNT/A binds GT1b on the plasma membrane of unstimulated neuron (blue). Step 2 Membrane depolarization, elicited in cultured cells using elevated extracellular potassium, triggers the opening of voltage-gated calcium channels, allowing influx of calcium. Step 3 Intracellular calcium binds synaptotagmin I/II, located in isolation and in complex with SV2, which signals for fusion of synaptic vesicles to the plasma membrane. Vesicle fusion exposes L4 loop of SV2, the protein receptor for BoNT/A. The HCR binds GT1b and SV2 simultaneously. Step 4 Complexes of synaptic vesicle proteins are endocytosed to be recycled. Step 5 The vacuolar ATPase acidifies the lumen of the synaptic vesicle. Step 6 The acidic environment triggers insertion of the HCT domain which facilitates translocation of a partially unfolded LC (red) through a channel made by HCT (green). Once in the cytoplasm, LC cleaves SNAP-25.
How BoNT/C and BoNT/D enter neurons is less clear. The dependence of gangliosides for entry has been demonstrated for BoNT/C and BoNT/D [46-48]. Recently an unidentified synaptic vesicle protein was suggested as a receptor for BoNT/C based on depolarization-dependent toxicity, while BoNT/D has been proposed to utilize two carbohydrates for entry, although it, too, responds to depolarization [46, 48]. This review will describe the history and our current understanding of the entry of BoNT/C and BoNT/D into neurons.
BoNT/C and BoNT/D
BoNT/C and BoNT/D are not typically associated with human intoxications [49]. Although not toxic to humans following ingestion [50], BoNT/C is toxic for human tissues and cleaves SNAP25 and syntaxin in human neurons [51]. BoNT/C was initially isolated in 1922 and determined to be responsible for avian botulism [52-54]. A role for gangliosides in BoNT/C intoxication was supported by studies with mouse knockouts that were deficient in complex gangliosides that were more resistant to BoNT/C intoxication than wild type mice and by the observation that BoNT/C directly bound gangliosides GD1b and GT1b [47] . Conversely, binding of BoNT/C to neuronal cell lysates is insensitive to proteinase K [43, 47, 48]. A co-receptor for BoNT/C has yet to be identified, although Rummel et al demonstrated an increase in toxicity upon stimulation, which indicates entry may be synaptic-vesicle specific. Furthermore, BoNT/C competed with BoNT/E and F in a mouse hemidiaphragm paralysis experiment, although whether these serotypes compete for a protein or ganglioside is still unclear [48].
BoNT/D-1873 was initially observed in 1929 in cattle and remains associated with animal botulism [55]. Human intoxication by BoNT/D has not been reported, and one study indicated toxicity in human tissues was not observed at concentrations sufficient for BoNT/A toxicity [51]. A discrepancy arose regarding BoNT/D and ganglioside interactions due to an early study demonstrating gangliosides competing BoNT/D toxicity [56], while later studies did not detect BoNT/D binding to gangliosides, but reported the direct binding of BoNT/D to phosphatidylethanolamine derivatives [47]. The requirement of gangliosides was most recently demonstrated through the use of hemidiaphragm preparations from mice lacking β-1,4-N-acetylgalactosamine transferase and GD3 synthetase, which were partially resistant to toxicity [46]. The current model by Binz and colleagues proposes two carbohydrate binding sites [46]; the role of phospholipid binding remains unclear. In addition, BoNT/D toxicity is increased with neuronal stimulation, suggesting at least one receptor is specific to synaptic vesicles [48].
In addition to the BoNT prototypic serotypes, BoNT/C-Stockholm and BoNT/D-1873 mosaic toxins have been reported, which comprise D-C and C-D structural organization. These mosaic toxins appear to have originated from recombination events, presumably through a phage-mediated mechanism, since the genes encoding BoNT/C and BoNT/D are located within phage [55]. BoNT/D-South Africa (BoNT/D-SA) is a D-C mosaic toxin that has attracted interest with the observation that mice immunized with HCR/C were only partially protected from BoNT/D-SA challenge, while immunization with HCR/D did not protect from BoNT/D-SA challenge [57-60]. Thus, the study on BoNT/D-SA may provide information on the immunological protection elicited by the HCR domains.
The primary amino acid homology among BoNT/C-Stockholm, BoNT/D-1873 and BoNT/D-SA are shown in Table 1. This alignment showed that BoNT/D-SA is a mosaic composed of the LC and HCT of BoNT/D and the HCR of BoNT/C. While the LC and HCT showed a high amount of homology between the respective domains of BoNT/D-SA and BoNT/C and BoNT/D, there was limited identity between the C-terminal sub-domains of the HCRs of BoNT/D-SA with BoNT/C (62%). This indicated that BoNT/D-SA and BoNT/C had undergone considerable genetic drift since the generation of BoNT/D-SA. [61, 62], The genetic divergence within the C terminus of BoNT/D-SA supports a role for the HCR in eliciting a protective immune response to BoNT intoxication which is consistent with the inability of HCR/C or HCR/D vaccination to completely protect against challenge by BoNT/D-SA [57].
Table 1.
Amino acid identity among BoNT/C, BoNT/D, and BoNT/D-SA
| Identity with BoNT/D-SA (%)A | ||||
|---|---|---|---|---|
| LC | HCT | HCRN | HCRC | |
| BoNT/C | 47 | 70 | 90 | 62 |
| BoNT/D | 98 | 95 | 50 | 24 |
Structures of HCR domains of BoNT/C, BoNT/D, and BoNT/D-SA
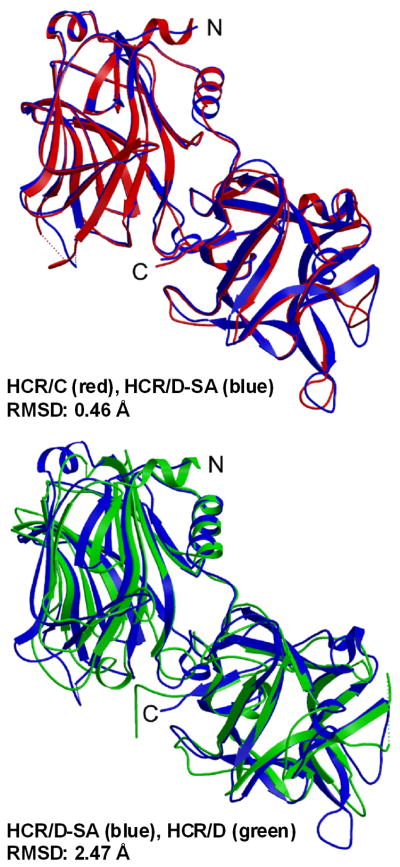
The crystal structures of HCR/C, HCR/D, and HCR/D-SA (Figure 3) [62] show a conservation of structure among each other as well as among other BoNT serotypes [4, 8, 26, 63, 64]. The HCRs are organized into two sub-domains, an N-terminal jelly-roll domain and a C-terminal β-trefoil domain. The root mean square deviation (RMSD) was 2.5 Å for HCR/D-SA and HCR/D and 0.5 A for HCR/D-SA and HCR/C. The greater RMSD for HCR/D-SA and HCR/D was due to the different bent angle between the N-terminal jelly-roll domain and C-terminal β-trefoil domain, which perturbed the calculated RMSD values between the two HCR. The majority of structural divergence between HCR/D-SA and HCR/C and HCR/D is primarily within the C-terminal sub-domain, specifically within the loops of the C-terminal β-trefoil domain, which includes the Ganglioside Binding Pocket (GBP) described within other BoNT serotypes [28].
Figure 3. Crystal structures of HCR/C, HCR/D, and HCR/D-SA.
Shown are overlays of the crystal structure of the HCR/D-SA (blue) with HCR/C (left panel, red) RMSD: 0.46 Δ and HCR/D-SA (blue) with HCR/D (right panel, green) RMSD: 2.47Δ. PDB: HCR/C, 3N7K; HCR/D, 3N7J; HCR/D-SA, 3N7L. Reproduced from [62] with permission.
A structure based alignment showed that the overall C-terminal β-trefoil domain was conserved among BoNT/C, BoNT/D, and BoNT/A. Using HCR/A and the corresponding amino acid residues the main chain can be traced. From the C terminus, the main chain of the HCRs proceeds from a conserved internal Trp-Phe towards the N terminus, upon emerging from the interior, the main chain forms the helical conformation of the GBP (Figure 4). Three residues, Tyr1267, Trp1266, and Ser1264 (HCR/A), are present on the N-terminal side of the GBP helix and contribute to ganglioside binding, while Glu1203 (HCR/A) also participates in ganglioside binding [65]. As the main chain continues towards the N terminus, a β-hairpin loop is formed and continues into an anti-parallel-β-sheet which aligns adjacent to Trp1266 of the GBP and includes His1253 (HCR/A), which also contributes to the GBP. The regions within HCR/C and HCR/D that are analogous to the GBP have an overall structural similarity to HCR/A, however, neither HCR/D nor HCR/C contain the conserved Trp, Ser, or His. Thus, while the main chain organization of the GBP is conserved, residues that contribute to ganglioside binding are absent in BoNT/C and BoNT/D. This implicated a unique mechanism for ganglioside binding by BoNT/C and BoNT/D. The structure based alignment shows that, following the GBP, HCR/C and HCR/D form Trp-containing β-loops similar in size the hydrophobic loop in HCR/B. This loop represents a novel ganglioside binding region termed the Ganglioside Binding Loop (GBL) (Figure 5) [62]. While each BoNT serotype contains a loop that corresponds to the GBL, the loops vary in size and composition. For example, HCR/A has a β-finger (i.e., a two-residue turn) with several hydrophobic residues toward the end of the finger followed by two consecutive Asn at the tip of the finger which is exposed to solvent, and HCR/B has a loop more structurally analogous to HCR/C and HCR/D, but lacking a Trp.
Figure 4. Ganglioside Binding Pocket of HCR/A overlay with HCR/D-SA.
HCR/D-SA (blue) includes conserved internal Phe1280 and Trp1282 (grey), and corresponding residues that represent the Ganglioside Binding Pocket of HCR/A (green). Enlarged views of the Ganglioside Binding Pocket of HCR/A (lower) and the corresponding region of HCR/D-SA (upper) are shown. Residues that contribute to ganglioside binding of HCR/A (Glu1203, His1253, Ser1264, Trp1266 and Tyr1267) and corresponding residues within HCR/D-SA are shown. Reproduced from [62] with permission.
Figure 5. Ganglioside Binding Loops of HCR/C, HCR/D, and HCR/D-SA overlay with HCR/A and HCR/B.
HCR/D-SA (blue, upper) includes conserved Phe1280 and Trp1282 (black), and the Ganglioside Binding Loop (GBL) is enlarged, rotated, (lower) and aligned with the structurally analogous β-hairpin loops of BoNT/A (purple), /B (orange), /C (red), /D (green). HCR/C and /D-SA loops. Note, BoNT/B has an extended β-hairpin loop like HCR/C, HCR/D, and HCR/D-SA, but lacks a tryptophan residue. BoNT/A, in contrast, does not have an extended β-hairpin loop. Reproduced from [62] with permission.
Ganglioside binding by HCR/C and HCR/D-SA
Early studies showed that HCR/C bound GD1b and GT1b [47]. Quantitative binding assays showed that HCR/C bound GD1b with the highest affinity, followed by GT1b, GD1a, and GM1a, while HCR/D-SA displayed a unique binding preference for GM1a, followed by GD1a with a lower affinity for b-series gangliosides. Directed mutagenesis experiments showed that HCR/C (Trp1258Ala) had reduced binding to GD1b, while HCR/D-SA(Trp1252Ala) binding to GM1a was also reduced. This supports a role for the Trp within the GBL as contributing to the coordination of ganglioside binding by HCR/C and HCR/D-SA. Thus, HCR/C and HCR/D-SA utilize the GBL for ganglioside binding.
The GBLs of BoNT/C, BoNT/D and BoNT/D-SA represent a gain of function of ganglioside binding with a loss of function at the prototypical GBP. The overlap of the Trp locations within the GBL of HCR/C and HCR/D-SA indicates that while Trp is required for ganglioside binding, this residue does not contribute to specificity and other residues within the GBL may contribute to ganglioside binding specificity. Tsukamoto and colleagues reported that the mutation Trp1258Ala reduced the ability of HCR/C to compete with BoNT/C for synaptosome binding [66], also implicating a role for this Trp in cell recognition. In addition to Arg1253, HCR/C has two Args flanking either side of the GBL (R1251, R1260), which may provide contacts for sialic acid residues in b-series gangliosides. The GBL loop of HCR/D-SA also contains Asp1249, which may repel the sialic acid carboxylate of b-series gangliosides. Future studies will determine which residues make contact with gangliosides and how gangliosides interact with this novel GBL. Cell binding experiments showed that HCR/C and HCR/D-SA bound neurons, while neither HCR/C(Trp1258Ala) nor HCR/D-SA(Trp1252Ala) bound neurons. Unlike HCR/C and D-SA, HCR/D did not show detectable cell binding, suggesting HCR/D binding and affinity varied from HCR/C and HCR/D-SA. Under similar conditions, HCR/A binding to neurons is also not detected unless neurons are depolarized [40]. Thus, high-affinity binding of HCR/D may require the presence of a synaptic vesicle co-receptor.
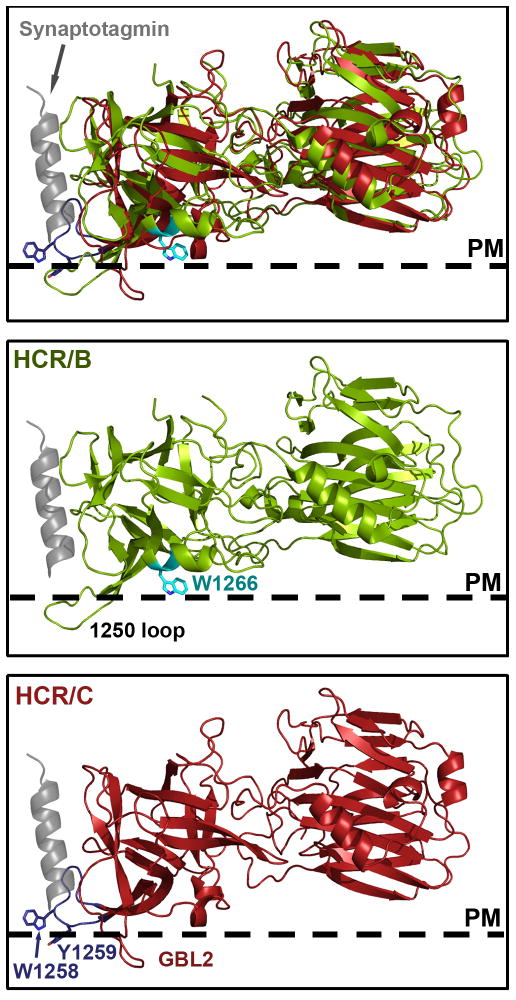
Using receptor bound HCR/B as a model, a prediction for how the GBL can align with the host plasma membrane is shown in Figure 6. Plasma membrane orientation was achieved by aligning the indol rings of BoNT/B Trp1266 and BoNT/C Trp1258 parallel to the plasma membrane. Binding to ganglioside and synaptotagmin simultaneously positions HCR/B on the membrane so that the 1250 loop may contact the lipid bilayer. When HCR/C is modeled in the same orientation, the synaptotagmin peptide occupies a region on the C terminus of the HCR and TrpW1258 of the GBL is positioned to interact with PM embedded ganglioside. Additional interaction with the plasma membrane may be accomplished by HCR/C though Tyr1259 and the loop residues Met1183-Iso1198.
Figure 6. Alignment of HCR/C to the HCR/B-Synaptotagmin complex.
A. Crystal structure of HCR/B (green) bound to synaptotagmin peptide (grey, PBD: 2NM1 [26], aligned with HCR/C (red, PDB: 3N7K). Trp1266 and Tyr1267 of the ganglioside binding pocket (GBP) are shown in cyan. The ganglioside binding loop (GBL1) of HCR/C is in purple with Trp1258 shown. Structures were aligned so that Trp1266 of HCR/B and Trp1258 of HCR/C are parallel with the plasma membrane (dashed line, PM). B. The 1250 loop described by Stevens and coworkers [64] potentially penetrates the PM while the synaptotagmin peptide fits into a crevice within the C terminus of HCR/B. C. Same alignment as in A. except HCR/B was omitted for clarity. Tyr1273 maps to the GBP and is shown in cyan. Trp1258 is positioned to interact with PM embedded ganglioside and along with Tyr1259 may penetrate the PM lipid bilayer, with another loop, GBL2, also membrane associated.
Vaccines against Botulism
The potency and duration of paralysis in humans place the BoNTs as category A select agents. BoNT/C and BoNT/D cause paralysis in human neuromuscular preparations [51, 67] and have been implicated as agents for human therapy [68]. In addition, there is a need to develop vaccines that neutralize all BoNT serotypes and variants. Traditional vaccination strategies use formaldehyde-inactivated BoNT, which eliminates toxicity and retains immunogenicity, but these are complicated to produce [69]. Recombinant HCRs are an alternative vaccine strategy, since the HCRs can be produced in large quantities free of neurotoxin contamination [70]. In addition, mice immunized with a cocktail of the seven prototypical serotypes (HCR/A-G) were resistant to challenge by each neurotoxin (BoNT/A-G) demonstrating the efficacy of this strategy [71]. Anti-sera from mice immunized with the hepta-serotype HCR vaccine blocked binding of HCRs to gangliosides in vitro. This indicates that neutralizing antibodies interfere with receptor recognition that is located adjacent to known human immune reactive epitopes. Humans do not appear to produce antibodies against to the region comprising the GBP, which suggests that the GBP may not be immunogenic [72, 73]. Unlike the GBP, the GBL of BoNT/C and BoNT/D-SA is a β-hairpin loop which protrudes from the HCR. The lack of cross protection observed by mice immunized with HCR/C upon challenge with BoNT/D-SA indicates the neutralizing epitopes are not conserved between these two BoNT subtypes and thus, the β-loop may be a potential site to elicit serotype specific neutralizing antibodies. Consistent with this region contributing to immune stimulation is the recent observation by Fairweather and coworkers, who reported that deletion of the GBL homologous region of HCR/TeNT reduced the capacity to elicit a neutralizing immune response [74]. Studies are underway to determine the role of the GBL in eliciting a protective response against botulism.
Future Perspectives
BoNT/C, BoNT/D, and the related mosaic toxins are a cluster in which ganglioside recognition has deviated from the mechanism utilized by BoNT/A, B, E-G, and TeNT. This evolution in protein function may have occurred as a gain of function process where the GBL acquired the ability to bind ganglioside, allowing a loss of function by the GBP for ganglioside binding while maintaining tertiary structure. Unanswered questions remain regarding the unusual ganglioside specificity of BoNT/D-South Africa and potential co-receptors for BoNT/C and BoNT/D cluster. BoNT/C and BoNT/D may use SV cycling to enter neurons [48], but thus far BoNT/C has not been found to interact with any known SV proteins [43], and potential secondary interactions for BoNT/D remains unclear. Identifying ganglioside specificity and entry mechanisms for the BoNT/C and BoNT/D cluster will expand the known capabilities of BoNT entry strategies. Furthermore, the discovery of basis for BoNT/D-SA evasion of both HCR/C and HCR/D immunization may provide a better understanding of how immunization leads to neutralization of BoNT intoxication.
Acknowledgments
JTB and JJK acknowledge membership and support from National Institutes of Health Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research Program, Great Lakes Regional Center of Excellence Award NIH-NIAID 1-U54-AI-057153. MRB acknowledges support from NIH-NINDS NS061763.
Abbreviations
- BoNT
botulinum neurotoxin
- HCR
heavy chain receptor binding domain of BoNT
- VAMP2
vesicle-associated membrane protein 2
- SNAP25
synaptosomal-associated protein of 25 kDa
- SNARE
soluble NSF attachment receptors
References
- 1.Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Mol Biol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh BR, Gimenez JA, DasGupta BR. Comparative molecular topography of botulinum neurotoxins from Clostridium butyricum and Clostridium botulinum type E. Biochim Biophys Acta. 1991;1077:119–126. doi: 10.1016/0167-4838(91)90533-6. [pii] [DOI] [PubMed] [Google Scholar]
- 3.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, et al. Genetic diversity among Botulinum Neurotoxin-producing clostridial strains. J Bacteriol. 2007;189:818–832. doi: 10.1128/JB.01180-06. JB.01180-06 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Mol Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 5.Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. nature03123 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Mol Biol. 2000;7:693–699. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- 7.Fischer A, Garcia-Rodriguez C, Geren I, Lou J, Marks JD, Nakagawa T, Montal M. Molecular Architecture of Botulinum Neurotoxin E Revealed by Single Particle Electron Microscopy. Journal of Biological Chemistry. 2008;283:3997–4003. doi: 10.1074/jbc.M707917200. [DOI] [PubMed] [Google Scholar]
- 8.Kumaran D, Eswaramoorthy S, Furey W, Navaza J, Sax M, Swaminathan S. Domain Organization in Clostridium botulinum Neurotoxin Type E Is Unique: Its Implication in Faster Translocation. Journal of Molecular Biology. 2009;386:233–245. doi: 10.1016/j.jmb.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, Camilli PD, Sudhof TC, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- 10.Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal R, Schmidt JJ, Stafford RG, Swaminathan S. Mode of VAMP substrate recognition and inhibition of Clostridium botulinum neurotoxin F. Nat Struct Mol Biol. 2009;16:789–794. doi: 10.1038/nsmb.1626. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Wan HY. Molecular mechanisms of substrate recognition and specificity of botulinum neurotoxin serotype F. Biochemical Journal. 2010;433:277–284. doi: 10.1042/bj20101310. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal R, Swaminathan S. SNAP-25 Substrate Peptide (Residues 180–183) Binds to but Bypasses Cleavage by Catalytically Active Clostridium botulinum Neurotoxin E. Journal of Biological Chemistry. 2008;283:25944–25951. doi: 10.1074/jbc.M803756200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Barbieri JT. Engineering botulinum neurotoxin to extend therapeutic intervention. Proceedings of the National Academy of Sciences. 2009;106:9180–9184. doi: 10.1073/pnas.0903111106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer A, Montal M. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proceedings of the National Academy of Sciences. 2007;104:10447–10452. doi: 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer A, Mushrush DJ, Lacy DB, Montal M. Botulinum Neurotoxin Devoid of Receptor Binding Domain Translocates Active Protease. PLoS Pathog. 2008;4:e1000245. doi: 10.1371/journal.ppat.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koriazova LK, Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat Struct Mol Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- 18.Lacy DB, Stevens RC. Recombinant Expression and Purification of the Botulinum Neurotoxin Type A Translocation Domain. Protein Expression and Purification. 1997;11:195–200. doi: 10.1006/prep.1997.0772. [DOI] [PubMed] [Google Scholar]
- 19.Halpern JL, Loftus A. Characterization of the receptor-binding domain of tetanus toxin. Journal of Biological Chemistry. 1993;268:11188–11192. [PubMed] [Google Scholar]
- 20.Umland TC, Wingert LM, Swaminathan S, Furey WF, Schmidt JJ, Sax M. Structure of the receptor binding fragment HC of tetanus neurotoxin. Nat Struct Mol Biol. 1997;4:788–792. doi: 10.1038/nsb1097-788. [DOI] [PubMed] [Google Scholar]
- 21.Herreros J, Lalli G, Schiavo G. C-terminal half of tetanus toxin fragment C is sufficient for neuronal binding and interaction with a putative protein receptor. Biochem J. 2000;347:199–204. [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S, Zhou Y, Singh BR. Cloning, expression, and purification of C-terminal quarter of the heavy chain of botulinum neurotoxin type A. Protein Expression and Purification. 2006;45:288–295. doi: 10.1016/j.pep.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 23.Lalli G, Herreros J, Osborne S, Montecucco C, Rossetto O, Schiavo G. Functional characterisation of tetanus and botulinum neurotoxins binding domains. J Cell Sci. 1999;112:2715–2724. doi: 10.1242/jcs.112.16.2715. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Fotinou C, Black I, Fairweather NF, Charles IG, Watts C, Hewitt E, Isaacs NW. The Structures of the HC Fragment of Tetanus Toxin with Carbohydrate Subunit Complexes Provide Insight into Ganglioside Binding. Journal of Biological Chemistry. 2000;275:8889–8894. doi: 10.1074/jbc.275.12.8889. [DOI] [PubMed] [Google Scholar]
- 25.Fotinou C, Emsley P, Black I, Ando H, Ishida H, Kiso M, Sinha KA, Fairweather NF, Isaacs NW. The Crystal Structure of Tetanus Toxin Hc Fragment Complexed with a Synthetic GT1b Analogue Suggests Cross-linking between Ganglioside Receptors and the Toxin. Journal of Biological Chemistry. 2001;276:32274–32281. doi: 10.1074/jbc.M103285200. [DOI] [PubMed] [Google Scholar]
- 26.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 27.Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Molecular Microbiology. 2004;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 28.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal Structure of Botulinum Neurotoxin Type A in Complex with the Cell Surface Co-Receptor GT1b—Insight into the Toxin–Neuron Interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Fu Z, Kim J-JP, Barbieri JT, Baldwin MR. Gangliosides as High Affinity Receptors for Tetanus Neurotoxin. Journal of Biological Chemistry. 2009;284:26569–26577. doi: 10.1074/jbc.M109.027391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayaraman S, Eswaramoorthy S, Kumaran D, Swaminathan S. Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin. Proteins: Structure, Function, and Bioinformatics. 2005;61:288–295. doi: 10.1002/prot.20595. [DOI] [PubMed] [Google Scholar]
- 31.Montecucco C. How do tetanus and botulinum toxins bind to neuronal membranes? Trends in Biochemical Sciences. 1986;11:314–317. [Google Scholar]
- 32.Nishiki T-i, Tokuyama Y, Kamata Y, Nemoto Y, Yoshida A, Sato K, Sekiguchi M, Takahashi M, Kozaki S. The high-affinity binding of Clostridium botulinum type B neurotoxin to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Letters. 1996;378:253–257. doi: 10.1016/0014-5793(95)01471-3. [DOI] [PubMed] [Google Scholar]
- 33.Nishiki T-i, Tokuyama Y, Kamata Y, Nemoto Y, Yoshida A, Sekiguchi M, Takahashi M, Kozaki S. Binding of botulinum type B neurotoxin to Chinese hamster ovary cells transfected with rat synaptotagmin II cDNA. Neuroscience Letters. 1996;208:105–108. doi: 10.1016/0304-3940(96)12557-x. [DOI] [PubMed] [Google Scholar]
- 34.Kozaki S, Kamata Y, Watarai S, Nishiki T-i, Mochida S. Ganglioside GT1b as a complementary receptor component for Clostridium botulinumneurotoxins. Microbial Pathogenesis. 1998;25:91–99. doi: 10.1006/mpat.1998.0214. [DOI] [PubMed] [Google Scholar]
- 35.Nishiki T, Kamata Y, Nemoto Y, Omori A, Ito T, Takahashi M, Kozaki S. Identification of protein receptor for Clostridium botulinum type B neurotoxin in rat brain synaptosomes. Journal of Biological Chemistry. 1994;269:10498–10503. [PubMed] [Google Scholar]
- 36.Rummel A, Karnath T, Henke T, Bigalke H, Binz T. Synaptotagmins I and II Act as Nerve Cell Receptors for Botulinum Neurotoxin G. Journal of Biological Chemistry. 2004;279:30865–30870. doi: 10.1074/jbc.M403945200. [DOI] [PubMed] [Google Scholar]
- 37.Simpson LL. Kinetic studies on the interaction between botulinum toxin type A and the cholinergic neuromuscular junction. Journal of Pharmacology and Experimental Therapeutics. 1980;212:16–21. [PubMed] [Google Scholar]
- 38.Hughes R, Whaler BC. Influence of nerve-ending activity and of drugs on the rate of paralysis of rat diaphragm preparation by Cl. botulinum type A toxin. The Journal of Physiology. 1962;160:221–233. doi: 10.1113/jphysiol.1962.sp006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162:1293–1303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 Is the Protein Receptor for Botulinum Neurotoxin A. Science. 2006;312:592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 41.Dong M, Liu H, Tepp WH, Johnson EA, Janz R, Chapman ER. Glycosylated SV2A and SV2B Mediate the Entry of Botulinum Neurotoxin E into Neurons. Mol Biol Cell. 2008;19:5226–5237. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Z, Chen C, Barbieri JT, Kim J-JP, Baldwin MR. Glycosylated SV2 and Gangliosides as Dual Receptors for Botulinum Neurotoxin Serotype F. Biochemistry. 2009;48:5631–5641. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baldwin MR, Barbieri JT. Association of Botulinum Neurotoxin Serotypes A and B with Synaptic Vesicle Protein Complexes. Biochemistry. 2007;46:3200–3210. doi: 10.1021/bi602396x. [DOI] [PubMed] [Google Scholar]
- 44.Bennett MK, Calakos N, Kreiner T, Scheller RH. Synaptic vesicle membrane proteins interact to form a multimeric complex. The Journal of Cell Biology. 1992;116:761–775. doi: 10.1083/jcb.116.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Südhof TC. The Synaptic Vesicle Cycle. Annual Review of Neuroscience. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 46.Strotmeier J, Lee K, Völker AK, Mahrhold S, Zong Y, Zeiser J, Zhou J, Pich A, Bigalke H, Binz T, et al. Botulinum neurotoxin serotype D attacks neurons via two carbohydrate-binding sites in a ganglioside-dependent manner. Biochemical Journal. 2010;431:207–216. doi: 10.1042/bj20101042. [DOI] [PubMed] [Google Scholar]
- 47.Tsukamoto K, Kohda T, Mukamoto M, Takeuchi K, Ihara H, Saito M, Kozaki S. Binding of Clostridium botulinum Type C and D Neurotoxins to Ganglioside and Phospholipid. Journal of Biological Chemistry. 2005;280:35164–35171. doi: 10.1074/jbc.M507596200. [DOI] [PubMed] [Google Scholar]
- 48.Rummel A, Hafner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–1954. doi: 10.1111/j.1471-4159.2009.06298.x. JNC6298 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Oguma K, Yokota K, Hayashi S, Takeshi K, Kumagai M, Itoh N, Tachi N, Chiba S. Infant botulism due to Clostridium botulinum type C toxin. The Lancet. 1990;336:1449–1450. doi: 10.1016/0140-6736(90)93157-k. [DOI] [PubMed] [Google Scholar]
- 50.Gangarosa EJ, Donadio JA, Armstrong RW, Meyer KF, Brachman PS, Dowell VR. Botulism in the United States, 1899–1969. Am J Epidemiol. 1971;93:93–101. doi: 10.1093/oxfordjournals.aje.a121239. [DOI] [PubMed] [Google Scholar]
- 51.Coffield JA, Bakry N, Zhang RD, Carlson J, Gomella LG, Simpson LL. In vitro characterization of botulinum toxin types A, C and D action on human tissues: Combined electrophysiologic, pharmacologic and molecular biologic approaches. Journal of Pharmacology and Experimental Therapeutics. 1997;280:1489–1498. [PubMed] [Google Scholar]
- 52.Neimanis A, Gavier-Widen D, Leighton F, Bollinger T, Rocke T, Morner T. An outbreak of type C botulism in herring gulls (Larus argentatus) in southeastern Sweden. J Wildl Dis. 2007;43:327–336. doi: 10.7589/0090-3558-43.3.327. [DOI] [PubMed] [Google Scholar]
- 53.Davletov B, Bajohrs M, Binz T. Beyond BOTOX: advantages and limitations of individual botulinum neurotoxins. Trends in Neurosciences. 2005;28:446–452. doi: 10.1016/j.tins.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Brand CJ, Schmitt SM, Duncan RM, Cooley TM. An outbreak of type E botulism among common loons (Gavia immer) in Michigan's upper peninsula. J Wildl Dis. 1988;24:471–476. doi: 10.7589/0090-3558-24.3.471. [DOI] [PubMed] [Google Scholar]
- 55.Oguma K, Iida H, Shiozaki M, Inoue K. Antigenicity of converting phages obtained from Clostridium botulinum types C and D. Infect Immun. 1976;13:855–860. doi: 10.1128/iai.13.3.855-860.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simpson LL, Rapport MM. Ganglioside inactivation of botulinum toxin. Journal of Neurochemistry. 1971;18:1341–1343. doi: 10.1111/j.1471-4159.1971.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 57.Webb RP, Smith TJ, Wright PM, Montgomery VA, Meagher MM, Smith LA. Protection with recombinant Clostridium botulinum C1 and D binding domain subunit (Hc) vaccines against C and D neurotoxins. Vaccine. 2007;25:4273–4282. doi: 10.1016/j.vaccine.2007.02.081. [DOI] [PubMed] [Google Scholar]
- 58.Moriishi K, Syuto B, Kubo S, Oguma K. Molecular diversity of neurotoxins from Clostridium botulinum type D strains. Infect Immun. 1989;57:2886–2891. doi: 10.1128/iai.57.9.2886-2891.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oguma K, Syuto B, Agui T, Iida H, Kubo S. Homogeneity and Heterogeneity of Toxins Produced by Clostridium botulinum Type C and D Strains. Infect Immun. 1981;34:382–388. doi: 10.1128/iai.34.2.382-388.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oguma K, Syuto B, Iida H, Kubo S. Antigenic similarity of toxins produced by Clostridium botulinum type C and D strains. Infect Immun. 1980;30:656–660. doi: 10.1128/iai.30.3.656-660.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moriishi K, Koura M, Abe N, Fujii N, Fujinaga Y, Inoue K, Ogumad K. Mosaic structures of neurotoxins produced from Clostridium botulinum types C and D organisms. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1996;1307:123–126. doi: 10.1016/0167-4781(96)00006-1. [DOI] [PubMed] [Google Scholar]
- 62.Karalewitz AP, Kroken AR, Fu Z, Baldwin MR, Kim JJ, Barbieri JT. Identification of a unique ganglioside binding loop within botulinum neurotoxins C and D-SA. Biochemistry. 2010;49:8117–8126. doi: 10.1021/bi100865f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 64.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 65.Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Molecular Microbiology. 2004;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 66.Tsukamoto K, Kozai Y, Ihara H, Kohda T, Mukamoto M, Tsuji T, Kozaki S. Identification of the receptor-binding sites in the carboxyl-terminal half of the heavy chain of botulinum neurotoxin types C and D. Microbial Pathogenesis. 2008;44:484–493. doi: 10.1016/j.micpath.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 67.Kalandakanond S, Coffield JA. Cleavage of intracellular substrates of botulinum toxins A, C, and D in a mammalian target tissue. J Pharmacol Exp Ther. 2001;296:749–755. [PubMed] [Google Scholar]
- 68.Eleopra R, Tugnoli V, Rossetto O, Montecucco C, De Grandis D. Botulinum neurotoxin serotype C: a novel effective botulinum toxin therapy in human. Neuroscience Letters. 1997;224:91–94. doi: 10.1016/s0304-3940(97)13448-6. [DOI] [PubMed] [Google Scholar]
- 69.Aoki KR, Smith LA, Atassi MZ. Mode of action of botulinum neurotoxins: current vaccination strategies and molecular immune recognition. Crit Rev Immunol. 2010;30:167–187. doi: 10.1615/critrevimmunol.v30.i2.50. [pii] [DOI] [PubMed] [Google Scholar]
- 70.Smith LA, Jensen MJ, Montgomery VA, Brown DR, Ahmed SA, Smith TJ. Roads from vaccines to therapies. Mov Disord. 2004;19(Suppl 8):S48–52. doi: 10.1002/mds.20009. [DOI] [PubMed] [Google Scholar]
- 71.Baldwin MR, Tepp WH, Przedpelski A, Pier CL, Bradshaw M, Johnson EA, Barbieri JT. Subunit vaccine against the seven serotypes of botulism. Infect Immun. 2008;76:1314–1318. doi: 10.1128/IAI.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dolimbek BZ, Steward LE, Aoki KR, Atassi MZ. Immune recognition of botulinum neurotoxin B: antibody-binding regions on the heavy chain of the toxin. Mol Immunol. 2008;45:910–924. doi: 10.1016/j.molimm.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 73.Dolimbek BZ, Aoki KR, Steward LE, Jankovic J, Atassi MZ. Mapping of the regions on the heavy chain of botulinum neurotoxin A (BoNT/A) recognized by antibodies of cervical dystonia patients with immunoresistance to BoNT/A. Mol Immunol. 2007;44:1029–1041. doi: 10.1016/j.molimm.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Qazi O, Sesardic D, Tierney R, Soderback Z, Crane D, Bolgiano B, Fairweather N. Reduction of the ganglioside binding activity of the tetanus toxin HC fragment destroys immunogenicity: implications for development of novel tetanus vaccines. Infect Immun. 2006;74:4884–4891. doi: 10.1128/IAI.00500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]