Abstract
Mutant p53 (mutp53) cancers are surprisingly dependent on their hyperstable mutp53 protein for survival, identifying mutp53 as a potentially significant clinical target. However, exploration of effective small molecule therapies targeting mutp53 has barely begun. Mutp53 hyperstabilization, a hallmark of p53 mutation, is cancer cell-specific and due to massive upregulation of the HSP90 chaperone machinery during malignant transformation. We recently showed that stable complex formation between HSP90 and its mutp53 client inhibits E3 ligases MDM2 and CHIP, causing mutp53 stabilization. Histone deacetylase (HDAC) inhibitors (HDACi) are a new class of promising anti-cancer drugs, hyperacetylating histone and non-histone targets. Currently, suberoylanilide hydroxamic acid (SAHA) is the only FDA-approved HDACi. We show that SAHA exhibits preferential cytotoxicity for mutant, rather than wild-type and null p53 human cancer cells. Loss/gain-of-function experiments revealed that although able to exert multiple cellular effects, SAHA's cytotoxicity is caused to a significant degree by its ability to strongly destabilize mutp53 at the level of protein degradation. The underlying mechanism is SAHA's inhibition of HDAC6, an essential positive regulator of HSP90. This releases mutp53 and enables its MDM2- and CHIP-mediated degradation. SAHA also strongly chemosensitizes mutp53 cancer cells for chemotherapy due to its ability to degrade mutp53. This identifies a novel action of SAHA with the prospect of SAHA becoming a centerpiece in mutp53-specific anticancer strategies.
Keywords: SAHA, mutant p53, HDAC6, activated Hsp90 chaperone, MDM2, CHIP
For wild-type p53 (wtp53) harboring cancers, several small molecule-based anticancer therapies such as Nutlin and RITA aiming at non-genotoxic activation of wtp53 by release from its negative regulation by MDM2 are currently intensely investigated. However, over 50% of human tumors harbor mutant p53 (mutp53). Ninety percent are missense point mutations in the p53 DNA-binding domain. Mutp53 knock-in mice (KI), modeling the human tumor-derived point mutations, established that mutp53 alleles can drive tumor formation, invasion and metastasis via dominant negative inhibition of wtp53 and via a true gain-of-function (GOF), so-called ‘neomorphic' oncogenic activities, compared with p53 null alleles.1, 2 In addition to their role in tumor formation and progression, mutp53 proteins, whose hallmark is aberrant constitutive hyperstabilization in tumor cells (but not in normal cells),1, 2 are clinically significant because their expression renders tumor cells resistant to chemotherapeutic drugs.3, 4, 5 Importantly, mutp53 KI mice also clearly established that p53 mutation alone does not confer hyperstability of mutp53 protein, but reflects a tumor-specific activation of p53-stabilizing pathways.6 We recently showed that stable complex formation between HSP90, which is highly and ubiquitously upregulated specifically in cancer cells, and mutp53 inhibits the responsible E3 ligases MDM2 and CHIP, and constitutes the major underlying mechanism for mutp53 stabilization.7 Importantly, experimental interference with mutp53 function in tumor cells either by expression of a dominant negative minipeptide or by RNAi-mediated knockdown of elevated levels of mutp53 protein, severely attenuated tumor growth in vitro and in xenografts, effectively decreased invasion and metastasis8, 9 and restored chemotherapy-induced apoptosis.5, 10
Histone deacetylase inhibitors (HDACi) are one of the most promising classes of new anticancer drugs and currently under intense investigation. HDACi were initially thought to simply ‘normalize' the widespread transcriptional silencing mediated by the aberrant cancer cell epigenome. However, with increasing understanding of the acetylome and its regulation, a growing number of non-histone targets of HDACs and thus novel targets of HDACi are being identified. Non-histone targets of HDACs are transcription factors (e.g., p53, E2F1, STAT1, NF-κB). In wtp53 tumor cells, HDACi induce p53 hyperacetylation that stabilizes and transcriptionally activates p53 for pro-apoptotic targets. Other important non-histone targets of HDACs are the specific cytoplasmic proteins α-tubulin, HSP90 and Ku70. The α-tubulin and HSP90 are specific targets of HDAC6 only.11 HDAC6 is an obligate positive regulator of the multi-component HSP90 chaperone machinery that is required for proper folding of many oncoproteins including mutp53. Acetylation of its core component, Hsp90 protein, by HDAC6 knockdown or HDACi inactivates its chaperone activity and lead to degradation of its clients HER2, ErbB1, ErbB2, Akt, c-Raf, BCR–ABL and FLT3.12, 13, 14, 15
HDACi show pleiotropic anti-cancer effects. These comprise tumor-cell intrinsic effects of inducing (typically p53 independently) apoptosis, cell cycle arrest and differentiation, and extrinsic effects of inhibiting invasion, tumor vasculature and enhancing the host immune response. Encouragingly, HDACi show a big therapeutic window in cytotoxicity, with transformed cells at least 10 × more sensitive than normal cells. For example, HDACi selectively induce apoptosis in acute promyelocytic leukemic cells but not in normal bone marrow or preleukemic cells that already express the PML-RARα fusion oncoprotein.11 Thus, HDACi exhibit an excellent toxicity profile. Currently there are over 70 mostly early phase clinical trials ongoing for 11 different HDACi compounds, either alone or in combination with other drugs, covering a broad range of liquid and solid tumors. Suberoylanilide hydroxamic acid (SAHA) (Vorinostat) is the first and currently only FDA-approved HDACi drug (since 2006 for cutaneous T-cell lymphoma). SAHA inhibits class I, II and IV HDACs, including the cytoplasmic HDAC6, a member of class IIb, and therefore can have multiple cellular effects. In cancer cells, SAHA activates apoptosis by Bid cleavage, upregulation of BH3-only proteins like Bim and Bmf, accumulation of ROS, suppression of Bcl2 and ROS scavengers and TNFα family activation.16
For mutp53 harboring tumors, we hypothesized that destabilizing mutp53 may provide a novel therapeutic strategy of clinical significance. However, pharmacologically effective mutp53-targeting small molecule approaches have not been properly explored. Here we show that SAHA shows preferential cytotoxicity in mutp53 cancer cells by destabilizing mutp53 through inhibition of the HDAC6-Hsp90 chaperone axis. In sum, this data provides encouraging evidence for the feasibility of mutp53-targeted anticancer therapy using a well-tolerated small molecule inhibitor that is already in late clinical trials.
Results
SAHA downregulates mutp53 but not wtp53
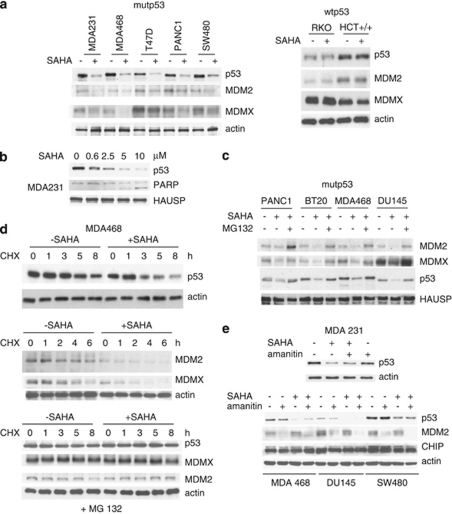
This effect occurs at the level of protein degradation but not at the level of transcription. To analyze whether SAHA has an effect on p53 expression, a panel of human tumor cell lines harboring either mutant or wild-type p53 were analyzed by immunoblots. As shown in Figure 1a, SAHA strongly downregulates the various aberrantly accumulated mutp53 proteins in all cases, but does not alter levels of wtp53 protein. Also, SAHA treatment decreases the levels of MDM2 and MDMX, all bona fide physiologic substrates of MDM2 in mutant, but not wtp53 cells (Figure 1a). Moreover, SAHA-mediated downregulation of mutp53 by low micromolar concentrations is dose-dependent and correlates with induction of PARP cleavage in cancer cells (Figure 1b). Furthermore, consistent with mutant-specific downregulation, SAHA induces ubiquitination of mutant but not wtp53 (Supplementary Figure 1). Importantly, proteasome inhibition by MG132 completely rescues SAHA-mediated downregulation of mutp53, MDM2 and MDMX, indicating that SAHA regulates their stability on the post-transcriptional level (Figure 1c). To further explore the mechanism of SAHA-induced destabilization of mutp53, we next performed cycloheximide (CHX) chase experiments. As shown in Figure 1d, SAHA dramatically decreases the half-life not only of mutp53 protein, but also of MDMX and of MDM2 itself. Moreover, proteasome inhibiton completely rescues SAHA-induced destabilization of mutp53, MDM2 and MDMX after CHX treatment (Figure 1d). Conversely, mutp53 cells were treated with SAHA plus α-amanitin, a potent transcriptional inhibitor that blocks RNA polymerase II. However, transcriptional inhibition does not interfere with SAHA-mediated downregulation of mutp53 in all tumor lines tested (Figure 1e). Taken together, our results indicate that SAHA's effect is specific for mutp53, and is largely posttranslational at the level of protein degradation.
Figure 1.
SAHA downregulates mutp53 but not wtp53. This effect occurs at the level of protein degradation but not at the level of transcription. (a) SAHA (5 μM) strongly downregulates mutant p53, MDM2 and MDMX protein levels. In contrast, SAHA does not alter levels of wild-type p53 protein. A panel of human tumor cell lines harboring mutant or wild-type p53 were analyzed by immunoblot as indicated. Actin, loading control. (b) SAHA-mediated downregulation of mutant p53 is dose-dependent and correlates with induction of PARP cleavage. (c) Proteosome inhibitor MG132 (5 μM) rescues SAHA-mediated downregulation of mutp53, MDM2 and MDMX. Cells were simultaneously treated with SAHA (5 μM) and MG132 (5 μM) for 16 h. HAUSP was used as a loading control. (d) SAHA (5 μM) dramatically decreases the half-life of mutant p53, MDM2 and MDMX, all bona fide physiologic substrates of MDM2. CHX chase for the indicated times. Actin, loading control. Bottom, proteasome inhibitor MG132 (5 μM) rescues SAHA-induced destabilization of mutp53, MDM2 and MDMX after CHX treatment. (e) The SAHA-mediated downregulation of mutant p53 does not occur at the level of transcription. Mutp53 cells were treated with SAHA (5 μM) and/or α-amanitin (10 μg/ml) for 16 h, a potent and specific transcriptional inhibitor of RNA polymerase II
SAHA-induced degradation of mutp53 is mediated by reactivation of MDM2 and CHIP E3 ligases
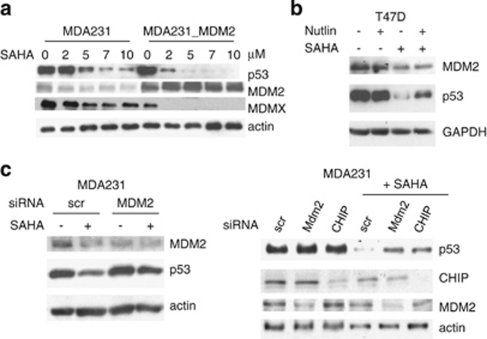
Next we sought to identify the specific E3 ubiqutin ligase(s) that are responsible for SAHA's effect on mutp53. We focused on MDM2 and CHIP, as CHIP was identified as a mutp53 E3 ligase17, 18 and the functional inactivation of these two ligases is the cause for aberrant stabilization of mutp53 in cancer cells.7 Stable overexpression of MDM2 enhances SAHA-induced degradation of mutp53 (and of MDMX) compared with vector controls (Figure 2a). Moreover, the specific MDM2 inhibitor Nutlin partially prevents SAHA-induced destabilization of mutp53 (Figure 2b). Furthermore, direct siRNA-mediated knockdown of MDM2 and CHIP partially rescues SAHA-induced destabilization of mutp53 (Figures 2c and 5e). Taken together, these data imply that mechanistically SAHA causes mutp53 degradation by inducing functional reactivation of MDM2 and CHIP.
Figure 2.
SAHA-induced degradation of mutp53 is mediated by reactivation of MDM2 and CHIP E3 ligases. (a) Overexpression of MDM2 enhances SAHA-induced degradation of mutp53 and MDMX. The cell system used here is described by Li et al.7 wherein we show that in human cancer cells endogenous mutant p53 – despite its ability to interact with MDM2 – suffers from a profound lack of ubiquitination as the root cause of its degradation defect. Multiple lines of evidence indicate the functional impairment of MDM2 in mutp53 cancer cells by the HSP90 chaperone. We found that in contrast to transiently overexpressed MDM2, physiologically tolerated, stably overexpressed MDM2 is silent and fails to affect ubiquitination and mutp53 levels, indicating the presence of selective pressure against active MDM2 in mutp53 cancer cells. Immunoblot of parental MDA231 and cells stably overexpressing MDM2. Actin as loading control. (b) Nutlin partially prevents SAHA-induced destabilization of mutp53, indicating MDM2 reactivation upon SAHA (5 μM). GAPDH, loading control. (c) siRNA-mediated knockdown of MDM2 and CHIP partially rescues SAHA-induced (5 μM) destabilization of mutp53
HDAC6 inhibition destabilizes mutp53 by inactivating HSP90. SAHA destabilizes mutp53 by inactivating HDAC6
Mutp53 KI mouse models definitively clarified that p53 missense mutations alone do not confer aberrant p53 protein stability, as normal tissues of these mice exhibit low, wild-type like p53 levels.1, 2 Thus, they showed that hyperstability is not due to loss of transcriptional activation of the p53 target gene MDM2, the long favored proposed mechanism for hyperstability. Instead mutp53 stabilization reflects a tumor-specific activation of a p53-stabilizing pathway. The identity of this pathway seems to be linked to the HSP90 chaperone machinery, which is highly and ubiquitously upregulated specifically in cancer cells. Mutp53 proteins are damaged in their conformation-sensitive core domain. In fact, in tumor cells mutp53 forms stable complexes with Hsp90. These large stable heterocomplexes were previously proposed to inhibit proteasome-mediated degradation of mutp53.19
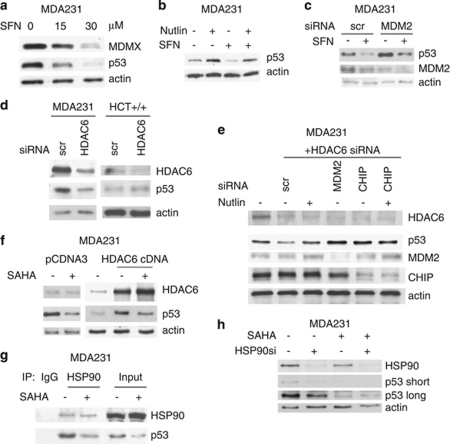
HSP90 chaperone activity is regulated by acetylation/deacetylation of Hsp90. HSP90 acetylation at K294 inactivates its chaperone activity by inhibiting its cyclical binding to client proteins and cochaperones. HDAC6 is the positive regulator of the HSP90 chaperone activity by mediating K294 deacetylation.20, 21 Indeed, HDAC6 pharmacological inhibition or HDAC6 siRNA-mediated knockdown leads to degradation of mutp53. As shown in Figure 3a, HDAC6 inhibition by Sulfaraphane (SFN)22 destabilizes mutp53 (and Mdmx). Conversely, Nutlin (Figure 3b) and MDM2 siRNA (Figure 3c) partially prevent SFN-induced degradation of mutp53, again implicating MDM2 in SFN-mediated degradation. More importantly, HDAC6 inhibition by siRNA specifically destabilizes mutp53 (but not wtp53, that is in HCT116 cells) (Figure 3d) and this siHDAC6 effect is again rescued by Nutlin or siRNA against MDM2 and CHIP (Figure 3e). Conversely, HDAC6 overexpression stabilizes mutp53 (Figure 3f, middle lane). Importantly, this stabilization is largely rescued by concomitant treatment with SAHA (Figure 3f, third lane). Furthermore, SAHA targets all HDAC isoforms including HDAC6.16 Importantly, SAHA releases mutp53 by inhibiting the complex between Hsp90 and its p53 client (Figure 3g). To further support the fact that SAHA specifically works through Hsp90, we examined the effect of Hsp90 silencing. Indeed, although downregulation of Hsp90 by siRNA destabilizes mutp53 (Figure 3h, compare lanes 1 and 2), SAHA does not induce further destabilization in HSP90-ablated cells (Figure 3h, compare lanes 3 with 4). Thus, our aggregate results of Figures 1, 2, 3 concerning the mechanism of SAHA-induced degradation of mutp53 strongly suggest the following chain of events: in the context of mutp53, to a large (albeit not exclusive) extent SAHA works by inhibiting HDAC6 deacetylation. This leads to inactivation of Hsp90 and release of mutp53 that in turn leads to reactivation of MDM2 and CHIP E3 ligases and p53 degradation.
Figure 3.
Inhibition of HDAC6 inhibits HSP90 and destabilizes mutp53 by reactivating MDM2 and CHIP. (a) HDAC6 inhibitor SFN destabilizes mutp53 and MDMX. Nutlin (b) and MDM2 siRNA (c) partially prevent SFN-induced degradation of mutp53. (d) Likewise, HDAC6 inhibition by siRNA also destabilizes mutp53 (in MDA231) but does not destabilize wtp53 (in HCT116). (e) siRNA-mediated knockdown of HDAC6 results in degradation of mutp53 that is rescued by Nutlin (lane 3) or by siRNA against MDM2 and CHIP. Scr, scrambled siRNA. Immunoblot. (f) Conversely, HDAC6 overexpression stabilizes mutp53, which is significantly rescued by concomitant SAHA. Immunoblot. (g) SAHA (5 μM) inhibits the complex between Hsp90 and mutp53. Lysates from MDA231 cells left untreated or treated with SAHA were immunoprecipitated with IgG or anti-Hsp90. (h) In HSP90-ablated cells, SAHA does not induce further destabilization of mutp53. MDA231 cells were transfected with siHSP90 or scrambled siRNA control. After 24 h, cells were treated with 2.5 μM SAHA for an additional 24 h followed by immunoblot
SAHA shows preferential cytotoxicity for mutp53 tumor cells
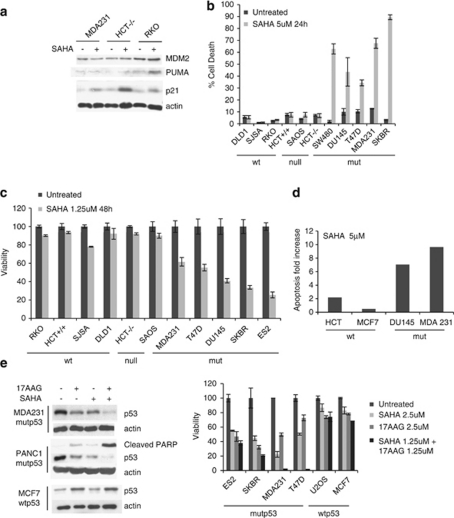
Our results so far suggest that mutp53-harboring tumor cells might be especially sensitive to SAHA's anti-tumor effects. To test this notion, we employed multiple death assays to examine side-by-side randomly selected panels of human tumor cell lines representing different p53 mutational status. One study had previously reported that mutp53 was associated with increased cytotoxicity to the HDAC inhibitors FR901228 (aka FK-228 or depsipeptide) and trichostatin A (TSA).23 However, both drugs are of different chemical classes than SAHA and were reported to somehow ‘pharmacologically rescue' mutp53 by inducing a wild-type p53-like transcriptional response, indicated by p21 and MDM2 target gene induction. Because mutp53 is highly expressed, a sudden restoration of wild-type p53 functions was proposed to be the basis of cytotoxicity for these two HDACi.23 However, in contrast to FR901228 and TSA, SAHA does not restore wild-type p53-like functions in mutp53. In SAHA-treated MDA231 cells, MDM2 fails to be induced by SAHA. p21, although mildly increased, is induced in a p53-independent manner, as indicated by the highest p21 induction in p53 null cells (HCT116−/−), and comparable (to mutp53 cells) p21 induction in wtp53 cells (RKO) (Figure 4a). SAHA is well known to induce p21 (CDKN1A).11, 24, 25 Furthermore, in contrast to wtp53 cells we did not observe upregulation of Puma, Noxa and Bax in mutp53 cells following SAHA treatment (Figure 4a and Supplementary Figure 3).
Figure 4.
SAHA shows preferential cytotoxicity for mutp53 tumor cells. (a) SAHA does not pharmacologically rescue mutant p53 to assume wild type function. MDM2 is not induced. Although p21 is slightly induced, it appears to be in a p53-independent manner, based on p53-deficient HCT116 cells (HCT−/−). SAHA was used at 5 μM. (b–d) SAHA shows strong cytotoxic cell killing towards mutp53 tumor cells, as measured by trypan blue exclusion assay (b), cell viability CTB assay (c) and subG1 FACS (d). In contrast, wtp53 and p53 null tumor cells show only a minimal cytotoxic response to SAHA. Random panel of human tumor cells. (e) SAHA and 17AAG can synergize to induce preferential apoptosis of mutp53 cancer cells. Co-treatment of SAHA and 17AAG causes a synergistic loss of cell viability specifically in MDA231 and T47D. Combined efficacy correlates with the degree of mutp53 destabilization and PARP cleavage (right immunoblots). This synergism is due to complementary drug targets within the HSP90 chaperone machinery
Concerning anti-tumor effects, SAHA indeed shows preferential cytotoxicity in mutp53 cancer cells. SAHA induced strong cell killing in human tumor cells harboring mutp53, as measured by trypan blue exclusion assays (Figure 4b), cell viability assays (Figure 4c) and subG1 fractions in FACS analysis (Figure 4d). In contrast, wtp53 or p53 null human tumor cells show only minimal cytotoxic responses to SAHA (Figures 4b–d). This result was further supported by determination of IC50s (inhibitory concentrations at which 50% killing is achieved), which was profoundly lower in mutp53 cancer cells (Table 1 and Supplementary Figure 2). Moreover, as SAHA is an effective HDAC6 inhibitor, which is a positive regulator of the HSP90 chaperone complex, we predicted that in the context of mutp53, SAHA might synergize with a direct Hsp90 inhibitor. Highly specific Hsp90 inhibitors such as the geldanamycin and its clinically active derivative, the prototype 17AAG, competitively bind to the N-terminal ATP-binding pocket and stop the chaperone cycle, leading to client protein degradation.26, 27 Indeed, 17AAG alone shows similar preferential efficacy in mutp53 tumor cells as SAHA alone. Moreover, co-treatment of SAHA plus 17AAG causes a synergistic loss of cell viability in some mutp53 cancer cells (MDA231), but much less so or not at all in wtp53 cancer cells. This is very obvious in MDA231 and T47D cells that were killed with 100% efficiency by the drug combination (Figure 4e, right). This synergistic efficacy correlates with the degree of mutp53 destabilization and PARP cleavage in mutant but not wild-type cells (Figure 4e, left). Of note, SAHA alone induces mutp53 destabilization at least as efficiently as 17AAG alone. Thus, molecularly the synergism of SAHA+17AAG is explained by complementary drug targets against distinct components of the HSP90 chaperone machinery, leading to direct and indirect (HDAC6) inhibition of the complex's ability to hyperstabilize mutp53.
Table 1. IC50 of SAHA.
| Cell line | IC50 (μM) | p53 status |
|---|---|---|
| RKO | 393.0 | wt |
| HCT+/+ | 128.0 | wt |
| T47D | 1.732 | mut |
| MDA231 | 1.105 | mut |
| ES2 | 1.980 | mut |
Causality – SAHA's preferential cytotoxic effect on mutp53-harboring cancer cells is to a significant degree due to its ability to degrade mutp53
Our data so far shows that SAHA preferentially kills mutp53 tumor cells and that its cytotoxicity completely correlates with SAHA's ability to degrade hyperstable mutp53 protein, even under conditions of strong transcriptional inhibition. Therefore we asked whether the anti-tumor effect of SAHA is dependent on HSP90-mediated mutp53 degradation and if so, to what degree. To test for dependence, we used complementary gain- and loss-of-function approaches of mutp53 cancer cells and interrogated them in an array of functional assays that included cell death, clonogenicity, invasion and chemosensitization.
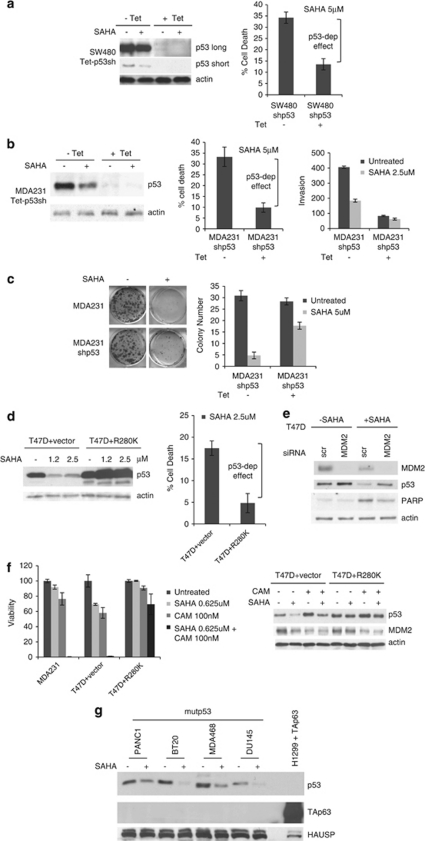
As shown in Figure 5, SAHA loses more than 50% of its killing efficacy in mutp53-harboring cancer cells when its target (mutp53) is knocked down by Tet-inducible shp53 RNAi, as shown in SW480 and MDA231 cells (Figures 5a and b). In both cases, SAHA did retain a small remnant efficacy despite p53 downregulation, which might be due to incomplete p53 elimination, and/or to a p53-independent component of the anti-tumoral SAHA effect. These results were confirmed by long-term clonogenicity assays (Figure 5c). Furthermore, SAHA-induces an invasion block in MDA231 cells that correlates with its degradation of mutp53 (−Tet, Figure 5b, left and right panels). Conversely, the SAHA-induced invasion block is virtually eliminated when mutp53 is downregulated (+Tet, Figure 5b, left and right panels). This argues that the invasion block in p53-proficient control MDA231 cells is at least in part due to SAHA-induced HSP90-mediated mutp53 degradation.
Figure 5.
Causality – SAHA's preferential cytotoxic effect on mutp53-harboring cancer cells is to a significant degree due to its ability to degrade mutp53. (a–c) Pseudo-null lines were generated to eliminate mutp53 as a target of SAHA. Cells were pretreated with tetracycline to achieve ‘isogenicity'. Cells with high (control) and very low levels of mutp53 were then treated with SAHA. In both MDA231 and SW480, SAHA partially loses cytotoxicity when mutp53 is very low. (a and b) In mutp53-harboring cancer cells SAHA loses over 50% killing efficacy when mutp53 is knocked down. Left, immunoblots of (a) SW480 and (b) MDA231 cells stably harboring Tet-inducible shp53. Middle, trypan blue exclusion assays. Percent increase in cell death of SAHA-treated cells relative to their respective untreated controls without SAHA. (b, right) Knockdown of mutp53 by SAHA or/and Tet- inducible shRNA inhibits invasion. (b, left) Inhibition of invasion directly correlates with the extent of mutp53 destabilization. Aliquots were used for the Matrigel Boyden chambers and corresponding immunoblot. (c) Clonogenicity assay of MDA231 cells harboring Tet-inducible shp53 in response to SAHA. Right, quantitation. For experiments in a–c, cells were pretreated with tetracycline for 48 h and then seeded for subsequent SAHA treatment (5 μM) for an additional 24 h. (d) Likewise, SAHA loses killing efficacy when its ability to degrade mutp53 is overwhelmed by excess ectopic mutant p53. Left, immunoblots of empty vector or mutp53 R280K overexpressing T47D. Right, increase in cell death of SAHA-treated cells relative to their respective untreated controls without SAHA. Trypan blue exclusion assay. (e) siRNA-mediated knockdown of MDM2 partially rescues SAHA-induced destabilization of mutp53 and inhibits the cytotoxic effect of SAHA, indicated by reduced PARP cleavage. Cells were transfected with siMDM2 or scrambled siRNA, followed by SAHA treatment (5 μM) for 16 h. Immunoblot, actin as loading control. (f) SAHA strongly chemosensitizes mutp53 cancer cells and this is due to its ability to degrade mutp53. Left, viability of MDA231, T47D vector and T47D cells overexpressing mutp53 R280K (see Figure 5d) after low-dose camptothecin (100 nM) and SAHA (625 nM) treatment alone and in combination. CTB assays. Right, corresponding immunoblots of the T47D set. (g) SAHA fails to induce TAp63 protein in mutant p53 cancer cells. Despite multiple forced attempts (overloading, overexposure), we were unable to detect SAHA-induced TAp63 protein levels in any of the cell lines. Immunoblot with a pan-p63 antibody (H137) of total cell lysates (20 μg per lane) from mutp53 cancer cells grown in the absence or presence of SAHA (5 μM) for 24 h. As control, 2 μg of total cell lysate from H1299 cells transfected with a TAp63α plasmid was loaded
In further support of a causal link for SAHA targeting mutp53, SAHA loses killing efficacy when its ability to degrade mutp53 is overwhelmed by excess amounts of ectopically expressed mutp53 (‘overstuffed'). Excess ectopic R280K mutp53 that has exhausted SAHA's ability to degrade it at the concentration used also squelches SAHA's ability to induce cell death in T47D and SW480 cells by more than 50% (Figure 5d and data not shown). As expected and already seen with the knockdown systems (Figures 5a–c), SAHA retains some remnant efficacy, suggesting a partial p53-independent component of SAHA action. Most importantly, however, in both experimental conditions cells become partially resistant to SAHA because SAHA is no longer able to downregulate mutp53 levels.
As predicted and already seen above, knockdown of MDM2 by siRNA partially rescues SAHA-induced destabilization of mutp53 (Figures 2c, 5e) and inhibits the cytotoxic effect of SAHA, indicated by reduced PARP cleavage (Figure 5e).
One possible mechanism of SAHA-induced apoptosis in mutp53 cells might be TAp63 activation. Although such an effect was shown to be wtp53-dependent,28 at least in principle SAHA might also activate TAp63 in our mutant p53 system in a transcription-dependent and independent way: first, massive transcriptional induction of TAp63 mRNA (over 3000 fold), leading to p63 protein induction from undetectable to highly detectable levels, was reported for HDAC inhibitors TSA28, 29 and SAHA29 in wtp53-harboring cancer cells. However, we do not see TAp63 mRNA and protein induction in mutp53 cells (Supplementary Figure 3 and Figure 5g). On the other hand, destabilization of mutp53 by SAHA might release TAp63 from a proposed mutp53-TAp63-Smad inhibitory complex and empower its apoptotic function.8 This follow-up question is currently under active investigation in our lab.
SAHA strongly chemosensitizes mutp53 cancer cells and this is due to its ability to degrade mutp53
RNAi-mediated mutp53 knockdown in human cancer cells was previously shown to cause chemosensitization towards an array of conventional genotoxic drugs,5, 10, 30 hinting that such cancer cells are addicted to the continuous expression of mutp53. Thus, we tested whether pharmacological degradation of mutp53 via HSP90 targeting by SAHA can also mediate chemosensitization in response to conventional genotoxic drugs such as topoisomerase inhibitor camptothecin. As shown in Figure 5f, this is indeed the case. Although low doses of camptothecin (100 nM) and SAHA (625 nM) alone induces only modest cell killing of 10 and 25%, respectively, in MDA231 cells, SAHA markedly synergizes with camptothecin to cause 100% killing. A similar situation is seen with T47D cells. Of note, this chemosensitization is due to SAHA's ability to degrade mutp53, as excess ectopic mutp53, which overwhelms SAHA, completely squelches this effect (Figure 5f).
Discussion
There is growing evidence that the highly accumulated mutp53 protein, a hallmark of almost 50% of human tumors, is a clinically relevant target for intervention. It is the hyperstability of mutp53 that is the basis for its GOF and dominant-negativity (over wtp53 in case of heterozygosity) that promotes malignancy and chemoresistance.31 Aberrant accumulation of mutp53 does not occur in normal cells1, 2 but is tumor-specific due to massive upregulation of the multi-component HSP90 chaperone machinery that almost ubiquitously accompanies malignant transformation.7, 26, 32, 33 RNAi-mediated knockdown of mutp53 was shown to decrease tumor cell proliferation in vitro and in xenografts, inhibit invasion and metastasis, and sensitize tumor cells towards genotoxic therapy.5, 6, 8, 9 However, exploration of pharmacologically effective small molecule therapy that targets mutp53 degradation has barely begun. Our results here provide significant support for the hypothesis that destabilization of mutp53 is indeed an effective strategy for treating this large group of human cancers and that this can be achieved by pharmacological means that are clinically already well developed. Thus, these data carry some tangible translational importance.
We show here that HDAC6, a cytoplasmic non-histone HDAC that deacetylates Hsp90 and functions as an obligate positive regulator of the HSP90 chaperone machinery,20, 21 is critically important in enabling aberrant stability of mutp53. Inhibition of HDAC6, known to maintain Hsp90 chaperone activity towards other cancer-related client proteins such as androgen receptor, estrogen receptor and ErbB2,34 destabilizes mutp53 by inactivating Hsp90 (Figure 3). Furthermore, SAHA treatment leads to strong acetylation of HSP90, interfering with its chaperone activity.35 Importantly, we could demonstrate for the first time that the HDAC inhibitor SAHA is able to effectively and specifically downregulate mutp53 by promoting its degradation, while having no effect on wtp53 (Figure 1). Mechanistically, SAHA acts by inhibiting HDAC6, thereby disturbing the physical interaction between Hsp90 and mutp53 that is the basis of mutp53 hyperstability. This in turn enables the reactivation of the endogenous ubiquitin ligases MDM2 and CHIP to mediate mutp53 degradation, thus depriving these tumors of a crucial survival factor (Figure 2, see model in Figure 6). As a result, we find that SAHA shows preferential cytotoxicity for mutp53 tumor cells, whereas wtp53 and p53 null tumor cells are much less sensitive (Figure 4).
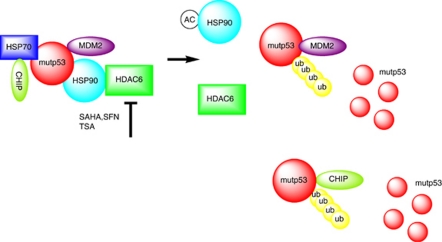
Figure 6.
Proposed model of SAHA-mediated destabilization of mutant p53 by inhibiting the HDAC6-Hsp90 chaperone axis. Based on results from us and others, we propose the following model. Normal tissues that harbor missense mutp53 are able to efficiently control their mutp53 levels despite the fact that their MDM2 levels are only supported by constitutive P1 promoter-derived transcription. In contrast, tumor-specific stabilization of mutp53 proteins, which contributes to driving the tumor phenotype, depends on a second alteration these cells undergo upon malignant transformation. This is their addiction to support from the activated heat shock machinery for survival. In contrast to wtp53, the aberrant conformation of mutp53 proteins requires permanent heat shock support, thus mutp53 is stably engaged in complexes with the highly activated HSP90 chaperone to prevent aggregation. Intimately linked to this conformational stabilization, however, is the fact that this interaction also acts as a large protective ‘cage' against degradation, thereby enabling mutp53's GOF. The E3 ligases MDM2 and CHIP, which in principle are capable of degrading mutp53, might also be trapped in this complex in an inactive state. As mutp53 is fully competent to bind to MDM2, HSP90 likely binds to pre-existing mutp53-MDM2 complexes. Alternatively, chaperone-bound mutp53 could recruit MDM2. HDAC6 is a cytoplasmic non-histone HDAC that deacetylates Hsp90 and an obligate positive regulator of the HSP90 chaperone activity.21, 22 Inhibiting HDAC6 by SAHA and related drugs leads to hyperacetylation and inhibition of Hsp90. This destroys the complex, releases mutp53 and enables MDM2/CHIP-mediated degradation
Our result is important on several accounts. First, it was previously thought that SAHA-induced tumor cell death was independent of their p53 status. This conclusion, however, needs to be updated as it was largely based on equal efficacy between p53-null and wtp53 tumor cells.16, 36, 37 Our study, which is the first systematic side-by-side comparison that includes a random selection of established human mutp53 tumor cells, clearly shows a significantly increased responsiveness in mutp53 tumors. Moreover, SAHA is a pleiotropic drug that traditionally is thought to act primarily at the epigenetic transcriptional level.11, 16 However, our gain-and loss-of function studies demonstrate that in the case of mutp53 tumor cells, SAHA's effect is mainly posttranslational. Its cytotoxicity is to a substantial – albeit not exclusive – degree directly and causally linked to its ability to destabilize mutp53 at the protein level.
SAHA induces mutp53 destabilization at least as efficiently as 17AAG, the prototype of direct Hsp90 inhibitors that until now were the only other class of mutp53 destabilizing agents.7 Moreover, a synergistic effect of both drugs is seen in some mutant lines that correlates with further decrease of mutp53 levels (Figure 4e). We propose that inhibiting HDAC6 by SAHA, that in turn causes hyperacetylation of HSP90, further lowers the threshold of inhibiting HSP90 by 17AAG, resulting in enhanced ubiquitination of HSP90 client proteins including mutp53. Cancer cells that overexpress mutp53 are generally highly resistant to conventional chemotherapeutic drugs. Of note, using gain/loss-of-function manipulation, we demonstrate that SAHA, by virtue of depleting mutp53, is able to dramatically chemosensitize mutp53 cells to genotoxic stress agents such as camptothecin (Figure 5).
Encouragingly, SAHA is being used in the clinic since several years and found to be well tolerated. Overall our data identifies a novel mutp53-directed action of SAHA and carries the important translational prospect of SAHA becoming a centerpiece in mutp53-specific anticancer strategies. They provide the rationale to next test SAHA and other promising HDACi under development in preclinical in vivo models, currently under way in our laboratory.
Materials and Methods
Human cancer cells
Breast cancer MDA231 (p53R280K), MDA468 (p53R273K), T47D (p53L194F) and SKBR3 (p53R175H), colon cancer SW480 (p53 R273H and P309S), prostate cancer DU145 (p53P223L and V274F), pancreatic cancer PANC1 and ovarian cancer EB2 cell lines harbor mutp53. Conversely, MCF7 (breast), RKO, HCT 116 and DLD1 (colon), U2OS and SJSA (osteosarcoma) contain functional wtp53. SAOS2 and HCT116−/− cells are p53 null. Stable mutant Tet-On SW480 (p53 R273H/P309S) inducibly express shp53 under the control of a tetracycline-regulated promoter when tetracyclin is added into the culture medium (1.0 mg/ml).28 T47D and SW480 cells stably overexpressing excess ectopic p53R280K were generated by transfection and selection. All cells were cultured in 10% FCS/DMEM. Where indicated, cells were treated with CHX (50 μg/ml, Sigma, St. Louis, MO, USA), MG132 (5 μM) or α-amanitin (10 μg/ml, Sigma) added to the medium. Treatment with SAHA was for 24 h at the indicated concentrations. SFN (20 μM, Sigma) was used for 24 h, 17AAG (5 μM, 17-allylamino-17-demethoxygeldanamycin, LC Laboratories, Woburn, MA, USA) was used for 24 h, 5 μM Camptothecin for 3 h and Nutlin (20 μM, Sigma) for 24 h. Cell death was determined by trypan blue exclusion, subG1 fractions in propidium iodide-stained flow cytometry, CellTiter-Blue cell viability assays (Promega, Fitchburg, WI, USA; 96-well format with 10 000 cells/well seeded 24 h before) and standard clonogenicity assays. Invasion was determined by standard Matrigel Boyden chambers. All cell viability assays were done using CellTiter-Blue Cell Viability Assay (Promega, 96-well format with 5000 cells/well seeded 24 h before). Cells were treated for 48 h in various concentrations of drug used. Florescence was detected by SPECTRAmax M2 (Molecular Devices, Sunnyvale, CA, USA). IC50 was calculated using GraphPad Prism 5, 2 sites-Fit logIC50 program (GraphPad Prism, La Jolla, CA, USA).
Plasmids
pCMV-MDM2 and pcDNA3.3-mutp53 plasmids carrying a Neomycin resistance gene were transfected with Lipofectamine (Invitrogen, Carlsbad, CA, USA). Stably transfected clones were selected in 700 mg/ml G418 (Gibco, Carlsbad, CA, USA).
RNA interference
Pools of four different siRNA duplexes specific for human HDAC6 (Dharmacon, Lafayette, CO, USA), MDM2 (Ambion, Austin, TX, USA) and CHIP (Dharmacon) or scrambled control duplexes were transfected with Lipofectamine 2000. Cells were harvested 48 h later for analysis. For Hsp90 silencing, MDA231 cells were transfected with 10 pmol of Silencer Select siRNAs (Ambion).
Immunoblots and immunoprecipitations
For immunoblots, equal total protein of crude cell lysates (2.5–5 μg) were loaded. Antibodies were FL393 and DO1 for p53, SMP14 for MDM2 (Santa Cruz, Santa Cruz, CA, USA), MDMX (Bethyl Lab, Montgomery, TX, USA), CHIP (Calbiochem, Darmstadt, Germany), HDAC6 (Santa Cruz) HAUSP (Calbiochem), cleaved PARP (Cell Signaling, Beverly, MA, USA), Hsp90, E-cadherin, pan-p63 H137 (all Santa Cruz), actin and rabbit IgG (Sigma). For detecting endogenous complexes, crude lysates were immunoprecipitated with 1 μg antibody for 2 h. Beads were washed three times with SNNTE plus 2 × RIPA (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% Na deoxycholate, pH 7.4) before immunoblotting.
Acknowledgments
This work was funded by grants from the National Cancer Institute (CA0664 and CA93853 to UMM), Deutsche Krebshilfe (108173 to UMM), Deutsche Forschungsgemeinschaft (MO 1998/1-1 to UMM) and the Carol Baldwin Breast Cancer Research Fund (to NDM). We also thank Sulan Xu, Alisha Yalowitz and Chia-Ying Yang (Stony Brook University) for their technical assistance.
Glossary
- wtp53
wild-type p53
- mutp53
mutant p53
- GOF
gain-of-function
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- CHX
cycloheximide
- KI
knock-in
- SFN
Sulforaphane
- SAHA
suberoylanilide hydroxamic acid
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- Li R, Sutphin PD, Schwartz D, Matas D, Almog N, Wolkowicz R, et al. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene. 1998;16:3269–3277. doi: 10.1038/sj.onc.1201867. [DOI] [PubMed] [Google Scholar]
- Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–309. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Marchenko N, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Bali P, George P, Cohen P, Tao J, Guo F, Sigua C, et al. Superior activity of the combination of histone deacetylase inhibitor LAQ824 and the FLT-3 kinase inhibitor PKC412 against human acute myelogenous leukemia cells with mutant FLT-3. Clin Cancer Res. 2004;10:4991–4997. doi: 10.1158/1078-0432.CCR-04-0210. [DOI] [PubMed] [Google Scholar]
- Fuino L, Bali P, Wittmann S, Donapaty S, Guo F, Yamaguchi H, et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol Cancer Ther. 2003;2:971–984. [PubMed] [Google Scholar]
- Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, et al. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res. 2003;63:5126–5135. [PubMed] [Google Scholar]
- Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, et al. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Esser C, Scheffner M, Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Sutphin P, An WG, Schulte T, Blagosklonny MV, Neckers L. Geldanamycin-stimulated destabilization of mutated p53 is mediated by the proteasome in vivo. Oncogene. 1997;14:2809–2816. doi: 10.1038/sj.onc.1201120. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A, Schwartzman J, Deng V, Alumkal J. Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6. Proc Natl Acad Sci USA. 2009;106:16663–16668. doi: 10.1073/pnas.0908908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Trostel S, Kayastha G, Demidenko ZN, Vassilev LT, Romanova LY, et al. Depletion of mutant p53 and cytotoxicity of histone deacetylase inhibitors. Cancer Res. 2005;65:7386–7392. doi: 10.1158/0008-5472.CAN-04-3433. [DOI] [PubMed] [Google Scholar]
- Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshochi R, Halim F, Sunters A, Alao JP, Madureira PA, Hart SM, et al. ICI182,780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor alpha from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J Biol Chem. 2005;280:3185–3196. doi: 10.1074/jbc.M408063200. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH. The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1998;18:1517–1524. doi: 10.1128/mcb.18.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayan BS, Yang AL, Conforti F, Bernardini S, Tucci P, Vasa-Nicotera M, et al. Induction of Tap63 by histone deacetylase inhibitors. Biochem Biophys Res Commun. 2010;391:1748–1751. doi: 10.1016/j.bbrc.2009.12.147. [DOI] [PubMed] [Google Scholar]
- Beyer U, Moll-Rocek J, Moll UM, Dobbelstein M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci USA. 2011;108:3624–3629. doi: 10.1073/pnas.1016201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Liu G, Scoumanne A, Chen X. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res. 2008;68:6789–6796. doi: 10.1158/0008-5472.CAN-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223:116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- King FW, Wawrzynow A, Hohfeld J, Zylicz M. Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J. 2001;20:6297–6305. doi: 10.1093/emboj/20.22.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S, Neckers L. Extracellular heat shock protein 90: a role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 2007;98:1536–1539. doi: 10.1111/j.1349-7006.2007.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Wei W, Wang SY, Du ZY, Xu YJ, Yu XD. Histone deacetylase inhibitor SAHA induces ERalpha degradation in breast cancer MCF-7 cells by CHIP-mediated ubiquitin pathway and inhibits survival signaling. Biochem Pharmacol. 2008;75:1697–1705. doi: 10.1016/j.bcp.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Muhlenberg T, Zhang Y, Wagner AJ, Grabellus F, Bradner J, Taeger G, et al. Inhibitors of deacetylases suppress oncogenic KIT signaling, acetylate HSP90, and induce apoptosis in gastrointestinal stromal tumors. Cancer Res. 2009;69:6941–6950. doi: 10.1158/0008-5472.CAN-08-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L, et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci USA. 2007;104:8071–8076. doi: 10.1073/pnas.0702294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.