Abstract
Objective
Two functional single nucleotide polymorphisms (SNP) in the PTPN22 gene (rs24746601 and rs33996649) have been associated with autoimmunity. The aim of this study was to investigate the role of the R263Q SNP for the first time and to re-evaluate the role of the R620W SNP in the genetic predisposition to systemic sclerosis (SSc) susceptibility and clinical phenotypes.
Methods
3422 SSc patients (2020 with limited cutaneous SSc and 1208 with diffuse cutaneous SSc) and 3638 healthy controls of Caucasian ancestry from an initial case--control set of Spain and seven additional independent replication cohorts were included in our study. Both rs33996649 and rs2476601 PTPN22 polymorphisms were genotyped by TaqMan allelic discrimination assay. A meta-analysis was performed to test the overall effect of these PTPN22 polymorphisms in SSc.
Results
The meta-analysis revealed evidence of association of the rs2476601 T allele with SSc susceptibility (pFDRcorrected=0.03 pooled, OR 1.15, 95% CI 1.03 to 1.28). In addition, the rs2476601 T allele was significantly associated with anticentromere-positive status (pFDRcorrected=0.02 pooled, OR 1.22, 95% CI 1.05 to 1.42). Although the rs33996649 A allele was significantly associated with SSc in the Spanish population (pFDRcorrected=0.04, OR 0.58, 95% CI 0.36 to 0.92), this association was not confirmed in the meta-analysis (p=0.36 pooled, OR 0.89, 95% CI 0.72 to 1.1).
Conclusion
The study suggests that the PTPN22 R620W polymorphism influences SSc genetic susceptibility but the novel R263Q genetic variant does not. These data strengthen evidence that the R620W mutation is a common risk factor in autoimmune diseases.
Systemic sclerosis (SSc) is a complex disease with an autoimmune origin in which extensive fibrosis, vascular alterations and autoantibodies against various cellular antigens are among the principal features.1 There are two major subgroups in the actual classification of SSc: limited cutaneous (lcSSc) and diffuse cutaneous (dcSSc).2 In lcSSc, fibrosis is mainly restricted to the hands, arms and face. Anticentromere antibodies (ACA) occur in 50–90% of lcSSc patients. Conversely, dcSSc is a rapidly progressing disorder that affects a large area of skin and compromises one or more internal organs. Antitopoisomerase I antibodies (ATA) are more frequently associated with this form of SSc.1,2
SSc occurs in genetically predisposed individuals who have encountered specific environmental factors and/or other stochastic factors.1-3 Similar to other autoimmune disorders, the most consistent and reproducible genetic association with SSc corresponds to the major histocompatibility complex.3 Genes encoding molecules involved in immune function have also recently been associated with susceptibility to SSc, such as IRF5, STAT4 genes and the C8orf13-BLK region.4-9 In spite of these findings, the complete genetic background of SSc, the nature of its genetic determinants and how they contribute to SSc susceptibility and clinical manifestations are still poorly understood.1,3
The protein tyrosine phosphatase non-receptor 22 (PTPN22) gene encodes the protein tyrosine phosphatase lymphoid tyrosine phosphatase, which is a critical gatekeeper of T-cell receptor (TCR) signalling. In T cells, lymphoid tyrosine phosphatase potently inhibits signalling through dephosphorylation of several substrates, including the Src family kinases Lck and Fyn, as well as ZAP-70 and TCRzeta.10-12 Interestingly, PTPN22 has emerged as an important genetic risk factor for human autoimmunity. In particular, two missense single nucleotide polymorphisms (SNP) are associated with autoimmune disorders. The R620W (C1858T, rs2476601) polymorphism in PTPN22 exon 14 was first associated with type 1 diabetes13 and subsequently with other autoimmune disorders such as rheumatoid arthritis (RA)14,15 and systemic lupus erythematosus (SLE)16 and others (reviewed in Lee et al).17 Interestingly, the role of the R620W polymorphism in SSc has also been investigated and shows a trend of association.18-21 Another polymorphism in PTPN22 that is associated with autoimmunity is R263Q (G788A; rs33996649) in exon 10, which alters an amino acid in the catalytic domain of the enzyme. The R263Q polymorphism is a protective factor to SLE.22 Both polymorphisms seem to have functional relevance in the immune response.13,22-26
In this study, we evaluated the role of the PTPN22 R263Q polymorphism in SSc for the first time and re-evaluated the influence of the R620W polymorphism in the genetic background of SSc and its clinical phenotypes.
MATERIALS AND METHODS
Patients
A total of 3422 SSc patients and 3638 controls was included in this study. First, we analysed an initial case–control set of 636 SSc patients (370 with lcSSc and 182 with dcSSc) and 1128 healthy controls of Spanish Caucasian ancestry. In addition, seven independent replication cohorts were analysed (Belgium 120 lcSSc, 58 dcSSc and 256 controls; England 344 lcSSc, 128 dcSSc and 373 controls; Germany 164 lcSSc, 128 dcSSc and 288 controls; Italy 292 lcSSc, 115 dcSSc and 371 controls; The Netherlands 131 lcSSc, 41 dcSSc and 277 controls; USA 607 lcSSc, 388 dcSSc and 693 controls; and Sweden 270 lcSSc, 191 dcSSc and 280 controls).
All of the patients fulfilled the 1980 American College of Rheumatology (ACR) classification criteria for SSc.27 In addition, patients were classified as having limited or diffuse SSc. When patients with SSc have cutaneous involvement distal to the elbows and knees, they fulfil definitions for limited sclero-derma.2 Those SSc patients with cutaneous changes proximal to the elbows and knees were classified as having diffuse SSc.28 In addition, the following clinical data were collected to ascertain the clinical SSc phenotype: age, gender, disease duration, the presence of SSc-specific autoantibodies and the presence of ACA and ATA (anti-Scl70). The methods used to determine the autoantibodies were the same in all contribution centres and have been described previously.20 Lung involvement was assessed according to international guidelines.29 Pulmonary fibrosis was assessed by a CT scan. Restrictive syndrome and diffusion capacity of the lungs was defined as a forced vital capacity of less than 75% of the predicted value and a diffusion capacity for carbon monoxide of less than 75% of the predicted value (based on age, sex, height and ethnic origin). All of the populations studied and their recruiting centres have been reported and described previously.6,8,9,20,30 The main clinical features of the SSc patients from all of the analysed case sets are summarised in table 1.
Table 1.
Main clinical features of SSc patients from the Spanish and the seven replication cohorts
| SSc patients population
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Spain | Belgium | England | Germany | Italy | Netherlands | USA | Sweden | |
| Female (%) | 89 | 77.6 | 83.5 | 87 | 95 | 72 | 88.4 | 78 |
| Limited phenotype (%) | 58.2 | 63.4 | 72.6 | 47.2 | 70.8 | 68.9 | 39.2 | 78.3 |
| ACA positivity (%) | 41.8 | 21.69 | 28.1 | 37 | 42 | 22.6 | 28.3 | 27.4 |
| Anti-Scl70 positivity (%) | 19.8 | 12.5 | 11.9 | 26.8 | 25.3 | 24.7 | 16.5 | 17.7 |
| Pulmonary fibrosis on CT scan (%) | 32 | – | 43 | 36 | 36.2 | 40 | – | 45 |
| Low FVC (<75% predicted) (%) | 28 | – | 30 | 18 | 18 | 24 | – | 35 |
| Low DLCO (<75% predicted) (%) | 45 | – | 11.4 | 50 | 50 | 20 | – | 24 |
Values are expressed in %
ACA, anticentromere antibody; DLCO, diffusion capacity for carbon monoxide; FVC, forced vital capacity; SSc, systemic sclerosis.
Clinical records about the co-occurrence of other autoimmune disorders in SSc patients were not available for all study cohorts. Therefore, to follow a homogeneous inclusion criteria in all case–control sets patients having a concomitant autoimmune condition were not excluded from the analyses. The control population consisted of unrelated, healthy individuals recruited in the same geographical region as the SSc patients and was matched by age, sex and ethnicity with the SSc patients groups. The study was approved by the local ethical committees from all of the participating centres, and written informed consent was obtained from both patients and controls.
PTPN22 genotyping
DNA from patients and controls was obtained using standard methods. Samples were genotyped for the rs33996649 PTPN22 polymorphism using TaqMan 5′ allelic discrimination assay technology, designed by Custom TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, California, USA). The rs2476601 PTPN22 polymorphism was genotyped using a pre-designed SNP genotyping assay provided by Applied Biosystems (part number: C__16021387_20). The PCR were performed as described previously.15
All samples were genotyped in the same centre to avoid inconsistencies. To verify genotyping accuracy, randomly selected samples were genotyped twice and showed 99% identical genotypes; the call rate was higher than 90% for all studied populations.
Statistical analysis
We tested Hardy–Weinberg equilibrium (HWE) for each case–control set using the program FINETI (http://ihg.gsf.de/cgi-bin/hw/hwa2.pl). Significance was calculated with 2×2 contingency tables and Fisher’s exact test to obtain p values, OR and 95% CI using PLINK V.1.07 (http://pngu.mgh.harvard.edu/purcell/plink/). p Values less than 0.05 were considered statistically significant. Multiple testing was corrected by false discovery rate control (pFDR). Linkage disequilibrium measurements (r2) between rs33996649 and rs2476601 were estimated by the expectation-maximisation algorithm using HAPLOVIEW version 4.1 (Broad Institute of MIT and Harvard).
A search of the literature was made using Medline citations to identify available articles in which the association of the rs2476601 PTPN22 (C1858T, R620W) polymorphism with SSc disease had been examined. The medical subject heading terms and text words used were ‘protein tyrosine phosphatase’, ‘PTPN22’, ‘scleroderma’ and ‘SSc’. A previously published study was included in the meta-analysis if (1) it was published by October 2009, (2) it was original data (independent among studies), (3) it provided enough data to calculate the OR and (4) it included SSc patients diagnosed by the 1980 ACR classification criteria for SSc.27 An article was excluded if (1) it contained overlapping data, (2) the number of null and wild genotypes could not be ascertained and (3) the patients and controls included were related. We obtained (via personal communication) the frequencies of the R620W polymorphism in the SSc subtypes and autoantibodies if they were not provided in the selected manuscripts. Dieudé et al19 thus provided a more complete dataset for their French cohort, and Gourh et al20 provided genotypic frequency distribution in the subtypes of SSc.
The analysis of the combined data from all populations was performed using Stats Direct version 2.6.6 (StatsDirect Ltd, Cheshire, UK) for global SSc, the clinical subgroups of the disease (lcSSc and dcSSc) and the autoantibody classification (ACA and ATA). Homogeneity of the OR among cohorts was calculated using Breslow–Day and Cochran’s Q test methods. Higgins’ test (I2) was used to determine if the percentage of total variation across the studies is due to heterogeneity rather than chance (I2 <25% low, I2 50% moderate and I2 >75% high).31 The pooled OR were calculated under a fixed-effects model (Mantel–Haenszel meta-analysis if I2 <25% or I2 50%) or random effects model (DerSimonian–Laird if I2 >75%).32 The estimation of the statistical power of the study was performed using the CaTS-Power Calculator (Andrew Skol and Gonçalo Abecasi, 2006).
RESULTS
The PTPN22 R263Q polymorphism is associated with SSc in the Spanish population
First, we conducted an association study in a case–controls set of Spanish Caucasian ancestry. The distribution of the allelic frequencies of the two studied polymorphisms, R263Q and R620W, was in HWE in both Spanish SSc patients and controls (table 2). The allele frequencies for the R620W variant in the Spanish population (minor allelic frequency (MAF) 0.07) were very similar to that reported previously in Caucasian populations, including the international HapMap project (MAF 0.1) (http://www.hapmap.org) and previous studies (MAF 0.05–0.1).13-17
Table 2.
Genotype and allele frequencies for rs33996649 and rs2476601 PTPN22 polymorphisms in healthy controls and patients with SSc from eight countries
| SNP | Population | GG | GA | AA | G | A | p Value* | OR | (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs33996649 | Spain | SSc (n=599) | 0.960 | 0.040 | 0 | 0.979 | 0.0211 | |||
| Controls (n=1128) | 0.932 | 0.068 | 0 | 0.966 | 0.0341 | 0.04¶ | 0.58 | 0.36 to 0.92 | ||
| Belgium | SSc (n=187) | 0.952 | 0.048 | 0 | 0.976 | 0.0241 | ||||
| Controls (n=236) | 0.949 | 0.051 | 0 | 0.975 | 0.0254 | 0.9 | 0.95 | 0.39 to 2.27 | ||
| England | SSc (n=477) | 0.956 | 0.042 | 0.002 | 0.977 | 0.0231 | ||||
| Controls (n=382) | 0.969 | 0.031 | 0 | 0.984 | 0.0157 | 0.28 | 1.48 | 0.73 to 3.01 | ||
| Germany | SSc (n=354) | 0.966 | 0.034 | 0 | 0.983 | 0.0169 | ||||
| Controls (n=285) | 0.958 | 0.042 | 0 | 0.979 | 0.0211 | 0.59 | 0.8 | 0.36 to 1.8 | ||
| Italy | SSc (n=419) | 0.945 | 0.055 | 0 | 0.973 | 0.0274 | ||||
| Controls (n=371) | 0.938 | 0.062 | 0 | 0.969 | 0.0310 | 0.68 | 0.88 | 0.49 to 1.59 | ||
| Netherlands | SSc (n=185) | 0.959 | 0.041 | 0 | 0.979 | 0.0207 | ||||
| Controls (n=263) | 0.951 | 0.049 | 0 | 0.975 | 0.0247 | 0.72 | 0.83 | 0.32 to 2.22 | ||
| USA | SSc (n=1050) | 0.947 | 0.053 | 0 | 0.973 | 0.0267 | ||||
| Controls (n=693) | 0.958 | 0.042 | 0 | 0.979 | 0.0209 | 0.28 | 1.28 | 0.81to 2.02 | ||
| Sweden | SSc (n=191) | 0.974 | 0.026 | 0 | 0.987 | 0.0131 | ||||
| Controls (n=280) | 0.946 | 0.054 | 0 | 0.973 | 0.0268 | 0.15 | 0.48 | 0.17 to1.34 | ||
| Pooled† | SSc(n=3422) | 0.954 | 0.045 | 0 | 0.977 | 0.0230 | ||||
| Controls (n=3638) | 0.947 | 0.053 | 0 | 0.973 | 0.0530 | |||||
| Fixed model | 0.36 | 0.89 | 0.72 to 1.12 | |||||||
|
| ||||||||||
| SNP | Population | CC | CT | TT | C | T | p Value* | OR | (95% CI) | |
|
| ||||||||||
| rs2476601 | Spain | SSc (n=636) | 0.857 | 0.137 | 0.006 | 0.925 | 0.075 | |||
| Controls (n=1128) | 0.857 | 0.136 | 0.007 | 0.925 | 0.075 | 0.98 | 0.99 | 0.77 to 1.29 | ||
| Belgium | SSc (n=189) | 0.788 | 0.212 | 0.000 | 0.894 | 0.106 | ||||
| Controls (n=256) | 0.859 | 0.129 | 0.012 | 0.927 | 0.073 | 0.08 | 1.51 | 0.95 to 2.42 | ||
| England | SSc (n=463) | 0.793 | 0.201 | 0.006 | 0.893 | 0.107 | ||||
| Controls (n=373) | 0.853 | 0.147 | 0.000 | 0.926 | 0.074 | 0.04¶ | 1.5 | 1.07 to 2.12 | ||
| Germany | SSc (n=343) | 0.802 | 0.175 | 0.023 | 0.889 | 0.111 | ||||
| Controls (n=288) | 0.774 | 0.212 | 0.014 | 0.880 | 0.120 | 0.62 | 0.92 | 0.65 to 1.3 | ||
| Italy | SSc (n=383) | 0.890 | 0.104 | 0.005 | 0.943 | 0.057 | ||||
| Controls (n=371) | 0.935 | 0.059 | 0.005 | 0.965 | 0.035 | 0.08¶ | 1.68 | 1.02 to 2.76 | ||
| Netherlands | SSc (n=190) | 0.805 | 0.189 | 0.005 | 0.900 | 0.100 | ||||
| Controls (n=277) | 0.819 | 0.177 | 0.004 | 0.908 | 0.092 | 0.68 | 1.1 | 0.7 to 1.71 | ||
| Sweden | SSc (n=175) | 0.789 | 0.200 | 0.011 | 0.889 | 0.111 | ||||
| Controls (n=279) | 0.789 | 0.190 | 0.022 | 0.884 | 0.116 | 0.82 | 0.95 | 0.62 to 1.45 | ||
| Dieudé et al19‡ | SSc (n=1018) | 0.820 | 0.172 | 0.008 | 0.906 | 0.094 | ||||
| Controls (n=1004) | 0.828 | 0.163 | 0.009 | 0.909 | 0.091 | 0.73 | 1.04 | 0.84 to 1.28 | ||
| Gourh et al20‡ | SSc (n=666) | 0.787 | 0.203 | 0.011 | 0.888 | 0.112 | ||||
| Controls (n=430) | 0.844 | 0.147 | 0.009 | 0.917 | 0.083 | 0.06¶ | 1.39 | 1.03 to 1.87 | ||
| Pooled§ | SSc (n=4063) | 0.819 | 0.173 | 0.009 | 0.905 | 0.095 | ||||
| Controls (n=4406) | 0.843 | 0.148 | 0.008 | 0.917 | 0.083 | |||||
| Fixed model | 0.03¶ | 1.15 | 1.03 to 1.28 | |||||||
p Value for the minor allele.
I2=17.6%, Breslow–Day p=0.281 Q=8.49 p=0.29; The statistical power for this pooled analysis was 97%.
The authors of those papers provided by personal communication the actualisation and specific data of their works.
I2=33.8%, Breslow–Day p=0.145 Q=12.08 p=0.15. The statistical power for this pooled analysis was 100%.
False discovery rate correction p value.
SNP, single nucleotide polymorphism; SSc, systemic sclerosis.
After comparing the genotypic and allelic frequencies of the R263Q and R620W PTPN22 genetic variants between SSc Spanish patients and healthy controls, we observed that the R263Q A allele was associated with SSc (p=0.02 (pFDRcorrected=0.04), OR 0.58, 95% CI 0.36 to 0.92). However, we did not observe any significant difference for the R620W polymorphism in this population (p=0.98, OR 0.99, 95% CI 0.77 to 1.29) (table 2).
In addition, we performed an analysis stratifying the patients according to their clinical outcome, that is, dcSSc, lcSSc, ACA or ATA-positive patients. Interestingly, we observed that the frequency of the A allele of the R236Q polymorphism was higher in healthy controls (3%) compared with lcSSc Spanish patients (2%) (p=0.02 (pFDRcorrected=0.04), OR 0.49, 95% CI 0.27 to 0.91) (table 3). However, no significant association was found between R620W and the subtypes of the disease or between the R263Q or R620W PTPN22 polymorphisms and ACA and ATA-positive subsets of SSc (tables 3 and 4).
Table 3.
Distribution of PTPN22 rs33996649 and rs2476601 polymorphisms in SSc subtypes and healthy controls
| SNP | Population | GG | GA | AA | G | A | p Value* | OR | (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs33996649 | Spain | Limited (n=351) | 0.966 | 0.034 | 0 | 0.983 | 0.0171 | 0.04†† | 0.49 | 0.27 to 0.91 |
| Diffuse (n=169) | 0.947 | 0.053 | 0 | 0.973 | 0.0266 | 0.47 | 0.77 | 0.38 to 1.56 | ||
| Controls (n=1128) | 0.932 | 0.068 | 0 | 0.966 | 0.0341 | |||||
| Belgium | Limited (n=119) | 0.950 | 0.050 | 0 | 0.975 | 0.0252 | 0.99 | 0.99 | 0.37 to 2.68 | |
| Diffuse (n=58) | 0.931 | 0.052 | 0 | 0.957 | 0.0259 | 0.96 | 1.04 | 0.29 to 3.73 | ||
| Controls (n=236) | 0.949 | 0.051 | 0 | 0.975 | 0.0254 | |||||
| England | Limited (n=344) | 0.962 | 0.035 | 0.003 | 0.980 | 0.0203 | 0.51 | 1.30 | 0.60 to 2.83 | |
| Diffuse (n=128) | 0.945 | 0.055 | 0 | 0.973 | 0.0273 | 0.23 | 1.76 | 0.69 to 4.52 | ||
| Controls (n=382) | 0.969 | 0.031 | 0 | 0.984 | 0.0157 | |||||
| Germany | Limited (n=164) | 0.970 | 0.030 | 0 | 0.985 | 0.0152 | 0.54 | 0.72 | 0.25 to 2.06 | |
| Diffuse (n=128) | 0.977 | 0.023 | 0 | 0.992 | 0.0078 | 0.35 | 0.55 | 0.15 to 1.97 | ||
| Controls (n=285) | 0.958 | 0.042 | 0 | 0.979 | 0.0211 | |||||
| Netherlands | Limited (n=109) | 0.954 | 0.046 | 0 | 0.977 | 0.0229 | 0.89 | 0.93 | 0.33 to 2.63 | |
| Diffuse (n=31) | 0.968 | 0.032 | 0 | 0.984 | 0.0161 | 0.67 | 0.65 | 0.08 to 5.03 | ||
| Controls (n=263) | 0.951 | 0.049 | 0 | 0.975 | 0.0247 | |||||
| Italy | Limited (n=292) | 0.952 | 0.062 | 0 | 0.976 | 0.0240 | 0.44 | 0.77 | 0.39 to 1.51 | |
| Diffuse (n=115) | 0.939 | 0.061 | 0 | 0.970 | 0.0304 | 0.97 | 0.98 | 0.42 to 2.32 | ||
| Controls (n=371) | 0.938 | 0.062 | 0 | 0.969 | 0.0310 | |||||
| USA | Limited (n=607) | 0.956 | 0.044 | 0 | 0.978 | 0.0222 | 0.82 | 1.06 | 0.63 to 1.81 | |
| Diffuse (n=388) | 0.938 | 0.062 | 0 | 0.969 | 0.0309 | 0.15 | 1.49 | 0.86 to 2.58 | ||
| Controls (n=693) | 0.958 | 0.042 | 0 | 0.979 | 0.0209 | |||||
| Sweden | Limited (n=270) | 0.493 | 0.015 | 0 | 0.984 | 0.0160 | 0.27 | 0.54 | 0.18 to 1.64 | |
| Diffuse (n=191) | 0.974 | 0.026 | 0 | 0.987 | 0.0131 | 0.28 | 0.34 | 0.04 to 2.60 | ||
| Controls (n=280) | 0.946 | 0.054 | 0 | 0.973 | 0.0268 | |||||
| Pooled†‡ | Limited (n=2020) | 0.959 | 0.040 | 0 | 0.980 | 0.0200 | 0.13 | 0.81 | 0.62 to 1.05 | |
| Diffuse (n=1208) | 0.951 | 0.049 | 0 | 0.975 | 0.0250 | 0.99 | 0.99 | 0.73 to 1.33 | ||
| Controls (n=3638) | 0.947 | 0.053 | 0 | 0.973 | 0.0530 | |||||
|
| ||||||||||
| SNP | Population | CC | CT | TT | C | T | p Value* | OR | (95% CI) | |
|
| ||||||||||
| rs2476601 | Spain | Limited (n=370) | 0.889 | 0.108 | 0.003 | 0.943 | 0.0568 | 0.09 | 0.74 | 0.52 to 1.05 |
| Diffuse (n=182) | 0.808 | 0.181 | 0.011 | 0.898 | 0.1016 | 0.08 | 1.40 | 0.96 to 2.03 | ||
| Controls (n=1128) | 0.857 | 0.136 | 0.007 | 0.925 | 0.0749 | |||||
| Belgium | Limited (n=120) | 0.775 | 0.225 | 0.000 | 0.888 | 0.1125 | 0.07 | 1.62 | 0.96 to 2.73 | |
| Diffuse (n=58) | 0.845 | 0.155 | 0.000 | 0.922 | 0.0776 | 0.85 | 1.08 | 0.50 to 2.30 | ||
| Controls (n=256) | 0.859 | 0.129 | 0.012 | 0.927 | 0.0725 | |||||
| England | Limited (n=336) | 0.774 | 0.217 | 0.009 | 0.882 | 0.1176 | 0.01†† | 1.67 | 1.17 to 2.40 | |
| Diffuse (n=122) | 0.852 | 0.148 | 0.000 | 0.926 | 0.0738 | 1.00 | 1.00 | 0.58 to 1.74 | ||
| Controls (n=373) | 0.853 | 0.147 | 0.000 | 0.926 | 0.0737 | |||||
| Germany | Limited (n=162) | 0.772 | 0.185 | 0.043 | 0.864 | 0.1358 | 0.49 | 1.16 | 0.77 to 1.73 | |
| Diffuse (n=120) | 0.817 | 0.175 | 0.008 | 0.904 | 0.0958 | 0.32 | 0.78 | 0.47 to 1.28 | ||
| Controls (n=288) | 0.774 | 0.212 | 0.014 | 0.880 | 0.1198 | |||||
| Netherlands | Limited (n=131) | 0.794 | 0.206 | 0.000 | 0.897 | 0.1031 | 0.62 | 1.13 | 0.69 to 1.85 | |
| Diffuse (n=41) | 0.854 | 0.122 | 0.024 | 0.915 | 0.0854 | 0.84 | 0.92 | 0.40 to 2.10 | ||
| Controls (n=277) | 0.819 | 0.177 | 0.004 | 0.908 | 0.0921 | |||||
| Italy | Limited (n=271) | 0.886 | 0.111 | 0.000 | 0.941 | 0.0590 | 0.09†† | 1.73 | 1.02 to 2.94 | |
| Diffuse (n=102) | 0.892 | 0.098 | 0.010 | 0.941 | 0.0588 | 0.13 | 1.72 | 0.85 to 3.47 | ||
| Controls (n=371) | 0.935 | 0.059 | 0.005 | 0.965 | 0.0350 | |||||
| Sweden | Limited (n=137) | 0.803 | 0.190 | 0.007 | 0.898 | 0.1022 | 0.54 | 0.86 | 0.54 to 1.38 | |
| Diffuse (n=175) | 0.789 | 0.200 | 0.011 | 0.889 | 0.1114 | 0.48 | 1.28 | 0.64 to 2.56 | ||
| Controls (n=279) | 0.789 | 0.190 | 0.022 | 0.884 | 0.1165 | |||||
| Dieudé et al19§ | Limited (n=641) | 0.811 | 0.181 | 0.008 | 0.902 | 0.0983 | 0.46 | 1.10 | 0.86 to 1.39 | |
| Diffuse (n=315) | 0.838 | 0.156 | 0.006 | 0.916 | 0.0841 | 0.62 | 0.93 | 0.68 to 1.28 | ||
| Controls (n=1004) | 0.828 | 0.163 | 0.009 | 0.909 | 0.0906 | |||||
| Gourh et al20§ | Limited (n=378) | 0.778 | 0.209 | 0.013 | 0.882 | 0.1177 | 0.04†† | 1.48 | 1.07 to 2.05 | |
| Diffuse (n=254) | 0.791 | 0.201 | 0.008 | 0.892 | 0.1083 | 0.11 | 1.35 | 0.94 to 1.96 | ||
| Controls (n=430) | 0.844 | 0.147 | 0.009 | 0.917 | 0.0826 | |||||
| Pooled¶** | Limited (n=2546) | 0.815 | 0.176 | 0.009 | 0.903 | 0.0970 | 0.12 | 1.18 | 0.96 to 1.44 | |
| Diffuse (n=1459) | 0.820 | 0.173 | 0.007 | 0.906 | 0.0940 | 0.28 | 1.09 | 0.94 to 1.26 | ||
| Controls (n=4406) | 0.843 | 0.148 | 0.008 | 0.917 | 0.0830 | |||||
p Value for the minor allele
Heterogeneity for systemic sclerosis (SSc) limited analysis I2=0.0%, Breslow–Day p=0.57 Q=5.59 p=0.58.
Heterogeneity for SSc diffused analysis I2=9.1%, Breslow–Day p=0.33 Q=7.70 p=0.36. The statistical power for this pooled analysis was 81%.
The authors of those papers provided by personal communication the actualisation and specific data of their works.
Heterogeneity for SSc limited analysis I2=59.79% Breslow–Day p=0.0102 Q=19.84 p=0.0109. The values showed for the limited form of the SSc meta-analysis correspond to the random effects model. The statistical power for this pooled analysis was 100%.
Heterogeneity for SSc diffused analysis I2=0%, Breslow–Day p=0.43 Q=7.91 p=0.44. The statistical power for this pooled analysis was 99%.
False discovery rate correction p value.
SNP, single nucleotide polymorphism.
Table 4.
Distribution of PTPN22 genetic variants according with scleroderma-specific autoantibody status
| SNP | Population | GG | GA | AA | G | A | p Value* | OR | (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs33996649 | Spain | ACA (n=253) | 0.964 | 0.036 | 0 | 0.982 | 0.0178 | 0.06 | 0.51 | 0.26 to 1.03 |
| ATA (n=112) | 0.964 | 0.036 | 0 | 0.982 | 0.0179 | 0.19 | 0.52 | 0.19 to 1.42 | ||
| Controls (n=1128) | 0.932 | 0.068 | 0 | 0.966 | 0.0341 | |||||
| Belgium | ACA (n=40) | 0.950 | 0.050 | 0 | 0.975 | 0.0250 | 0.98 | 0.98 | 0.22 to 4.48 | |
| ATA (n=32) | 0.938 | 0.063 | 0 | 0.969 | 0.0313 | 0.78 | 1.24 | 0.27 to 5.66 | ||
| Controls (n=236) | 0.949 | 0.051 | 0 | 0.975 | 0.0254 | |||||
| England | ACA (n=133) | 0.962 | 0.038 | 0 | 0.981 | 0.0188 | 0.73 | 1.20 | 0.42 to 3.44 | |
| ATA (n=56) | 0.982 | 0.018 | 0 | 0.991 | 0.0089 | 0.58 | 0.56 | 0.07 to 4.38 | ||
| Controls (n=382) | 0.969 | 0.031 | 0 | 0.984 | 0.0157 | |||||
| Germany | ACA (n=127) | 0.953 | 0.047 | 0 | 0.976 | 0.0236 | 0.82 | 1.13 | 0.42 to 3.03 | |
| ATA (n=97) | 0.979 | 0.021 | 0 | 0.990 | 0.0103 | 0.34 | 0.48 | 0.11 to 2.18 | ||
| Controls (n=285) | 0.958 | 0.042 | 0 | 0.979 | 0.0211 | |||||
| Netherlands | ACA (n=37) | 0.946 | 0.054 | 0 | 0.973 | 0.0270 | 0.91 | 1.10 | 0.24 to 4.96 | |
| ATA (n=32) | 0.938 | 0.063 | 0 | 0.969 | 0.0313 | 0.58 | 0.79 | 0.35 to 1.81 | ||
| Controls (n=263) | 0.951 | 0.049 | 0 | 0.975 | 0.0247 | |||||
| Italy | ACA (n=177) | 0.944 | 0.056 | 0 | 0.972 | 0.0282 | 0.80 | 0.91 | 0.43 to 1.93 | |
| ATA (n=114) | 0.939 | 0.061 | 0 | 0.969 | 0.0307 | 0.98 | 0.99 | 0.42 to 2.34 | ||
| Controls (n=371) | 0.938 | 0.062 | 0 | 0.969 | 0.0310 | |||||
| USA | ACA (n=296) | 0.956 | 0.044 | 0 | 0.978 | 0.0220 | 0.18 | 1.39 | 0.86 to 2.23 | |
| ATA (n=174) | 0.954 | 0.046 | 0 | 0.977 | 0.0230 | 0.15 | 1.49 | 0.86 to 2.58 | ||
| Controls (n=693) | 0.958 | 0.042 | 0 | 0.979 | 0.0209 | |||||
| Sweden | ACA (n=51) | 0.980 | 0.020 | 0 | 0.990 | 0.0098 | 0.30 | 0.36 | 0.05 to 2.75 | |
| ATA (n=33) | 0.939 | 0.061 | 0 | 0.970 | 0.0303 | 0.87 | 1.14 | 0.25 to 5.08 | ||
| Controls (n=280) | 0.946 | 0.054 | 0 | 0.973 | 0.0268 | |||||
| Pooled†‡ | ACA (n=1114) | 0.957 | 0.043 | 0 | 0.978 | 0.0225 | 0.46 | 0.87 | 0.63 to 1.21 | |
| ATA (n=650) | 0.957 | 0.043 | 0 | 0.978 | 0.0215 | 0.48 | 0.85 | 0.56 to 1.27 | ||
| Controls (n=3638) | 0.947 | 0.053 | 0 | 0.973 | 0.0265 | |||||
|
| ||||||||||
| SNP | Population | CC | CT | TT | C | T | p Value* | OR | (95% CI) | |
|
| ||||||||||
| rs2476601 | Spain | ACA (n=266) | 0.853 | 0.139 | 0.008 | 0.923 | 0.0771 | 0.87 | 1.03 | 0.72 to 1.47 |
| ATA (n=126) | 0.881 | 0.111 | 0.008 | 0.937 | 0.0635 | 0.51 | 0.84 | 0.49 to 1.42 | ||
| Controls (n=1128) | 0.857 | 0.136 | 0.007 | 0.925 | 0.0749 | |||||
| Belgium | ACA (n=41) | 0.805 | 0.195 | 0.000 | 0.902 | 0.0976 | 0.43 | 1.38 | 0.62 to 3.08 | |
| ATA (n=32) | 0.750 | 0.250 | 0.000 | 0.875 | 0.1250 | 0.14 | 1.83 | 0.81 to 4.12 | ||
| Controls (n=255) | 0.863 | 0.129 | 0.008 | 0.927 | 0.0725 | |||||
| England | ACA (n=130) | 0.777 | 0.223 | 0.000 | 0.888 | 0.1115 | 0.06 | 1.58 | 0.98 to 2.53 | |
| ATA (n=55) | 0.891 | 0.091 | 0.018 | 0.936 | 0.0636 | 0.70 | 0.85 | 0.38 to 1.93 | ||
| Controls (n=373) | 0.853 | 0.147 | 0.000 | 0.926 | 0.0737 | |||||
| Germany | ACA (n=127) | 0.787 | 0.157 | 0.055 | 0.866 | 0.1339 | 0.57 | 1.14 | 0.73 to 1.76 | |
| ATA (n=92) | 0.804 | 0.196 | 0.000 | 0.902 | 0.0978 | 0.42 | 0.80 | 0.46 to 1.38 | ||
| Controls (n=288) | 0.774 | 0.212 | 0.014 | 0.880 | 0.1198 | |||||
| Netherlands | ACA (n=43) | 0.791 | 0.209 | 0.000 | 0.895 | 0.1047 | 0.71 | 1.15 | 0.55 to 2.44 | |
| ATA (n=47) | 0.851 | 0.149 | 0.000 | 0.926 | 0.0745 | 0.58 | 0.79 | 0.35 to 1.81 | ||
| Controls (n=277) | 0.819 | 0.177 | 0.004 | 0.908 | 0.0921 | |||||
| Italy | ACA (n=161) | 0.888 | 0.106 | 0.006 | 0.941 | 0.0590 | 0.07 | 1.73 | 0.94 to 3.17 | |
| ATA (n=97) | 0.907 | 0.093 | 0.000 | 0.954 | 0.0464 | 0.46 | 1.34 | 0.62 to 2.91 | ||
| Controls (n=371) | 0.935 | 0.059 | 0.005 | 0.965 | 0.0350 | |||||
| Sweden | ACA (n=48) | 0.792 | 0.188 | 0.021 | 0.885 | 0.1146 | 0.96 | 0.98 | 0.50 to 1.94 | |
| ATA (n=31) | 0.613 | 0.355 | 0.032 | 0.790 | 0.2097 | 0.08†† | 2.01 | 1.04 to 3.91 | ||
| Controls (n=279) | 0.789 | 0.190 | 0.022 | 0.884 | 0.1165 | |||||
| Dieudé et al19§ | ACA (n=372) | 0.812 | 0.183 | 0.005 | 0.903 | 0.0968 | 0.62 | 1.08 | 0.81 to 1.44 | |
| ATA (n=247) | 0.834 | 0.158 | 0.008 | 0.913 | 0.0870 | 0.80 | 0.97 | 0.69 to 1.37 | ||
| Controls (n=1004) | 0.828 | 0.163 | 0.009 | 0.909 | 0.0906 | |||||
| Gourh et al20§ | ACA (n=188) | 0.761 | 0.229 | 0.011 | 0.875 | 0.1250 | 0.04†† | 1.60 | 1.08 to 2.35 | |
| ATA (n=107) | 0.710 | 0.271 | 0.019 | 0.846 | 0.1542 | 0.005†† | 2.05 | 1.32 to 3.19 | ||
| Controls (n=430) | 0.844 | 0.147 | 0.009 | 0.917 | 0.0826 | |||||
| Pooled¶** | ACA (n=1376) | 0.815 | 0.174 | 0.011 | 0.902 | 0.0981 | 0.02†† | 1.22 | 1.05 to 1.42 | |
| ATA (n=834) | 0.824 | 0.168 | 0.008 | 0.908 | 0.0923 | 0.26 | 1.17 | 0.89 to 1.55 | ||
| Controls (n=4126) | 0.847 | 0.145 | 0.007 | 0.920 | 0.0800 | |||||
p Value for the minor allele.
Heterogeneity for systemic sclerosis (SSc) patients with anticentromere antibody (ACA)-positive and healthy controls analysis I2=0%, Breslow–Day p=0.61 Q=5.01 p=0.66. The statistical power for this pooled analysis was 69%.
Heterogeneity for SSc patients with antitopoisomerase antibody (ATA)-positive and healthy controls analysis I2=0.0%, Breslow–Day p=0.89 Q=2.82 p=0.90. The statistical power for this pooled analysis was 42%.
These data were provided by personal communication by the authors, because they did not showed the complete frequencies in the previous work.
Heterogeneity for SSc patients with ACA-positive and healthy controls analysis I2=0%, Breslow–Day p=0.60 Q=6.36 p=0.61. The statistical power for this pooled analysis was 99%.
Heterogeneity for SSc patients with ATA-positive and healthy controls analysis I2=49.9%, Breslow–Day p=0.04 Q=15.96 p=0.04. The values showed for the ATA-positive status correspond to the random effects model. The statistical power for this pooled analysis was 90%, in this case the values correspond to the random effect model.
False discovery rate correction p value.
SNP, single nucleotide polymorphism.
A replication study and meta-analysis showed that the R620W polymorphism is associated with SSc and ACA-positive patients
In view of the interesting findings observed in the Spanish population, we conducted a large replication study including seven independent populations with Caucasian ancestry. All analysed control populations were in HWE for both PTPN22 R263Q and R620W genetic variants. As previously reported,22 no linkage disequilibrium between the PTPN22 R263Q and R620W genetic variants in any population was observed (r2<0.03 for all studied populations).
None of the seven replication cohorts showed a significant association of the R263Q PTPN22 polymorphism with SSc susceptibility (table 2), with the disease subtypes (lcSSc and dcSSc) (table 3), or with ACA and ATA status (table 4). Neither did the meta-analysis confirm the significant association of the R263Q genetic variant and SSc, observed in the Spanish population (p=0.36, pooled OR 0.89, 95% CI 0.72 to 1.12) (figure 1A and table 2).
Figure 1.
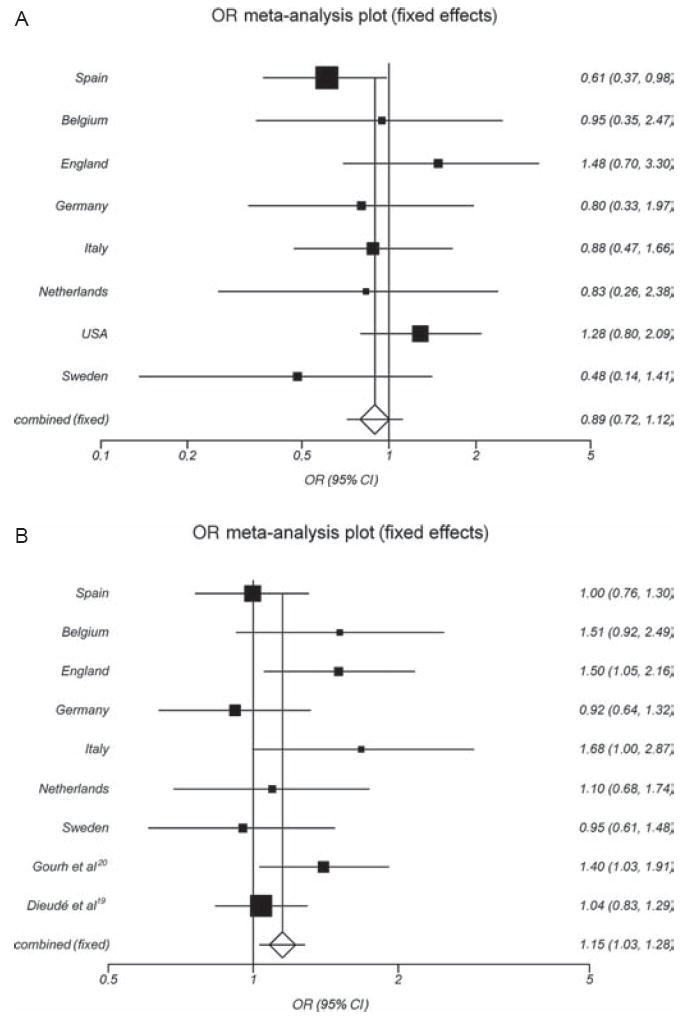
(A) Forest plot for the meta-analysis of the PTPN22 R263Q (G788A; rs33996649) polymorphism in systemic sclerosis in eight Caucasian cohorts. (B) Forest plot of the PTPN22 R620W (C1858T; rs2476601) polymorphism and SSc in seven Caucasian cohorts and two previous studies.
The replication study of the PTPN22 R620W polymorphism showed a slightly increased frequency of the T allele in SSc patients compared with controls in most cohorts (table 2). However, this difference was statistically significant after FDR correction only in the English population (p=0.02 (pFDRcorrected=0.04), OR 1.50, 95% CI 1.07 to 2.12) (table 2, figure 1B). In this regard, the frequency of the T allele was significantly increased in lcSSc English patients (11%) compared with healthy controls (7%) (p=0.005 (pFDRcorrected=0.01), OR 1.67, 95% CI 1.17 to 2.40) (table 3). To evaluate the overall effect of the classic PTPN22 R620W genetic variant in SSc susceptibility, we performed a meta-analysis including the seven case–control sets analysed in this study (from Spain, Belgium, England, Germany, Italy, The Netherlands and Sweden), together with previously published reports. Four studies that analysed the PTPN22 R620W polymorphism in SSc were identified through a Medline search (published in October 2009),18-21 but two of these studies were excluded due to overlapping data,21,18 The authors of the selected manuscripts were contacted to obtain more detailed data information (see Materials and methods).
Heterogeneity was not observed, and the inconsistency was low in the meta-analysis of the R620W PTPN22 polymorphism and SSc (Q=12.08, p=0.15, I2=33.8%). Accordingly, in this pooled analysis, the T allele was significantly associated with susceptibility to develop SSc (p=0.010 (pFDRcorrected=0.03), OR 1.15, 95% CI 1.03 to 1.28) (figure 1B and table 2). Moreover, the meta-analysis of the T allele of PTPN22 R620W and the lcSSc form showed a significant association under the fixed effects model (p=0.02, OR 1.15, 95% CI 1.02 to 1.31). However, heterogeneity was detected (Q=19.84, p=0.01, I2=59.8%), and the final result was based on the random effects model that showed no significant association between the variant allele and the lcSSc subtype (p=0.12, OR 1.18, 95% CI 0.96 to 1.44) (figure2A and table 3). Neither the meta-analysis for the R620W polymorphism nor for the dcSSc form showed significant association (p=0.10, pooled OR 1.15, 95% CI 0.97 to 1.35) (table 3). However, the meta-analysis performed after stratifying SSc patients according to autoantibody status showed that the T allele of the PTPN22 R620W polymorphism was a risk factor for the ACA-positive subset of SSc (p=0.01 (pFDRcorrected=0.02), pooled OR 1.22, 95% CI 1.05 to 1.42) (figure 2B and table 4). In this line, we performed a meta-analysis for the PTPN22 R620W polymorphism between the ACA-positive versus ACA-negative patients in the available data cohorts and we observed the same direction of the OR but the results did not reach significant difference (p=0.16, pooled OR 1.15, 95% CI 0.95 to 1.39, see supplementary figure 1, available online only).
Figure 2.
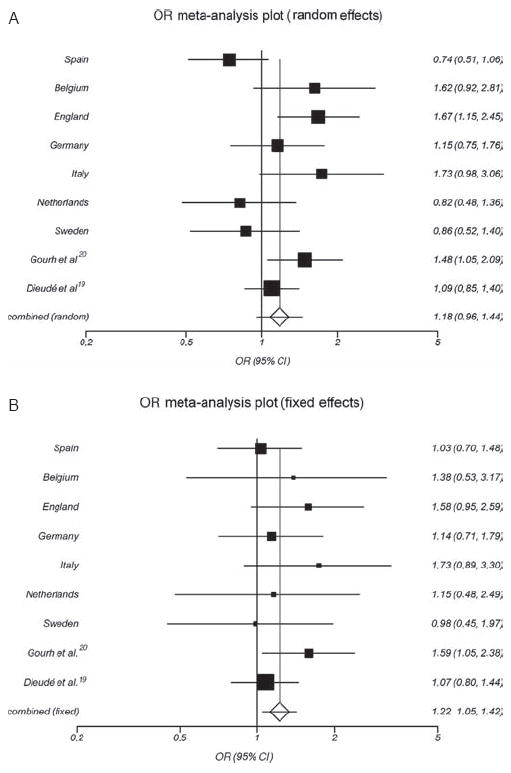
(A) Forest plot for the meta-analysis of the PTPN22 the R620W (C1858T; rs2476601) polymorphism and limited cutaneous systemic sclerosis subtype, in seven Caucasian cohorts and previous studies. (B) Forest plot for the meta-analysis of the PTPN22 the R620W (C1858T; rs2476601) polymorphism and anticentromere antibody-positive autoantibody status of systemic sclerosis, in seven Caucasian cohorts and previous studies.
DISCUSSION
A loss-of-function PTPN22 variant (R263Q, G788A, rs33996649), which is less effective in reducing TCR signalling, was recently associated with SLE.22 Recent findings also demonstrate an overlap in the genetic factors involved in both SLE and SSc, with consistent associations of genetic variants in the STAT4, IRF5 and BANK1 genes.6-9,20,30,33,34 This fact prompted us to analyse the possible influence of the R263Q PTPN22 mutation in the genetic component of SSc and its clinical manifestations. We studied eight Caucasian cohorts and performed a pooled analysis. Although we observed a significant association with the 788A allele in SSc and its lcSSc subtype in the Spanish cohort, this was not confirmed in the pooled analysis that included seven Caucasian ancestry replication cohorts. Considering the statistical power reached by the pooled analysis (97%), we suggest that the R263Q PTPN22 polymorphism does not influence the genetic background of SSc. In spite of the lack of significant association, the pooled OR shows a protective trend (0.87) in the meta-analysis for the R263Q variant and SSc, the same direction observed for the association of this polymorphism in SLE.22 In order to determine with greater accuracy if this polymorphism is implicated in the clinical outcome of SSc or the ACA and ATA autoantibody subsets (the statistical power for these analyses range between 42% and 81%), further studies are needed.
Conversely, in the present study, we wanted to re-evaluate the role of the PTPN22 R620W (C1858T, rs24746601) polymorphism in SSc genetic susceptibility by taking advantage of our large Caucasian cohort and performing a meta-analysis, which is a robust tool for resolving contradictory results and increasing the statistical power in genetic association studies.35-37 Our meta-analysis had high statistical power (100% for the global SSc disease, 100% for SSc in the lcSSc clinical subgroup, 99% for dcSSc, 99% for SSc ACA autoantibody status and 97% for ATA) and showed evidence of association of the 1858T allele with SSc, the ACA subset, and (to a lesser extent) the lcSSc form. Our data confirm and extend previous reports showing a trend of association of the R620W PTPN22 variant with SSc.19,20 For instance, Gourh et al20 observed an increased frequency of the T allele in SSc patients compared with controls in a Caucasian population (p=0.13, OR 1.25). Similarly, Dieudé et al19 conducted a meta-analysis including three Caucasian cohorts, and they observed a trend of association between the T allele and SSc (p=0.12, OR 1.18). Data from our more powerful meta-analysis showed a statistically significant association of the T allele with SSc (pFRDcorrected=0.03, OR 1.15), which highlights the need for large cohorts and a meta-analysis approach to detect minor associations in genetic studies.35-37 The major discrepancies between our study and previous reports are related to the association of the R620W PTPN22 variant with autoantibody status. Gourh et al20 showed a significant association between the 1858 T allele and both ACA and ATA-positive status. However, Dieudé et al19 observed a weaker effect (OR 1.08) between the CT/TT genotypes and ATA but not ACA-positive status. In our large study, we only observed an association between the 1858T allele and ACA-positive status. Interestingly, a meta-analysis for this allele between the ACA-positive versus ACA-negative status confirmed this tendency and showed that the OR maintains the risk direction (OR 1.15) but due to the statistical power (75%) we could not detect a significant difference (see supplementary figure 1, available online only). The discrepancies between these results could be explained partly by the clinical heterogeneity of the disease between populations.38 In addition, it is now clear that the R620W polymorphism displays a wide range of allele frequencies in normal Caucasian populations.37
The observed effect magnitude of the 1858T allele on genetic susceptibility to SSc (OR 1.15) seems to be weaker than that of other autoimmune diseases, such as SLE and RA, indicating that the PTPN22 gene contributes to a lesser extent to SSc genetic susceptibility. However, the specific immunological mechanisms of each disease may explain such results.37,39
Some limitations could be attributed to our study, as patients with SSc complicated by other autoimmune diseases could not be excluded from the analysis. Therefore, given that the R620W polymorphism is associated with multiple autoimmune phenotypes, it can be argued that our findings may result from the genetic background of those SSc patients presenting with another autoimmune disease associated with the PTPN22 gene. Although this possibility cannot be completely discounted, this seems not to be the case, because the most frequent co-autoimmune disease reported in SSc patients, Sjögren’s syndrome, is not associated with R620W in Caucasian populations.17,40 In this regard, it is worth mentioning that Sjögren’s syndrome frequently presents concomitantly with other autoimmune diseases. This association is well described for RA or SLE in which the PTPN22 gene is an important genetic factor.41
Conversely, SLE and SSc fit within the same spectrum of interferon-mediated diseases. A subset of SSc patients shows a ‘lupus-like’ high interferon-inducible gene expression pattern,33 and recently Kariuki and Niewold24 demonstrated skewing of serum cytokine profiles in SLE patients carrying the 1858T risk allele towards high serum IFN-α. This implies that the 1858T allele could be a heritable risk factor for SSc through the interferon pathway. On the other hand, other functional studies have shown that primary T cells from patients with autoimmunity (type 1 diabetes and RA) carrying the W620 allele exhibited reduced IL-2 response to TCR engagement.42,43 IL-2 is known as one of the molecules that shapes immune responses and tolerance.44 All this together points out the pathways by which the R620W PTPN22 variant influence autoimmunity, but further functional genetics studies are needed to solve this completely. In conclusion, our results suggest that the R263Q PTPN22 variant is not associated with SSc, in contrast to the R620W polymorphism that is a known susceptibility factor for SSc and the ACA-positive subset. Our results indicate that compared with the predisposition conferred by the R620W variant in autoimmunity, the protective effect of R263Q appears to be weaker. One possible explanation for these observations is that the loss-of-function effect on TCR signalling due to the R263Q variant may be more easily compensated for than a gain-of-function effect caused by the R620W variant.22
Supplementary Material
Acknowledgments
The authors would like to thank Sofia Vargas and Sonia Garcia for their excellent technical assistance, the patients and control donors for their essential collaboration. The authors also thank Dieudé et al and Gourh et al, for kindly providing an update of their results to do a more complete analysis in the present study.
Funding This work was supported by grants SAF2009-11110, Junta de Andalucía, grants CTS-4977 and CTS-180 and by the RETICS Program, RD08/0075 (RIER) from the Instituto de Salud Carlos III (ISCIII). BR was supported by the I3P CSIC programme funded by the ‘Fondo Social Europeo’. LMDG was supported by COLFUTURO and the ‘Ayudas Predoctorales de Formación en Investigación en Salud (PFIS – FI09/00544)’ from the Instituto de Salud Carlos III. The USA contributors were supported by NIH/NIAMS grants P50AR054144, N01-AR-0-2251 and NIH/NCRR 3UL1RR024148.
Footnotes
Additional data (supplementary figure 1) are published online only. To view these articles please visit the journal online (http://ard.bmj.com).
Competing interests None.
Patient consent Obtained.
Ethics approval This study was conducted with the approval of each ethics committee of the participating hospitals.
Provenance and peer review Not commissioned; externally peer reviewed
References
- 1.Gabrielli A, Avvedimento EV, Krieg T. Scleroderma. N Engl J Med. 2009;360:1989–2003. doi: 10.1056/NEJMra0806188. [DOI] [PubMed] [Google Scholar]
- 2.LeRoy EC, Medsger TA., Jr Criteria for the classification of early systemic sclerosis. J Rheumatol. 2001;28:1573–6. [PubMed] [Google Scholar]
- 3.Agarwal SK, Tan FK, Arnett FC. Genetics and genomic studies in scleroderma (systemic sclerosis) Rheum Dis Clin North Am. 2008;34:17–40. v. doi: 10.1016/j.rdc.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Dieudé P, Guedj M, Wipff J, et al. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 2009;60:225–33. doi: 10.1002/art.24183. [DOI] [PubMed] [Google Scholar]
- 5.Ito I, Kawaguchi Y, Kawasaki A, et al. Association of a functional polymorphism in the IRF5 region with systemic sclerosis in a Japanese population. Arthritis Rheum. 2009;60:1845–50. doi: 10.1002/art.24600. [DOI] [PubMed] [Google Scholar]
- 6.Rueda B, Broen J, Simeon C, et al. The STAT4 gene infl uences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. 2009;18:2071–7. doi: 10.1093/hmg/ddp119. [DOI] [PubMed] [Google Scholar]
- 7.Dieudé P, Guedj M, Wipff J, et al. STAT4 is a genetic risk factor for systemic sclerosis having additive effects with IRF5 on disease susceptibility and related pulmonary fibrosis. Arthritis Rheum. 2009;60:2472–9. doi: 10.1002/art.24688. [DOI] [PubMed] [Google Scholar]
- 8.Gourh P, Agarwal SK, Martin E, et al. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J Autoimmun. 2010;34:155–62. doi: 10.1016/j.jaut.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radstake TR, Gorlova O, Rueda B, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–9. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 1999;189:111–21. doi: 10.1084/jem.189.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Katrekar A, Honigberg LA, et al. Identification of substrates of human protein-tyrosine phosphatase PTPN22. J Biol Chem. 2006;281:11002–10. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 12.Mustelin T, Rahmouni S, Bottini N, et al. Role of protein tyrosine phosphatases in T cell activation. Immunol Rev. 2003;191:139–47. doi: 10.1034/j.1600-065x.2003.00014.x. [DOI] [PubMed] [Google Scholar]
- 13.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–8. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 14.Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orozco G, Sánchez E, González-Gay MA, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52:219–24. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 16.Kyogoku C, Langefeld CD, Ortmann WA, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75:504–7. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YH, Rho YH, Choi SJ, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases – a meta-analysis. Rheumatology (Oxford) 2007;46:49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 18.Balada E, Simeón-Aznar CP, Serrano-Acedo S, et al. Lack of association of the, PTPN22 gene polymorphism R620W with systemic sclerosis. Clin Exp Rheumatol. 2006;24:321–4. [PubMed] [Google Scholar]
- 19.Dieudé P, Guedj M, Wipff J, et al. The PTPN22 620W allele confers susceptibility to systemic sclerosis: findings of a large case–control study of European Caucasians and a meta-analysis. Arthritis Rheum. 2008;58:2183–8. doi: 10.1002/art.23601. [DOI] [PubMed] [Google Scholar]
- 20.Gourh P, Tan FK, Assassi S, et al. Association of the PTPN22 R620W polymorphism with anti-topoisomerase I- and anticentromere antibody-positive systemic sclerosis. Arthritis Rheum. 2006;54:3945–53. doi: 10.1002/art.22196. [DOI] [PubMed] [Google Scholar]
- 21.Wipff J, Allanore Y, Kahan A, et al. Lack of association between the protein tyrosine phosphatase non-receptor 22 (PTPN22)*620W allele and systemic sclerosis in the French Caucasian population. Ann Rheum Dis. 2006;65:1230–2. doi: 10.1136/ard.2005.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orrú V, Tsai SJ, Rueda B, et al. A loss-of-function variant of PTPN22 is associated with reduced risk of systemic lupus erythematosus. Hum Mol Genet. 2009;18:569–79. doi: 10.1093/hmg/ddn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang WY, Ströbel P, Belharazem D, et al. The PTPN22 gain-of-function + 1858T(+) genotypes correlate with low IL-2 expression in thymomas and predispose to myasthenia gravis. Genes Immun. 2009;10:667–72. doi: 10.1038/gene.2009.64. [DOI] [PubMed] [Google Scholar]
- 24.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155:109–17. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanford SM, Mustelin TM, Bottini N. Lymphoid tyrosine phosphatase and autoimmunity: human genetics rediscovers tyrosine phosphatases. Semin Immunopathol. 2010;32:127–36. doi: 10.1007/s00281-010-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zikherman J, Hermiston M, Steiner D, et al. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol. 2009;182:4093–106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Preliminary criteria for the classification of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 28.LeRoy EC, Black C, Fleischmajer R, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15:202–5. [PubMed] [Google Scholar]
- 29.Matucci-Cerinic M, D’Angelo S, Denton CP, et al. Assessment of lung involvement. Clin Exp Rheumatol. 2003;21(3 Suppl 29):S19–23. [PubMed] [Google Scholar]
- 30.Rueda B, Gourh P, Broen J, et al. BANK1 functional variants are associated with susceptibility to diffuse systemic sclerosis in Caucasians. Ann Rheum Dis. 2010;69:700–5. doi: 10.1136/ard.2009.118174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 33.Assassi S, Mayes MD, Arnett FC, et al. Systemic sclerosis and lupus: points in an interferon-mediated continuum. Arthritis Rheum. 2010;62:589–98. doi: 10.1002/art.27224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dieudé P, Wipff J, Guedj M, et al. BANK1 is a genetic risk factor for diffuse cutaneous systemic sclerosis and has additive effects with IRF5 and STAT4. Arthritis Rheum. 2009;60:3447–54. doi: 10.1002/art.24885. [DOI] [PubMed] [Google Scholar]
- 35.Attia J, Ioannidis JP, Thakkinstian A, et al. How to use an article about genetic association: B: are the results of the study valid? JAMA. 2009;301:191–7. doi: 10.1001/jama.2008.946. [DOI] [PubMed] [Google Scholar]
- 36.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–9. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 37.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–91. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker UA, Tyndall A, Czirják L, et al. Geographical variation of disease manifestations in systemic sclerosis: a report from the EULAR Scleroderma Trials and Research (EUSTAR) group database. Ann Rheum Dis. 2009;68:856–62. doi: 10.1136/ard.2008.091348. [DOI] [PubMed] [Google Scholar]
- 39.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 40.Avouac J, Airò P, Dieudé P, et al. Associated autoimmune diseases in systemic sclerosis define a subset of patients with milder disease: results from 2 large cohorts of European Caucasian patients. J Rheumatol. 2010;37:608–14. doi: 10.3899/jrheum.090815. [DOI] [PubMed] [Google Scholar]
- 41.Theander E, Jacobsson LT. Relationship of Sjögren’s syndrome to other connective tissue and autoimmune disorders. Rheum Dis Clin North Am. 2008;34:935–47. viii–ix. doi: 10.1016/j.rdc.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Aarnisalo J, Treszl A, Svec P, et al. Reduced CD4+ T cell activation in children with type 1 diabetes carrying the PTPN22/Lyp 620Trp variant. J Autoimmun. 2008;31:13–21. doi: 10.1016/j.jaut.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Rieck M, Arechiga A, Onengut-Gumuscu S, et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–10. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 44.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–65. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


