Abstract
There is growing evidence that the p53 tumor suppressor protein not only can function to activate gene transcription but also to repress the expression of specific genes. Although recent studies have implicated the transcriptional repression function of p53 in the pathway of apoptosis, the molecular basis of this activity remains poorly understood. This study takes a first step toward elucidating this mechanism. We report that trichostatin A (TSA), an inhibitor of histone deacetylases (HDACs), abrogates the ability of p53 to repress the transcription of two genes that it negatively regulates, Map4 and stathmin. Consistent with this finding, we report that p53 physically associates in vivo with HDACs. This interaction is not direct but, rather, is mediated by the corepressor mSin3a. Both wild-type p53 and mSin3a, but not mutant p53, can be found bound to the Map4 promoter at times when this promoter preferentially associates with deacetylated histones in vivo. Significantly, inhibition of p53-mediated transcriptional repression with TSA markedly inhibits apoptosis induction by p53. These data offer the first mechanistic insights for p53-mediated transcriptional repression and underscore the importance of this activity for apoptosis induction by this protein.
Keywords: p53 protein, gene transcription, transcriptional repression, apoptosis induction
The p53 tumor suppressor gene continues to hold distinction as the most frequently mutated gene in human cancer. The accumulated data suggest that the high frequency of p53 mutations in cancer reflects the ability of this protein to induce programmed cell death, or apoptosis. p53 is a transcription factor that can be activated by stimuli such as DNA damage, hypoxia, or viral and cellular oncogenes. Following such stimuli, p53 functions to up-regulate a set of p53-response genes and down-regulate another set of genes (for review, see White 1996; Ko and Prives 1996; Levine 1997). The transactivation activity of p53 is significantly better understood. However, data have accumulated indicating that p53 can induce apoptosis independent of its function as a transactivator. For example, p53 can induce apoptosis in the presence of inhibitors of new RNA or protein synthesis (Caelles et al. 1994; Wagner et al. 1994). Synthetic mutants of p53 that are defective in transactivation have been shown to induce apoptosis when overexpressed in tumor cells (Haupt et al. 1995). Additionally, in the spleen of mice, p53-dependent apoptosis has been shown to occur in the absence of transactivation of endogenous or transgenic p53-response genes (MacCallum et al. 1996; Gottlieb et al. 1997).
Several studies have implicated a role for transcriptional repression in p53-dependent apoptosis. For example, there are at least three proteins that can inhibit p53-dependent apoptosis; these are BCL2, its adenoviral homolog E1B–19K, and the Wilms' tumor suppressor gene WT-1. Each of these proteins has been shown to inhibit p53-mediated transcriptional repression, whereas transactivation and G1 arrest remain unaffected (Shen and Shenk 1994; Maheswaran et al. 1995; Sabbatini et al. 1995; Murphy et al. 1996). Certain human tumor-derived mutant forms of p53 that are incapable of inducing apoptosis have been found to be defective at transrepression but not transactivation (Ryan and Vousden 1998). Similarly, deletion of the proline-rich region of p53 renders it defective at apoptosis induction and transrepression, but not transactivation (Walker and Levine 1996; Sakamuro et al. 1997; Venot et al. 1998). Notably, during p53-dependent apoptosis induced by hypoxia, p53 does not transactivate p53-response genes like bax and p21/waf1 but continues to function as a transcriptional repressor (Koumenis et al. in prep.). Our studies have focused on the transrepression activity of p53, both in the context of identifying p53-repressed genes and in elucidating the mechanism of this activity.
Previously we identified Map4 as a p53-repressed gene (Murphy et al. 1996). Map4 mRNA is down-regulated at the level of transcriptional initiation in a p53-dependent manner in multiple cell lines (Murphy et al. 1996; Zhang et al. 1998, 1999). In contrast, this gene is not down-regulated during apoptosis or growth arrest that is induced independent of p53 (Murphy et al. 1996). We have found that the Map4 promoter can confer transcriptional repression by p53 to a heterologous gene; this repression occurs even when this gene is stably transfected into cells (W.H. Hoffman and M. Murphy, in prep.). Other genes found to be negatively regulated following p53 induction include DNA topoisomerase IIα, DP-1, wee1, presenilin-1, and others (Wang et al. 1997; Gopalkrishnan et al. 1998; Leach et al. 1998; Roperch et al. 1998). Overexpression of Map4 and presenilin-1 proteins markedly inhibits apoptosis (Murphy et al. 1996; Roperch et al. 1998), underscoring the physiological relevance of p53-mediated repression to the process of apoptosis. We recently identified the gene stathmin (oncoprotein 18), as one that, like Map4, is transcriptionally repressed by p53 (Ahn et al. 1999). Interestingly, like Map4, the Stathmin protein product influences microtubule polymerization dynamics.
In this study we have used the Map4 and stathmin genes as tools to probe the mechanism of transcriptional repression by wild-type p53. Recently, the work of several groups has established an evolutionarily conserved role for histone deacetylases (HDACs) in the mechanism of repression by transcription factors, such as Mad/Max, Rb, and the nuclear hormone receptors (Hassig et al. 1997; Laherty et al. 1997; Nagy et al. 1997; Luo et al. 1998). In this study we show that inhibition of HDAC activity abrogates the ability of p53 to repress the expression of endogenous p53-target genes like Map4 and stathmin. We report the identification of p53 in complexes with the corepressor mSin3a, along with HDAC. Only wild-type p53, and not transrepression-inactive mutant forms of this protein, can be found bound to the promoter of Map4 in vivo. Additionally, mSin3a binds to the Map4 promoter only in the presence of wild-type p53, and when these proteins are bound, the endogenous Map4 promoter shows decreased association with acetylated histone H3. The combined data place p53 in a bona fide transcriptional repression complex and provide the first indication that p53 may use an evolutionarily conserved mechanism for transcriptional repression—selective targeting of mSin3a, coupled with a HDAC, to the regulatory regions of specific p53-repressed genes.
Results
Trichostatin A inhibits p53-mediated repression of endogenous target genes
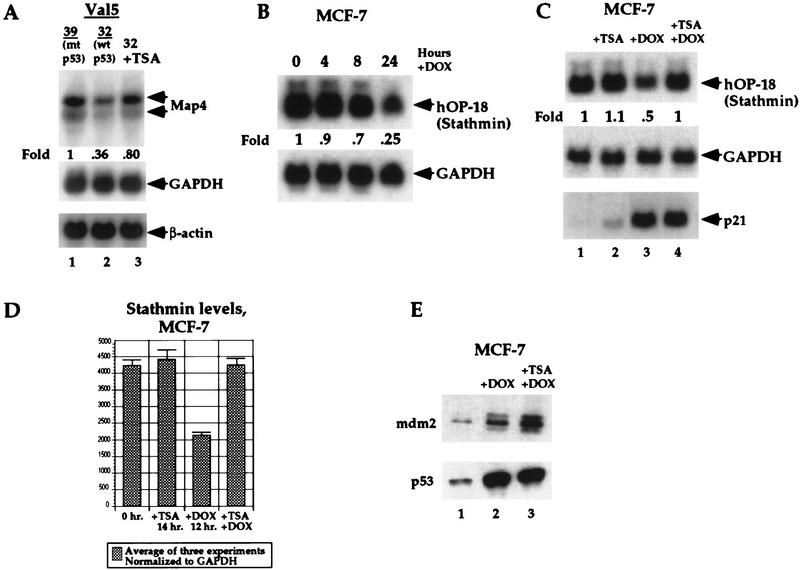
To address the possibility that transcriptional repression of Map4 and other p53-repressed genes involves a recruitment of histone deacetylases to these promoters, we tested the ability of trichostatin A (TSA) to inhibit repression of these genes following p53 induction. TSA is a potent and specific inhibitor of HDAC activity and is active in nanomolar concentrations (Yoshida et al. 1990). Initially for these analyses we utilized the murine cell line Val5, which harbors a temperature-sensitive p53 protein (Martinez et al. 1991; Wu et al. 1993). p53 exists in a mutant (inactive) conformation in this cell line at 39°C; temperature shift to 32°C results in wild-type p53 conformation and activity. Val5 cells were grown at 39°C, or temperature-shifted to 32°C, in the presence or absence of 100 nm TSA; similar concentrations of TSA have been shown to alleviate transcriptional repression of Mad/Max and Rb-repressed genes (Laherty et al. 1997; Luo et al. 1998). Induction of wild-type p53 following temperature shift of Val5 cells to 32°C resulted in an approximately threefold reduction of Map4 mRNA, consistent with our previous reports (Fig. 1, lane 2). In the presence of TSA, however, this repression was inhibited, and Map4 RNA levels maintained ∼80% of starting levels (lane 3). In contrast, neither GAPDH nor β-actin levels were altered by temperature shift, or by incubation with TSA (Fig. 1). Flow cytometric analyses indicated that TSA treatment did not alter the cell cycle distribution of Val5 cells cultured at 32°C (data not shown).
Figure 1.
Transcriptional repression of Map4 and Stathmin by p53 is inhibited by the HDAC inhibitor TSA. (A) Northern analysis of Map4 mRNA levels in Val5 cells at 39°C (lane 1, mutant p53) and following temperature shift to 32°C for 12 hr [wild-type (wt) p53, G1 arrest] in the absence (lane 2) and presence (lane 3) of 100 nm TSA. The decrease of Map4 mRNA is largely alleviated by incubation with TSA during temperature shift and p53 induction (lane 3). In contrast, neither temperature shift nor TSA affects the levels of the housekeeping genes GAPDH or β-actin. (B) Northern analysis of Stathmin levels in MCF-7 cells treated with 0.5 μg/ml DOX for 0, 4, 8, and 24 hr indicates that repression of Stathmin expression is evident as early as 4 hr, but is greatest after 24 hr of DOX treatment. (C) Northern analysis of Stathmin levels in MCF-7 cells treated with 0.5 μg/ml DOX for 12 hr indicates that DOX treatment leads to 50% repression of Stathmin levels (lane 3); TSA treatment alone does not repress Stathmin levels (lane 2). The 50% repression of Stathmin by DOX treatment is effectivelyabrogated by TSA treatment at 100 nm (lane 4). In contrast, TSA treatment does not affect p53-mediated transactivation of the p53 response gene p21/waf1. (D) The averaged data from three independent Northern analyses, described above, using probes of comparable specific activity and following normalization to the levels of GAPDH. The error bars mark standard deviations. (E) Western analysis of p53 and MDM2 protein levels in MCF-7 cells treated with adriamycin (+ DOX, lane 2) or adriamycin and TSA (+ DOX + TSA, lane 3). These data indicate that TSA does not affect the post-translational stabilization of p53 following DOX treatment nor does it inhibit the induction of MDM2 protein. Equal microgram amounts of protein were loaded in each lane, and the blot was Coomassie-stained following protein detection to verify equal loading.
To extend these observations, the human tumor-derived cell lines MCF7 (breast carcinoma) and A2780 (ovarian carcinoma) were analyzed following treatment with adriamycin to activate the endogenous, wild-type p53 protein. Because of difficulties detecting human Map4 message, we analyzed the mRNA levels of stathmin (also called oncoprotein 18). Recently, we identified stathmin as a p53-repressed gene (Ahn et al. 1999). As shown in Figure 1B, a time course of treatment of MCF-7 cells with the DNA-damaging agent adriamycin (doxorubicin, DOX) results in ∼30% reduction of stathmin RNA levels after 8 hr and 75% reduction after 24 hr (Fig. 1B). We chose to focus further on the 12-hr time point of adriamycin treatment, where 50% repression of Stathmin was evident (Fig. 1C, lane 3), to limit the toxicity sometimes associated with TSA (Yoshida et al. 1990). Significantly, although incubation with TSA alone had undetectable effects on Stathmin levels (lane 2), TSA was able to completely abrogate the reduction of stathmin levels following adriamycin treatment (lane 4). The results of three independent experiments are plotted in Figure 1D; these data indicate that the repression of Stathmin expression evident after 12 hr of adriamycin treatment is completely abrogated by incubation with TSA. In contrast, neither the post-translational stabilization of p53 by DOX nor the induction of the p53 response genes mdm2 and p21/waf1 were affected by TSA (Fig. 1C,E). Likewise, neither TSA nor adriamycin had any effect on Stathmin levels in the H1299 human lung adenocarcinoma, which is null for p53 protein (data not shown). Therefore, TSA specifically inhibits the repression of stathmin following p53 induction. We have performed identical experiments in A2780 cells (ovarian carcinoma, wild-type p53). In these studies, adriamycin treatment led to ∼50% repression of stathmin expression after 12 hr; coincubation with TSA was able to abrogate completely this repression (data not shown). The combined data indicate that HDAC activity may be an integral component of the p53-dependent repression of Map4 and stathmin.
p53 associates with HDAC-1 and mSin3a in vivo
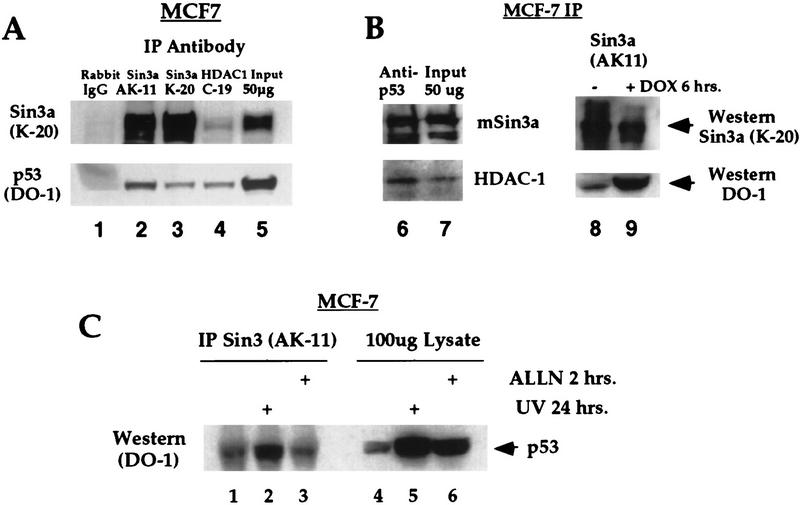
To address the possibility that p53 utilizes HDACs to repress transcription, we tested immunocomplexes from MCF-7 cells for the presence of a complex containing HDAC and p53. Immunoprecipitation (IP) of MCF-7 extract with antisera specific to HDAC-1 (Santa Cruz Biotechnology) consistently revealed the presence of p53 in these immunocomplexes, as ascertained by Western analysis (Fig. 2, lane 4). These complexes persisted under stringent washing conditions (RIPA buffer washes; see Materials and Methods) and were not present using control antisera (rabbit IgG, lane 1). However, in vitro binding reactions indicated that p53 was unable to bind to HDAC-1 directly (data not shown; see, e.g., Fig. 5, lane 8, below). Therefore, we tested the possibility that p53 utilizes a corepressor protein, like mSin3a, SMRT, or NCoR to couple it to HDAC. Immunoprecipitation of MCF-7 lysate using two different antibodies to mSin3a reproducibly demonstrated the presence of p53 in mSin3a complexes (Fig. 2, lanes 2,3). In contrast, we found no evidence for an interaction between p53 and the corepressor SMRT (data not shown). Significantly, both HDAC-1 and mSin3a were present in immunocomplexes generated using p53 antisera (p53 polyclonal, Fig. 2B, lane 6, and mAb 421, data not shown). These data are a strong indication that a complex among p53, mSin3a, and HDAC-1 exists in vivo.
Figure 2.
Wild-type p53 associates with HDAC1 and mSin3a in vivo. (A) IP–Western analysis of MCF-7 cell extract with antisera specific for rabbit IgG (Sigma, negative control), mSin3a (AK11 and K20 antibodies, Santa Cruz Biotechnology) and HDAC1 (C19, Santa Cruz Biotechnology), followed by Western analysis for p53 protein (DO-1 antibody, Calbiochem). Western analysis indicates the consistent presence of p53 in both HDAC1 (lane 4) and mSin3a (lanes 2,3,8) immunocomplexes. (B) IP with polyclonal antisera to p53 (p53 FL1–393, Santa Cruz Biotechnology) reveals the presence of both mSin3a and HDAC1 in a complex with this protein (lane 6). The amount of p53 present in mSin3a complexes increases approximately fivefold following post-translational stabilization of p53 protein mediated by 6 hr treatment in DOX (lane 9). (C) IP of MCF-7 cell lysate from untreated cells (lanes 1,4), cells treated with 4 J/m2 UV irradiation (lanes 2,5), and cells treated with ALLN (lanes 3,6). In lanes 1–3, lysate was immunoprecipitated with antisera for mSin3a (AK-11, Santa Cruz Biotechnology) and Western blotted with DO-1 (Calbiochem). Lanes 4–6 represent Western blots of straight lysate from the indicated samples.
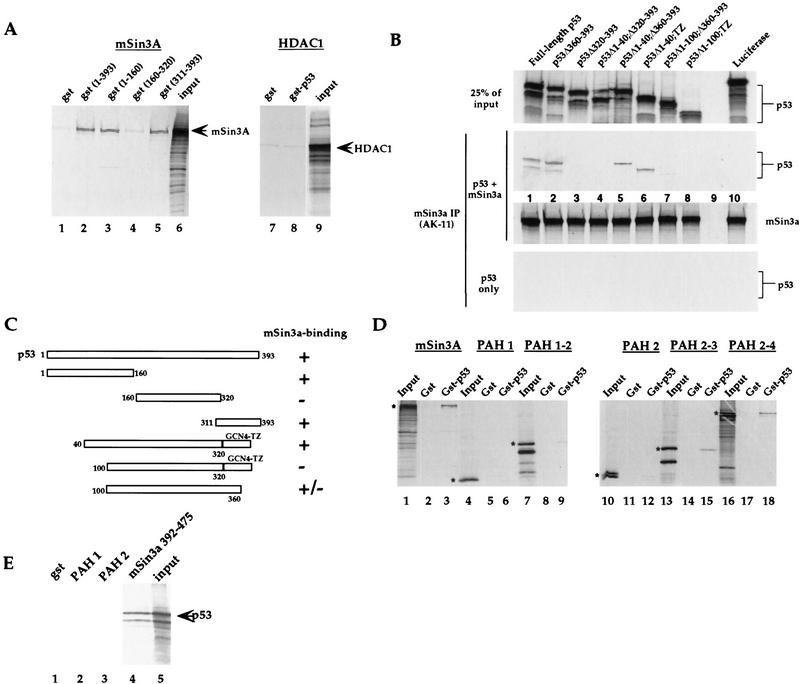
Figure 5.
p53 interacts with mSin3a in vitro. (A) GST fusion proteins of p53, which include the p53 amino acids listed, were assayed for binding to the in vitro-translated radiolabeled proteins listed above each panel. Input lanes include 50% of the radiolabeled input protein, unless noted otherwise. Arrows mark the major in vitro-translated product. These data indicate that p53 amino acids 1–160 (lane 3) and 311–393 (lane 5) interact with mSin3a in this assay. Negative controls include GST alone (lanes 1,7), and in vitro-translated HDAC1 (lanes 7,8). (B) Binding assays of in vitro-translated mSin3a (100% input in bottom panel) with in vitro-translated synthetic p53 mutants (25% of input in top panel). Although deletion of amino acids 360–393 of p53 does not inhibit interaction with mSin3a (middle panel, lane 2), deletion of amino acids 320–393 (the oligomerization domain, lane 3) abolished binding. Replacement of the p53 oligomerization domain with a modified leucine zipper tetramerization domain from GCN4 (TZ) allows for mSin3a binding (data not shown), and deletion of p53 amino acids 1–40 does not inhibit this binding (Δ1–40;TZ, lane 6). In contrast, deletion of amino acids 1–100 (Δ1–100;TZ) inhibits the ability of p53 to complex with mSin3a (middle panel, lane 8). As negative controls, luciferase showed undetectable binding to either mSin3a or p53, and mSin3a antiserum was unable to immunoprecipitate the p53 mutants incubated alone (bottom panel). (C) Summary of data obtained from in vitro binding assays; the combined data indicate that p53 amino acids 40–160 comprise the amino-terminal binding domain for mSin3a; amino acids 320–360 comprise the carboxy-terminal binding domain. (D) Binding of GST alone or a GST fusion protein encoding full-length p53 (GST–p53) with in vitro-translated radiolabeled full-length mSin3a, or mSin3a deletion mutants containing the paired amphipathic helical (PAH) domains listed above each panel. The mSin3a construct encoding PAH domains 2–3 constitutes the minimal p53 binding region. (E) A GST fusion protein containing mSin3a amino acids 392–475, which comprises the domain in between PAH domains 2 and 3, is sufficient to bind to in vitro-translated p53.
Next, we analyzed the p53–mSin3a interaction following post-translational stabilization of p53 mediated by DNA-damaging agents. We found that the amount of p53 in mSin3a complexes was approximately fivefold greater following treatment of MCF-7 cells with doxorubicin (Fig. 2B, lane 9). The significance of this increased association was unclear, however, given that p53 levels are also increased under conditions of DNA damage. To address the question of whether the avidity or affinity of p53 for mSin3a increases following DNA damage, we compared the amount of p53–mSin3a complexes following DNA damage (UV irradiation, 4 J/m2) to that following treatment of cells with the calpain I inhibitor ALLN. This inhibitor has been shown to stabilize p53 protein significantly but not activate it for DNA binding (Kubbutat and Vousden 1997). As depicted in Figure 2C, treatment of MCF-7 cells with 4 J/m2 of UV radiation results in a four- to fivefold increase in p53 protein levels (Fig. 2C, lane 5) and a concomitant increase in p53 associated with mSin3a (lane 2). Because of the stringent washing conditions we perform on these immunoprecipitations, this increased p53–mSin3a complex is not entirely reflective of the overall p53 increase. Treatment of cells with ALLN led to an approximately fourfold increase in p53 protein but an undetectable increase in the p53–mSin3a complex (lane 3). These data have been reproducible with both UV and adriamycin treatment, and support the conclusion that agents of DNA damage cause an increased affinity and/or avidity of p53 for mSin3a.
mSin3a binds both wild-type and mutant p53 proteins
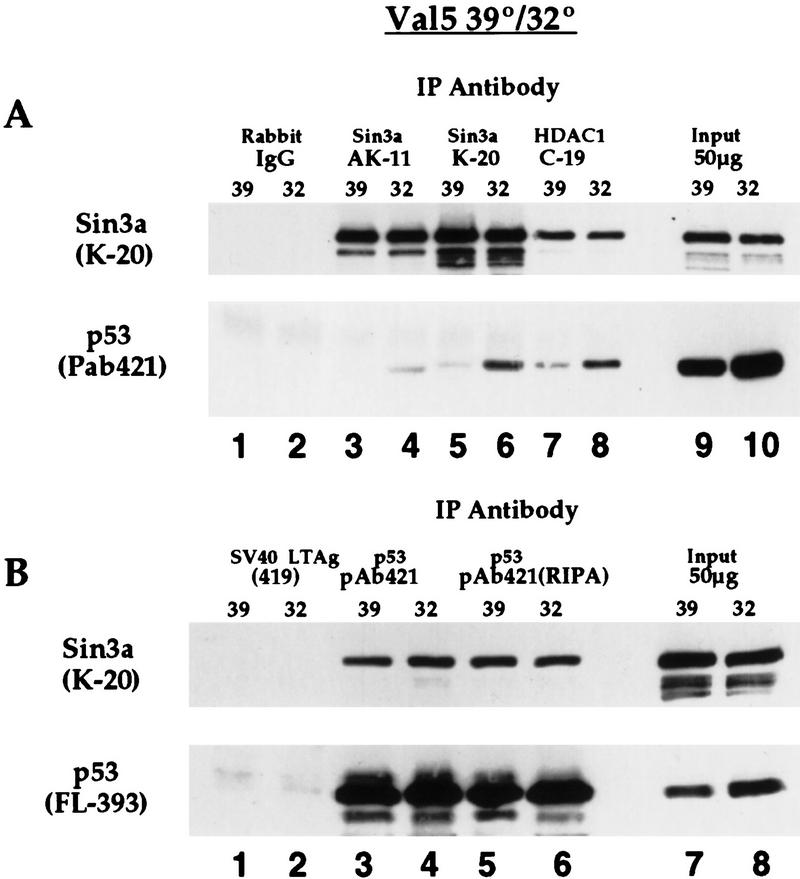
Several proteins that interact with wild-type p53 also interact with tumor-derived missense mutant alleles of this protein; these proteins include MDM2, TafII31, and the large T antigen of SV40 virus (Levine 1997). To determine if mSin3a complexes with both mutant and wild-type conformations of p53 protein, we performed IP–Western analyses in Val5 cells. Immunoprecipitation of Val5 cells grown at 39°C (mutant p53) with two different Sin3a antisera, or antisera specific to HDAC1, revealed the reproducible presence of mutant p53 in all three immunocomplexes (Fig. 3A, lanes 3,5,7). The presence of wild-type p53 in these complexes is also clearly detectable, even following washes of the immune complex in RIPA buffer (Fig. 3A, lanes 4,6,8). Similar results were obtained following IP with p53 antisera (mAb 421, detects both wild-type and mutant conformation) followed by Western analysis for mSin3a (Fig. 3B), supporting the finding that mSin3a can interact with both wild-type and mutant p53 protein. Consistent with this, we have found that the human tumor-derived p53 mutant R175H is able to interact with mSin3a in vitro (data not shown). These data indicate that like other p53-binding proteins such as MDM2, TafII31, and SV40 large T antigen, mSin3a can interact with both mutant and wild-type p53.
Figure 3.
mSin3a interacts with both mutant and wild-type conformations of p53 protein. IP–Western analysis of Val5 cell extract (temperature-sensitive p53 protein) at 39°C (mutant p53) and following temperature shift to 32°C for 24 hr (wild-type p53). (A) Both wild-type and mutant forms of p53 protein are present in mSin3a immunocomplexes (top, lanes 3–6), and in HDAC1 immunocomplexes (lanes 7,8). No p53 is immunoprecipitated with irrelevant antisera (rabbit IgG, lanes 1,2). Fifty micrograms of total protein from each sample is loaded as a control (lanes 9,10). (B) (Top) mSin3a is present in p53 immunocomplexes precipitated by mAb 421, which detects both mutant and wild-type p53, (bottom), even following extensive washes in RIPA buffer (see Materials and Methods). Western analysis of the bottom panel was performed using polyclonal antisera raised against full-length p53 (FL-393, Santa Cruz Biotechnology).
Wild-type but not mutant p53 associates with mSin3a at the Map4 promoter
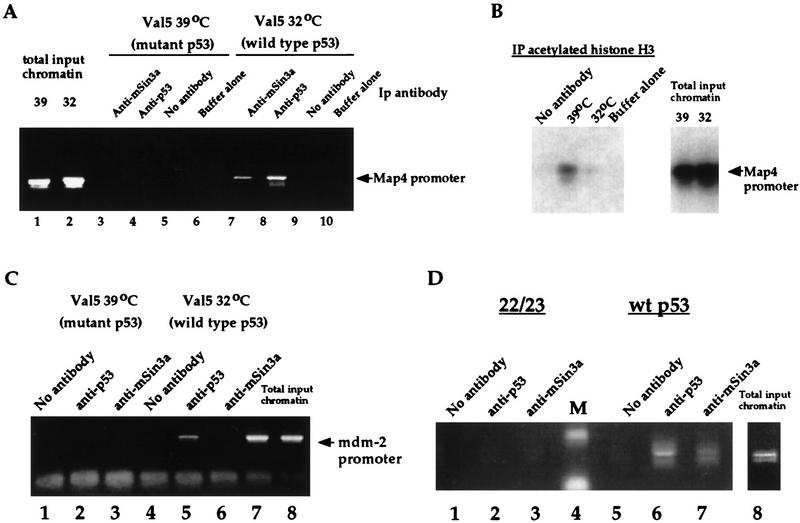
Although it is clear from these studies that p53 associates in vivo in a complex containing mSin3a and HDAC and that HDACs play a role in p53-mediated repression, it became important to assess the overall contribution of this complex to the repression of genes like Map4. Therefore, we performed chromatin immunoprecipitations (ChIPs) of the Map4 promoter using antisera specific for p53, mSin3a, and acetylated histone H3 (as per Boyd et al. 1998). Initially these studies were performed in the cell line Val5, described above (Wu et al. 1993). Immunoprecipitation of DNA using polyclonal antisera generated against full-length p53 or mSin3a was performed on formaldehyde–cross-linked extract from cells grown at 39°C (mutant p53) and 32°C (wild-type p53). Following reversal of cross-links and proteinase K digestion, a fragment corresponding to nucleotides −70 to +350 of the Map4 promoter was amplified by PCR (see Materials and Methods). These studies revealed that this fragment of the Map4 promoter is reproducibly present in a complex with p53 and mSin3a. However, this binding is detectable only when p53 is wild type and not when this protein exists in a mutant conformation (Fig. 4, cf. lanes 3 and 4 with 7 and 8). These data indicate that although mutant p53 can interact with mSin3a, only wild-type p53 and mSin3a are present at the promoter of Map4 in vivo. Therefore, like the p53 coactivator TafII31 (Lu and Levine 1995), the mSin3a corepressor associates with both mutant and wild-type forms of p53 protein, but only the latter interaction is accompanied by DNA binding and is therefore functionally consequential.
Figure 4.
Wild-type p53 and mSin3a interact with the Map4 promoter in vivo, resulting in decreased association of this promoter with acetylated histones. (A) ChIPs assay of the Map4 promoter (nucleotides −70 to +350, where +1 represents that start site of transcription) in Val5 cells cultured at 39°C (mutant p53) or 32°C (wild-type p53). IP of formaldehyde–cross-linked lysate was performed using 10 μg of anti-p53 or anti-mSin3a, followed by PCR using oligonucleotides specific for the Map4 promoter. As negative controls, lysis buffer alone was added to an anti-p53 IP (lanes 6,10), or lysate was incubated without antibody (lanes 5,9). The doublet band represents an occasional internal priming event seen with the Map4 reverse primer. For total input chromatin, 1 μl of a 1:300 dilution of DNA was used for the PCR. (B) ChIPs analysis of the Map4 promoter in Val5 cells using antisera specific for acetylated histone H3 (Upstate Biotechnology, Inc.). Following immunoprecipitation of Val5 extract and PCR of eluted chromatin, the agarose gel was blotted and hybridized to the full-length Map4 promoter. As negative controls, no antisera or 5 μg of anti-acetylated histone H3 was used for the IP of extract and binding buffer alone, respectively. (C) Wild-type p53, but not mutant p53 or mSin3a, associates with the mdm2 promoter in vivo. Immunoprecipitated chromatin from the experiment in A was used for PCR of the murine mdm2 promoter, using oligonucleotides that flank the p53-binding sites in this promoter. Prior to PCR for 25 cycles, immunoprecipitated chromatin was diluted 1:1000. For total input chromatin, 1 μl of a 1:300 dilution was used. (D) ChIPs assays were performed as above using extract from H1299 cells (human lung adenocarcinoma, p53 null) transfected with either human wild-type p53 (lanes 5–7) or human p53 containing mutations of amino acids 22 and 23 (lanes 1–3). Ten micrograms of polyclonal antisera to p53 or mSin3a (Santa Cruz Biotechnology), or no antibody, was used for immunoprecipitations, followed by PCR of the Map4 promoter. Western analysis indicated that equivalent levels of wild-type p53 and the 22/23 mutant were expressed in the transfected cells.
We extended these studies to include an analysis of the acetylation status of nucleosomes at the Map4 promoter in Val5 cells. Immunoprecipitation of Val5 cells using antisera specific for acetylated histone H3 (Upstate Biotechnology, Inc.) revealed that the Map4 promoter was preferentially associated with acetylated histone H3 when p53 was in the mutant conformation (39°C, Fig. 4B). Temperature shift and wild-type p53 induction caused a significant decrease in the association of this promoter with acetylated histone H3 (Fig. 4B, 32°C). The simplest interpretation of these data is that histones at the Map4 promoter are deacetylated in the presence of wild-type p53. As a positive control for these studies, we analyzed by ChIPs assay the interaction of wild-type p53 with the mdm2 promoter. Here, only p53 in the wild-type conformation was able to immunoprecipitate the mdm2 promoter (Fig. 4C, lane 5). Antisera to mSin3a was unable to immunoprecipitate the mdm2 promoter at either temperature (Fig. 4C, lanes 3,6). The mSin3a interaction thus appears specific to the promoter of the p53-repressed gene Map4.
Like human tumor-derived mutants of p53, the synthetic p53 double mutant at amino acids 22 and 23 (Lin et al. 1994) is also unable to repress transcription of genes like Map4 (Sang et al. 1994; Murphy et al. 1996). Interestingly, like mutant conformations of p53, we found that the 22/23 mutant of p53 was unable to immunoprecipitate with the Map4 promoter, under conditions where wild-type p53 was able to bind (Fig. 4D, lanes 2,6). Following transfection of p53 into H1299 cells (p53 null), antisera to mSin3a was likewise able to immunoprecipitate the Map4 promoter only in the presence of wild-type p53 and not the 22/23 mutant (Fig. 4D, lanes 3,7). These data indicate that there is a consistent correlation between repression-defective p53 mutants (valine 135 and 22/23) and an inability to assemble with mSin3a at the Map4 promoter.
p53 and mSin3a binding domains
To identify protein domains required for the p53–mSin3a interaction, we performed in vitro binding assays using GST fusion proteins of p53 and mSin3a. We first tested for the ability of a GST fusion protein of full-length p53 for the ability to associate with in vitro-translated, [35S-]methionine-labeled mSin3a. As shown in Figure 5A, GST p53 (amino acids 1–393) but not GST alone was able to interact with radiolabeled mSin3a following precipitation of the complex using glutathione–Sepharose and extensive washing (lanes 1,2). In contrast, neither GST nor GST–p53 showed an interaction with HDAC1 (Fig. 5A, lanes 7–9). These results suggest that p53 recruits HDAC1 through a physical association with mSin3a, in a manner similar to that reported for the Mad/Max heterodimer (Laherty et al. 1997). Next, we wished to determine which region(s) of p53 mediate the association with mSin3a. Similar in vitro binding assays were performed using GST fusion proteins comprised of truncated versions of p53 (corresponding to amino acids 1–160, 160–320, and 311–393). Interestingly, two distinct regions of the p53 protein, amino acids 1–160 (Fig. 5A, lane 3) and amino acids 311–393 (lane 5) were both found to interact with mSin3a. Therefore, like SMRT, p53 appears to possess two discrete regions that interact with mSin3a (Nagy et al. 1996). It is of note that these same two mSin3a-binding domains of p53 (1–160 and 311–393) overlap with two regions shown previously to function as autonomous transrepression domains (Sang et al. 1994; Horikoshi et al. 1995).
To more finely map the amino-terminal region of p53 necessary for the interaction with mSin3a, we extended these studies to include mutants of p53 for which the tetramerization domain of p53 has been replaced with that of GCN4 (Waterman et al. 1996). These studies were necessary because our data indicate that the carboxy-terminal mSin3a interaction domain of p53 overlaps with the tetramerization domain of p53 (amino acids 320–360; M. Murphy et al., unpubl.). Deletion of this domain from p53 (necessary to map the amino-terminal interaction domain), however, significantly inhibited the ability of the amino-terminal domain of p53 to interact with mSin3a (data not shown). Therefore, in vitro binding assays were performed using p53 synthetic mutants containing the GCN4 tetramerization domain (TZ) in place of the p53 oligomerization domain. These studies indicated that both full-length p53 containing the GCN tetramerization domain and a deletion mutant in which the first 40 amino acids of p53 are deleted (Δ1–40;TZ) were capable of interacting with mSin3a (Fig. 5B, lane 6; data not shown). In contrast, deletion of amino acids 1–100 of p53 rendered the TZ mutant unable to bind (Δ1–100;TZ, Fig. 5B, lane 8). A conservative interpretation of these data is that amino acids 40–160 of p53 are necessary for the amino-terminal interaction with mSin3a. A summary of results from in vitro binding assays is included in Figure 5C.
To localize the region in mSin3a mediating the association with p53, similar in vitro interaction experiments were performed with GST p53 and truncated versions of mSin3a corresponding to amino acids 112–192 (paired amphipathic helix 1, PAH1), 112–386 (PAH1–2), 297–386 (PAH 2), 297–529 (PAH2–3) and 297–965 (PAH2–4). As shown in Figure 5D, neither PAH1 nor PAH2 (lanes 6,9) was observed to interact with GST p53. In contrast, both PAH2–3 and PAH2–4 (lanes 15,18) associated with GST p53 in a manner similar to that of the full-length mSin3a protein (lane 3). Further delineation of the p53 binding site of mSin3a revealed that this binding domain is actually located between the second and third PAH domains of mSin3a (amino acids 392–475) (Fig. 5E, lane 4). The specificity of this interaction is supported by a failure of the 392–475 mutant of mSin3a to associate with the corepressor SMRT (data not shown), which has been shown to require the PAH4 domain to bind mSin3a (Nagy et al. 1997). These results indicate that p53 interacts with mSin3a in a region that does not include a PAH domain but, rather, is located in between PAH domains 2 and 3 (amino acids 392–475).
TSA inhibits p53-mediated apoptosis
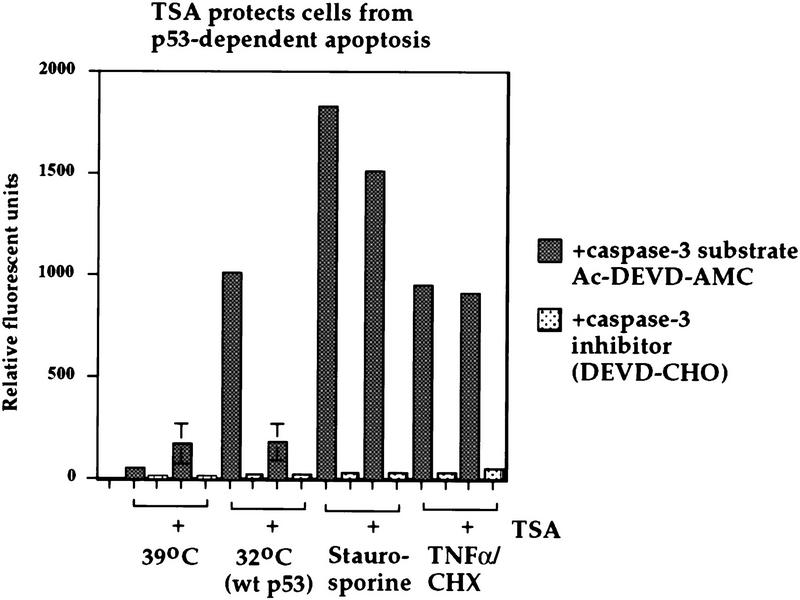
To assess the contribution of p53-mediated repression to apoptosis, we utilized the cell line Vm10. This murine cell line contains a stably transfected p53 gene encoding a temperature-sensitive protein, as well as a constitutively expressed c-myc proto-oncogene; the characteristics of this line, and its ability to undergo apoptosis at 32°C, have been described (Chen et al. 1996). Temperature shift and wild-type p53 induction of these cells results in a significant increase in caspase-3-dependent AMC–DEVD hydrolysis, a readout for apoptosis, as measured in a spectrofluorometric assay (see Fig. 6). In contrast, lysate from cells cultured at 39°C, or following addition of the caspase-3 inhibitor Ac–DEVD–CHO, showed negligible AMC cleavage (Fig. 6). Culture of Vm10 cells at 32°C (wild-type p53) in the presence of 100 nm TSA resulted in a greater than fourfold inhibition of caspase-3-dependent AMC hydrolysis. The remaining caspase-3 activity was not significantly different from Vm10 cells cultured at 39°C (mutant p53) and treated with TSA, indicating that this residual activity stems from toxicity associated with TSA treatment, rather than from p53 induction. We have also performed flow cytometry of Vm10 cells cultured at 39°C and 32°C in the presence and absence of TSA and found that 100 nm TSA reduces the sub-G1 population of Vm10 cells cultured at 32°C (wild-type p53; data not shown). TSA treatment was unable to inhibit apoptosis significantly (quantitated as caspase 3-dependent proteolysis of AMC–DEVD) induced in Vm10 cells cultured at 39°C (mutant p53), and treated with either staurosporine or tumor necrosis factor α(TNFα; Figure 6). Therefore, the ability of TSA to inhibit apoptosis is specific for p53-dependent apoptosis.
Figure 6.
p53-induced apoptosis in the Vm10 cell line is inhibited by TSA. Caspase-3 activity was analyzed in the Vm10 cell extracts indicated; cells were cultured at 39°C (mutant p53) or 32°C [wild-type (wt) p53, apoptosis] in the presence or absence of 100 nm TSA for 24 hr. As positive controls, Vm10 cells were cultured at 39°C in the presence of 100 nm staurosporine (Calbiochem), TNFα (10 ng/ml, RD Systems) plus cycloheximide (CHX, 40 μg/ml), or dilution vehicle alone (DMSO, 39°C). The activity of caspase-3 is measured as the ability of cell extract to catalyze the cleavage of AMC–DEVD and release the AMC fluorochrome. Readings were performed in duplicate; in the case of temperature-shifted cells, the data plotted are the average results of three independent experiments, with standard deviations plotted on the error bars. For staurosporine and TNFα-treated cells, the data plotted are the averages of two independent experiments. As a negative control, an inhibitor specific for caspase-3 (Ac–DEVD–CHO) was also added to the cell extract (light stippled bars).
The p53/mSin3a complex persists following DNA damage
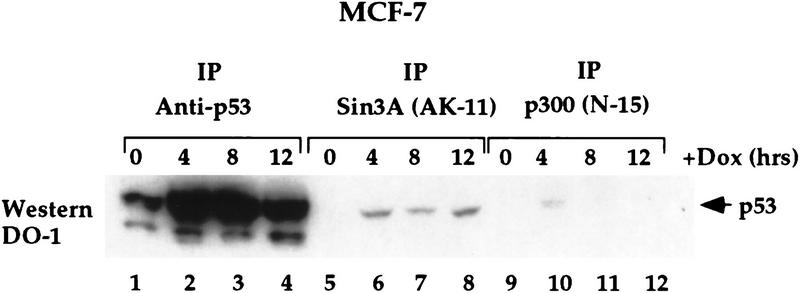
Next we analyzed the time course of the p53–mSin3a interaction following exposure of cells to the DNA-damaging agent DOX. Both p53–mSin3a and p53–p300 complexes were monitored by IP–Western analysis in a time course following adriamycin treatment of MCF-7 cells. As a control, p53 was immunoprecipitated using polyclonal antisera generated against full-length p53 (Santa Cruz Biotechnology). p53 levels were also assessed at each time point by Western analyses (data not shown). As depicted in Figure 7, DOX treatment of MCF-7 cells resulted in a significant increase in total p53 protein after 4 and 8 hr of treatment; this level decreased somewhat after 12 hr (Fig. 7, lanes 1–4). IP with two different antisera to mSin3a revealed a similar increase in the amount of p53 coimmunoprecipitating with mSin3a antisera at 4, 8, and 12 hr after DOX treatment (Fig. 7, lanes 5–8; data not shown). The p53–p300 complex was consistently more difficult to detect, and typically was present only transiently, at 4 hr after DOX treatment (Fig. 7, lane 10). These results were consistent in over six independent experiments, using two different antibodies to p300, both of which immunoprecipitated similar amounts of p300 (data not shown). These data indicate that the time course of p53 association with mSin3a and p300 may be distinct following DNA damage; this might reflect the ability of p300 to serve as a template for the degradation of p53, mediated by MDM2 (Grossman et al. 1998).
Figure 7.
p53-mSin3a complexes exist for extended periods of time following DNA damage. IP of MCF7 cells treated with 0.5 μg/ml DOX for the indicated time points with antibodies to p53 (rabbit polyclonal to p53, p53, FL393, Santa Cruz Biotechnology), mSin3a (AK11, Santa Cruz Biotechnology), and p300 (N15, Santa Cruz Biotechnology), followed by Western analysis for p53 (DO-1, Calbiochem). The depicted data are representative of six independent experiments and were reproducible with two different antibodies to p300, which detected similar amounts of total p300 protein (see Materials and Methods).
Discussion
It has been clear for some time that wild-type p53 can repress gene transcription (Ko and Prives 1996; Murphy and Levine 1998). Attempts to understand this activity have been complicated by the ability of p53 to repress gene transcription in a non-sequence-specific manner, particularly in reporter assays using transient transfections where p53 and target promoters are greatly overexpressed. Therefore, the relevance of transcriptional repression by p53, and elucidation of the underlying mechanism, has awaited the reliable identification of endogenous p53-repressed genes. Several groups have recently reported the existence of genes whose endogenous expression is decreased following wild-type p53 induction; these include the stathmin (Ahn et al. 1999) and Map4 genes (Murphy et al. 1996). Nuclear run-on analyses indicate that Map4 is repressed at the level of transcriptional initiation following p53 induction (Murphy et al. 1996). How p53 can repress the expression of genes like Map4 has been unclear to date. Our data place p53 in a complex with a known corepressor and HDAC-binding protein, mSin3a, at the promoter of the Map4 gene. A decreased association of the Map4 promoter with acetylated histones accompanies the p53–mSin3a interaction. Mutant p53 proteins defective at transcriptional repression (valine 135 and human 22/23) are likewise defective at associating with mSin3a at the Map4 promoter. These data constitute a powerful argument that sequence-specific binding is necessary for p53–mediated repression, and further that the p53–mSin3a–HDAC1 complex has a role in transcriptional repression of gene expression mediated by p53.
How p53 is targeted to the promoters of p53-repressed genes is currently an active area of study by several groups. We have cloned and characterized the Map4 promoter and shown that a specific fragment of this promoter interacts with wild-type p53, but not mutant forms of this protein, in a sequence-specific manner (W.H. Hoffman and M. Murphy, in prep.). Notably, however, this promoter lacks a canonical p53-response element, so this binding activity apparently involves a novel DNA-binding site. Additionally, although p53 immunopurified from mammalian nuclear extract is capable of interacting with the Map4 promoter, p53 protein purified from baculovirus p53-infected insect cells is not (W.H. Hoffman and M. Murphy, in prep.). It is possible that at least one other protein, not present in insect cells, is required to target p53 to the promoter of Map4 and perhaps to other repressed promoters as well. Recently, two groups have identified DNA-binding elements involved in p53-mediated transcriptional repression. One group found that a p53 half-site (two palindromic pentamers of consensus 5′-PuPuPuCA/TT/AGPyPyPy-3′) allows p53 to repress the hepatitis B virus (HBV) enhancer (Ori et al. 1998). Similarly, Lee and colleagues (1999) found that a perfect half site, possibly coupled with an imperfect pentamer, competes with HNF-3 and represses the α-fetoprotein gene. As the Map4 promoter contains neither a perfect nor an imperfect pentamer site, it is likely that an entirely novel binding element for repression exists for this promoter. However, the mechanism of repression (coupling of mSin3a and HDAC) may well be conserved for all three of these promoters.
Our in vitro data indicate that there is most likely a direct interaction between p53 and mSin3a. However, as these studies used in vitro-translated proteins, we cannot exclude the formal possibility that a protein present in reticulocyte lysate mediates this interaction. We have mapped the domain necessary for the p53–mSin3a interaction to two regions of p53 protein, from amino acids 40–160 and 320–360 (human p53). Notably, these two domains of p53 overlap with those previously implicated in transcriptional repression by p53 (Sang et al. 1994; Horikoshi et al. 1995). We have also shown that nanomolar concentrations of TSA can effectively inhibit p53-dependent apoptosis, but do not detectably affect p53-dependent transactivation nor apoptosis induced by staurosporine or TNFα. These data argue for a role of p53-dependent transrepression in apoptosis induction by this protein, and are consistent with reports that MDM2 protein which inhibits transrepression by p53 (Chen et al. 1995), also inhibits p53-dependent apoptosis in the Vm10 cell line (Chen et al. 1996). These data are further supported by a recent report from Koumenis et al. (manuscript submitted). In this study these investigators found that during p53 induction mediated by hypoxia, p53 functions as a transcriptional repressor of genes like Map4 and stathmin but does not detectably activate the expression of genes like bax and p21/waf1. Furthermore, uncoupling of the mSin3a–HDAC1 complex markedly inhibits apoptosis induced by p53 in hypoxic cells (Koumenis et al., in prep.).
The p53–HDAC interaction may have further functional significance. Recently it was demonstrated that p53 interacts with the coactivator p300, as well as the closely related protein CBP (Avantaggiati et al. 1997; Lill et al. 1997). Both of these proteins have histone acetyltransferase (HAT) activity and also interact with another HAT protein, pCAF-1 (Blanco et al. 1998). The p53–p300 interaction has been shown to be critical for both transactivation (Avantaggiatti et al. 1997; Lill et al. 1997) and apoptosis induction (Avantaggiatti et al. 1997) by p53. Additionally, however, p53 was shown to be a substrate for acetylation by this protein (Gu and Roeder 1997). Acetylation of p53 on lysine 382 has been demonstrated to relieve part of the negative regulation of p53 conferred by the carboxyl terminus of this protein (Gu and Roeder 1997; Sakaguchi et al. 1998) and may represent an important post-translational modification of p53 following DNA damage (Giaccia and Kastan 1998). Our data indicate that in addition to associating with HATs, p53 also associates with HDACs. These data raise the formal possibility that in addition to contributing to transrepression of genes like stathmin and Map4, the HDAC–mSin3a–p53 association may also result in the deacetylation of p53 and have a role in the regulation of this protein. This hypothesis awaits further testing. The data presented here support the hypothesis that mSin3a plays a role in p53-mediated transcriptional repression by tethering p53 in a repressor complex with HDACs. How the formation of this complex is regulated and targeted to the selected promoters of p53-repressed genes remain the next questions to be addressed.
Materials and methods
Cell culture
MCF-7 cells were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin and streptomycin. The A2780 ovarian carcinoma cell line, kindly provided by Tom Hamilton (Fox Chase Cancer Center), was maintained in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin and streptomycin, 0.3 mg/ml glutamine, and 0.25 U/ml porcine insulin. The Val5 and Vm10 murine embryo fibroblast cell lines were maintained as described (Wu et al. 1993; Chen et al. 1996). All cells were grown at 37°C (unless noted otherwise) in a 5% CO2 humidified atmosphere.
Cell treatment/p53 induction
Subconfluent cultures of MCF-7 and A2780 cells were treated with 100 nm TSA (Sigma) for 2 hr, followed by incubation with 0.5 μg/ml DOX (Sigma) for the indicated time points. Val5 cells grown at 39°C were treated with 100 nm TSA for 2 hr, followed by temperature shift to 32°C for 12 hr. p53 induction was monitored by Western analysis of 100 μg of protein as described (Murphy et al. 1996), using 0.1 μg/ml DO-1 (Ab-6, Calbiochem) or 421 (Ab-1, Calbiochem). MDM2 induction was monitored using mAb 2A10, kindly provided by Arnold Levine (Rockefeller University, New York, NY). For UV treatments, cells were irradiated with 4 J/m2 of UV-C with a Spectroline X series UV lamp, and output was monitored with a traceable UV light meter (Fisher Scientific). For ALLN treatment, MCF-7 cells were treated with 50 μm ALLN (Sigma) made in DMSO (Sigma) or with DMSO alone for 2 hr prior to harvesting.
Northern analysis
Total RNA was isolated from cells using CsCl purification (Murphy et al. 1996) or using Trizol, as per the manufacturer (GIBCO/BRL). Northern analyses were performed as described (Murphy et al. 1996). Probes for Northern blots were radiolabeled using random primers (Prime-It-II, Stratagene) and [α-32P]dCTP (NEN). Autoradiographs were quantitated using NIH image software. The average values from three independent experiments were plotted onto bar graphs following normalization to GAPDH.
Immunoprecipitation and Western analysis
Subconfluent cultures of cells were harvested and lysed in NP-40 buffer supplemented with protease inhibitors (1 mm PMSF, 10 μg/ml pepstatin A, 10 μg/ml aprotinin, and 5 μg/ml leupeptin). Protein concentrations were determined by the method of Bradford (Bio-Rad). Equal microgram amounts of protein (between 2.0 and 3.0 mg) were immunoprecipitated with 1 μg of each antibody, except for Val5 immunoprecipitations, which used 1000 μg of protein, as described (Murphy et al. 1996). For p300, antibody N15 (Santa Cruz Biotechnology) and AB-1 (Calbiochem) yielded similar results. Each IP was washed twice in NP-40 buffer (Wu et al. 1993) followed by two washes in RIPA buffer (50 mm Tris at pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 1% Na deoxycholate). IPs were run on 7.5%–10% SDS–polyacrylamide gels and transferred overnight onto Immuno-Blot PVDF membrane (Bio-Rad). Western blots were blocked and incubated in antibody in PBS/0.2% Tween-20/5% nonfat dry milk. Blots were incubated with 1 μg/ml antibody for 1 hr at room temperature, followed by washing in PBS/0.2% Tween-20 and incubation in peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories), as well as chemiluminescence detection (NEN).
ChIPs
ChIPs assays were performed essentially as described (Boyd et al. 1998), except that lysate from 1 × 107 cells was diluted eightfold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris at pH 8.1, 167 mm NaCl), and bound complexes were collected with protein A–agarose containing salmon sperm DNA and BSA (Upstate Biotechnology, Inc.). For ChIPs assays using antisera specific to acetylated histone H3, the acetyl–histone H3 ChIP assay protocol was performed exactly as specified by the manufacturer (Upstate Biotechnology, Inc.). PCR of the Map4 promoter was performed on immunoprecipitated chromatin using oligonucleotides 5′-AGGTGGCCGCTTCCTCGTCG-3′ (forward) and 5′-CTTCTCAACTTGGTCCAGC-3′ (reverse), as described (Boyd et al. 1998). PCR of the mdm2 promoter was performed using the oligonucleotides 5′-CGGGTCGCGCTGGCTCGTTG-3′ and 5′-ATGCATTTACGAAGGAGACA-3′. To ensure that PCR was in the linear range, 1:300 (Map4) and 1:1000 (mdm2) dilutions of DNA were amplified for a maximum of 30 cycles. For transfection experiments, asynchronously growing H1299 cells (human lung adenocarcinoma, p53-null) were transfected using lipofectin (GIBCO/BRL) with plasmids encoding the murine Map4 promoter in pGL2-basic (10 μg) plus 2 μg of either pRc/CMV human wild-type p53, or pRc/CMV human p53 22/23 (courtesy of Jiayuh Lin and Arnold Levine). Thirty-six hours following transfection, lysate from 106 cross-linked cells was immunoprecipitated with antisera to p53 (rabbit polyclonal, Santa Cruz Biotechnology) or mSin3a (AK-11, Santa Cruz Biotechnology) using the method outlined above, and PCR was performed as above. In some cases, PCR products were run on 2% agarose, blotted by the method of Southern, and hybridized with the full-length Map4 promoter. Equivalent expression of the wild-type p53 and 22/23 mutant in transfections was monitored by Western analysis of transfected cells.
In vitro interaction assays
GST fusion proteins were prepared from Escherichia coli cells as described (Murphy et al. 1996). One microgram of GST or fusion protein was bound to glutathione–Sepharose beads and incubated with 10 μl of [35S]methionine-labeled, in vitro-translated proteins generated in a coupled transcription/translation system (TNT, Promega). Binding occurred in 300 μl of binding buffer (20 mm HEPES at pH 7.9, 150 mm KCl, 1 mm EDTA, 4 mm MgCl2, 1 mm DTT, 0.02% NP-40, 10% glycerol) supplemented with protease inhibitors. Proteins were bound for 30 min at 4°C and bound complexes were washed three times in wash buffer with constant vortexing (20 mm HEPES at pH 7.9, 150 mm KCl, 1 mm EDTA, 4 mm MgCl2, 1 mm DTT, 0.1% NP-40, 10% glycerol). Bound proteins were eluted in 1× Laemmli sample buffer and separated by 7.5% or 10% SDS-PAGE. For interaction assays using only in vitro-translated proteins, 15 μl of radiolabeled mSin3a was added to 15 μl of nonlabeled protein in 300 μl of NP-40 buffer. After 30 min at 4°C, 0.2 μg of rabbit polyclonal antisera to p53 or mSin3a (Santa Cruz Biotechnology) and 30 μl of 50% (vol/vol) protein A–Sepharose were added and incubated for 30 min longer at 4°C. Bound complexes were washed twice in NP-40 buffer, once in RIPA buffer, and resolved by SDS-PAGE.
Construction of p53 deletion mutants
Deletion mutants containing the GCN4 tetramerization domain were generated by PCR of the plasmid pGEMhp53TZ334NR (kindly provided by Thanos Halazonetis) using forward oligonucleotides initiating at amino acid 40 or amino acid 100. In all cases, sequence analysis of the cloned PCR products revealed wild-type p53 sequence.
Apoptosis assays
Apoptosis was quantitated using the AMC–caspase assay specific for caspase-3 (Pharmingen). Vm10 cells were grown at 39°C or temperature-shifted to 32°C for 24 hr in the presence or absence of 100 nm TSA. Vm10 cells at 39°C were treated with staurosporine (Calbiochem) at 100 nm, dilution vehicle (DMSO), or TNFα plus cycloheximide (10 ng/ml TNFα, RD Systems, plus 40 μg/ml cycloheximide, Sigma). Cells were harvested and lysed, and lysate was monitored for the caspase-3-dependent hydrolysis of the fluorogenic substrate Ac–DEVD–AMC, according to protocols derived from the manufacturer (Pharmingen). As a control for nonspecific proteolysis, lysates from 39°C and 32°C were also assayed in the presence of the caspase-3 inhibitor Ac–DEVD–CHO (N-acetyl-Asp-Glu-Val-Asp-aldehyde, Pharmingen). Fluorescence emissions were quantified on a spectrofluorometer (VersaFluor Cuvette Fluorometer) with excitation wavelength of 360 nm and emission of 460 nm. The data represent duplicate readings of the averaged values from two (TNFα and staurosporine) or three (temperature shift) independent experiments.
Acknowledgments
We thank Donald Ayer for the mSin3a construct, Thanos Halazonetis for some of the p53 constructs utilized in this study, Laszlo Nagy for GST–Sin3 constructs, Julie Malchiodi for technical assistance, and Susan Law and Siham Biade for reagents. We thank Peter Adams, Elisabeth Hopper, and Jack Zilfou for critical reading of the manuscript, and Costas Koumenis and Amato Giaccia for communicating results prior to publication. R.M.E. is an Investigator of the Howard Hughes Medical Institute, and March of Dimes Chair in Molecular and Developmental Biology. K.W. is a fellow of the Leukemia Society of America. This work was supported by National Institutes of Health grants CA66741 (D.L.G.), CA80854 (M.M.), and by a generous grant from the W.W. Smith Charitable Trust (M.M.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ME_Murphy@FCCC.edu; FAX (215) 728-4333.
References
- Ahn, J., M. Murphy, S. Kratowicz, A. Wang, A.J. Levine, and D.L. George. 1999. Down-regulation of the stathmin/Op18 and FKBP25 genes following p53 induction. Oncogene (in press). [DOI] [PubMed]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes & Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- Chen J, Lin J, Levine AJ. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med. 1995;1:142–152. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wu X, Lin J, Levine AJ. Mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: Emerging patterns from divergent signals. Genes & Dev. 1998;12:2973–2981. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Gopalkrishnan RV, Lam EWF, Kedinger C. The p53 tumor suppressor inhibits transcription from the TATA-less mouse DP1 promoter. J Biol Chem. 1998;273:10972–10978. doi: 10.1074/jbc.273.18.10972. [DOI] [PubMed] [Google Scholar]
- Gottlieb E, Haffner R, King A, Asher G, Gruss P, Lonai P, Oren M. Transgenic mouse model for studying the transcriptional activity of the p53 protein: Age- and tissue-dependent changes in radiation-induced activation during embryogenesis. EMBO J. 1997;16:1381–1390. doi: 10.1093/emboj/16.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, Howley PM, Livingston DM. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3a. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes & Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- Horikoshi N, Usheva A, Chen J, Levine AJ, Weinmann R, Shenk T. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol Cell Biol. 1995;15:227–234. doi: 10.1128/mcb.15.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kubbutat MHG, Vousden KH. Proteolytic cleavage of human p53 by calpain: A potential regulator of p53 stability. Mol Cell Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Yang W-M, Sun J-M, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Leach SD, Scatena CD, Keefer CJ, Goodman HA, Song SY, Yang L, Pietenpol JA. Negative regulation of wee1 expression and cdc2 phosphorylation during p53-mediated growth arrest and apoptosis. Cancer Res. 1998;58:3231–3236. [PubMed] [Google Scholar]
- Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2, and the adenovirus E1B 55–kD protein. Genes & Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Lu H, Levine AJ. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Hupp TR, Midgely CA, Stuart D, Campbell SJ, Harper A, Walsh FS, Wright EG, Balmain A, Lane DP, Hall PA. The p53 response to ionizing radiation in adult and developing murine tissues. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- Maheswaran S, Englert C, Bennett P, Heinrich G, Haber DA. The WT1 gene product stabilizes p53 and inhibits p53-mediated apoptosis. Genes & Dev. 1995;9:2143–2156. doi: 10.1101/gad.9.17.2143. [DOI] [PubMed] [Google Scholar]
- Martinez J, Georgoff I, Martinez J, Levine AJ. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes & Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- Murphy M, Hinman A, Levine AJ. Wild type p53 negatively regulates the expression of a microtubule-associated protein. Genes & Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- Murphy M, Levine AJ. The role of p53 in apoptosis. In: Potten CS, Booth C, Wilson JW, editors. Apoptosis regulatory genes. New York, NY: Thompson Science; 1998. [Google Scholar]
- Nagy L, Kao H-Y, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3a, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Ori A, Zauberman A, Doitsh G, Paran N, Oren M, Shaul Y. p53 binds and represses the HBV enhancer: An adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544–553. doi: 10.1093/emboj/17.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roperch JP, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron MC, Israeli D, Dausset J, Oren M, Amson R, Telerman A. Inhibition of presenilin 1 expression is promoted by p53 and p21WAF1 and results in apoptosis and tumor suppression. Nat Med. 1998;4:835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- Ryan KM, Vousden KH. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18:3692. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatini P, Chiou S-K, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by adenovirus E1B 19K protein. Mol Cell Biol. 1995;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Apella E. DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes & Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D, Sabbatini P, White E, Prendergast GC. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- Sang B-C, Chen J-Y, Minna J, Barbosa MS. Distinct regions of p53 have a different role in transcriptional activation and repression functions. Oncogene. 1994;9:853–859. [PubMed] [Google Scholar]
- Shen Y, Shenk TE. Relief of p53-mediated transcriptional repression by the adenovirus E1B-19K protein or the cellular bcl2 protein. Proc Natl Acad Sci. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AJ, Kokontis JM, Hay N. Myc-mediated apoptosis requires wild type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes & Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zambetti GP, Suttle DP. Inhibition of DNA topoisomerase IIα gene expression by the p53 tumor suppressor. Mol Cell Biol. 1997;17:389–397. doi: 10.1128/mcb.17.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman MJF, Waterman JLF, Halazonetis TD. An engineered four-stranded coiled coil substitutes for the tetramerization domain of wild-type p53 and alleviates transdominant inhibition by tumor-derived p53 mutants. Cancer Res. 1996;56:158–163. [PubMed] [Google Scholar]
- White E. Life, death, and the pursuit of apoptosis. Genes & Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson DC, Levine AJ. The p53–mdm2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijimia M, Akita M, Beppu T. Potent and specific inhibition of histone deacetylase both in vitro and in vivo by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- Zhang CC, Yang J-M, White E, Murphy M, Levine AJ, Hait WN. The role of Map4 expression in the sensitivity to paclitaxel and resistance to vinca alkaloids in p53 mutant cells. Oncogene. 1998;16:1617–1624. doi: 10.1038/sj.onc.1201658. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Yang JM, Bash-Babula J, White E, Murphy M, Levine AJ, Hait WN. DNA damage increases sensitivity to vinca alkaloids and decreases sensitivity to taxanes through p53-dependent repression of microtubule-associated protein 4. Cancer Res. 1999;59:3663–3670. [PubMed] [Google Scholar]