Summary
Morphology evolves often through changes in developmental genes, but the causal mutations, and their effects, remain largely unknown. The evolution of naked cuticle—rather than trichomes—on larvae of Drosophila sechellia resulted from changes in five transcriptional enhancers of shavenbaby, a gene encoding a transcription factor that governs trichome morphogenesis. Here we show that the function of one of these enhancers evolved through multiple single nucleotide substitutions that altered both the timing and level of shavenbaby expression. The consequences of these nucleotide substitutions on larval morphology were quantified with a novel functional assay. We found that each substitution had a relatively small phenotypic effect, and that many nucleotide changes account for this large morphological difference. In addition, we observed that the substitutions displayed non-additive effects to generate a large phenotypic change. These data provide unprecedented resolution of the phenotypic effects of substitutions and show how individual nucleotide changes in a transcriptional enhancer have caused morphological evolution.
The genetic mechanisms underlying morphological evolution remain largely unknown1,2. Comparative studies suggest that changes in the timing (heterochrony), location (heterotopy), and level of gene expression have caused much of morphological evolution3–8. But, with a few exceptions9–11, we do not know the specific DNA changes responsible for altered expression, leaving several important questions unanswered. How many genetic changes underlie new morphologies12? Do multiple substitutions have independent effects or do they contribute instead to epistasis, where the effects of one change are dependent on other changes13–15? Do the changes that cause morphological evolution have minimal pleiotropic effects, as has been predicted16–18? Does transcriptional regulation evolve through deletion and de novo creation of enhancers, or through subtle modification of existing cis-regulatory modules19–21?
Here we identify the molecular changes in a transcriptional enhancer underlying a case of morphological evolution. To shed light on the interplay between gene expression divergence and morphological evolution, we evaluated the effects of these changes on timing and level of expression and also determined their effects on the resulting phenotype.
Modular enhancers regulate svb transcription
Drosophila melanogaster larvae are decorated with a complex pattern of microtrichia (hereafter called “trichomes”) resulting from the differentiation of epidermal cells (Fig. 1a, b). We focus on the dorso-lateral epidermis that differentiates quaternary trichomes in D. melanogaster and in most related species22 (Fig. 1b, c). Evolution of cis-regulatory regions of the shavenbaby (svb) gene, which encodes a transcription factor that orchestrates trichome morphogenesis23,24, cause D. sechellia larvae to differentiate smooth cuticle, rather than quaternary trichomes25 (Fig. 1c). This derived phenotype resulted from the specific loss of svb expression in quaternary cells (Fig. 1d,e), while svb expression is conserved in other epidermal cells, such as those that produce the ventral stout trichomes, called denticles22.
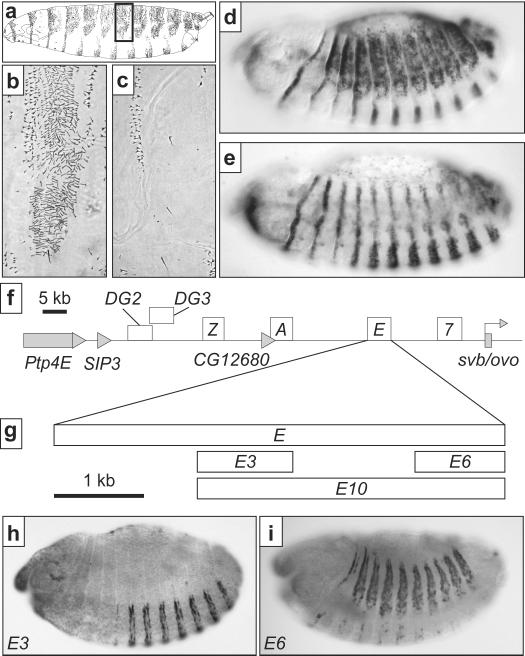
Figure 1.
The pattern of trichomes has evolved between Drosophila species due to changes in the enhancers of the svb gene. (a) Lateral view drawing of a first instar larva of D. melanogaster. The dark rectangle indicates the region shown in b and c. (b,c) The pattern of dorso-lateral trichomes on the fourth abdominal segment of D. melanogaster (b) and D. sechellia (c). Some of the dorso-lateral cells differentiate thin “quaternary” trichomes in D. melanogaster and naked cuticle in D. sechellia. (d, e) Pattern of svb RNA expression in stage 14 embryos of D. melanogaster (d) and D. sechellia (e). (f) Diagram illustrating the location of the six enhancers of svb (open boxes). The enhancers 7, E and, A were referred as proximal, medial, and distal, respectively, in ref. 25. Genes in the region are indicated with gray boxes and only the first exon of svb is shown. (g) Summary of the dissection of the E enhancer in D. melanogaster. Boxes indicate the enhancer constructs discussed in the text. (h) The E3 region drives expression in ventral stripes. (i) The E6 region drives expression in quaternary cells.
Through systematic dissection of the ~110 kb D. melanogaster svb locus, we identified six embryonic enhancers of ~5 kb25,26 (Fig. 1f). In D. sechellia, five of these six enhancers have evolved reduced activity in quaternary cells25,26. One of these enhancers, E, drives strong expression in quaternary cells and in the ventral denticle cells of D. melanogaster embryos25. The orthologous E region from D. sechellia drives greatly diminished expression in quaternary cells, which directly contributed to trichome pattern evolution25, while expression driven by this enhancer in ventral cells is conserved25. The E cis-regulatory element thus represents an attractive target for identifying the individual genetic changes that have contributed to morphological evolution in D. sechellia.
We found that the ventral and dorso-lateral expression driven by E are encoded in two distinct regions, each ~1 kb in length, that are separated by ~1.2 kb (Fig. 1g, Supp Fig 1). The first region, E3, drives expression in ventral cells that differentiate denticles (Fig. 1h) and the second region, E6, drives mostly dorso-lateral expression (Fig. 1i). No smaller constructs from the E6 region displayed equivalent activity; E6 sub-fragments drove expression that was either strongly reduced, partial, or ectopic (Supp. Fig. 1). The D. melanogaster E region thus comprises two cis-regulatory modules, E3, which drives expression in ventral cells, and E6, the minimal region that can drive a coherent pattern of expression in quaternary cells.
A svb enhancer evolved by level and timing changes
To assay the evolutionary modification of E activity between D. melanogaster and D. sechellia, for each species we generated E10 constructs, which included both the evolving E6 region and the conserved E3 region. The E3 region provided an internal control of conserved expression (Fig 2e, f). The D. melanogaster E10 construct (mel_E10) drove expression in dorsal cells beginning at stage 12–13 (Figs. 2a, c). This pattern strengthened and spread to more lateral cells in later stages (Figs. 2e, g). In stage 16 embryos, mel_E10 expression persisted in many dorsal and lateral cells (Fig. 2i), while endogenous svb mRNA is not present at this stage (data not shown). These constructs therefore produce artificially high levels of mRNA in late stage embryos. This experimental artifact allowed discovery of the surprising fact that, while the D. sechellia E10 (sec_E10) does not drive expression before stage 14 (Figs. 2b, d, f), it does drive expression in quaternary cells in late stage embryos (Figs. 2h, j), albeit at a much lower level than does mel_E10. In a separate set of experiments, we confirmed that the D. sechellia E6 region indeed drives this late dorsal expression (data not shown) indicating that it retains some weak and heterochronic expression. In contrast, the ventral expression driven by sec_E10 matched the timing and levels driven by mel_E10. These data therefore show that conserved ventral expression and divergent dorsal expression of the E10 regions from D. melanogaster and D. sechellia is correlated with the patterns of trichomes produced by each species, further localizing evolutionary changes to within the E6 region.
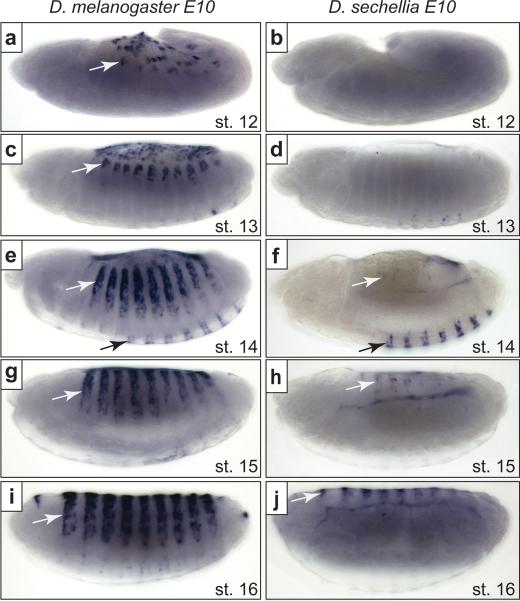
Figure 2.
D. sechellia E6 displays decreased and delayed expression relative to D. melanogaster E6. (a, c, e, g, i) The D. melanogaster E10 construct drives expression that is detected first in the most dorsal cells of stage 12 embryos (a). This expression strengthens and spreads laterally through stages 13 (c), 14 (e), 15 (g) and 16 (i). (b, d, f, h, j) The D. sechellia E10 construct does not drive detectable expression in stage 12 (b) or 13 (d) embryos. Dorsal expression (white arrows) is detected in only some stage 14 embryos (f) and is clearly observable in stage 15 and 16 embryos (h, j). Both the D. melanogaster and D. sechellia E10 constructs drive similar expression in ventral cells (black arrows) (e, f).
The E6 enhancer evolved at an accelerated rate
We next attempted to identify the DNA changes that caused the evolutionary shift in E6 function. We compared the sequences of the E6 region between D. sechellia and five closely related species, all of which, like D. melanogaster, produce dense quaternary trichomes. Multiple sequence alignment allowed us to identify thirteen substitutions and one single bp deletion that are unique to D. sechellia (Fig. 3, Supp. Fig. 2). These D. sechellia-specific substitutions are located in a region of ~ 500 bp (the “focal region”) of otherwise high sequence conservation, even in D. sechellia (Fig. 3a).
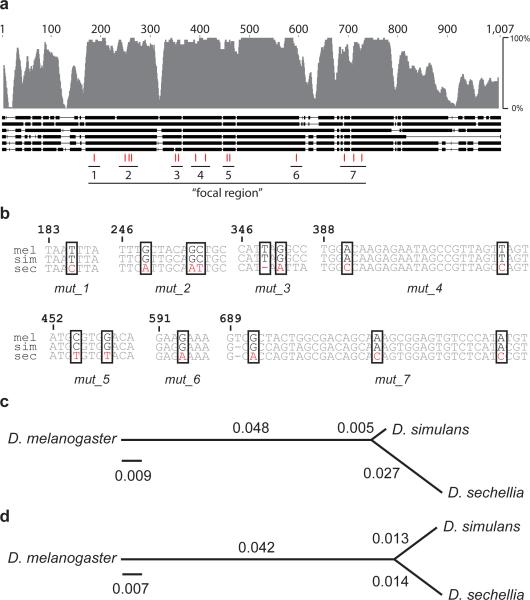
Figure 3.
Sequence conservation of the E6 region and location of the D. sechellia-specific substitutions. (a) The aligned E6 sequences from D. melanogaster, D. simulans, D. mauritiana, D. sechellia, D. yakuba, and D. erecta are represented as thick horizontal lines, with thin regions indicating gaps in the alignments. (Full alignment is provided as Supp. Fig. 2.) Sequence conservation over a 10 bp sliding window is represented above by the height of the gray bars. The positions of D. sechellia-specific substitutions are indicated with vertical red lines, the seven clusters of substitutions are indicated below the red lines, and the “focal region” is labeled. (b) Sequences of the seven regions containing the D. sechellia-specific substitutions (enclosed in rectangles) with the aligned sequences from D. melanogaster (mel), D. simulans (sim), and D. sechellia (sec). (c, d) Evolutionary trees of the E6 focal region (c) and 9 kb outside of the focal region (d), where branch lengths are proportional to the substitution rate.
Given the functional importance of E6, we examined whether this apparent clustering of substitutions within a highly conserved block represented an unusual substitution rate. We sequenced the E6 focal region from eight additional isolates of D. sechellia. All nine D. sechellia sequences were identical (data not shown), which is consistent with the low levels of polymorphism detected in other regions of the D. sechellia genome27,28. The absence of polymorphism in the E6 region in D. sechellia prevented us from employing commonly used tests of selection that rely on allele frequencies29. Instead, we analyzed substitution rates in the D. sechellia and D. simulans lineages, using D. melanogaster as an outgroup30. We observed a significant increase in D. sechellia divergence, compared to D. simulans, in the focal region of E6 (Fig. 3c; Tajima's relative rate test, χ2=6.25, P=0.012, 503 bases). To determine whether this pattern of accelerated divergence reflects simply an accelerated evolutionary rate of substitution at this genomic locus in D. sechellia, we sequenced ~9000 bp of DNA flanking the focal region, which does not include any of the other evolved enhancers, both from D. sechellia and from D. simulans. The ~9000 bp region has not evolved at significantly different rates in the two lineages (Fig. 3d; Tajima's relative rate test, χ2 = 0.56, P = 0.45, 7072 alignable bases). In the D. sechellia lineage, the focal region experienced a significantly higher substitution rate (4.8 times higher) than did the flanking regions (Fisher's exact test, two-tailed P = 0.016). Therefore, when compared to neighboring regions, the focal region of E6 evolved at a faster rate in the D. sechellia lineage, suggesting that it has evolved under positive selection31, or relaxed constraints32, or both.
Substitutions in E6 altered enhancer function
To assay the effect of the D. sechellia-specific substitutions in E6 on enhancer activity, we introduced all of these substitutions into mel_E10. We also performed the reciprocal experiment by reversing the D. sechellia-specific substitutions to the D. melanogaster sequence in sec_E10. To enable trichome rescue experiments, the mutated E10 versions were placed upstream of a svb cDNA that contained a heterologous tag in the 3' UTR, which allowed is to differentiate expression driven by the transgene from expression driven by the endogenous svb gene.
In stage 14 embryos, the D. melanogaster E10 construct carrying all of the D. sechellia-specific substitutions (mel_mut_All) drove substantially weaker expression in quaternary cells than did mel_E10 (Fig. 4a–c). Conversely, the D. sechellia E10 carrying all of the “reverse” substitutions to the D. melanogaster state (sec_mut_All) drove substantially stronger dorsal expression than did sec_E10 (Fig. 4b, d). These manipulated enhancers did not perfectly reproduce the temporal and spatial differences between mel_E10 and sec_E10 (Fig. 4), indicating that at least one other substitution in E10 contributed to the functional divergence of these enhancers. All together, these results confirm that at least one of the D. sechellia-specific substitutions in the E6 region caused most of the species difference in E6 function.
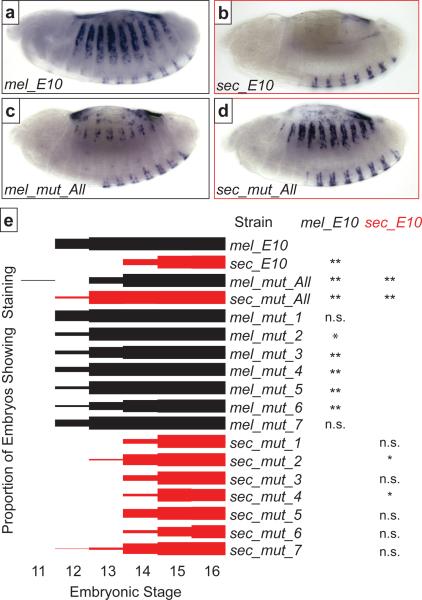
Figure 4.
Evolutionary engineering of the E10 enhancer reveals the role of evolved substitutions in altering the levels and timing of expression. (a,b) Reporter gene expression driven by the D. melanogaster E10 (a) and D. sechellia E10 (b) constructs in st. 14 D. melanogaster embryos. (c) Introducing all seven clusters of D. sechellia-specific substitutions into a mel_E10 construct (mel_mut_All) strongly reduces dorsal expression in st. 14 embryos. (d) Introducing the respective D. melanogaster nucleotides into a sec_E10 construct (sec_mut_All) almost completely restores dorso-lateral expression in st. 14 embryos. (e) The onset of expression driven by the E10 and mut enhancers was quantified by counting the proportion of embryos showing dorso-lateral expression at each of six embryonic stages. The mel_mut_All and sec_mut_All show strong changes in the onset of expression compared with the respective wild type constructs. Five of the D. melanogaster mut lines also show delayed onset of expression compared with mel_E10 construct. Two of the sec_mut lines show significant differences in the onset of expression compared with sec_E10. The E10 and mut_All comparisons were made at stage 13 and the individual cluster mel_mut and sec_mut comparisons were made at stages 12 and 14, respectively. Sequential Bonferroni test P values: * P < 0.05; ** P < 0.01; n.s.=not significant.
Many substitutions caused morphological evolution
We asked next which of the D. sechellia-specific substitutions caused the altered function of E6 in D. sechellia. Since the D. sechellia-specific substitutions in the E6 enhancer appeared clustered in seven regions (Fig 3a), we mutated separately these seven clusters of nucleotides (Fig. 3b) from the D. melanogaster to the D. sechellia sequence in mel_E10. We also performed the reverse experiment, separately mutating each of seven clusters from the D. sechellia to the D. melanogaster sequence in sec_E10. Some of the D. melanogaster constructs with individual mutated clusters displayed weaker lateral expression in stage 14 embryos than mel_E10 did (data not shown). Quantification of the onset of expression revealed further that five of seven of the D. melanogaster mutated enhancers drove significantly delayed expression when compared to mel_E10 (Fig. 4e, Suppl. Table 1). In the reciprocal experiments, some sec_E10 constructs with clusters of D. melanogaster substitutions drove slightly stronger dorso-lateral expression in quaternary cells than did sec_E10 (data not shown). Some of these sec_mut constructs drove a significantly altered onset of expression than did sec_E10, but these differences were not of large magnitude (Fig. 4e, Suppl. Table 1). Most importantly, no single cluster of substitutions in either direction recapitulated the temporal onset of expression observed when all substitutions were introduced together (Fig. 4e).
These results suggest that at least five of the D. sechellia specific substitutions in the E6 region contributed to the functional divergence of this enhancer. We therefore quantified the ability of these constructs to rescue trichomes in an embryo that lacked endogenous svb activity (Fig. 5b). We tested first whether mel_E10 and sec_E10 could rescue the production of trichomes with normal morphology in the correct spatial domains (Fig. 5a, c). mel_E10 rescued many, but not all, of the quaternary trichomes (Fig. 5c, m, n) and recovered many ventral trichomes (Suppl. Fig 3). The incomplete rescue of both dorsal and ventral trichomes was expected, because multiple svb enhancers together contribute to the complete pattern of svb expression26. sec_E10 rescued ventral trichomes as well as mel_E10 did (Suppl Fig. 3), but recovered only a few dorsal trichomes (Fig. 5i, m), consistent with the conserved and evolved functions of E10. Therefore, this rescue assay provides a reliable readout of the normal function of svb enhancers.
Figure 5.
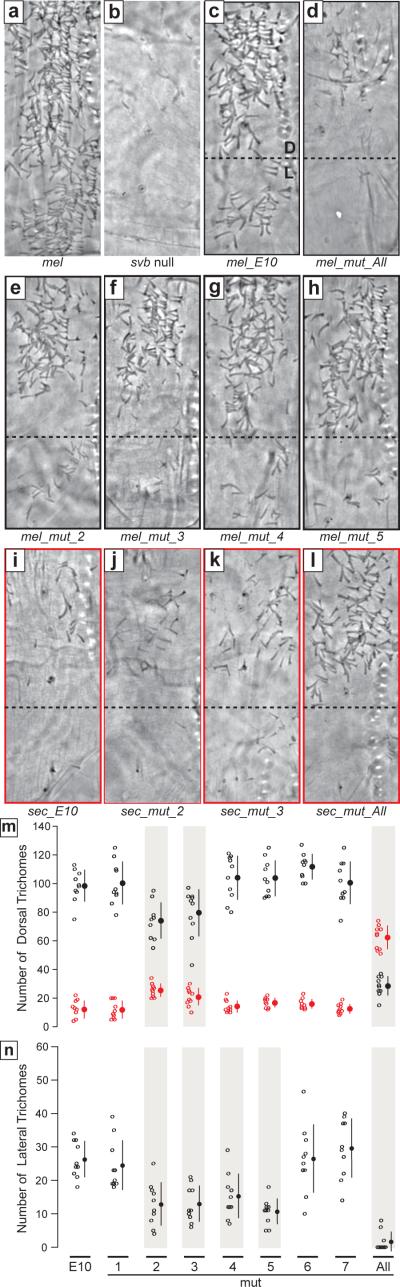
Effect of the engineered substitutions on trichome rescue in dorsal and lateral regions of the sixth abdominal segment of first-instar larvae. (a) Wild type D. melanogaster. (b) svb null. (c) mel_E10 in a svb null background. The dorsal (D) and lateral (L) regions where trichomes were counted are delimited with a dashed line. (d–h) mel_E10 constructs carrying all D. sechellia substitutions (d), or cluster 2 (e), 3 (f), 4 (g), or 5 (h) substitutions in a svb null background. (i) mel_E10 in a svb null background. (j–l) sec_E10 constructs carrying cluster 2 (e), 3 (f), or all D. melanogaster substitutions (l). (m, n) Number of trichomes rescued by the mel (black) and sec (red) constructs in the dorsal (m) and lateral (n) regions. All larvae carrying sec_mut constructs differentiated zero trichomes in the lateral region, and for clarity these data are not shown in n. Open circles represent counts for each individual. Closed circles and lines indicate the means and standard deviations, respectively. Grey shading encompasses the constructs with trichome counts that were significantly different from the E10 construct of the respective species (P < 0.05, Dunnet's test).
Since the D. sechellia-specific substitutions in E6 are sufficient to almost completely recapitulate the differences in expression patterns between the species, we asked whether these changes were sufficient to modify trichome patterning. Introduction of all of the D. sechellia-specific substitutions from E6 into mel_E10, mel_mut_All, caused larvae to produce many fewer trichomes than did mel_E10, and thus to look more like D. sechellia (Fig. 5d, m, n). Conversely, larvae carrying the reversed substitutions in a D. sechellia background (sec_mut_All) looked more like D. melanogaster larvae (Fig. 5l, m).
To determine how many substitutions cause this species difference in enhancer activity, we tested whether each cluster of substitutions influenced trichome patterns. In mel_mut_2, mel_mut_3, mel_mut_4, and mel_mut_5, the D. melanogaster to D. sechellia substitutions reduced the number of trichomes produced by 4.6–33.5 % (Fig. 5e–h, m, n, Suppl. Table 3). In contrast, in only sec_mut_2 and sec_mut_3 did the D. sechellia to D. melanogaster substitutions increase the number of trichomes by 9.9–14.6% (Fig. 5j–k, m, n, Suppl. Table 3).
Larvae carrying mel_mut_All differentiated significantly more trichomes than did larvae carrying sec_E10. The opposite is also true; sec_mut_All did not rescue as many trichomes as did mel_E10. Thus, additional substitutions within E10, other than those we tested, might also have contributed to the morphological difference between D. melanogaster and D. sechellia.
The functional rescue experiments show that at least four clusters of substitutions in E6 can alter trichome patterning on their own. Both the onset of expression data and the trichome rescue data indicate that the D. sechellia-specific substitutions display epistasis with respect to each other and with respect to the remaining E10 sequence. Indeed, the magnitude of the effect of mutating all seven clusters of substitutions together on trichome patterning is not recapitulated by summing up the effects of all clusters acting alone (Fig. 5m,n, Suppl. Table 3). The impact of each substitution on larval morphology is thus partly dependent on which other substitutions are already present.
Note, there is not perfect congruence between the analysis of gene expression patterns and the functional readout of trichome number. For example, mel_mut_6 altered expression timing, but not trichome number. This suggests that subtle expression differences may not always correctly predict the effects of genetic changes on morphological evolution.
Discussion
We have identified molecular changes in a cis-regulatory region that contributed to a morphological difference between closely related species. We found that, taken individually, each genetic change in a transcriptional enhancer had a relatively small effect on gene expression and on the final phenotype, but that when they were combined, they produced a large morphological difference. It is impossible to know the actual order in which these substitutions occurred nor whether all of the mutations went to fixation independently or whether some co-segregated. We thus focused on the effects of individual clusters of substitutions in the background of the parental species.
Our results strongly suggest that at least five substitutions in the E10 region—at least four in the mutated clusters and at least one other site—contributed to altered function of the E6 enhancer in D. sechellia. The substitutions that contributed to morphological evolution exhibited substantial epistasis, both with respect to the background E10 construct and with respect to the other substitutions in E6. Similarly, a study of pigmentation differences among D. melanogaster populations showed that multiple polymorphisms of small effect in enhancers of the gene ebony account for large phenotypic differences10. We hypothesize that enhancer structure influences the patterns of genetic change. When the function of a cis-regulatory module relies on multiple transcription factor binding sites, each with a small effect in expression, evolution may require changes of a large number of such sites to cause a significant phenotypic change.
Detecting the action of natural selection on specific non-coding genomic regions remains a major challenge for evolutionary genetics33–35. The accelerated substitution rate that we observed in the D. sechellia E6 focal region suggests that this region experienced either positive selection36, relaxation of purifying selection37, or both. In addition, none of the D. melanogaster to D. sechellia mutations led to a significant increase in trichome number, and none of the reciprocal mutations led to a decrease in trichome number. That is, along the lineage leading to D. sechellia, the E6 enhancer appears to have accumulated only substitutions that decrease trichome number. These observations also are consistent with the action of directional selection36, unless random mutations in this enhancer preferentially cause loss of expression.
When we reverted the D. sechellia-specific substitutions to the ancestral state, the D. sechellia E10 construct regained most of the functionality present in the D. melanogaster E10 construct. Thus, in principle, descendants of modern D. sechellia could re-evolve at least some trichomes through the accumulation of single nucleotide substitutions in an existing enhancer. Our results contrast with other recent studies of cis-regulatory evolution that have discovered large deletions in transcriptional enhancers9. For example, the wholesale deletion of an enhancer caused the loss of pelvic structures in some stickleback populations11. While this is a striking result, large deletions may contribute to morphological evolution only rarely. For example, enhancer deletions may have deleterious pleiotropic effects, since many single enhancer “modules” in fact encode expression in multiple domains1,38–40. In addition, new expression patterns may sometimes evolve through modification of existing enhancers21,41,42. Widespread deletion of cis-regulatory DNA may thus reduce the evolutionary potential of existing enhancers. It is worth noting that the stickleback populations with different pelvic structures diverged less than 10,000 years ago11. Our study focuses on morphological differences between species that diverged approximately 500,000 years ago. The dramatically different genetic architecture discovered in these two cases may indicate that different kinds of mutations are selected over different evolutionary timescales2
Our results suggest an additional explanation for the predominance of single nucleotide substitutions that have altered E6 function. Some constructs carrying large deletions of the E6 element generated ectopic expression (Supp. Fig. 1). This may be a general feature of enhancers that require multiple activation and repressive activities to define a precise spatio-temporal pattern of expression43,44. In such cases, large insertions or deletions may result in ectopic expression and, potentially, in dominant pleiotropic effects. In contrast, single nucleotide substitutions within activator and repressor binding sites may result in subtle changes in expression with minimal pleiotropic effects. For example, substitutions that lead to heterochrony in enhancer activity can modify a transcriptional program without deleterious effects on development. Such a heterochronic shift in enhancer activity could result from either downregulation of enhancer activity or from a temporal delay in the initiation of enhancer activation. Either or both kinds of events may have occurred in the D. sechellia lineage.
Methods Summary
Embryos were collected and fixed using standard conditions and ß-Gal expression was detected with immuno-histochemistry using a rabbit anti-βGal antibody (Cappel) used at 1:2000 and an anti-rabbit antibody coupled to HRP (Santa Cruz Biotech), also used at 1:2000. Staining was developed with DAB/Nickel.
To detect the expression of transgenic svb transcripts, we made a RNA probe complementary to the lacZ and SV40 sequence in the 3' UTR of the svb cDNA using the Dig RNA labeling kit (Roche). We tested for heterochronic changes in the onset of transgene expression by comparing the proportion of embryos showing staining between constructs at a single stage. We then tested for differences in the proportions of stained embryos with the Barnard test using a sequential Bonferroni correction for multiple tests.
For trichome rescue experiments, we cloned D. melanogaster and D. sechellia E10 into pRSQsvb26. Mutant plasmids were generated using site-directed mutagenesis (Genescript USA Inc.). Constructs were integrated into the attP site of line M(3xP3-RFP.attP)ZH- 86Fb; M(vas-int.Dm)ZH-2A. Males homozygous for the transgene were crossed to svb−/FM7c;twi∷GFP females. Non-fluorescent first instar larvae from this cross were mounted on a microscope slide in a drop of Hoyer's:lactic acid (1:1). Cleared cuticles were imaged with phase-contrast microscopy. Dorsal and lateral regions were defined using morphological landmarks and programmed as macros in Image J software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2009). Trichomes were counted using the cell-counter option of Image J. We performed pairwise comparisons of trichome numbers between the wild type construct and each mutated construct and statistical significance of comparisons was determined with Dunnet's test.
Additional experimental methods are available as Supplementary Online Material.
Supplementary Material
Acknowledgements
We thank Greg Davis, Purak Parikh, and Philippe Valenti for assistance with cloning and the Drosophila Species Stock Center for fly stocks. This work was supported by the Pew Charitable Trusts Latin American Fellows Program in the Biomedical Sciences Fellowship to N.F., a Ruth L. Kirschstein National Research Service Award to DE (F32 GM 83546-02), Agence Nationale de la Recherche (Blanc 2008, Netoshape) to F.P., and NIH (GM063622-06A1) and NSF (IOS-0640339) grants to D.L.S.
Footnotes
Author contributions N.F., D.F.E., A.P.M., and D.L.S. designed the experiments and analyzed the data. N.F., D.F.E., A.P.M., S.W. and F.P. performed the experimental work. N.F. and D.L.S. wrote the manuscript. D.F.E., A.P.M. and F.P. commented on the manuscript at all stage.
References
- 1.Monteiro A, Podlaha O. Wings, horns, and butterfly eyespots: how do complex traits evolve? PLoS Biol. 2009;7:e37. doi: 10.1371/journal.pbio.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern DL. Evolution, Development, & The Predictable Genome. Roberts & Co.; 2010. [Google Scholar]
- 3.Stern DL, Orgogozo V. The Loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 5.Erezyilmaz DF, Riddiford LM, Truman JW. The pupal specifier broad directs progressive morphogenesis in a direct-developing insect. Proc Natl Acad Sci U S A. 2006;103:6925–6930. doi: 10.1073/pnas.0509983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson EH. The regulatory genome: Gene regulatory networks in development and evolution. Academic Press; 2006. [Google Scholar]
- 7.Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: Molecular genetics and the evolution of animal design. Blackwell Science; 2001. [Google Scholar]
- 8.Wilkins AS. The Evolution of Developmental Pathways. Sinauer Associates; 2002. [Google Scholar]
- 9.Jeong S, et al. The Evolution of Gene Regulation Underlies a Morphological Difference between Two Drosophila Sister Species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadeau NJ, Jiggins CD. A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet. 2010 doi: 10.1016/j.tig.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerke J, Lorenz K, Cohen B. Genetic interactions between transcription factors cause natural variation in yeast. Science. 2009;323:498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution Int J Org Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- 16.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 17.Akam M. Hox genes, homeosis and the evolution of segment identity: no need for hopeless monsters. Int J Dev Biol. 1998;42:445–451. [PubMed] [Google Scholar]
- 18.Stern DL. Perspective: Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 19.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig MZ, et al. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 22.Sucena E, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proceedings of the National Academy of Sciences, USA. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 2006;4:e290. doi: 10.1371/journal.pbio.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payre F, Vincent A, Carreno S. ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature. 1999;400:271–275. doi: 10.1038/22330. [DOI] [PubMed] [Google Scholar]
- 25.McGregor AP, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 26.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kliman RM, et al. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics. 2000;156:1913–1931. doi: 10.1093/genetics/156.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legrand D, et al. Species-wide genetic variation and demographic history of Drosophila sechellia, a species lacking population structure. Genetics. 2009;182:1197–1206. doi: 10.1534/genetics.108.092080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 30.Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo T, Ikeo K, Gojobori T. Large-scale search for genes on which positive selection may operate. Mol Biol Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- 32.Baines JF, Chen Y, Das A, Stephan W. DNA sequence variation at a duplicated gene: excess of replacement polymorphism and extensive haplotype structure in the Drosophila melanogaster bicoid region. Mol Biol Evol. 2002;19:989–998. doi: 10.1093/oxfordjournals.molbev.a004179. [DOI] [PubMed] [Google Scholar]
- 33.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 34.Haygood R, Babbitt CC, Fedrigo O, Wray GA. Contrasts between adaptive coding and noncoding changes during human evolution. Proc Natl Acad Sci U S A. 2010;107:7853–7857. doi: 10.1073/pnas.0911249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moses AM. Statistical tests for natural selection on regulatory regions based on the strength of transcription factor binding sites. BMC Evol Biol. 2009;9:286. doi: 10.1186/1471-2148-9-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casillas S, Barbadilla A, Bergman CM. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol. 2007;24:2222–2234. doi: 10.1093/molbev/msm150. [DOI] [PubMed] [Google Scholar]
- 38.Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev Cell. 2010;18:359–370. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klingler M, Soong J, Butler B, Gergen JP. Disperse versus compact elements for the regulation of runt stripes in Drosophila. Dev Biol. 1996;177:73–84. doi: 10.1006/dbio.1996.0146. [DOI] [PubMed] [Google Scholar]
- 40.Howard KR, Struhl G. Decoding positional information: regulation of the pair-rule gene hairy. Development. 1990;110:1223–1231. doi: 10.1242/dev.110.4.1223. [DOI] [PubMed] [Google Scholar]
- 41.Wittkopp PJ. Evolution of cis-regulatory sequence and function in Diptera. Heredity. 2006;97:139–147. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- 42.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuh C-H, Bolouri H, Davidson EH. Genomic Cis-Regulatory Logic: Experimental and Computational Analysis of a Sea Urchin Gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 44.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO Journal. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.