Abstract
The volume of publicly available data in biomedicine is constantly increasing. However, these data are stored in different formats and on different platforms. Integrating these data will enable us to facilitate the pace of medical discoveries by providing scientists with a unified view of this diverse information. Under the auspices of the National Center for Biomedical Ontology (NCBO), we have developed the Resource Index—a growing, large-scale ontology-based index of more than twenty heterogeneous biomedical resources. The resources come from a variety of repositories maintained by organizations from around the world. We use a set of over 200 publicly available ontologies contributed by researchers in various domains to annotate the elements in these resources. We use the semantics that the ontologies encode, such as different properties of classes, the class hierarchies, and the mappings between ontologies, in order to improve the search experience for the Resource Index user. Our user interface enables scientists to search the multiple resources quickly and efficiently using domain terms, without even being aware that there is semantics “under the hood.”
Keywords: semantic Web, ontology-based indexing, semantic annotation, data integration, information mining, information retrieval, biomedical data, biomedical ontologies
1. Introduction
Researchers in biomedicine produce and publish enormous amounts of data describing everything from genomic information and pathways to drug descriptions, clinical trials, and diseases. These data are stored on many different databases accessible through Web sites, using idiosyncratic schemas and access mechanisms. Our goal is to enable a researcher to browse and analyze the information stored in these diverse resources. Then, for instance, a researcher studying allelic variations in a gene can find all the pathways that the gene affects, the drug effects that these variations modulate, any disease that could be caused by the gene, and the clinical trials that involve the drug or diseases related to that specific gene. The information that we need to answer such questions is available in public biomedical resources; the problem is finding that information.
The research community agrees that terminologies and ontologies are essential for data integration and translational discoveries to occur [1, 2, 3]. However, the metadata that describe the information in data resources are usually unstructured, often come in the form of free-text descriptions, and are rarely labelled or tagged using terms from ontologies that are available for the domains. Users often prefer labels from ontologies because they provide a clear point of reference during their search and mining tasks [4, 5, 6]. For example, researchers and curators widely use the Gene Ontology to describe the molecular functions, cellular location, and biological processes of gene products. These annotations enable the integration of the descriptions of gene products across several model organism databases [7].
However, besides these examples, semantic annotation of biomedical resources is still minimal and is often restricted to a few resources and a few ontologies [8]. Usually, the textual content of these online resources is indexed (e.g., using Lucene) to enable querying the resources with keywords. However, there are obvious limits to keyword-based indexing, such as the use of synonyms, polysemy, lack of domain knowledge. Furthermore, having to perform keyword searches at each Web site individually makes the navigation and aggregation of the available information extremely cumbersome, if not impractical. Search engines, like Entrez (www.ncbi.nlm.nih.gov/Entrez), facilitate search across several resources, but they do not currently use as many of the available and relevant biomedical ontologies.
The National Center for Biomedical Ontology (NCBO) Resource Index addresses these two problems by (1) providing a unified index of and access to multiple heterogeneous biomedical resources; and (2) using ontologies and the semantic representation that they encode to enhance the search experience for the user. The NCBO BioPortal—an open library of more than 200 ontologies in biomedicine [9]—serves as the source of ontologies for the Resource Index. We use the terms from these ontologies to annotate, or “tag,” the textual descriptions of the data that reside in biomedical resources and we collect these annotations in a searchable and scalable index (Figure 1). The key contributions to the field are (i) to build the search system for such an important number of ontologies and resources and (ii) to use the semantics that the ontologies encode.
Figure 1. NCBO Resource Index overview.
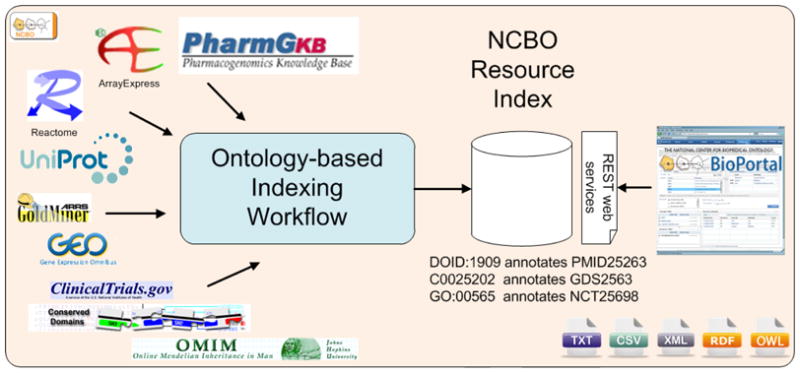
We process each bio-medical resource using the ontology-based indexing workflow. We store the resulting annotations in a database and make them available in several formats via REST Web services. BioPortal provides user-friendly interfaces to search and navigate the Resource Index.
In the context of our research, we call data element any identifiable entity or record (e.g., document, article, experimentation report) which belongs to a biomedical data resource (e.g., database of articles, experiments, trials). Usually, an element has an identifier and can be linked by a URL. For instance, the trial NCT00924001 is an element of the ClinicalTrials.gov data resource that can be accessed with: http://clinicaltrials.gov/ct2/show/NCT00924001. We call annotation—a central component— a link from an ontology term to a data element, indicating that the data element refers to the term either explicitly or not [10, 11]. We then use these annotations to “bring together” the data elements.
We currently index 22 resources, which are maintained by a variety of different institutions, with terms from more than 200 ontologies included in BioPortal (A). As of January 2011, our 1.5Tb MySQL database, which stores the annotations in the Resource Index, contains 11 Billion annotations, 3.3 Million ontology concepts, and 3.2 Million data elements. The user interface is available at http://bioportal.bioontology.org/resources.
A preliminary version of the system was presented in [12]. In this paper, we illustrate use case scenarios (Section 2), describe the system implementation (Section 3) and the details of the indexing workflow (Section 3.3), and the different means to access the Resource Index (Section 3.4). We demonstrate how semantic technologies enable information retrieval and mining scenarios that were not possible otherwise (Section 4).
2. Use case scenarios
We will describe the functionality of the Resource Index through three use case scenarios.
Scenario 1: Multiple-term search across resources
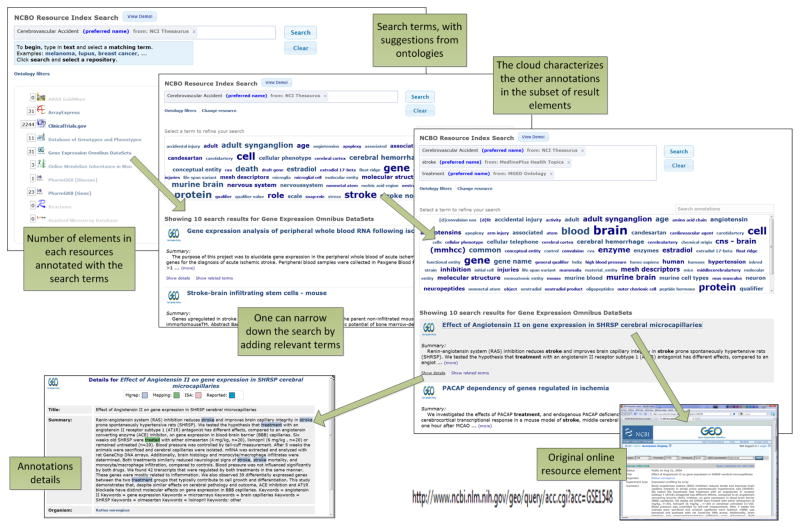
The user is interested in the role of tumor protein p53 in breast cancer. He can search the Resource Index for “Tumor Protein p53” AND “Breast Carcinoma” as defined in the NCI Thesaurus (Figure 2). The search results summarize the number of elements per resources annotated with both terms. The user can see there is relevant data linking p53 to breast cancer in such resources as ArrayExpress, ClinicalTrials.org, Gene Expression Omnibus (GEO), Stanford Microarray Database (SMD) and others. He can access the data elements within each resource quickly and navigate between resources.
Figure 2. Resource Index user interface.
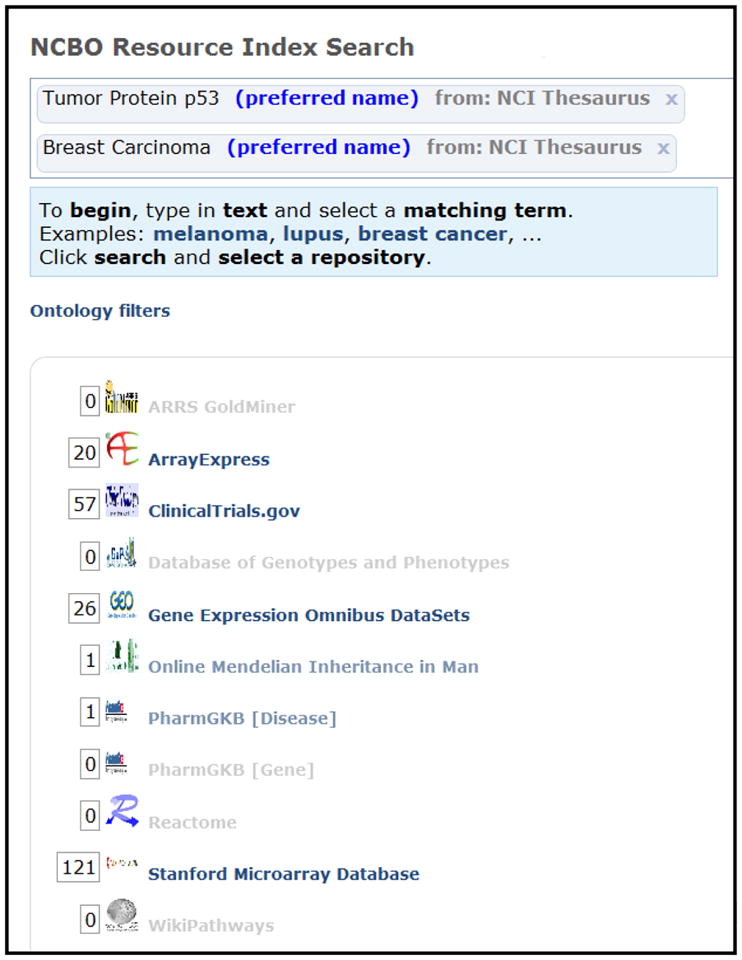
The search for resources that contain both “Tumor Protein p53” AND “Breast Carcinoma.”
Scenario 2: Exploratory search across resources
A researcher studying the causes and treatments for stroke in humans is interested in learning more about the genetic basis of the response to related conditions by searching the literature. She already knows that some related conditions such as stroke, transient is-chemic attack, and cerebral bleeding fall under the general category of cerebrovascular accidents (Figure 3). Therefore, she starts by typing “cerebr” and immediately gets feedback in the form of suggested terms from various ontologies. She selects and initiates a search for Cerebrovascular Accident from the National Cancer Institute (NCI) Thesaurus. She notices a number of hits from several resources and drills down to read more about the data elements from both the GEO and Database of Genotypes and Phenotypes (dbGAP) resources. She focuses on GEO: the tag cloud emphasizes other terms that are ranked highly in these 31 elements. Thus, she can get an idea of what these elments are about. She selects “Stroke” in the tag cloud, then “Treatment,” and gets to the 12 elements that are annotated with the three previous terms. A similar series of steps on dbGAP leads her to two elements annotated with “Cerebrovascular Accident,” “Stroke,” and “Physiology.” As a result of her search, she has quickly located gene-expression data (from rats) that is connected to genotype-phenotype data (from humans). In rats, researchers studied the gene-expression level response to both stroke and to drugs used to treat stroke. In humans, researchers studied genotypes that predispose humans to stroke and affect the physiology of the outcome.
Figure 3. Searching the Resource Indexin BioPortal.
The user searches for resources on “cerebrovascular accidents” and finds gene-expression data that are relevant to different types of cerebrovascular accidents, such as stroke.
Scenario 3: Semantically enriched search across resources
The user wants to search gene expression data about “retroperitoneal neoplasms.” A direct keyword search with “retroperitoneal neoplasm” on the GEO Web site will return no results. However, there are several datasets in GEO about “pheochromocytoma” and “renal cell cancer” both of which are retroperitoneal neoplasms and thus relevant to the previous search. When our user queries the Resource Index with “retroperitoneal neoplasm,” he will get the results that use the hierarchy represented in the BioPortal ontologies. Specifically, the NCI Thesaurus defines “pheochromocytoma” as a subclass of “retroperitoneal neoplasm.” Thus, the user will get all data elements that are annotated with “pheochromocytoma” as a response to the query on “retroperitoneal neoplasm,” including the relevant resources in GEO. Furthermore, he also gets results from ArrayExpress and SMD, which are other repositories of gene expression data also indexed in the Resource Index.
In the next section, we describe the implementation of the Resource Index, which enables these use cases.
3. The NCBO Resource Index
To create the Resource Index, we process metadata describing data elements in a variety of heterogeneous resources to create semantic annotations of these meta-data. We use the publicly available biomedical ontologies in BioPortal as a source of terms, their synonyms, and the relations between terms (Section 3.1). We use resource-specific access tools to process metadata that describe data elements in different resources (Section 3.2). We use an off-the-shelf concept-recognition tool to identify terms from BioPortal ontologies within the textual metadata and annotate, or tag, the corresponding element with the recognized terms. We expand these annotations using available ontology knowledge (Section 3.3). Finally, the Web services and user interface provide users with fast and scalable access to this index and support different use cases such as information retrieval and mining (Section 3.4).
3.1. Ontologies in the NCBO BioPortal
BioPortal, an open library of biomedical ontologies [9], provides uniform access to the largest collection of publicly available biomedical ontologies. At the time of this writing, there are 245 ontologies in this collection. BioPortal users can browse, search, visualize, and comment on ontologies both interactively, through a Web interface, and programmatically, via Web services. The majority of BioPortal ontologies were contributed by their developers directly to BioPortal. A number of ontologies come from Open Biomedical Ontologies (OBO) Foundry [13], a collaborative effort to develop a set of interoperable ontologies for biomedicine. Bio-Portal also includes publicly available terminologies from the Unified Medical Language System (UMLS), a set of terminologies which are manually integrated and distributed by the United States National Library of Medicine [14]. BioPortal includes ontologies that are developed in a variety of formats, including OWL, RDF(S), OBO (which is popular with many developers of biomedical ontologies), and RRF (which is used to distribute UMLS terminologies). BioPortal provides a uniform set of REST Web services to access basic lexical and structural information in ontologies represented in these heterogeneous formats.
We use the BioPortal REST services to traverse the ontologies and to create a dictionary of terms to use for direct annotations of data elements in biomedical resources. We use preferred name and synonym properties of classes for this dictionary. Some ontology formats have preferred name and synonym properties as part of the format (e.g., OBO and RRF). For OWL, ontology developers can either use the relevant SKOS properties to represent this information, or specify in the ontology metadata which are the properties that they use for preferred names (e.g., rdfs:label) and synonyms. Currently, our dictionary contains 6,835,997 terms, derived from the 3,349,338 concepts from 206 ontologies (the subset of BioPortal ontologies that are usable for annotation). We identify each concept by a URI defined in the original ontology or provided by NCBO.
3.2. Accessing biomedical resources
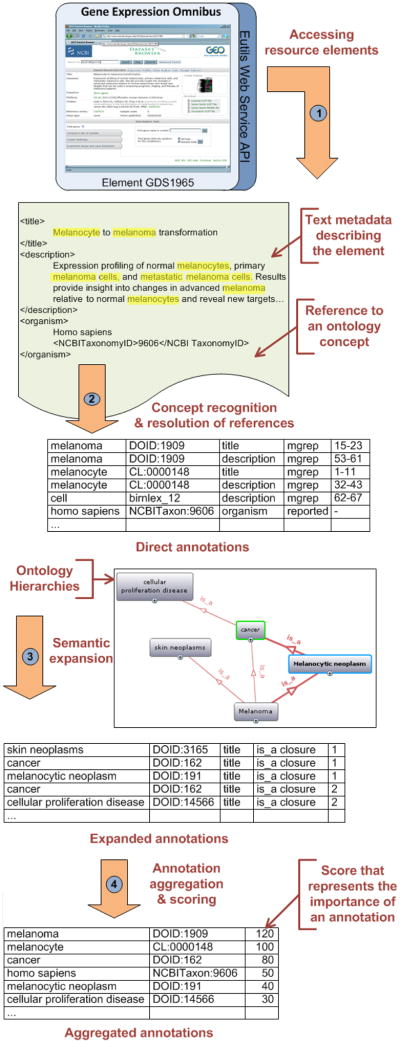
In addition to the ontology terms, the data elements from the biomedical resources are another major source of information for the Resource Index (Figure 1). As of January 2011, we have indexed 22 public biomedical resources of different sizes (up to 3.2 Million elements and 1.4Gb of data). We provide a list of sample resources in A. Data resources provide their data in idiosyncratic formats (often XML) and offer different means of access (often Web services). To access the information in the resources, we build a custom wrapper for each resource. The wrapper extracts the fields describing the data elements within a resource as illustrated in step 1 of Figure 4. In developing each wrapper, we work with a subject matter expert to determine which textual metadata fields (later called contexts) we must process (e.g., title, description). We also assign each context a weight [0,1] representing the importance of the field. We later use this weight to score annotations.2 For example, we may give annotations appearing in the title a higher weight based on the expert’s recommendation for that resource. In some cases, resources already tag elements with ontology terms, so the wrapper directly extracts the curated annotations and applies an appropriate weight. We call these annotations reported annotations. For example, the description of gene-expression data in GEO contains an organism field where a domain expert manually puts a term from the National Center for Biotechnology Information taxonomy, which refers to the relevant organism.
Figure 4. Example of annotations generated for a GEO element.
Direct annotations are generated from textual metadata and already existing ontology references of the data element. Then, expanded annotations are created using the ontology is a hierarchy. Finally, all the annotations are aggregated and scored taking into consideration their frequency and context.
Our resource-specific wrappers access the data elements incrementally, enabling us to process only the data elements that were added to the resource since the last time that we processed the resource.
3.3. Ontology-based annotation
After we access the data elements describing the resource, we perform the following steps to create annotations for the data elements in the resource: (a) direct annotation with ontology terms; (b) semantic expansion of annotations; (c) aggregation and scoring of annotations (Figure 4).
a. Creation of direct annotations
We process each textual metadata using a concept-recognition tool that detects the presence of concepts in text. Our workflow accepts different concept recognizers ranging from simple string matching techniques to advanced natural language processing algorithms. We currently use Mgrep [15, 16] which enables fast and efficient exact matching against a very large set of input strings (however without any advanced natural language processing (e.g., stemming, permutation, morphology)). Concept recognizers usually use a dictionary. The dictionary (or lexicon) is a list of strings that correspond to preferred names and synonyms of ontology concepts. At this step, Mgrep uses the 6.8 Million terms dictionary built before. In the example in Figure 4, the recognizer identifies the terms melanoma, melanocyte, and cell and creates a set of direct annotations with the corresponding concepts in the Human Disease, Cell type, and BIRNLex ontologies. We preserve the identified term, the context in whitch it appears, and its character position as provenance information about the annotation.
b. Semantic expansion of annotations
After direct annotations step, several semantic-expansion components leverage the knowledge in the ontologies to create expanded annotations from the direct annotations.
First, the is a transitive closure component traverses an ontology subclass–superclass hierarchy using a customized algorithm to create new annotations with superclasses of the classes that appear in direct annotations. We used the subclass transitive relation as defined by the original ontology e.g., is a (OBO), rdfs:subClassOf (OWL) and abstracted by BioPortal to compute the transitive closure on the whole ontology graph. For instance, we will expand a direct annotation of a data element with the concept melanoma from NCI Thesaurus, to annotations with melanocytic neoplasm, cancer, and cellular proliferation disease because NCI Thesaurus defines melanoma as a subclass of melanocytic neoplasm, which in turn is a subclass of cellular proliferation disease (Figure 4). We preserve the shortest ancestor level (direct parent, grandparent, etc.) as provenance information to use for scoring annotations. Naturally, the farther away the ancestor term is from the term in the direct annotation, the less relevant the corresponding expanded annotation is.
Second, the ontology-mapping component creates new annotations based on existing mappings between ontologies. BioPortal provides point-to-point mappings between terms in different ontologies. Some of these mappings were defined manually and some were created automatically using various mapping algorithms [17].3 We use the mappings that BioPortal stores and provides to expand our annotations and we do not follow them transitively. For instance, if a text is directly annotated with the concept treatment in Medical Subject Headings (MeSH), the mapping component will generate a new annotation with the concept therapeutic procedure from Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) because there is a mapping between these two terms in BioPortal. We preserve the type of mapping as provenance information to use for scoring annotations. It allows to score those expanded annotations proportionally to the mapping confidence (e.g., owl:sameAs, skos:exactMatch, skos:closeMatch, manually curated or automatically generated).
c. Annotation aggregation and scoring
We use the provenance information that we collect in creating direct and expanded annotations to assign each annotation a weight from 0 to 10 representing its relevance. For example, a match based on a preferred label gets a weight of 10 versus a synonym, which gets an 8; a match originating from a mapping gets a weight of 7 whereas one from an is a relationship get a diminishing weight based on ancestor level. Because several annotations with the same concept but with different provenance and context can co-exist we aggregate all those annotations of an element to a unique pair [concept-element], called aggregated annotation, to which a score is assigned. Those are the annotations used for searches. The scoring algorithm takes into account frequency, provenance and context of the annotation by doing the sum of the weights assigned to each annotation normalized by the weights of the original contexts.
At each step, the annotation workflow populates several relational tables and stores the detailed (direct & expanded) and aggregated annotations. Because both ontologies and resources are changing often, we need to automatically update the Resource Index tables regularly. The workflow handles (i) resource updates (i.e., incremental processing of new elements added to resources) using wrappers that pull only the data elements that have not been processed yet and (ii) ontology updates (i.e., incremental processing of new ontologies and new ontology versions) because BioPortal provides version specific identifiers for ontologies. For simplicity, when a new ontology version is added to BioPortal, the previous annotations associated with the ontology are removed from the Resource Index and new ones are added. The indexing workflow has been specifically optimized for this to occur rapidly [18]. We run these two different updates respectively weekly and monthly.
3.4. Accessing the NCBO Resource Index
The annotation and the scores that we described in the previous section constitute the Resource Index. The index contains 3 Billion aggregated annotations and 11 Billion detailed annotations (10% direct, 90% expanded) as illustrated by Figure 5. We provide both a Web service access to the index and a special-purpose easy-to-use graphical user interface, which enables domain experts to explore and analyze the information in the Resource Index.
Figure 5.
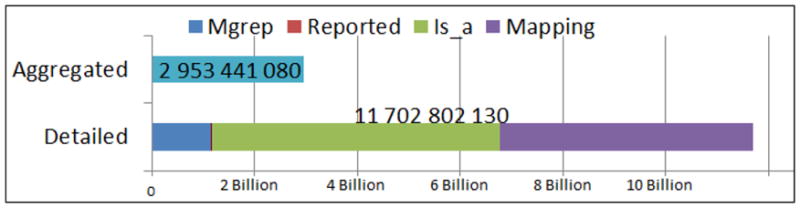
Number and types of annotations in the Resource Index.
The main Resource Index user interface, illustrated in Figures 2 and 3, is a search-based interface geared towards biomedical end-users. Users do not even need to be aware that semantic technologies are driving the user interface, and can use it through a simple search-box mechanism. As the user types in terms that she is interested in, she gets a list of auto-complete suggestions for the search terms and the source ontologies for these terms. Users can search data elements using AND and OR constructs.4 She is presented with a list of search results (as snippets) as well as a tag cloud of related terms (selected in the top 10 results) to help refine her search further. For each identified element, a user can see the details of the annotations highlighted in the original text and link back to the URLs of the original data elements.
Users can retrieve the content of the Resource Index programmatically by calling a Web service and specifying either ontology concepts or specific data elements that they are looking for. Specifically, we provide the following services:
For a given concept, obtain the set of elements in one or several resources annotated with this concept (e.g., GEO and ArrayExpress elements annotated with concept DOID:1909)
For several concepts, obtain the union or intersection of the set of elements annotated with these concepts (e.g., GEO and ArrayExpress elements annotated with both DOID:1909 and CL:0000148).
For a given data element, obtain the set of concepts in one or several ontologies annotating this element (e.g., NCI Thesaurus concepts annotating the GEO dataset GDS1965).
The first two information-retrieval services offer a unique endpoint to query several heterogeneous data resources and facilitate data integration (defined as view integration in Goble & Stevens [3]). The third service supports the type of exploration that the original resource may have never supported. This use case enables users to gather more information about a data element that they have already identified.
When retrieving annotations for a given element, users can filter out annotations using several mechanisms, such as limiting results to annotations with specific UMLS semantic types, using only results that match the whole word in the query, disabling the results obtained by matching synonyms, or selecting the type of mapping used for expanding annotations. Users can retrieve annotations in several formats (text, tab delimited, XML, RDF and OWL). The results are ordered by the scores assigned during the indexing phase.
4. Discussion and related work
The Resource Index provides semantically-enabled uniform access to a large set of heterogeneous biomedical resources. It leverages the semantics expressed in the ontologies in several different ways:
Preferred names and synonyms
Many biomedical ontologies specify, as class properties, not only labels (preferred names) but also synonyms for the class names, which we use during annotation. For example, a keyword search of caNanoLab resource with “adriamycin” would normally obtain no results. However, because the ontologies that we use have defined “doxorubicin” as a synonym for “adriamycin,” the Resource Index retrieves all caNanoLab elements annotated with the term “doxorubicin.”
Auto-complete
As users type a term into the search box, they receive immediate feedback giving both preferred names and synonyms for matching classes from different ontologies.
Hierarchies
We use subclass relations to traverse ontology hierarchies to create expanded annotations, therefore improving the recall of search on general terms. For example, a search with “retroperitoneal neoplasm,” will retrieve data annotated with “pheochromocytoma” (Section 2). Notice that subclass relationships are present in all ontologies thus enable to provide the same feature for all ontologies. Specific ontology relationships are not considered, although we acknowledge there are often useful on a per-ontology approach.
Mappings
We use BioPortal mappings to expand the set of annotations. For example, a search with the concept “treatment” from MeSH retrieves the elements annotated with “therapeutic procedure” in SNOMED-CT because there is a mapping between these two concepts in BioPortal.5
The use of ontologies significantly enhances recall of searches (i.e., more relevant data elements are retrieved) without affecting precision of the top results. Our aggregation and scoring addresses the issue of precision by ranking relevant results for the user e.g., the algorithm ranks the direct matches higher over the ones obtained via semantic expansion. Semantic disambiguation is not handled yet e.g., someone searching elements for Cell in NCI Thesaurus will obtain the elements mentioning the word cell as the abbreviation of cell phone. However, given the characteristics of the resources indexed (biomedical databases as opposed to general Web sites) the issue has not come up in practice.
Because the goal of the Resource Index is to improve runtime information retrieval and data-mining tasks, we decided to pre-compute inferences with ontologies (i.e., is a and mapping expansion) rather than to implement semantic query-expansion algorithms [19] that would have computed inferences dynamically but would have required longer response time. Our technical decisions in terms of design and architecture were often driven by benchmarking analysis and metrics [18]. The indexing workflow execution times range from a couple of minutes for the small resources to more than a week for the biggest one. Because it is impossible to include in the Resource Index all possible biomedical resources, NCBO provides the ontology-based annotation work-flow as a Web service [8], the NCBO Annotator, which allows researchers to annotate their text data automatically and get the annotations back. They can use this service to develop their own semantic-search applications. Researchers at the Medical College of Wisconsin have already created one such application for mining associations between gene expression levels and phenotypic annotations for microarray data from GEO (cf. http://gminer.mcw.edu).
Semantic annotation is an important research topic in the Semantic Web community [10]. Tools vary along with the types of documents that they annotate (e.g., image annotation [20]). For an overview and comparison of semantic annotation tools the reader may refer to the study by Uren and colleagues [11].
As we have mentioned earlier, our annotation work-flow can be configured to use any concept-recognition tool. A number of publicly available concept recognizers identify entities from ontologies or terminologies in text. These recognizers include IndexFinder [21], SAPHIRE [22], CONANN [23], and the University of Michigan’s Mgrep [15]. The National Library of Medicine (NLM)’s MetaMap [24], which identifies UMLS Metathesaurus concepts in text, is generally used as the gold standard for evaluating tools in the biomedical domain. Many of these tools are not under active development and are restricted to a particular ontology or the UMLS.
Related tools in the biomedical domain include Terminizer [25], which is an annotation service similar to the NCBO Annotator. Terminizer recognizes concept names and synonyms and their possible permutations but only for OBO ontologies. Terminizer does not allow any automatic semantic expansion of the annotations but allows refining annotations using broader or narrower terms in the user interface. Whatizit [26], which is a set of text mining Web services that can recognize several types of entities such as protein and drug names, diseases, and gene products. Reflect [27], which highlights gene, protein, and small-molecule names and can perform the recognition in HTML as well as PDF and MS Word documents. The originality of Reflect, when used in a Web browser, is that the tool links the identified terms to corresponding entries in biomedical resources e.g., UniProt, DrugBank. However, the tool is not driven by ontologies and does not execute any semantic expansion.
We have conducted a comparative evaluation of two concept recognizers used in the biomedical domain—Mgrep and MetaMap—and found that Mgrep has clear advantages in large-scale service oriented applications, specifically addressing flexibility, speed and scalability [8]. The precision of concept recognition varies depending on the text in each resource and type of entity being recognized: from 93% for recognizing biological processes in descriptions of gene expression experiments to 60% in clinical trials, or from 88% for recognizing disease terms in descriptions of gene expression experiments to 23% for PubMed abstracts [8]. Other studies reported similar results [28, 29]. The average precision is approximately 73%, average recall is 78%.
Most of the other annotation tools do not perform any semantic expansion, which gives the Resource Index and the Annotatora significant advantage. There are however other tools in the biomedical domain that use semantics internally including MedicoPort [30], which uses UMLS semantics to expand user queries; the work of Moskovitch and colleagues [4], who use ontologies for annotation (concept based search) and demonstrate the importance of the context (context-sensitive search) when annotating structured documents. Health-CyberMap [31] uses ontologies and semantic distances for visualizing biomedical resources information. Essie [32] shows that a judicious combination of exploiting document structure, phrase searching, and concept based query expansion is useful for domain optimized information retrieval. Finally, other studies such as Khelif and colleagues [33] illustrate the annotation of a specific resource with specific ontologies (the GeneRIF resource annotated with UMLS and Galen in this case).
Currently, we create annotations based only on textual fields. However, we can extend our approach to other kinds of documents (i.e., images, sounds) by changing the tool that we use for concept recognition. We currently process only text meta-data in English. However, as BioPortal now contains ontologies in multiple languages, we can start using concept recognizers for other languages in the future.
5. Challenges and future plans
We are currently working on expanding the Resource Index to include more resources. Our goal is to index up to 100 public resources, including PubMed, which provides access to all research articles in biomedicine (approximately 20 Million elements). We have analyzed the metrics on ontologies in order to re-structure the database backend for the Resource Index. This restructuring has enabled us to reduce the processing time for one of our larger datasets from one week to one hour [18]. With this type of optimizations, we can now annotate extremely large datasets such as PubMed. We have already indexed the last five years of it (20%). We note that since 2010, changes in MetaMap allow it to be deployed with ontologies outside of UMLS. We are investigating the possibility of including MetaMap as an alternative concept recognizer in the annotation work-flow.
One limiting factor in increasing the number of resources that we index is the need to develop custom access tools for most resources. However, most resource access tools follow the same principles, so we have built templates that enable our collaborators to build them easily and quickly to process their own datasets and to include them in the Resource Index.
Our next challenge is to evaluate the user interface and to understand what works best for domain experts. We have performed small-scale formative evaluations, but will need to work on larger scale evaluation, with different groups of users.
6. Conclusions
We have presented an ontology-based workflow to annotate biomedical resources automatically as well as an index constructed using this workflow. Ontology-based indexing is not new in biomedicine, however it is usually restricted to indexing a specific resource with a specific ontology (vertical approach). We adopt a horizontal approach, accessing annotations for many important resources using a large number of ontologies. This approach follows the translational bioinformatics and Semantic Web vision to discover new knowledge by recombining already existing knowledge (i.e., resources and ontologies) in a manner that the knowledge providers have not previously envisaged.
The Resource Index enables domain experts to search heterogeneous, independently developed resources. While we use ontologies and semantics “under the hood” to improve the quality of the results and to simplify the user interaction, the users are not aware of this complexity. They use a simple search-box interface and can drill down on the specific resources that contain their terms of interest or any other relevant terms.
A. Lists of ontologies and resources
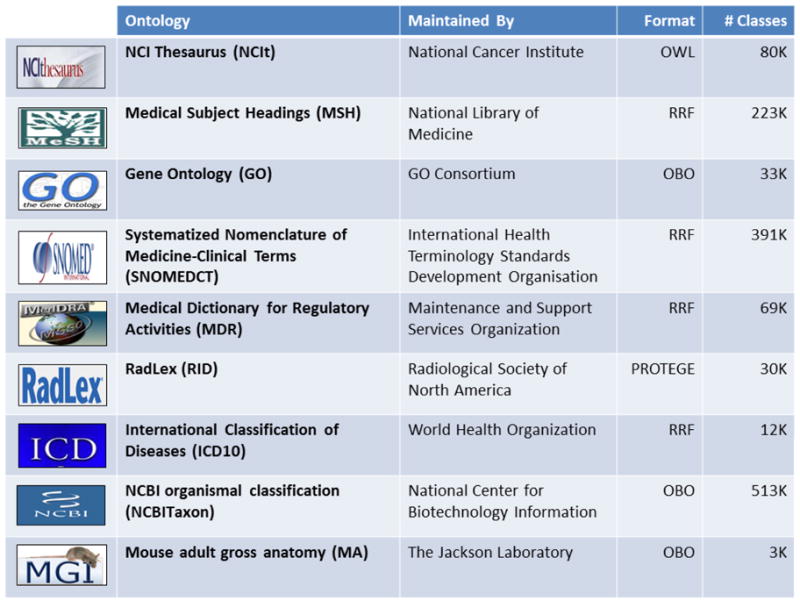
Table 1. A sample of ontologies included in the Resource Index.
Please refer to http://bioportal.bioontology.org/ontologies for a complete listing.
|
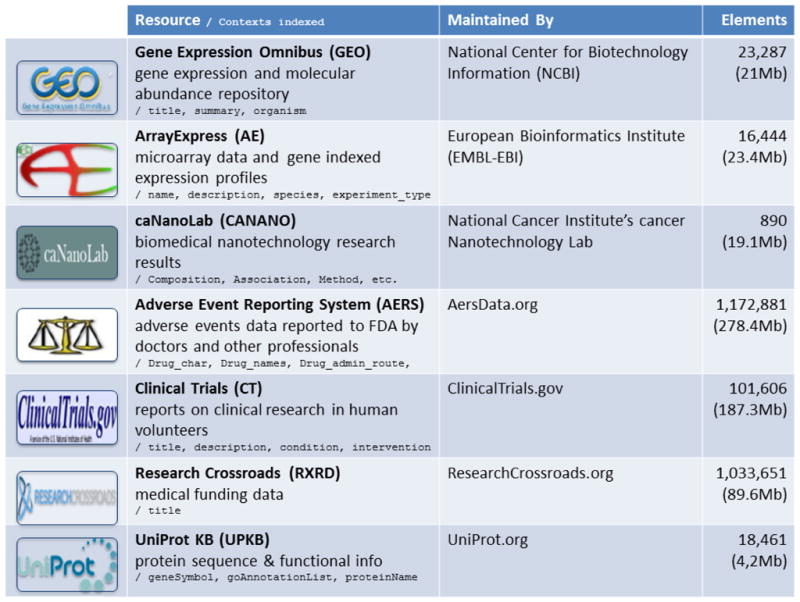
Table 2. A sample of resources included in the Resource Index.
Please refer to http://rest.bioontology.org/resource_index/resources/list/ for a complete listing
|
Acknowledgments
This work was supported in part by the National Center for Biomedical Ontology, under roadmap-initiative grant U54 HG004028 from the National Institutes of Health. The NCBO Resource Index won the First prize in the Semantic Web Challenge 2010 (http://challenge.semanticweb.org/).
Footnotes
Researchers have previously demonstrated the influence and importance of the original context in which a term appears on information retrieval [4].
In this work we assume mappings between ontologies already exists, the creation of biomedical mappings is discussed in numerous other papers.
The OR construct is currently available only through Web service; it is not available through the graphical user interface.
Notice there is no composition of the semantic expansion components e.g., mapping ancestors are not used for annotations.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bodenreider O, Stevens R. Bio-ontologies: Current Trends and Future Directions. Briefing in Bioinformatics. 2006;7(3):256–274. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butte AJ, Chen R. Finding disease-related genomic experiments within an international repository: first steps in translational bioinformatics. American Medical Informatics Association Annual Symposium, AMIA’06; Washington DC, USA. 2006. pp. 106–110. [PMC free article] [PubMed] [Google Scholar]
- 3.Goble C, Stevens R. State of the nation in data integration for bioinformatics. Biomedical Informatics. 2008;41(5):687–693. doi: 10.1016/j.jbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Moskovitch R, Martins SB, Behiri E, Weiss A, Shahar Y. A Comparative Evaluation of Full-text, Concept-based, and Context-sensitive Search. American Medical Informatics Association. 2007;14(2):164–174. doi: 10.1197/jamia.M1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spasic I, Ananiadou S, McNaught J, Kumar A. Text mining and ontologies in biomedicine: making sense of raw text. Briefing in Bioinformatics. 2005;6(3):239–251. doi: 10.1093/bib/6.3.239. [DOI] [PubMed] [Google Scholar]
- 6.Sneiderman CA, Demner-Fushman D, Fiszman M, Ide NC, Rindflesch TC. Knowledge-based Methods to Help Clinicians Find Answers in Medline. American Medical Informatics Association. 2007;14(6):772–780. doi: 10.1197/jamia.M2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nature Reviews Genetics. 2008;9:509–515. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- 8.Shah NH, Bhatia N, Jonquet C, Rubin DL, Chiang AP, Musen MA. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics. 10(9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith NB, Jonquet C, Rubin DL, Storey M-A, Chute CG, Musen MA. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research. 2009;37(web server):170–173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handschuh S, Staab S, editors. Frontiers in Artificial Intelligence and Applications. Vol. 96. IOS Press; 2003. Annotation for the Semantic Web. [Google Scholar]
- 11.Uren V, Cimiano P, Iria J, Handschuh S, Vargas-Vera M, Motta E, Ciravegna F. Semantic annotation for knowledge management: Requirements and a survey of the state of the art. Web Semantics: Science, Services and Agents on the World Wide Web. 2006;4(1):14–28. [Google Scholar]
- 12.Shah NH, Jonquet C, Chiang AP, Butte AJ, Chen R, Musen MA. Ontology-driven Indexing of Public Datasets for Translational Bioinformatics. BMC Bioinformatics. 10(2):S1. doi: 10.1186/1471-2105-10-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Consortium TO, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah NH, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32:267–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xuan W, Dai M, Mirel B, Athey B, Watson SJ, Meng F. Interactive Medline Search Engine Utilizing Biomedical Concepts and Data Integration. BioLINK: Linking Literature, Information and Knowledge for Biology, SIG, ISMB’08; Vienna, Austria. 2007. pp. 55–58. [Google Scholar]
- 16.Dai M, Shah NH, Xuan W, Musen MA, Watson SJ, Athey BD, Meng F. An Efficient Solution for Mapping Free Text to Ontology Terms. American Medical Informatics Association Symposium on Translational BioInformatics, AMIA-TBI’08; San Francisco, CA, USA. 2008. [Google Scholar]
- 17.Noy NF, Griffith NB, Musen MA. Collecting Community-Based Mappings in an Ontology Repository. 7th International Semantic Web Conference, Vol. 5318 of LNCS; Karlsruhe, Germany: Springer; 2008. pp. 371–368. [Google Scholar]
- 18.LePendu P, Noy NF, Jonquet C, Alexander PR, Shah NH, Musen MA. Optimize First, Buy Later: Analyzing Metrics to Ramp-up Very Large Knowledge Bases. 9th International Semantic Web Conference, Vol. 6496 of LNCS; Shanghai, China: Springer; 2010. pp. 486–501. [Google Scholar]
- 19.Bhogal J, Macfarlane A, Smith P. A review of ontology based query expansion. Information Processing and Management. 2007;43:866–886. [Google Scholar]
- 20.Hollink L, Schreiber G, Wielemaker J, Wielinga B. Semantic Annotation of Image Collections. Knowledge Markup and Semantic Annotation Workshop; Sanibel, FL, USA. 2003. [Google Scholar]
- 21.Zou Q, Chu WW, Morioka C, Leazer GH, Kangarloo H. IndexFinder: A Method of Extracting Key Concepts from Clinical Texts for Indexing. American Medical Informatics Association Annual Symposium, AMIA’03; Washington DC, USA. 2003. pp. 763–767. [PMC free article] [PubMed] [Google Scholar]
- 22.Hersh WR, Greenes RA. SAPHIRE - an information retrieval system featuring concept matching, automatic indexing, probabilistic retrieval, and hierarchical relationships. Computers and Biomedical Research. 1990;23(5):410–425. doi: 10.1016/0010-4809(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 23.Reeve LH, Han H. CONANN: An Online Biomedical Concept Annotator. 4th International Workshop Data Integration in the Life Sciences, Vol. 4544 of LNCS; Philadelphia, PA, USA: Springer-Verlag; 2007. pp. 264–279. [Google Scholar]
- 24.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. American Medical Informatics Association Annual Symposium, AMIA’01; Washington, DC, USA. 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock D, Morrison N, Velarde G, Field D. Terminizer –Assisting Mark-Up of Text Using Ontological Terms. 3rd International Biocuration Conference; Berlin, Germany. 2009. [Google Scholar]
- 26.Rebholz-Schuhmann D, Arregui M, Gaudan S, Kirsch H, Jimeno A. Text processing through Web services: Calling Whatizit. Bioinformatics. 2008;24(2):296–298. doi: 10.1093/bioinformatics/btm557. [DOI] [PubMed] [Google Scholar]
- 27.Pafilis E, O’Donoghue SI, Jensen LJ, Horn H, Kuhn M, Brown NP, Schneider R. Reflect: augmented browsing for the life scientist. Nature Biotechnology. 2009;27:508–510. doi: 10.1038/nbt0609-508. [DOI] [PubMed] [Google Scholar]
- 28.Simon JS, Twigger N, Geiger Joey. Using the NCBO Web Services for Concept Recognition and Ontology Annotation of Expression Datasets. Workshop on Semantic Web Applications and Tools for Life Sciences, SWAT4LS’09, Vol. 559 of CEUR Workshop Proceedings; Amsterdam, The Netherlands. 2009. [Google Scholar]
- 29.Sarkar IN. Leveraging Biomedical Ontologies and Annotation Services to Organize Microbiome Data from Mammalian Hosts. American Medical Informatics Association Annual Symposium, AMIA’10; Washington DC., USA. 2010. pp. 717–721. [PMC free article] [PubMed] [Google Scholar]
- 30.Can AB, Baykal N. MedicoPort: A medical search engine for all. Computer Methods and Programs in Biomedicine. 2007;86(1):73–86. doi: 10.1016/j.cmpb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Kamel-Boulos MN. A first look at HealthCyberMap medical semantic subject search engine. Technology and Health Care. 2004;12:33–41. [PubMed] [Google Scholar]
- 32.Ide NC, Loane RF, Demner-Fushman D. Essie: A Concept-based Search Engine for Structured Biomedical Text. American Medical Informatics Association. 2007;14(3):253–263. doi: 10.1197/jamia.M2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khelif K, Dieng-Kuntz R, Barbry P. An ontology-based approach to support text mining and information retrieval in the biological domain. Universal Computer Science, Special Issue on Ontologies and their Applications. 2007;13(12):1881–1907. [Google Scholar]