Abstract
The human single-stranded DNA-binding replication A protein (RPA) is involved in various DNA-processing events. By comparing the affinity of hRPA for artificial DNA hairpin structures with 3′- or 5′-protruding single-stranded arms, we found that hRPA binds ssDNA with a defined polarity; a strong ssDNA interaction domain of hRPA is positioned at the 5′ side of its binding region, a weak ssDNA-binding domain resides at the 3′ side. Polarity appears crucial for positioning of the excision repair nucleases XPG and ERCC1–XPF on the DNA. With the 3′-oriented side of hRPA facing a duplex ssDNA junction, hRPA interacts with and stimulates ERCC1–XPF, whereas the 5′-oriented side of hRPA at a DNA junction allows stable binding of XPG to hRPA. Our data pinpoint hRPA to the undamaged strand during nucleotide excision repair. Polarity of hRPA on ssDNA is likely to contribute to the directionality of other hRPA-dependent processes as well.
Keywords: Replication protein A, nucleotide excision repair, ERCC1–XPF, XPG, DNA-binding, polarity
Human replication protein A (hRPA) is a single-stranded DNA (ssDNA)-binding protein composed of three subunits of ∼70, 32, and 14 kD (Fairman and Stillman 1988; Wood et al. 1988). It is involved in multiple DNA-metabolizing processes, including (SV40−) DNA replication, recombination, and repair (for review, see Wold 1997). At a certain stage during all these processes, complementary DNA strands are separated and action is required along ssDNA intermediates. This action always occurs with a defined directionality; DNA polymerases synthesize new DNA strands in a 5′ → 3′ direction, Rad51-catalyzed recombinational strand exchange proceeds in a 3′ → 5′ direction (Baumann and West 1997), and repair incisions are made by nucleases that specifically cleave either at the 3′ or the 5′ side of a lesion (O’Donovan et al. 1994a; Matsunaga et al. 1995; Sijbers et al. 1996). Two properties of hRPA have generally been acknowledged to have a role in these processes—first, hRPA’s high affinity for ssDNA, and second, its ability to specifically interact with a variety of proteins.
hRPA binds to ssDNA with an apparent association constant of 109–1011 m, which is at least three orders of magnitude higher than its affinity for double-stranded DNA (dsDNA) (Kim et al. 1992, 1994). Binding of a monomer of hRPA to ssDNA occurs through the 70-kD subunit, and two different binding modes have been identified; hRPA interacts with a minimal occluded binding site of 8–10 nucleotides (Blackwell and Borowiec 1994) and with a 100-fold more stable occluded binding site of ∼30 nucleotides (Kim et al. 1992, 1994; Blackwell et al. 1996). In general, the high affinity of hRPA for ssDNA is thought to contribute to the stabilization of ssDNA intermediates and the removal of secondary structure from single-stranded regions. More specifically, binding of hRPA to ssDNA is required for unwinding of (SV40) replication origins and elongation of replication forks (Wold et al. 1987; Kenny et al. 1989; Weinberg et al. 1990; Tsurimoto and Stillman 1991). In recombination, hRPA stimulates DNA strand exchange between Rad51-coated ssDNA and duplex DNA (Baumann and West 1997; Sung 1997; New et al. 1998; Shinohara and Ogawa 1998). In nucleotide excision repair (NER), binding of hRPA to ssDNA might help the formation and stabilization of a locally unwound DNA intermediate. Interestingly, the extent to which DNA is opened around a lesion corresponds to the 30-nucleotide optimal binding region of hRPA (Huang et al. 1992; Evans et al. 1997a). Most of the activities mentioned above are nonspecific, in the sense that other single-stranded binding (SSB) proteins can carry out these functions as well.
The specificity of hRPA’s contribution to the different DNA-metabolizing events comes from its ability to interact with other proteins. hRPA specifically interacts with DNA polymerase α, probably to recruit this DNA polymerase to the unwound replication origin to initiate DNA replication (Kenny et al. 1989; Dornreiter et al. 1992; Braun et al. 1997). Also, hRPA binds to hRad52, an interaction that seems to be essential for homologous recombination (Park et al. 1996). Furthermore, hRPA interactions have been described with the NER proteins XPA, XPG, and ERCC1–XPF (He et al. 1995; Matsunaga et al. 1996). A complex between hRPA and the damage-recognition protein XPA was reported to show cooperative binding to DNA lesions (He et al. 1995).
XPG and ERCC1–XPF are structure-specific endonucleases that cut the damaged strand, respectively, at the 3′ and 5′ side of the locally unwound NER intermediate (O’Donovan et al. 1994a; Matsunaga et al. 1995; Sijbers et al. 1996; Evans et al. 1997a). It is not known what directs the nucleases to the damaged strand and prevents incisions in the nondamaged strand during NER. hRPA is indispensable for NER incisions (Coverley et al. 1991), but its physical localization with regard to the DNA and the remainder of the repair machinery is not clear. Interactions of hRPA with XPG and ERCC1–XPF were found to modulate the activity of both NER nucleases. Controversy exists, however, as to whether hRPA has a positive or negative effect on cleavage (Matsunaga et al. 1996; Bessho et al. 1997; Evans et al. 1997a). Here, we tested the hypothesis that the orientation of hRPA with regard to the cleavage site determines this outcome.
Results
hRPA binds to ssDNA with a defined polarity
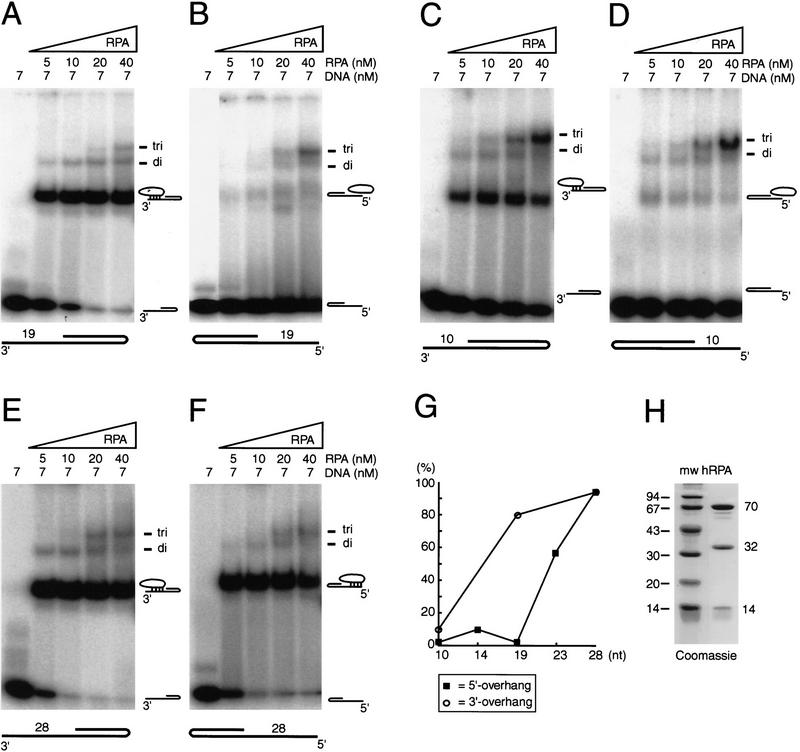
Several indications exist that the 8- to 10-nucleotide binding mode of hRPA is a precursor of the more stable 30-nucleotide binding mode (Blackwell and Borowiec 1994; Blackwell et al. 1996; Gomes et al. 1996). To investigate whether hRPA binds ssDNA with a defined polarity, we considered the possibility that initial DNA recognition occurs at a fixed position at one of the borders of the 30-nucleotide binding region. This would imply that, in the case of short (8–30 nucleotide) single-stranded arms protruding from duplex regions, hRPA shows a binding-preference for arms protruding in either the 3′ or the 5′ direction.
To study this, we used partially self-complementary oligonucleotides containing identical duplex regions with either 3′- or 5′-protruding single-stranded arms. The rationale to use hairpin DNA substrates, rather than combinations of two annealed oligonucleotides, is that their strong tendency to self-anneal counteracts the ability of hRPA to unwind duplex DNA regions (Georgaki et al. 1992; Treuner et al. 1996), which otherwise would complicate the analysis. As hRPA preferentially binds to single-stranded polypyrimidine tracts (Kim et al. 1992), the single-stranded overhangs of these substrates solely consisted of thymine residues, which also minimized the formation of secondary structures. Using gel-retardation assays, a dramatic difference in hRPA affinity for 3′- and 5′-protruding 19-nucleotide single-stranded arms [19-d(T) substrates] was observed. hRPA (Fig. 1H) bound very efficiently to the 3′ protruding 19-d(T) substrate, with almost 80% of the substrate complexed to hRPA monomers at approximately equimolar hRPA–DNA concentrations (Fig. 1A). In contrast, virtually no monomeric protein–DNA complexes were observed with the 5′-protruding substrate, not even when a more than threefold molar excess of hRPA was present (Fig. 1B). With these hRPA concentrations, only some hRPA trimers complexed to (possibly partially unwound) DNA hairpins were detected (Fig. 1B).
Figure 1.
RPA preferentially binds to 3′-protruding single-stranded arms. (A–F) Gel-retardation assays showing the binding characteristics of hRPA to 3′-protruding 19-d(T) substrate (A), 5′-protruding 19-d(T) substrate (B), 3′-protruding 10-d(T) substrate (C), 5′-protruding 10-d(T) substrate (D), 3′-protruding 28-d(T) substrate (E), 5′-protruding 28-d(T) substrate (F). (Di and tri) Dimeric and trimeric hRPA complexes bound to DNA, respectively. No glutaraldehyde was added to any of the reactions. (G) Graphic presentation showing DNA-binding efficiency of hRPA (in percentage of bound substrate at 10 nm hRPA, which is the hRPA concentration that is referred to in the text as the approximately equimolar concentration) vs. the size of the single-stranded overhang. The DNA-binding efficiencies presented here are not meant to give a quantative measure of hRPA’s affinity for the different substrates. (H) Coomassie-stained protein gel of purified hRPA.
The effect of the size of the single-stranded stretch on the hRPA-binding preference was examined. Using substrates with 10-nucleotide overhangs, essentially similar binding characteristics were observed as with the 19-nucleotide single-stranded overhangs. Very few protein–DNA complexes were formed with 5′-protruding 10-d(T) substrates, whereas hRPA–DNA complexes were clearly detectable with the 3′-protruding 10-d(T) substrate (Fig. 1C, D). The affinity of hRPA for the 3′-protruding 10-d(T) substrate, however, was reduced compared with its 3′-protruding 19-nucleotide counterpart, as at approximately equimolar concentrations, 10% instead of 80% of the substrate was complexed to an hRPA monomer (Fig. 1G).
Similar results were obtained with single-stranded overhangs composed of a random sequence instead of a poly(dT) tract, demonstrating that our results were not a peculiarity of specific oligonucleotides. Virtually no hRPA binding was observed to a 5′-protruding 13-d(N) substrate, whereas at approximately equimolar hRPA–DNA concentrations 30%–40% of a 3′-protruding 14-d(N) substrate was bound to hRPA monomers (data not shown). Changing the divalent ion concentration from 0.75 mm Mn2+ to 5 mm Mg2+ or adding 5 mm EDTA did not alter the affinity of hRPA for any of the substrates (data not shown).
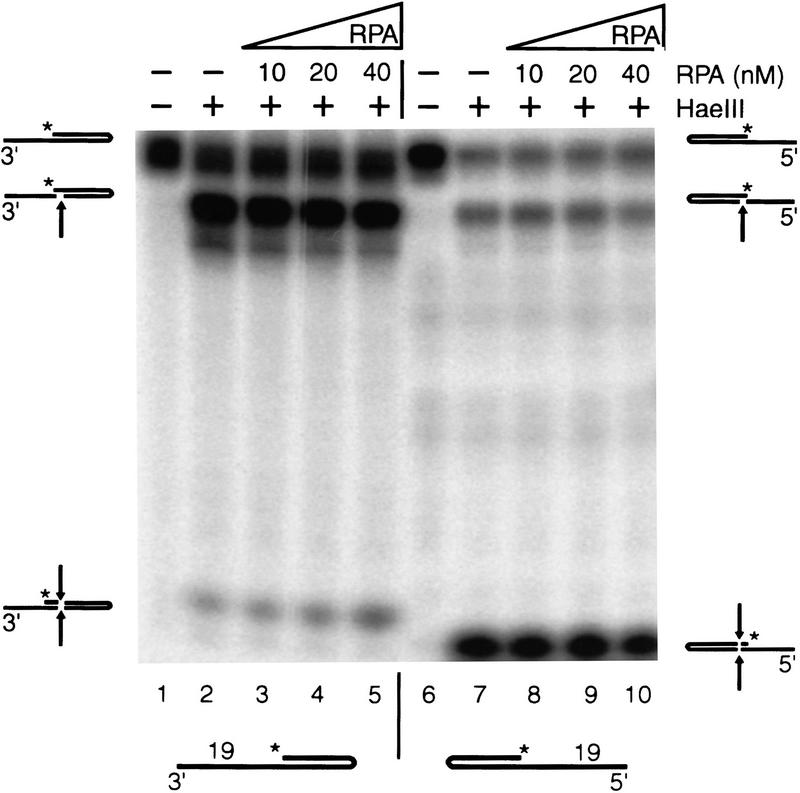
To find out whether unwinding of duplex DNA underlies the binding preference of hRPA for 3′-protruding single-strand regions, we performed a footprinting type of analysis with restriction enzymes. Cleavage of 3′- and 5′-protruding substrates by HaeIII, which recognizes the 4 bp immediately bordering the ssDNA portion of both substrates, was not affected by the presence of hRPA in the concentration range used above (Fig. 2). Similar results were obtained with BstB1, a restriction enzyme that recognizes 6 bp located near the turn of the hairpins (data not shown). These data demonstrated that the duplex region of these hairpins is not significantly unwound by hRPA monomers. Therefore, the observation that hRPA monomers only bind stably to 3′-protruding single-stranded arms and not to 5′-protruding single-stranded arms, at least when these arms range in size from 10–19 nucleotides, can only be explained by defined ssDNA-binding polarity of hRPA. Apparently, with arms protruding in the 5′ direction, the neighboring duplex DNA physically hinders the oriented interaction of hRPA with ssDNA.
Figure 2.
hRPA does not significantly unwind hairpin structures. Cleavage pattern of HaeIII in 3′- and 5′-protruding 19 nucleotide substrates is not altered when substrates are preincubated with hRPA (cf. lanes 3–5 with lane 2, and lanes 8–10 with lane 7, respectively). Note that, independent of the presence of hRPA, HaeIII is hardly able to cleave the nonprotruding strand of the 3′-protruding substrate (lanes 2–5). The dissimilar migration pattern of the released dinucleotides in lanes 2–5 vs. lanes 7–10 we attribute to different nucleotide compositions (CC vs. GG).
Next, we increased the size of the single-strand arms to that of the optimal hRPA-binding site. Equally efficient interactions were observed with both 3′- and 5′-protruding 28-d(T) substrates. More than 90% of both substrates were complexed to hRPA monomers at approximately equimolar hRPA–DNA concentrations (Fig. 1E–G). A partial loss of binding preference was already observed with a 23-nucleotide-long 5′-protruding substrate, which was bound with moderate affinity by hRPA (Fig. 1G). We conclude that hRPA preferentially binds 3′-protruding single-stranded arms, a phenomenon that is abolished when single-stranded overhangs reach the size of the optimal hRPA-binding site. The preference in affinity for single-stranded arms protruding in a certain direction implies that hRPA binds ssDNA with a defined polarity. We designate the strong ssDNA-binding side of hRPA as the “5′-oriented side” of hRPA, as it is located at the 5′ side of the bound ssDNA region (see Fig. 8, below). Conversely, the weak single-stranded interaction subdomain is defined as the “3′- oriented side” of hRPA. Obviously, the orientation of hRPA with respect to the ssDNA has important implications for the proteins with which it interacts.
Figure 8.
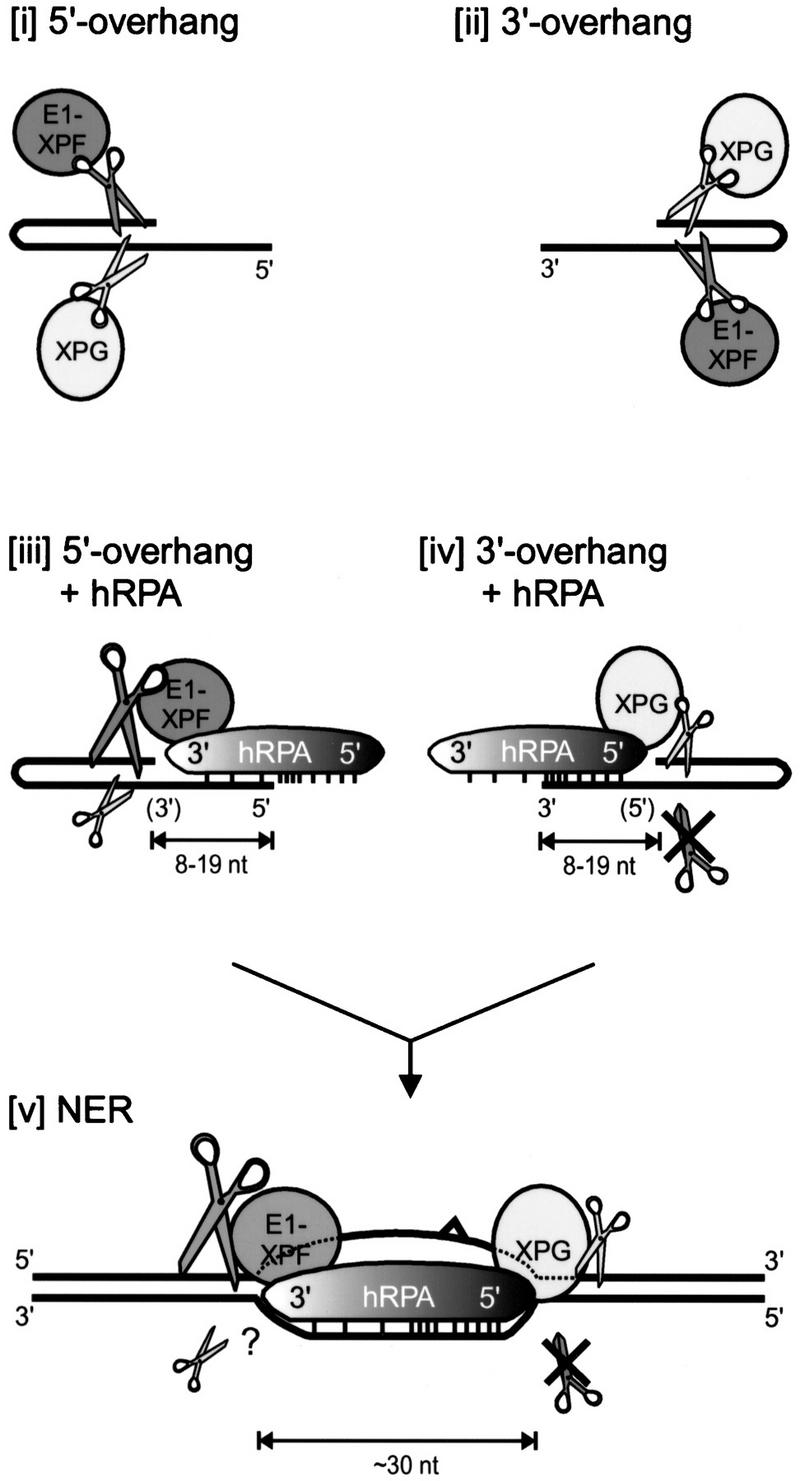
Model of hRPA binding to the undamaged strand during NER. (i,ii) ERCC1–XPF and XPG cleavage of 5′- and 3′-protruding substrates, respectively. (iii) On short 5′-protruding arms, hRPA weakly interacts with the single-stranded stretch, as bordering duplex DNA physically hinders stable binding of the strong DNA-interaction domain at the 5′-oriented side of hRPA. The 3′-oriented side of hRPA positions and strongly stimulates ERCC1–XPF-mediated incisions in the non-hRPA-bound strand, whereas it does not interact with XPG and slightly inhibits its activity. (iv) On short 3′-protruding arms, the strong binding domain of hRPA stably interacts with the single-stranded portion. This 5′-oriented side of hRPA positions XPG-mediated incisions (and slightly inhibits XPG-activity; see text), and completely blocks ERCC1–XPF-mediated incisions. (v) Extrapolation to nucleotide excision repair. On the formation of an opened DNA intermediate, which requires the activities of XPA, XPC–HHR23B, TFIIH, and hRPA (Evans et al. 1997b; Mu et al. 1997), an hRPA monomer is bound to the undamaged DNA strand and positions both nucleases onto the damaged strand; bound as such, it is able to stimulate strongly ERCC1–XPF incisions in the damaged strand and completely block ERCC1–XPF incisions in the undamaged strand. For clarity, XPA, XPC–HHR23B, and TFIIH are not depicted.
Defined ssDNA-binding polarity of hRPA modulates the activity of the excision repair nuclease ERCC1–XPF
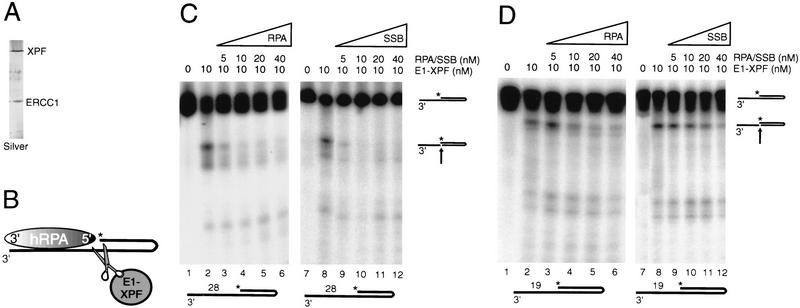
To investigate possible functional consequences of the defined ssDNA-binding polarity of hRPA, we focused on its role in NER. hRPA has been claimed to modulate the activity of XPG and the heterodimeric complex ERCC1–XPF (Matsunaga et al. 1996; Bessho et al. 1997; Evans et al. 1997a). Both are structure-specific NER endonucleases that direct their incisions to one strand of duplex DNA at ds-DNA–ssDNA junctions, with ERCC1–XPF cutting the strand that continues as a 3′-protruding single-stranded region and XPG cleaving the opposite strand (Matsunaga et al. 1996; Bessho et al. 1997; Evans et al. 1997a; see Figs. 3B, 4A, and 5B). Previously, we have shown that these enzymes require only one single-stranded arm protruding from duplex DNA to correctly position their incisions (de Laat et al. 1998). As such, the 3′- and 5′-protruding constructs used above are suitable substrates for cleavage by ERCC1–XPF and XPG. To determine whether the orientation of hRPA and the strand to which it binds influence the activity of the NER nucleases, we performed incision assays in the presence of hRPA. The data presented below were obtained with a homogeneous preparation of recombinant ERCC1–XPF purified from baculovirus-infected insect cells (Fig. 3A). The results of key experiments were verified with recombinant ERCC1–XPF purified from Escherichia coli to confirm that the observed nuclease activities were attributable to ERCC1–XPF and not a contaminating nuclease, and to exclude the possibility that other repair factors copurifying with ERCC1–XPF from insect cells obscured the outcome (for an example, see Fig. 4D).
Figure 3.
hRPA inhibits ERCC1–XPF endonuclease activity on 3′-protruding singles-stranded-arms. (A) Silver-stained protein gel of purified ERCC1–XPF. (B) Schematic presentation of ERCC1–XPF and hRPA acting on a 3′-protruding substrate. Asterisk indicates position of radioactive label. The 3′- and 5′-oriented side of hRPA, representing the weak and the strong ssDNA-binding side of hRPA, respectively, are indicated. (C,D) Denaturing polyacrylamide gels analyzing DNA incision products. (C) ERCC1–XPF nuclease assays on 3′-protruding 28-d(T) substrates, with increasing amounts of hRPA (lanes 3–6) and E. coli SSB protein (lanes 9–12). (D) ERCC1–XPF nuclease assays on 3′-protruding 19-d(T) substrates, with increasing amounts of hRPA (lanes 3–6) and E. coli SSB protein (lanes 9–12). Note that E. coli SSB protein concentration is given in nanomoles of tetramer per liter. The minor incision products visible in C and D do not correspond to known duplex single-stranded DNA junctions and probably arise from weak cutting activity near uncharacterized secondary structures in the DNA hairpin substrates.
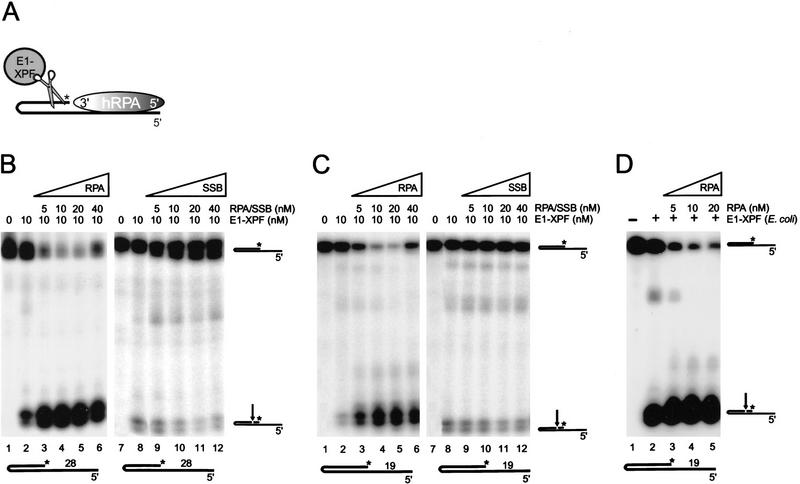
Figure 4.
hRPA stimulates ERCC1–XPF endonuclease activity on 5′-protruding single-stranded arms. (A) Schematic presentation of ERCC1–XPF and hRPA acting on a 5′-protruding substrate. Asterisk indicates position of radioactive label. The 3′- and 5′-oriented side of hRPA, representing the weak and the strong ssDNA-binding side of hRPA, respectively, are indicated. (B) ERCC1–XPF nuclease assays on 5′-protruding 28-d(T) substrates, with increasing amounts of hRPA (lanes 3–6) and E. coli SSB protein (lanes 9–12). (C) ERCC1–XPF nuclease assays on 5′-protruding 19-d(T) substrates, with increasing amounts of hRPA (lanes 3–6) and E. coli SSB protein (lanes 9–12). E. coli SSB protein concentration is given in nanomoles of tetramer per liter. (D) In C, lanes 1–5, but with ERCC1–XPF preparation purified from E. coli, to demonstrate that the effects of hRPA on ERCC1–XPF are not influenced by contaminants in the protein prep. Minor incision products visible in B–D do not correspond to known duplex ssDNA junctions and probably arise from weak cutting activity near uncharacterized secondary structures in the DNA hairpin substrates. Note that the ERCC1–XPF preparation obtained from E. coli (D) is more active, but yields similar incision products.
Figure 5.
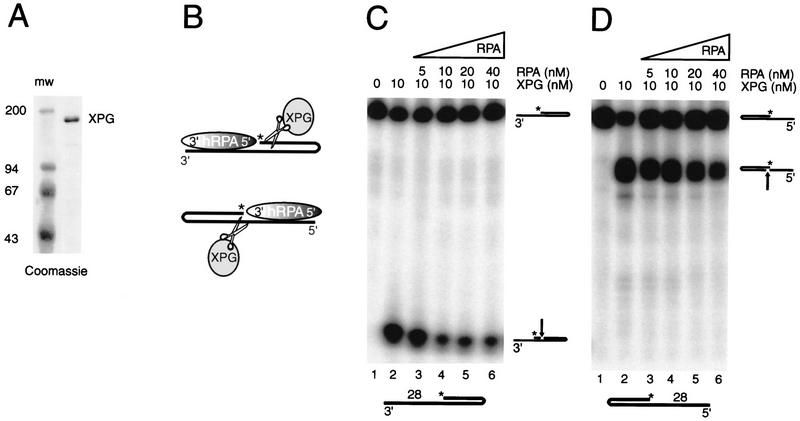
Polarity of hRPA on ssDNA has minor implications for XPG endonuclease activity. (A) Coomassie-stained protein gel of purified XPG. (B) Schematic presentation of XPG and hRPA acting on a 3′-protruding substrate (top) and a 5′-protruding substrate (bottom). Asterisks indicate positions of radioactive label. The 3′-oriented and 5′-oriented side of hRPA, representing the weak and the strong ssDNA-binding side of hRPA, respectively, are indicated. (C–D) XPG nuclease assays on C 3′-protruding 28-d(T) substrates, −/+ hRPA; (D) 5′-protruding 28-d(T) substrates, −/+ hRPA.
ERCC1–XPF cleaves 3′-protruding substrates in the strand that continues as a single-stranded arm, thereby removing the single-stranded portion (Fig. 3B). Preincubation of the 3′-protruding 28-d(T) substrate with increasing amounts of hRPA caused almost complete inhibition of ERCC1–XPF cleavage, a phenomenon also observed with the shorter 3′-protruding 19-d(T) substrates (Fig. 3C,D, lanes 1–6). To test whether this inhibitory effect was specific for hRPA, similar assays were performed with E. coli SSB protein. E. coli SSB protein blocked ERCC1–XPF activity approximately to the same extent as hRPA did (Fig. 3C,D, lanes 7–12), suggesting that hRPA-mediated inhibition of ERCC1–XPF cleavage on 3′-protruding DNA substrates does not require specific protein–protein interactions and is most likely attributable to steric hindrance.
Similarly, we tested 5′-protruding 28-d(T) DNA substrates. In the absence of hRPA, ERCC1–XPF cleaves these substrates near the junction in the nonprotruding strand, which results in extension of the single-stranded portion (Fig. 4A). In contrast to 3′ protrusions, preincubation of hRPA with the 5′-protruding 28-d(T) substrate had a strong stimulatory effect on ERCC1–XPF activity (Fig. 4B, lanes 1–6). No stimulation of ERCC1–XPF activity was observed with E. coli SSB protein (Fig. 4B, lanes 7–12), demonstrating that this effect was specific for hRPA and suggesting a direct protein–protein interaction between hRPA and ERCC1–XPF on 5′-protruding substrates. Suprisingly, even on 5′-protruding 19-d(T) substrates, for which bandshift experiments failed to reveal hRPA binding (see Fig. 1B), ERCC1–XPF cleavage was strongly stimulated by preincubation with hRPA (Fig. 4C). Apparently, hRPA does transiently interact with short 5′-protruding single-stranded arms, but this interaction is not stable enough to withstand bandshift conditions. ERCC1–XPF-mediated cleavage of 5′-protruding substrates yields a DNA product that can serve again as a substrate for cleavage (Fig. 4A), and with 5′-protruding hairpin substrates labeled at the 5′ end, we observed that in the presence of hRPA most of the incision products are incised again by ERCC1–XPF some nucleotides further away in the duplex (data not shown). The original positions of ERCC1–XPF incisions in both 3′- and 5′-protruding substrates were not altered by hRPA though (Figs. 3C,D and 4B,C), indicating again that hRPA binding does not significantly modify the hairpin structure. The specific stimulation of ERCC1–XPF activity on 5′-protruding single-stranded arms, not observed with 3′-protruding hairpin substrates, suggests orientation-specific interactions between hRPA and ERCC1–XPF.
hRPA-binding orientation has minor implications on XPG endonuclease activity
Similar incision assays were performed to determine the effect of differently oriented hRPA molecules on the activity of XPG, the complementary NER endonuclease. For this purpose, a homogeneous preparation of XPG protein was obtained from baculovirus-infected insect cells (Fig. 5A), using a standard purification protocol (with one additional purification step) that removes other repair factors and contaminating nucleases (O’Donovan et al. 1994b). XPG-mediated cleavage in 3′-overhanging hairpin substrates occurs in the nonprotruding strand, whereas hairpins with a 5′ single-stranded arm are cleaved in the protruding strand (Fig. 5B). On both the 3′- and 5′-protruding 28-d(T) substrates, we found an inhibitory effect of hRPA on XPG-incision activity (Fig. 5C,D). Inhibition, however, was not as pronounced as observed with ERCC1–XPF on 3′-protrusions, and was incomplete, even with an excess of hRPA. The partial inhibitory effect was also found with E. coli SSB replacing hRPA (data not shown). Again, hRPA did not change the position of XPG-mediated incisions in either substrate, once more confirming the footprinting analysis that demonstrated the structural integrity of these hairpins in the presence of hRPA. We conclude that hRPA, irrespective of its orientation with regard to the dsDNA–ssDNA junction, shows a weak and nonspecific modulation of XPG activity on the substrates tested.
hRPA–ERCC1–XPF complex formation occurs specifically on 5′-protruding substrates
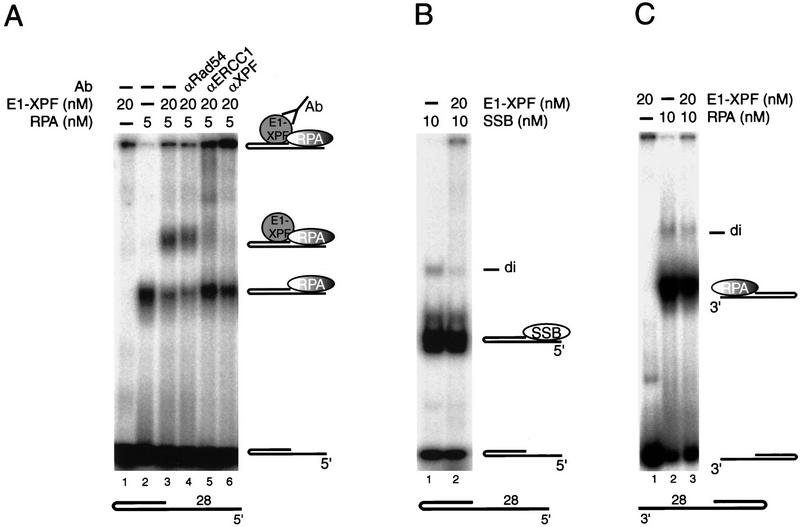
Obviously, the stimulatory effect of hRPA on ERCC1–XPF activity could imply that hRPA facilitates the positioning of this nuclease onto DNA, in line with the reported protein–protein interactions between them (Matsunaga et al. 1996; Bessho et al. 1997). Selective stimulation of ERCC1–XPF activity on 5′-protruding DNA substrates suggests that such a physical interaction strongly depends on the orientation of hRPA with regard to the dsDNA–ssDNA junction. To investigate this further, ERCC1–XPF binding to preformed DNA–hRPA complexes was monitored in gel-retardation assays. In the absence of hRPA, we could not detect DNA–ERCC1–XPF complexes with any of the substrates, neither in standard nuclease buffers nor in the presence of divalent ions such as Ca2+ (Fig. 6A, lane 1). After coincubation of ERCC1–XPF with hRPA and the 5′-protruding 28-d(T) substrate in standard nuclease buffers, a supershifted band was vaguely detectable, possibly representing a complex of DNA, hRPA, and ERCC1–XPF (data not shown). The strong stimulation by hRPA of ERCC1–XPF-mediated cleavage, however, is expected to hamper the trapping of transient complexes. To circumvent this problem, a similar coincubation was performed in a standard buffer containing 5 mm Ca2+ instead of Mn2+ or Mg2+. In the presence of calcium, which was shown previously to block ERCC1–XPF incision activity (de Laat et al. 1998), a clear supershifted complex was detectable in addition to the DNA–hRPA complex, which was dependent on the presence of DNA, hRPA, and ERCC1–XPF and originated from the DNA–hRPA complex (Fig. 6A, cf. lane 3 and lanes 1 and 2). Antibodies against ERCC1 and XPF further shifted this particular complex to the slot of the gel, whereas an unrelated antibody did not (Fig. 6A, lane 4–6). Therefore, the supershifted band represented a ternary complex, consisting of ERCC1–XPF and hRPA bound to the 5′-protruding DNA substrate. The use of EDTA instead of Ca2+ completely disrupted ternary complex formation but did not alter the binding of hRPA to DNA (data not shown), demonstrating that not only a direct protein interaction with hRPA, but also divalent cation-dependent DNA binding is a prerequisite for ERCC1–XPF to interact with hRPA–DNA complexes. Calcium, apparently, allows ERCC1–XPF to bind DNA but prevents further endonucleolytic processing. Ternary complex formation required specific interactions between ERCC1–XPF and hRPA, as E. coli SSB protein was not able to produce such a supershift (Fig. 6B). Therefore, it appears that hRPA stimulates ERCC1–XPF activity by facilitating, via direct protein–protein interactions, the positioning of this nuclease onto the DNA.
Figure 6.
hRPA–ERCC1–XPF complex formation occurs specifically on 5′-protruding substrates. (A–C) Gel-retardation assays; all incubations were performed in 5 mm CaCl2, in the presence of 0.05% glutaraldehyde. (A) hRPA–ERCC1–XPF complex formation on 5′-protruding 28-d(T) substrates. For clarity, only 5 mm hRPA was used; higher concentrations of hRPA induced some hRPA dimer formation on these substrates, which migrated similar to the ternary hRPA–ERCC1–XPF complexes. Ternary complex formation was also observed at 10 mm hRPA (data not shown). (B) No complex formation between E. coli SSB and ERCC1–XPF on 5′-protruding 28-d(T) substrates. (C) No hRPA–ERCC1-XPF complex formation on 3′-protruding 28-d(T) substrates.
Under the optimized conditions described above, ERCC1–XPF could not form a ternary complex on a 3′-protruding 28-d(T) substrate preincubated with hRPA (Fig. 6C), not even when twice the amount of enzyme was added (data not shown). This demonstrated that physical interaction among ERCC1–XPF, hRPA, and DNA is dependent on the orientation of hRPA with regard to the dsDNA–ssDNA junction. As on 5′-protruding substrates, the 3′-oriented side of hRPA is positioned toward the junction, we conclude that hRPA, bound to ssDNA, can only position and stimulate ERCC1–XPF-mediated incisions in duplex DNA that borders directly at its 3′-oriented side (see Fig. 8, below).
hRPA–XPG complex formation occurs specifically on 3′-protruding substrates
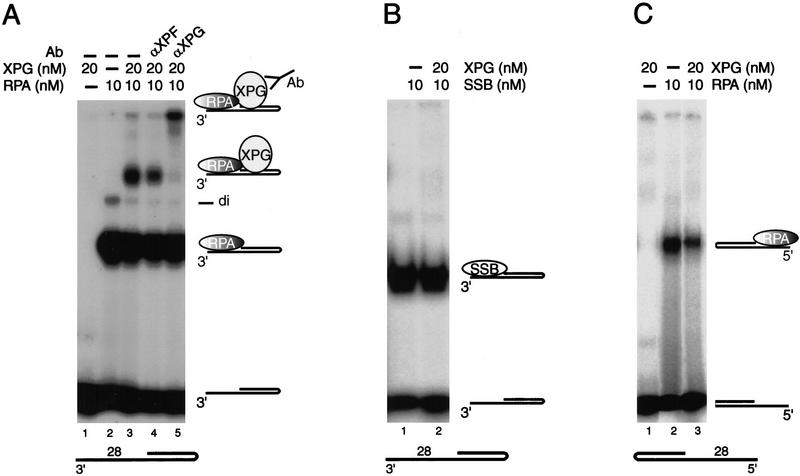
Although hRPA did not dramatically affect XPG activity under the conditions tested here, the possibility of a specific engagement when acting in close vicinity on DNA had to be considered. In the presence of Ca2+, no ternary complex was detected when XPG was coincubated with hRPA bound to either DNA substrate (data not shown). When binding was allowed in standard nuclease buffer containing 0.75 mm Mn2+, however, clearly a new, slowly migrating complex was observed. Its formation required the presence of hRPA, XPG, and the 3′-protruding 28-d(T) substrate (Fig. 7A, cf. lane 3 and lanes 1 and 2). Addition of an antibody against XPG specifically shifted this complex to the slot of the gel, whereas an unrelated antibody did not, demonstrating the presence of XPG in this complex (Fig. 7A, lanes 4,5). Complex formation could not be realized by E. coli SSB, showing the specificity of the XPG–hRPA interaction (Fig. 7B). Furthermore, no interaction between XPG and hRPA–DNA complexes was detected on 5′-protruding 28-d(T) substrates (Fig. 7C), revealing also that the interaction between XPG and hRPA depends on the orientation of hRPA with regard to the dsDNA–ssDNA junction. Therefore, hRPA can only specifically interact with XPG on DNA when the duplex region starts immediately at its 5′-oriented side, which is exactly opposite to what was found for ERCC1–XPF.
Figure 7.
hRPA–XPG complex formation occurs specifically on 3′-protruding substrates. (A–C) Gel-retardation assays; all incubations were performed in 0.75 mm MnCl2, in the presence of 0.05% glutaraldehyde. (A) hRPA–XPG complex formation on 3′-protruding 28-d(T) substrates. Note that at 10 mm hRPA some hRPA dimer formation, (di) occurs (lane 2). (B) No complex formation between E. coli SSB and XPG on 3′-protruding 28-d(T) substrates. (C) No hRPA–XPG complex formation on 5′-protruding 28-d(T) substrates.
Discussion
hRPA is a DNA-binding protein involved in multiple DNA transactions, including replication, recombination, repair, and possibly transcription (Wold 1997). A common denominator in all these processes is a ssDNA intermediate along which action takes place with a defined directionality. Two properties of hRPA have generally been acknowledged to have an important role in these processes. First, its high affinity for ssDNA is thought to be required for the stabilization of ssDNA intermediates and the removal of secondary structures from these regions. Second, its ability to specifically interact with many proteins, such as DNA polymerases and repair enzymes, is assumed to coordinate the action of these factors. Here, we introduced a new parameter important for hRPA functioning. Using a set of defined DNA structures, containing a duplex region with one single-stranded arm protruding either 3′ or 5′, we demonstrated that hRPA binds ssDNA with a defined polarity. A strong interaction domain of hRPA, required for initial ssDNA binding, is positioned at the 5′ side of the ssDNA-binding region and a weaker binding domain resides at the 3′ side. Functional implications were demonstrated for the NER system. The oriented binding of hRPA to ssDNA determines the positioning of the two NER nucleases on neighboring duplex DNA and modulates the activity of ERCC1–XPF. Below, we deduce a model for NER in which hRPA is situated on the nondamaged strand and directs incisions to the damaged strand. We anticipate that the defined ssDNA-binding polarity of hRPA contributes to the directionality of other hRPA-dependent processes as well.
hRPA binding to ssDNA initiates at the 5′ side of the prospective 30-nucleotide binding region
Of the three hRPA subunits, only RPA70 is able to interact stably with ssDNA on its own (Gomes et al. 1996; Gomes and Wold 1996). Within RPA70, two subdomains are present that together are required for binding to ssDNA (Pfuetzner et al. 1997). Cocrystallization of these domains with ssDNA revealed that they form a relatively straight channel that can bind stably to an 8-mer single-stranded oligonucleotide (Bochkarev et al. 1997). As binding of this hRPA fragment had a defined polarity, the interaction of the complete hRPA trimer with ssDNA was considered likely to have polarity as well (Bochkarev et al. 1997). Our finding that the affinity of hRPA for small single-stranded protrusions depends on their polarity strongly supports this hypothesis. Formally, the bandshift experiments in Figure 1 do not exclude the possibility that the observed binding preference is caused by the hydrogen-bonded DNA terminus rather than the polarity of the single strand. In this scenerio, a hydrogen-bonded 5′, but not 3′, end stabilizes the binding of hRPA molecules to short (3′) protrusions. Because we also find that modulation of the NER nucleases by hRPA strongly depends on the polarity of the single-stranded region and others have shown that the DNA-binding domain of hRPA binds ssDNA with a defined polarity (Bochkarev et al. 1997), we believe that polarity of the single strand is the main determinant of the observed binding preference. The fact that hRPA prefers short 3′ protrusions suggests that initial binding occurs at the 5′ side of the prospective 30-nucleotide ssDNA-binding region [see Fig. 8, (iii) and (iv)], and predicts a 5′- → 3′-directed progression from initial binding to the stable 30-nucleotide binding mode (Fig. 8). Stable binding may require a conformational change of hRPA or additional contacts between hRPA and the DNA.
In this study we used E. coli SSB protein to verify the specificity of protein–protein interactions. E. coli protein SSB and eukaryotic RPA carry out many of the same essential roles in different DNA-metabolizing processes but show little amino acid sequence similarity. The 19-kD E. coli SSB protein preferentially binds ssDNA as a homotetramer (Lohman and Overman 1985); RPA, on the other hand, is composed of three distinct subunits, that together form a stable ssDNA-binding monomer. A recent study revealed the presence of four potential ssDNA-binding domains (SBDs) within the three subunits of RPA, all showing weak sequence homology to the E. coli SSB protomer (Philipova et al. 1996). Two of the SBDs reside in the RPA70 subunit and correspond to the DNA-binding domains mentioned above, the others reside in RPA14 and RPA32. Therefore, binding of RPA to ssDNA may structurally resemble the E. coli SSB tetramer bound to ssDNA. This notion, combined with the fact that E. coli SSB cannot replace hRPA in human in vitro repair reactions (Coverley et al. 1991), makes E. coli SSB a proper control to determine the specificity of interactions between hRPA and human NER proteins.
hRPA is bound to the undamaged strand during NER
To investigate possible functional implications of the defined ssDNA-binding polarity of hRPA, we focused on its role in NER. During NER, the structure-specific endonucleases ERCC1–XPF and XPG cleave the damaged strand at the borders of a partially unwound DNA intermediate, respectively, at the 5′ and the 3′ side of the lesion (O’Donovan et al. 1994; Matsunaga et al. 1995; Sijbers et al. 1996; Evans et al. 1997a). Protein–protein interactions are thought to position the two nucleases specifically onto the damaged strand and prevent incisions in the undamaged strand. hRPA was reported to interact with both ERCC1–XPF (Matsunaga et al. 1996; Bessho et al. 1997) and XPG (He et al. 1995). Here, we demonstrate that the orientation of hRPA with regard to the ssDNA–dsDNA junction has a crucial role in the positioning of the two nucleases. On 5′-protruding substrates, the 3′-oriented side of hRPA faces the DNA junction. This allows a specific interaction with ERCC1–XPF, resulting in strong stimulation of ERCC1–XPF-mediated incisions in the DNA strand not bound by hRPA [Fig. 8 (iii)]. No physical interaction is observed in this orientation between hRPA and XPG, which cleaves these substrates in the same strand to which hRPA is bound [Fig. 8 (iii)]. Conversely, on 3′-protruding substrates the 5′- oriented side of hRPA is positioned toward the DNA junction. Now hRPA does not physically interact with ERCC1–XPF, but sterically blocks incisions by this nuclease in the strand to which hRPA is bound [Fig. 8 (iv)]. The 5′-oriented side of hRPA facing a DNA junction allows specific interaction with XPG, which cleaves the strand opposite the one that is bound by hRPA [Fig. 8 (iv)]. Therefore, hRPA positions both NER nucleases and even stimulates ERCC1–XPF activity, when cleavage is to occur in the strand opposite the one to which hRPA is bound. This implies that during NER hRPA is bound to the undamaged strand, and directs incisions to the damaged strand [Fig. 8 (v)]. Obviously, it is essential that incisions are restricted to the damaged strand, as the nondamaged strand later in the NER reaction has to serve as a template for gap-filling DNA synthesis. hRPA bound to the nondamaged strand even protects this strand from inadvertent incision by ERCC1–XPF, as we demonstrated that ERCC1–XPF is not able to make incisions in a DNA strand that is bound nearby by hRPA. On the other hand, inhibition of XPG-mediated incisions in hRPA-bound DNA strands was much less pronounced, indicating that possibly other factors are required to prevent XPG from cutting the template strand during NER. Curiously, hRPA-mediated positioning of XPG on the opposite strand does not result in stimulation of XPG incision activity, but rather in slight inhibition, which seems to obscure the functional relevance of this interaction. Possibly, this relevance becomes more apparent on less-defined DNA substrates, where hRPA’s ability to remove secondary structures from ssDNA may provide XPG with a better cleavable substrate. This notion is supported by the fact that stimulation of XPG-activity by hRPA was observed on substrates which are very poorly incised by XPG (Matsunaga et al. 1996). Alternatively, hRPA may be the more active profiteer of this interaction during NER by use of XPG as the 5′ border of its 30-nucleotide binding region. In this respect it is interesting to note that the distance from the lesion to the XPG-mediated 3′ incision, which is thought to preceed the ERCC1–XPF-mediated 5′ incision, varies in a lesion type-dependent manner, whereas the 30-nucleotide distance between the two incisions remains virtually constant (Huang et al. 1992; Mu et al. 1995; Moggs et al. 1996).
hRPA has been reported to bind to cisplatin-, AAF-, and UV-damaged DNA (Clugston et al. 1992; He et al. 1995; Burns et al. 1996). Together with the damage-recognition protein XPA, it showed a striking cooperativity in binding to DNA lesions (He et al. 1995). Our data pinpoint hRPA to the nondamaged strand during NER, strongly arguing that it is not the binding of hRPA to the damage per se, but rather local helical distortion and partial single-stranded character induced by the lesion that allows hRPA to bind to damaged DNA. The interaction of hRPA with XPA is likely to facilitate or stabilize the binding of the latter protein to the opposite strand containing the damage.
Implications of the defined ssDNA-binding polarity of hRPA for other DNA-metabolizing processes
Association of hRPA to ssDNA is likely to be most efficient when binding is initiated at the strong ssDNA affinity side of the molecule, which we defined as the 5′-oriented side of hRPA. This ssDNA-binding parameter of hRPA may well have an impact on other hRPA-dependent processes. In an in vitro recombination assay, binding of hRPA to ssDNA was found to inhibit the formation of Rad51 nucleoprotein filaments on that same DNA template, and inhibition could be alleviated by Rad52, involving specific Rad52–hRPA and Rad52–Rad51 protein–protein interactions (Sung 1997; Benson et al. 1998; New et al. 1998; Shinohara and Ogawa 1998). When hRPA was added after ssDNA–Rad51 nucleoprotein filaments had been allowed to form, however, it was found to stimulate Rad51-catalyzed strand exchange between a linear duplex and a circular ssDNA fragment (Shinohara and Ogawa 1998; Sung 1997; Baumann and West 1997; Benson et al. 1998; New et al. 1998). Strand exchange involved the transfer of the 5′ terminus of a strand of linear duplex DNA to circular ssDNA (Baumann and West 1997), and our data suggest that such hRPA-mediated stimulation can be explained by preferential binding of this protein factor to the released 3′-protruding single-strand arm from the linear duplex to prevent reannealing (Lavery and Kowalczykowski 1992; Baumann and West 1997).
After the incision stage of NER, hRPA has an additional involvement in gap-filling DNA repair synthesis, probably similar to its role during regular DNA replication (Shivji et al. 1995). Both DNA polymerase δ, which is stimulated by hRPA (Kenny et al. 1989), and DNA polymerase ε have been implicated in repair synthesis (Coverley et al. 1992). To allow synthesis of new DNA strands, hRPA has to leave the template strand. Interestingly, the DNA polymerase, proceeding in a 3′ → 5′ direction on the template strand, faces the weak ssDNA-binding side, the 3′-oriented side, of hRPA. It is probably more efficient to peel off a bound hRPA molecule from ssDNA starting from the side of the protein that is not so strongly attached to the template. DNA polymerase α, involved in the initiation of DNA replication, is also stimulated by hRPA. In contrast to DNA polymerase δ, however, this requires a specific interaction with hRPA (Kenny et al. 1989; Matsumoto et al. 1990; Braun et al. 1997). For this protein and other hRPA-interacting proteins that act with a defined directionality on DNA, like ERCC1–XPF and XPG, it will be interesting to see where the interacting subdomains reside in an hRPA molecule bound to ssDNA. Most likely, however, this awaits cocrystallization of the complete hRPA trimer on ssDNA.
Materials and methods
Purified proteins
Recombinant hRPA, expressed from the construct p11d–tRPA (a generous gift of Dr. M.S. Wold, University of Iowa, Iowa City), was purified from E. coli as described by Henricksen (1994). For biochemical assays, fresh dilutions were made from a 125 ng/μl purified hRPA stock in buffer A [25 mm Tris at pH 7.5, 1 mm EDTA, 0.3 m NaCl, 50% glycerol, 1 mm dithiothreitol (DTT), 0.2 μg/ml BSA]. Purification of recombinant human ERCC1–XPF complex from baculovirus-infected insect cells has been described previously (de Laat et al. 1998). Purified fractions, directly used for assays, contained ∼20 ng/μl ERCC1–XPF complex in buffer B (20 mm HEPES at pH 7.8, 50 mm kcl, 2 mm MgCl2, 1 mm EDTA, 10% glycerol, 2 mm DTT, 0.02% NP-40). Recombinant human XPG protein was purified from baculovirus-infected insect cells, using a protocol described by O’Donovan (1994b), with one additional purification step. After the hydroxyapatite column, the peak fractions were pooled, dialyzed against buffer C (25 mm Tris-HCl at pH 7.5, 1 mm EDTA, 0.01% NP-40, 10% glycerol) containing 0.1 m KCl, and loaded onto an HPLC Mono Q HR5/5 column, equilibrated in the same buffer. After washing, a gradient from 0.1 to 0.5 m KCl (20 ml) was applied at 0.5ml/min and 0.5 ml fractions were collected. XPG eluted between 0.4 m and 0.5 m KCl, and peak fractions contained ∼250 ng/μl of purified XPG protein. For biochemical assays, fresh dilutions of these fractions were made in buffer B.
DNA substrates
The sequences of the DNA substrates used, were as follows: 3′-protruding 10-d(T) substrate: 5′-GGCCGTGCTCTGAATTCCTGGATGTTCGAAAG/CTTTCGAACATCCAGGAATTCAGAGCACGGCC(T)10-3′; 5′-protruding 10-d(T) subtrate: 5′-(T)10GGCCGTGCTCTGAATTCCTGGATGTTCGAAAG/CTTTCGAACATCCAGGAATTCAGAGCACGGCC-3′; 3′-protruding 19-d(T) substrate: 5′-GGCCGTGCTCTCCTGGATGTTCGAAAG/CTTTCGAACATCCAGGAGAGCACGGCC(T)19-3′; 5′-protruding 19-d(T) substrate: 5′-(T)19GGCCGTGCTCTCCTGGATGTTCGAAAG/CTTTCGAACATCCAGGAGAGCACGGCC-3′; 3′-protruding 28-d(T) substrate: 5′-GGCCGTGCTCTGATGTTCGAAAG/CTTTCGAACATCAGAGCACGGCC(T)28-3′; 5′-protruding 28-d(T) substrate: 5′-(T)28GGCCGTGCTCTGATGTTCGAAAG/CTTTCGAACATCAGAGCACGGCC-3′; 3′-protruding 14-d(N) substrate: 5′-GTGCTCTCCTGGATGTTCGAAAGCTGGGCGT/ACGCCCAGCTTTCGAACATCCAGGAGAGCAC(AGACTTGGACCCGC)-3′; 5′-protruding 13-d(N) substrate: 5′-(CCTAGACTTAAGA)GGCCGTGCTCTCCTGGATGTTCGAAAGCTGGG/CCCAGCTTTCGAACATCCAGGAGAGCACGGCC-3′. Underlined sequences were filled in by Klenow fragment, using dGTP and (radio-labeled) dCTP. HaeIII restriction sites (GG/CC) and BstB1 restriction sites (TT/CGAA) are indicated in boldface type.
Nuclease assays
DNA oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis (DPAGE) and 150 of ng DNA substrate was 32P-labeled using polynucleotide kinase or Klenow fragment, followed by phenol extraction in 100 μl and G50–Sepharose column centrifugation. To allow self-annealing, oligonucleotides were heated for 3 min at 95°C and put on ice. Labeled substrates were kept at 4°C.
Nuclease assays (15 μl) were carried out in nuclease buffer D (50 mm Tris at pH 8.0, 0.75 mm MnCl2, 0.1 mg/ml BSA, 0.5 mm β-mercaptoethanol), containing ∼100 fmoles of DNA substrate. First, 1 μl with the indicated amount of hRPA (or 1 μl of buffer A) was preincubated with the DNA substrate in buffer D for 10 min at 37°C. Then 150 fmoles of ERCC1–XPF or 150 fmoles of XPG was added on ice and reactions were continued for 50 min at 37°C. Reactions were stopped by adding 90% formamide, heated for 3 min at 95°C and applied onto 10%–15% DPAGE. Reactions products were visualized by autoradiography or phosphorimager. In the case of the footprinting type of analysis, 10 units of HaeIII or 10 units of BstBI were added after 10 min hRPA preincubation, and incubation was proceded for another 4 hr at 37°C. Similar results, as shown in Figure 2, were obtained when HaeIII cleavage was allowed for 50 min at 37°C with 28-nucleotide hairpin substrates.
Gel-retardation assays
To analyze hRPA–DNA complex formation, the indicated amounts of hRPA were coincubated with 100 fmoles of DNA substrate and 1 μl of buffer B in buffer D (total reaction volume, 15 μl; final KCl concentration was ∼25 mm). No glutaraldehyde was added. After incubation for 1 hr at 37°C, reactions were put on ice, 3 μl of ice-cold 30% glycerol was added, and samples were loaded immediately onto a precooled 5% (1:40 acrylamide/bisacrylamide) native gel. Gels were run in 0.5× TBE for 2–3 hr at 150 V (20 må) and 4°C.
To analyze ternary complex formation, 100 fmoles of 3′ or 5′ protruding 28-d(T) substrate was preincubated with the indicated amounts of hRPA (or 1 μl buffer A) in buffer D containing either 0.75 mm MnCl2, 5 mm CaCl2, 5 mm MgCl2, or 5 mm EDTA. After 10 min at 30°C, the indicated amounts of ERCC1–XPF or XPG (or corresponding volumes of buffer B) were added on ice, and incubation was continued for 10 min at 30°C. Then, 1.5 μl of 0.5% glutaraldehyde (freshly diluted in 50 mm Tris at pH 7.5) was added at room temperature (final concentration: 0.05% in 15 μl reaction volume) and cross-linking was allowed for 40 min (30°C). In reactions containing antibody, after 25 min of glutaraldehyde-cross-linking, 2 μl of antibody was added and incubation was continued for another 15 min at 30°C. Reactions were stopped on ice, 3 μl of ice-cold 30% glycerol was added and samples were loaded immediately onto a precooled 5% (1:40) native PAGE (0.5× TBE) (see above). Products were visualized by autoradiography or PhosphorImager.
Acknowledgments
We thank Dr. M.S. Wold for generously providing the RPA expression construct p11d–tRPA, and Dr. Elizabeth Evans and Dr. Richard D. Wood for giving the XPG-containing baculovirus construct. We also thank D. Bootsma for stimulating interest, and the other members of our laboratory for advice and reagents. This work was supported by the Dutch Scientific Organisation (NWO, Foundation for Chemical Research), the Louis Jeantet Foundation, and the Biodesign Research Program grant from the Institute of Physical and Chemical Research (RIKEN).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hoeijmakers@gen.fgg.eur.nl; FAX 31-10-4360225.
References
- Baumann P, West SC. The human rad51 protein: Polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Bessho T, Sancar A, Thompson LH, Thelen MP. Reconstitution of human excision nuclease with recombinant XPF-ERCC1complex. J Biol Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA. Human replication protein A binds single-stranded DNA in two distinct complexes. Mol Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA, Masrangelo IA. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol Cell Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner R, Edwards A, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Braun K, Lao Y, He Z, Ingles C, Wold M. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- Burns J, Guzder S, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- Clugston C, McLaughlin K, Kenny M, Brown R. Binding of human single-stranded DNA binding protein to DNA damaged by the anticancer drug cis-diamminedichloroplatinum (II) Cancer Res. 1992;52:6375–6379. [PubMed] [Google Scholar]
- Coverley D, Kenny MK, Lane DP, Wood RD. A role for the human single-stranded DNA binding protein HSSB/RPA in an early stage of nucleotide excision repair. Nucleic Acids Res. 1992;20:3873–3880. doi: 10.1093/nar/20.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Kenny MK, Munn M, Rupp WD, Lane DP, Wood RD. Requirement for the replication protein SSB in human DNA excision repair. Nature. 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Jaspers NGJ, Hoeijmakers JHJ. DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- Dornreiter I, Erdile L, Gilbert I, von Winkler D, Kelly T, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997a;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Moggs JG, Hwang J, Egly J, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997b;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman MP, Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgaki A, Strack B, Podust VN, Hubscher U. DNA unwinding activity of replication protein A. FEBS Lett. 1992;308:240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- Gomes X, Henricksen L, Wold MS. Proteolytic mapping of human replication protein A: Evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry. 1996;35:5586–5595. doi: 10.1021/bi9526995. [DOI] [PubMed] [Google Scholar]
- Gomes X, Wold M. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision stepa of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- Henricksen L, Umbricht C, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Huang J C, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny MK, Lee SH, Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: Single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc Natl Acad Sci. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- Kim C, Snyder RO, Wold MS. Binding properties of replication protein-A from human and yeast cells. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery P, Kowalczykowski S. A postsynaptic role for single-stranded DNA-binding protein in recA protein-promoted DNA strand exchange. J Biol Chem. 1992;267:9315–9320. [PubMed] [Google Scholar]
- Lohman T, Overman L. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc Natl Acad Sci. 1990;87:9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Mu D, Park C-H, Reardon JT, Sancar A. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J Biol Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Park C-H, Bessho T, Mu D, Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Yarema KJ, Essigmann JM, Wood RD. Analysis of incision sites produced by human cells extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-Cisplatin adduct. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- Mu D, Park C-H, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- Mu D, Wakasugi M, Hsu D, Sancar A. Characterization of reaction intermediates of human excision repair nuclease. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- New J, Sugiyama T, Zaitseva E, Kowalczykowski S. Rad52 protein stimulates DNA strand exchange by Rad51 and replication. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- O’Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994a;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- O’Donovan A, Scherly D, Clarkson SG, Wood RD. Isolation of active recombinant XPG protein, a human DNA repair endonuclease. J Biol Chem. 1994b;269:15965–15968. [PubMed] [Google Scholar]
- Park MS, Ludwig DL, Stigger E, Lee S-H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- Pfuetzner R, Bochkarev A, Frappier L, Edwards A. Replication protein A. Characterization and crystallization of the DNA binding domain. J Biol Chem. 1997;272:430–434. doi: 10.1074/jbc.272.1.430. [DOI] [PubMed] [Google Scholar]
- Philipova D, Mullen J, Maniar H, Lu J, Gu C, Brill S. A hierarchy of SSB protomers in replication protein A. Genes & Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shivji MKK, Podust VN, Hubscher U, Wood RD. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei Y-F, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, Rademakers S, de Rooij J, Jaspers NGJ, Hoeijmakers JHJ, Wood RD. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between Replication Protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J Mol Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- Weinberg DH, Collins KL, Simancek P, Russo A, Wold MS, Virshup DM, Kelly TJ. Reconstitution of simian virus 40 DNA replication with purified proteins. Proc Natl Acad Sci. 1990;87:8692–8696. doi: 10.1073/pnas.87.22.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wold MS, Li JJ, Kelly TJ. Initiation of simian virus 40 DNA replication in vitro: Large-tumor-antigen- and origin-dependent unwinding of the template. Proc Natl Acad Sci. 1987;84:3643–3647. doi: 10.1073/pnas.84.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]