Abstract
Preventing or delaying progress through the cell cycle in response to DNA damage is crucial for eukaryotic cells to allow the damage to be repaired and not incorporated irrevocably into daughter cells. Several genes involved in this process have been discovered in fission and budding yeast. Here, we report the identification of human and mouse homologs of the Schizosaccharomyces pombe DNA damage checkpoint control gene rad1+ and its Saccharomyces cerevisiae homolog RAD17. The human gene HRAD1 is located on chromosome 5p13 and is most homologous to S. pombe rad1+. This gene encodes a 382-amino-acid residue protein that is localized mainly in the nucleus and is expressed at high levels in proliferative tissues. This human gene significantly complements the sensitivity to UV light of a S. pombe strain mutated in rad1+. Moreover, HRAD1 complements the checkpoint control defect of this strain after UV exposure. In addition to functioning in DNA repair checkpoints, S. cerevisiae RAD17 plays a role during meiosis to prevent progress through prophase I when recombination is interrupted. Consistent with a similar role in mammals, Rad1 protein is abundant in testis, and is associated with both synapsed and unsynapsed chromosomes during meiotic prophase I of spermatogenesis, with a staining pattern distinct from that of the recombination proteins Rad51 and Dmc1. Together, these data imply an important role for hRad1 both in the mitotic DNA damage checkpoint and in meiotic checkpoint mechanisms, and suggest that these events are highly conserved from yeast to humans.
Keywords: DNA damage, checkpoint control, synaptonemal complex, meiosis, cell cycle, DNA repair
Repair of damage to the genetic material is crucial for living organisms because they are continuously exposed to DNA-damaging agents, both physiological and environmental. In higher eukaryotes, for example, deficient DNA repair can lead to tumorigenesis and has been linked to certain degenerative disease states. Eukaryotic cells respond to DNA damage by delaying progress through the cell cycle and by repairing different types of damage with distinct sets of proteins. This cell cycle delay is called the DNA damage checkpoint and either causes irreversible withdrawal from the cell cycle (Di Leonardo et al. 1994), or allows the cell time to repair the damaged DNA before it is replicated or mitotic segregation takes place (Elledge 1996; Paulovich et al. 1997). The available evidence indicates that specialized protein complexes sense the DNA damage and trigger inhibitory signals that impinge on the cell cycle machinery. Many of the genes for these proteins were identified in fission and budding yeast through mutations that cause defective cell cycle checkpoints and hypersensitivity to DNA damaging agents. Conservation of some of these genes in distantly divergent yeasts indicates that DNA damage checkpoint mechanisms are highly conserved throughout evolution (for review, see Lehmann 1996).
Three key yeast checkpoint genes are Saccharomyces cerevisiae RAD17 (scRAD17; Lydall and Weinert 1995; Siede et al. 1996) and its Schizosaccharomyces pombe homolog rad1+ (sprad1+; Al-Khodairy and Carr 1992; Rowley et al. 1992; Long et al. 1994), scRAD24 and its homolog sprad17+ (Griffiths et al. 1995), and sprad3+ and its homologs scMEC1 and scTEL1 (Morrow et al. 1995; Paulovich and Hartwell 1995; Bentley et al. 1996). In addition to being involved in the DNA damage checkpoint, sprad1+ is also involved in another cell cycle checkpoint triggered in the presence of S-phase inhibitors (the S–M phase checkpoint; Al-Khodairy and Carr 1992; Rowley et al. 1992). The products of scRAD17 and sprad1+ show homology to Ustilago maydis Rec1p, a protein implicated in recombination, mutagenesis, and checkpoint control (Onel et al. 1996). Although the precise roles of these three factors in the DNA-damage checkpoint is unclear, Rec1 has been reported to possess 3′ → 5′ exonuclease activity (Thelen et al. 1994). Recently, spRad1 was found to interact with another DNA damage checkpoint protein, spHus1, supporting the hypothesis that these two proteins form part of a multisubunit DNA damage checkpoint complex (Kostrub et al. 1998).
Strains deficient in scRAD24/sprad17+ have similar phenotypes to scRAD17/sprad1+-deficient strains, and the products of these genes display homology with replication factor C subunits (Griffiths et al. 1995), which are involved in recruitment of replication factors onto primed templates during the initiation of DNA replication. Recently, scMec1p, scTel1p, and spRad3 have received much interest because they belong to a family of large proteins that are homologous to phosphatidylinositol 3-kinase (Jackson 1995; Hoekstra 1997). This family includes other proteins involved in DNA repair or checkpoint controls such as the human DNA-dependent protein kinase catalytic subunit (DNA-PKcs; Hartley et al. 1995) and ATR/FRP1, which is the mammalian homolog of spRad3 and scMec1p (Bentley et al. 1996; Cimprich et al. 1996). Notably, human ATR complements partially the UV sensitivity of a mec1 mutant S. cerevisiae strain (Bentley et al. 1996) and recently was shown to encode a component of the mammalian G2–M phase DNA damage checkpoint apparatus (Cliby et al. 1998). Also in this family of proteins is the human homolog of scTel1p, ATM (Savitsky et al. 1995), which functions in the G1–S, S–M, and G2–M checkpoints that are triggered after exposure to ionizing radiation (Lavin and Shiloh 1997). Other DNA damage checkpoint components that have been identified in mammalian systems are p53 and some of its downstream effectors, such as p21 and p16 (Levine 1997; Shapiro et al. 1998). In addition, hRad9, the human homolog of the S. pombe checkpoint protein Rad9 has been identified (Lieberman et al. 1996). hRad9 partially complements the checkpoint defect of a S. pombe rad9 mutant strain after exposure to ionizing radiation (Lieberman et al. 1996), supporting the notion that the fundamentals of checkpoint control are conserved from yeast to humans.
Recently, a link has been established between the mitotic DNA damage checkpoints and control of progression through meiosis. During meiotic prophase I, homologous chromosomes pair, synapse, recombine, then segregate (for a review, see Roeder 1997). Studies in S. cerevisiae indicate that one of the earliest events in recombination is the formation of DNA double-stranded breaks (Storlazzi et al. 1995; Keeney et al. 1997). The subsequent steps in this process require a group of proteins involved in DNA mismatch repair or homologous recombination (Shinohara and Ogawa 1995; Hassold 1996; Schwacha and Kleckner 1997). Rad51 and Dmc1, two eukaryotic homologs of the bacterial recombination factor RecA, are found in distinct foci in synapsed meiotic chromosomes during stages of prophase I progression in humans, rodents, and budding yeast, suggesting that they play important roles in meiotic recombination or its signaling (Bishop 1994; Barlow et al. 1997; Moens et al. 1997). During meiotic recombination in S. cerevisiae, a checkpoint control results in prophase I arrest if recombination is incomplete (Bishop et al. 1992). Several mitotic DNA damage checkpoint genes (scRAD24, scRAD17, and scMEC1) are involved in this process (Lydall et al. 1996), revealing important connections between the DNA damage and meiotic checkpoint controls. Furthermore, mutations in scMEC1 lead to defective meiosis (Kato and Ogawa 1994), and mutations in mei-41, the Drosophila homolog of sprad3+/scMEC1, cause reduced meiotic recombination frequencies and aberrant recombination nodules—the sites of recombination events (Hari et al. 1995). Recent results indicate that ATM and ATR also function in the control of meiotic progression. Thus, ATM-deficient humans have reduced fertility, ATM-deficient mice are infertile as a result of a failure to carry out meiotic prophase I during spermatogenesis (Xu et al. 1996; Barlow et al. 1998), and ATM and ATR were repoted to be located in synapsed and unsynapsed meiotic chromosomes, respectively (Keegan et al. 1996; Barlow et al. 1998). hChk1, the human homolog of the S. pombe checkpoint protein Chk1, is also located along unsynapsed chromosomes, and a possible interaction with ATR has been suggested (Flaggs et al. 1997). Taken together, these findings imply a dual role for certain proteins in DNA damage-induced mitotic checkpoints and in meiotic checkpoint control mechanisms. It will, therefore, be of great interest and importance to identify and characterize other proteins that function in these pathways. In this report, we describe the isolation of mammalian homologs of sprad1+. We show that the human gene complements substantially both the radiation sensitivity and checkpoint phenotypes of a sprad1 mutant strain. Furthermore, we find that expression of mammalian Rad1 is high in proliferating tissues and is transcriptionally up-regulated in testis. Finally, we report that Rad1 is associated with discrete foci on the chromosomes of mouse spermatocytes undergoing meiotic prophase I.
Results
Isolation and chromosomal location of human and mouse homologs of sprad1+
By screening expressed-sequence tag (EST) databases for sequences with similarity to sprad1+, we found a 480-bp cDNA sequence derived from human uterus (GenBank accession no. AA029300) whose translation product displayed significant homology to the carboxy-terminal region of the yeast protein. To clone the entire cDNA for this putative human RAD1 (HRAD1), primers that aligned with the 5′ or 3′ DNA strand of the EST were designed and used to carry out PCR on a B-cell cDNA plasmid library, in conjunction with plasmid primers. Two fragments corresponding to the 5′ and 3′ ends of the gene were thus amplified, cloned, and sequenced. Two further oligonucleotides were designed to allow cloning of the full-length cDNA from different libraries, and these were verified as sprad1+homologs by sequencing. After the full-length human cDNA sequence was obtained, we proceeded to isolate the mouse RAD1 homolog by designing various primers corresponding to the human cDNA and performing PCR with a mouse cDNA library as template (see Materials and Methods). The human and mouse cDNA clones were each found to contain a single long open reading frame. The predicted translation products of these are highly related to each other (91% identical), are homologous to spRad1 (∼30% identity, ∼55% similarity), and are also related to a slightly lesser degree to scRad17p (17% identity, 46% similarity) and Ustilago maydis Rec1 (umRec1; 30% identity, 54% similarity; Fig. 1). The mammalian proteins, however, are shorter than their yeast counterparts and lack the carboxy-terminal extensions that are present in the yeast proteins. Because umRec1 was reported to be a 3′ → 5′ exonuclease and certain residues (indicated by asterisks in Fig. 1) are known to be crucial for the activity of exonucleases (Thelen et al. 1994), it was of interest to determine whether these residues are conserved in the mammalian proteins. Notably, only two of the residues of umRec1, which were proposed to be important for catalysis, are found in the mouse or human sequences, perhaps suggesting that umRec1 functions differently than the mammalian proteins.
Figure 1.

Sequence comparison of the putative human and mouse Rad1 homologs. Multiple alignment of amino acid sequences of human (hRad1) and mouse Rad1 (mRad1) with Schizosaccharomyces pombe Rad1 (spRad1), Ustilago maydis Rec1 (umRec1), and Saccharomyces cerevisiae Rad17p (scRad17). Amino acid residues, which are identical, are shown by reverse shading and conservative substitutions are indicated by gray shading. Numbers indicate the amino acid position, and residues that align with those of umRec1, which have been proposed to define parts of a nuclease catalytic site (Thelen et al. 1994), are indicated by asterisks. The alignment was obtained with the PILEUP program from the GCG package, and shading was by the BOXSHADE program.
In light of the above, we decided to test whether hRad1 possesses nuclease activity. To this end, we expressed in Escherichia coli the full-length hRad1 protein fused to an amino-terminal hexahistidine tag, then purified the protein, which was present in high levels in the soluble fraction, to homogeneity using Ni2+-NTA affinity chromatography (see Materials and Methods). The resulting protein had no exonuclease activity when tested under various assay conditions with a variety of different DNA substrates (including blunt-ended DNA and DNA with 3′ or 5′ overhanging ends; data not shown). To rule out the possibility that the lack of exonuclease activity was attributable to the type of fusion protein used, we also cloned and expressed hRad1 as a GST fusion protein. Again, no exonuclease activity was detected (data not shown). Although we cannot rule out the possibility that hRad1 is folded incorrectly when produced in E. coli, these results suggest that hRad1 is not a nuclease. Alternatively, hRad1 may require an essential cofactor/partner for its exonuclease activity or may have an unusual substrate specificity.
To determine the chromosomal location of hRAD1, we used a PCR-based method using human–mouse and human–hamster hybrid cell lines that each contain a single human chromosome (Kelsell et al. 1995; see Materials and Methods). This revealed that HRAD1 maps to chromosome 5 (data not shown). Subsequent studies using the Genebridge cell hybrid panel (Gyapay et al. 1996) containing specific human chromosomal regions allowed mapping of HRAD1 to genomic markers close to chromosome 5 region p13. Interestingly, this localization places HRAD1 near a position known as a site of frequent deletion in lung and bladder carcinomas (Weiland and Bohm 1994; Weiland et al. 1996; Bohm et al. 1997), raising the possibility that loss of hRad1 can contribute to the generation of these types of cancer.
HRAD1 complements mutations in sprad1
The S. pombe rad1 mutant strain is sensitive to DNA-damaging agents such as UV- and γ-radiation, at least in part because it is unable to delay progress through the cell cycle when the DNA is damaged. To study whether or not HRAD1 might perform similar functions to the yeast gene, we expressed the HRAD1 cDNA in the S. pombe rad1 mutant strain and in the parental wild-type (WT) strain. To do this, HRAD1 was expressed in a S. pombe expression vector in which HRAD1 was under the control of the thiamine repressible nmt promoter (Maundrell 1990). Expression of the hRad1 protein was verified by Western blot using a hRad1-specific polyclonal antibody (see below). We also introduced the empty vector and a vector containing the sprad1+ gene into the sprad1 mutant strain. Sensitivity to UV irradiation was used to test whether the human gene complements the sprad1 mutant defect. Notably, when derepressed, HRAD1 reproducibly rescued the UV sensitivity of sprad1 mutant cells and, at higher UV doses, the viability of the strain expressing HRAD1 was ∼15-fold greater than that of the sprad1-deficient strain (Fig. 2A). However, we only observed subtle (∼threefold) complementation of the ionizing radiation sensitivity of the sprad1 mutant strain (Fig. 2B). Apart from functioning in the DNA damage checkpoint, sprad1+ is also involved in the S–M phase checkpoint that causes cell cycle arrest in S-phase when unreplicated DNA is present. This involvement is demonstrated by the hypersensitivity of sprad1 mutants to hydroxyurea (Al-Khodairy and Carr 1992; Rowley et al. 1992), which blocks DNA replication by inhibiting the de novo synthesis of deoxyribonucleotides. In contrast to the UV sensitivity complementation, expression of HRAD1 in the sprad1 mutant strain does not complement the hydroxyurea hypersensitive phenotype (data not shown). Taken together, these data indicate that HRD1 can complement some but not all of the phenotypes of the S. pombe rad1 mutant strain, and suggest strongly that hRad1 is indeed a functional homolog of spRad1.
Figure 2.
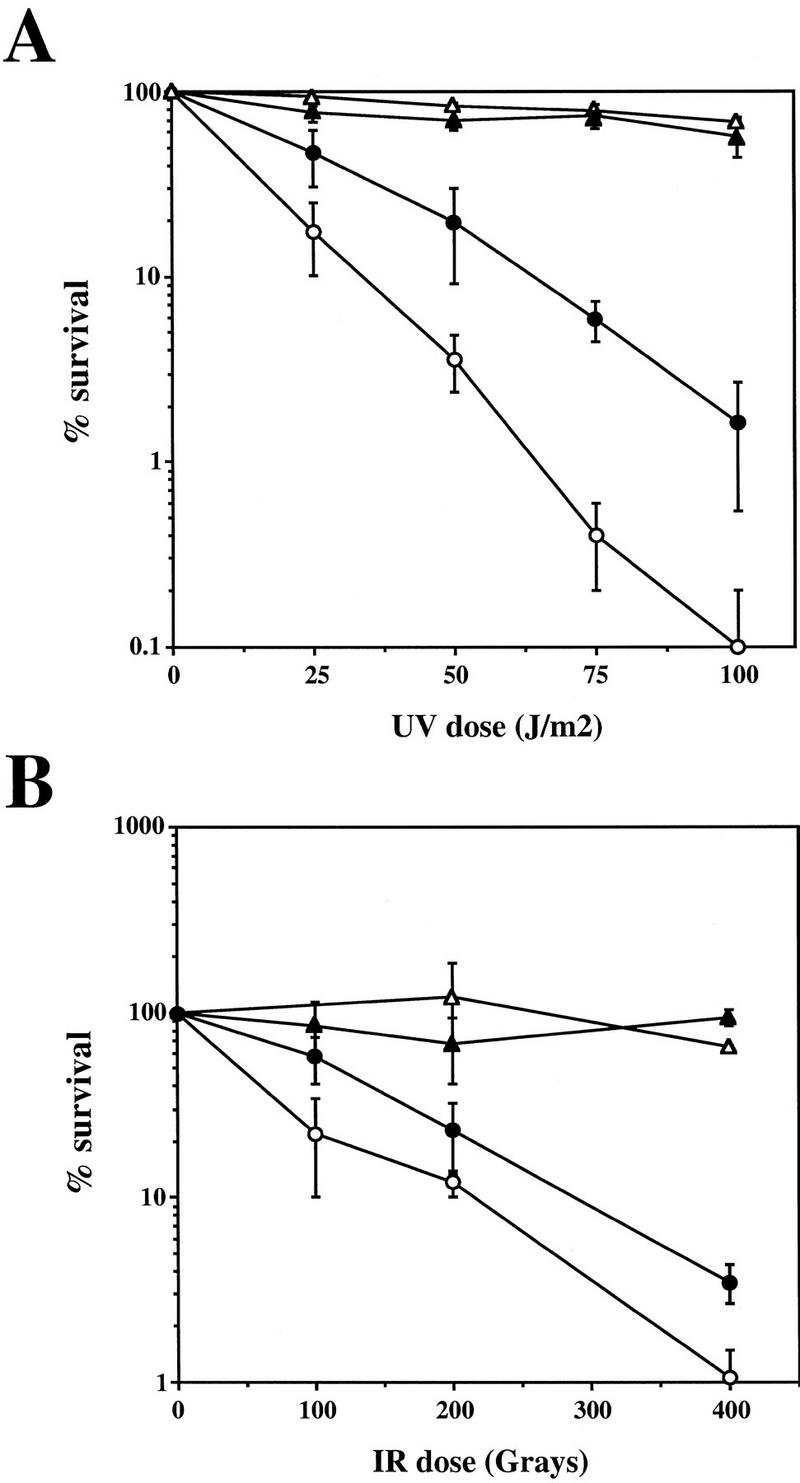
Complementation of radiation sensitivity of the S. pombe rad1 mutant by HRAD1. Viability of wild-type and rad1::ura4+ strains containing pREP3X (WT and rad1, respectively) and the rad1::ura4+ strain containing the pREP3X/sprad1+ plasmid (rad1/sprad1+) or pREP3X/HRAD1 (rad1/HRAD1) in response to different doses of UV light (A) or ionizing radiation (IR) (B) are shown. Each data point is the mean of at least three independent experiments. (▵) Wild type; (▴) rad1/sprad1+; (○) rad1; (•) rad1/HRAD1.
hRad1 can function in yeast cell cycle checkpoint control
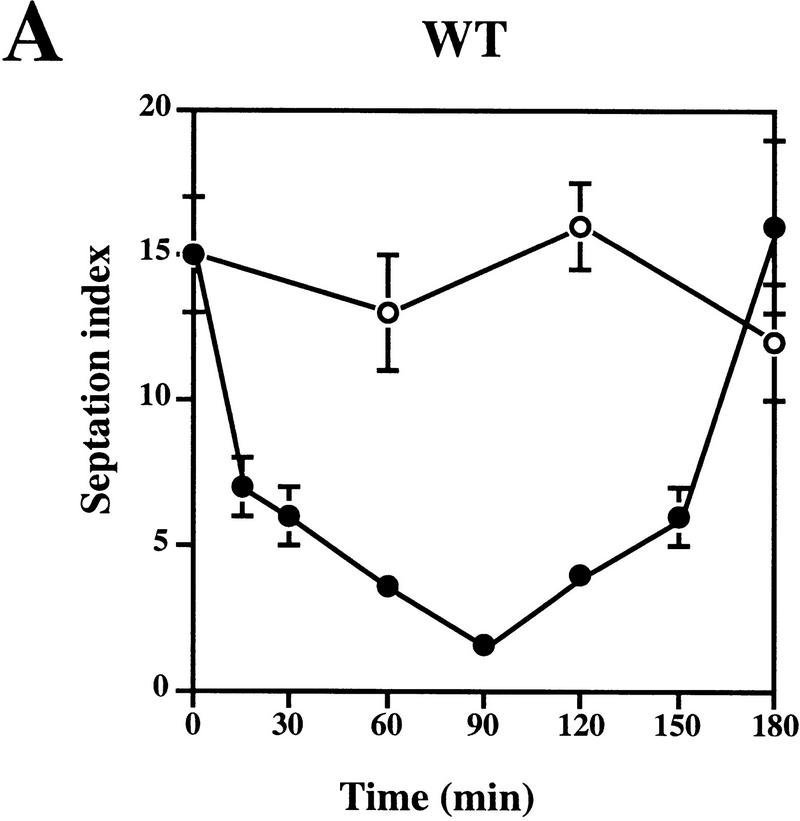
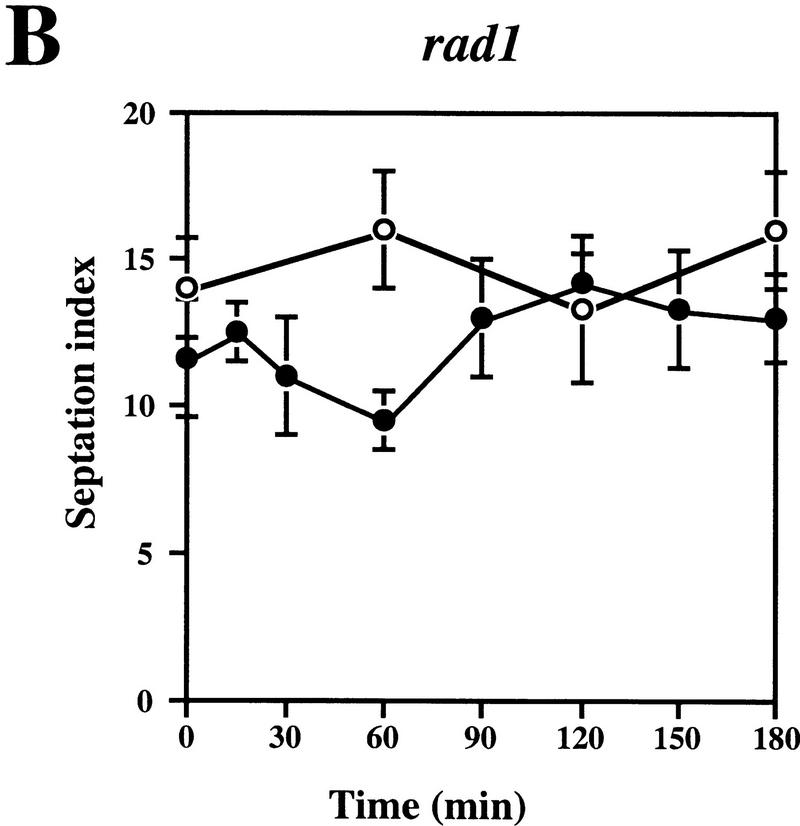
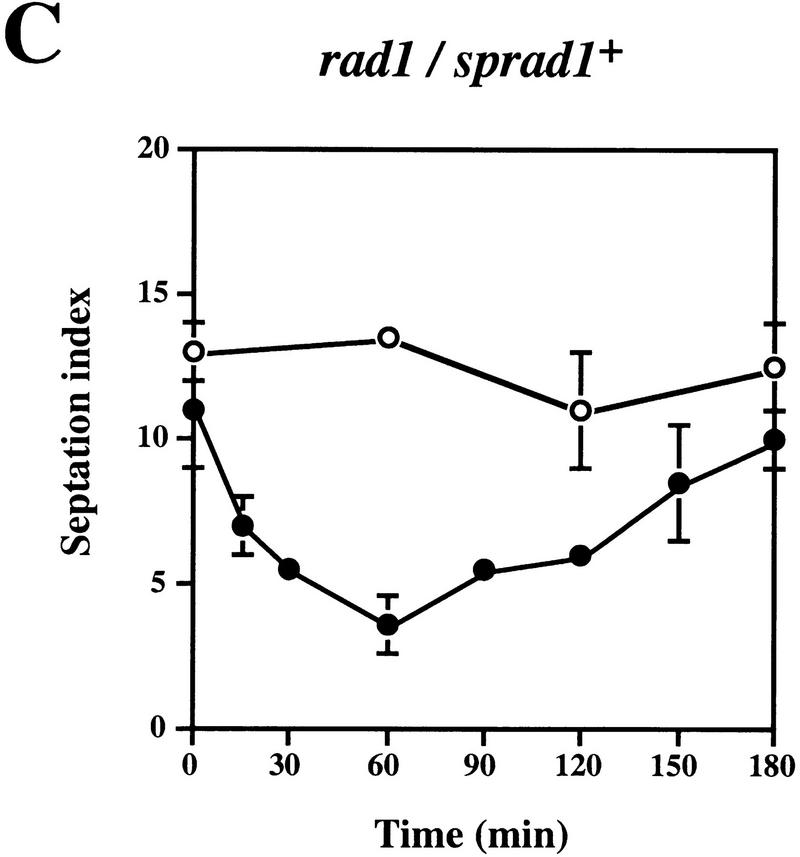
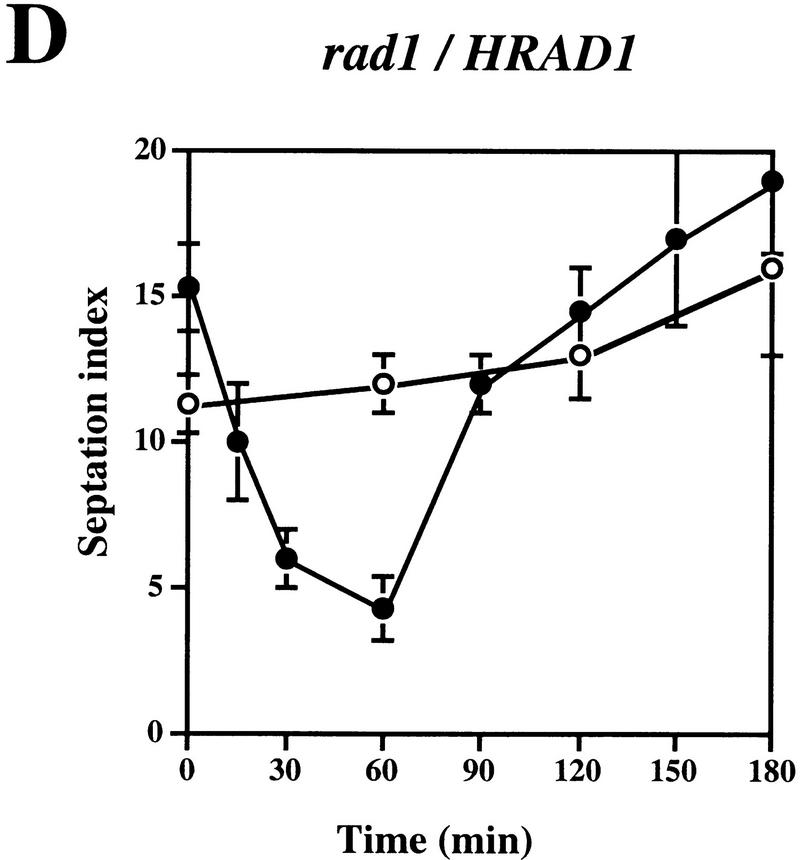
We then wished to determine whether complementation of the UV sensitivity of the sprad1-deficient strain by HRAD1 correlated with a correction of the cell cycle checkpoint after UV-induced DNA damage. To do this, we examined the G2–M phase checkpoint delay after DNA damage in the WT and rad1 mutant strains overexpressing either HRAD1 or sprad1+. The septation index of yeast cultures at various times after exposure to UV was measured as described previously (Edwards and Carr 1997; see Materials and Methods). This index reflects the proportion of cells that have just completed mitosis, and is an effective method for monitoring cell cycle progression. As reported previously, whereas the WT strain exhibits a checkpoint delay in response to 50 J/m2 UV irradiation, no similar delay is apparent in the rad1 mutant strain (Fig. 3A,B). Notably, the sprad1 strain expressing HRAD1 has a largely normal cell cycle checkpoint in response to this dose of UV (Fig. 3D), a dose at which we also observed significant complementation in viability assays. Entry into and exit from the G2–M phase checkpoint after irradiation followed similar kinetics in the wild-type and the rad1 mutant strains overexpressing sprad1+ (Fig. 3A,C). In the rad1 mutant strain expressing HRAD1, however, entry into the checkpoint appeared normal, whereas exit was advanced by ∼90 min (Fig. 3, cf. D with A). This effect is dominant as it also occurred when HRAD1 was overexpressed in the WT strain (data not shown). Taken together, these data provide further evidence that HRAD1 is a functional homolog of S. pombe rad1+ and indicate that HRAD1 encodes a functional checkpoint protein.
Figure 3.
Complementation of the S. pombe rad1 checkpoint defect by HRAD1. Strain designations are as in Fig. 2. Exponentially growing cultures of S. pombe strains [(A) Wild type (WT); (B) rad1; (C) rad1/sprad1+, (D) rad1/HRAD1] were either UV-irradiated with 50 J/m2 (•) or left untreated (○). Samples were taken at the indicated time points and the septation index was measured as described in Materials and Methods. Data shown are the mean ±s.d. of at least three independent experiments, each one conducted in duplicate.
hRad1 is mainly nuclear and is not induced by DNA-damaging agents
To study the expression of hRad1, we raised antibodies against an amino-terminal hexahistidine-tagged derivative of full-length hRad1 that had been expressed in E. coli then purified to essential homogeneity (see Materials and Methods). Western blot of HeLa cell extracts revealed that the resulting antisera recognize a polypeptide of ∼32 kD (Fig. 4A). Subsequently, the anti-hRad1 antibodies were affinity purified on a column containing immobilized recombinant hRad1, which resulted in improved specificity on Western blots. Titration studies revealed that the purified antibodies detected subnanogram quantities of recombinant hRad1 (Fig. 4B, lanes 1–3). Consistent with these antibodies recognizing hRad1 specifically, they were found to detect a protein of the appropriate size in extracts of the S. pombe strains described above that contain the HRAD1 expression plasmid, but did not detect this protein in extracts of strains containing the parental expression plasmid (Fig. 4B, lanes 4,5; as expected, this is slightly smaller than the recombinant protein, which bears the amino-terminal epitope tag). As further verification that the affinity-purified antibodies recognized hRad1 with high specificity, Western blots of extracts of human HeLa cells, or HeLa cells that had been transfected with a vector directing the expression of a Flag epitope-tagged version of full-length hRad1, were probed with these antibodies and also with a monoclonal antibody that recognizes the Flag–tag epitope. The presence of the Rad1–Flag–tag construct leads to a novel polypeptide being detected by anti-hRad1 antibodies that migrates slightly slower than the endogenous hRad1 protein (Fig. 4B, cf. lane 6 with lane 7). Unlike endogenous hRad1, this band is also detected by the anti-Flag antibody (lane 8) indicating that this novel species indeed corresponds to the Flag–hRad1 fusion. Together, these results indicate that the antibodies raised recognize hRad1 with high affinity and specificity.
Figure 4.
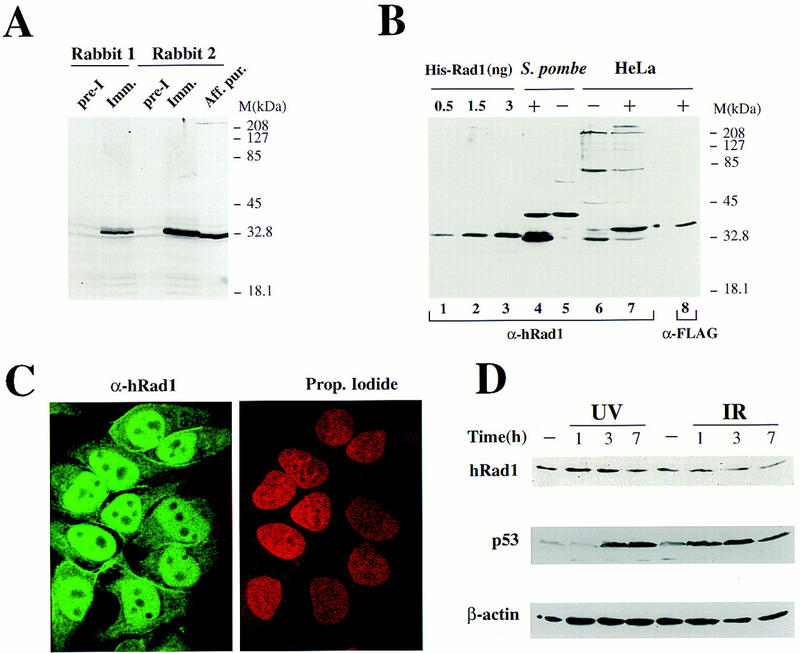
Generation of anti-hRad1 antibodies and their use in immunolocalization studies. (A) Western blot with two different antisera raised against hRad1. Each lane contains 60 μg of HeLa nuclear extract. The two antisera recognize a polypeptide of ∼32 kD. (B) Affinity-purified antibody 1 was used to probe Western blots of the indicated amounts of recombinant tagged bacterially expressed hRad1 (lanes 1–3); 10 μg of extract of the S. pombe rad1 strain expressing hRad1 (lane 4; +) and the same strain with empty vector (lane 5; −); 60 μg of HeLa whole cell extract (lane 6; −) and extract of HeLa cells transfected with Flag-tagged HRAD1 cDNA (lane 7; +). In addition, 60 μg of whole cell extract of HeLa cells transfected with Flag-tagged HRAD1 were also probed with monoclonal anti-Flag antibody (lane 8; +). (C) Immunofluorescence microscopy of HeLa cells with anti-hRad1 affinity-purified antibody using a fluorescein-conjugated secondary antibody (green; left), and staining of nuclear DNA with propidium iodide (red; right). Immunostaining reveals endogenous hRad1 in HeLa cells. (D) Western blot of U2OS cell extracts (50 μg per lane) after UV (50 J/m2) or IR (10 Gy) treatment. Western blots were probed with anti-hRad1, anti-β actin or anti-p53 antibodies, as indicated.
To determine the subcellular distribution of hRad1, we used the affinity-purified anti-hRad1 antibodies described above in immunofluorescence microscopy studies. This revealed that, despite the absence of any obvious nuclear localization sequences, the endogenous hRad1 protein is located mainly in the nucleus of HeLa cells but appears to be excluded from the nucleoli (Fig. 4C, left; the right panel shows nuclear DNA as revealed by propidium iodide staining). A low but significant signal was also detected in the cytoplasm, however, suggesting that a small proportion of hRad1 may be cytoplasmic. Similar staining with anti-Rad1 antibody was also obtained when other human cells were analyzed (human KB cells and human primary fibroblasts; data not shown). In agreement with these results, Western blotting of nuclear and cytoplasmic fractions confirmed that hRad1 is predominantly a nuclear protein (data not shown).
Because the levels of some proteins implicated in cell cycle checkpoint control are regulated in response to genotoxic insults or throughout the cell cycle, we tested whether this is the case for hRad1. No marked variation in hRad1 levels was observed at various times after exposing cells to UV or ionizing radiation (Fig. 4D). In contrast, the well-characterized induction of p53 by these agents is clearly evident (Fig. 4D). Similarly, no significant variation of hRad1 protein levels or hRad1 electrophoretic mobility on SDS–polyacrylamide gels was detected (data not shown). These results, together with the nuclear localization of hRad1, are consistent with hRad1 functioning as part of the DNA damage detection machinery rather than as an inducible downstream effector of DNA damage signaling pathways.
Expression pattern of Rad1 in various tissues
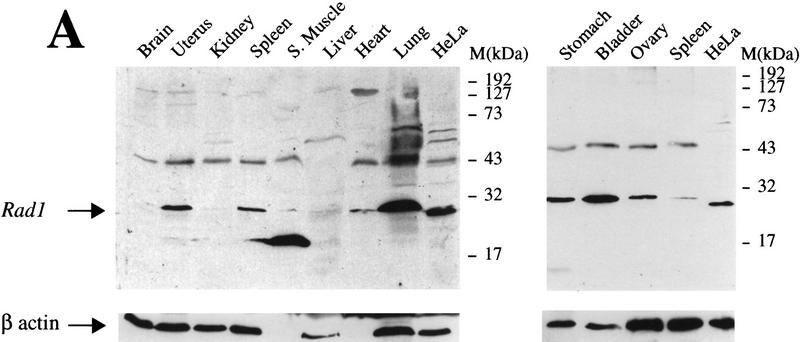
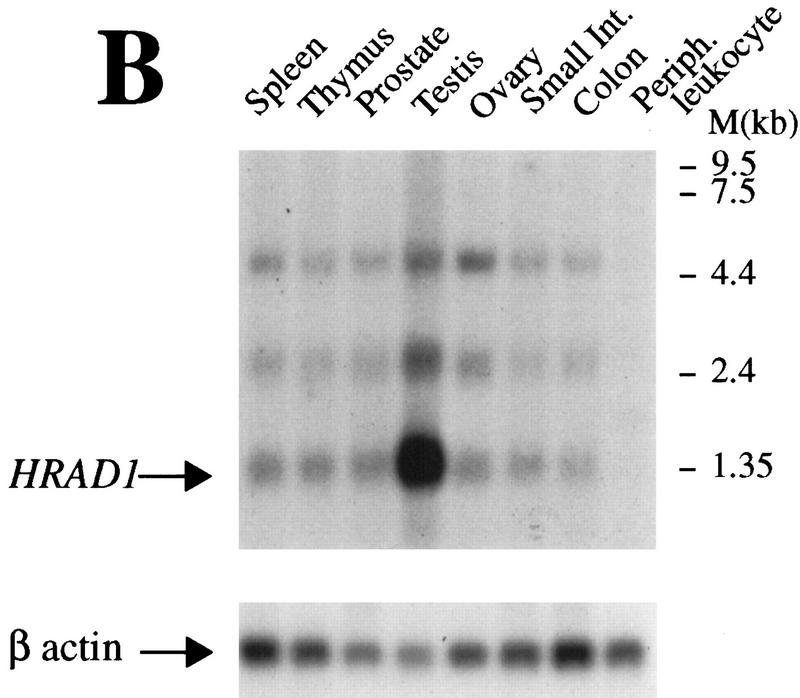
To gain further insights into hRad1 function, we performed Western blots using anti-hRad1 antiserum on extracts derived from various mouse tissues. As expected from the high degree of homology between human and mouse Rad1 (see Fig. 1), the anti-hRad1 antiserum recognizes the ∼32-kD mouse protein effectively (Fig. 5A). Furthermore, mouse Rad1 protein was detected in all tissues examined, although its levels varied in a manner that correlates with proliferative rate. Thus, relatively high levels of Rad1 protein were detected in proliferative tissues such as uterus, bladder, spleen, ovaries, and lung, whereas it was only just detectable in tissues with low proliferative capacity, such as brain and muscle (Fig. 5A). We then assessed the expression of the HRAD1 mRNA in various human tissues by Northern blot. This revealed that the major HRAD1 transcript (∼1.4 kb) is present at similar levels in all tissues examined, with the exception of testis, where expression levels appear to be ∼20-fold higher (Fig. 5B). In addition, the Northern blot revealed the existence of two additional HRAD1-hybridizing RNA species of ∼2.5 and 5 kb, raising the possibility that HRAD1-related genes or alternative versions of the HRAD1 mRNA may exist.
Figure 5.
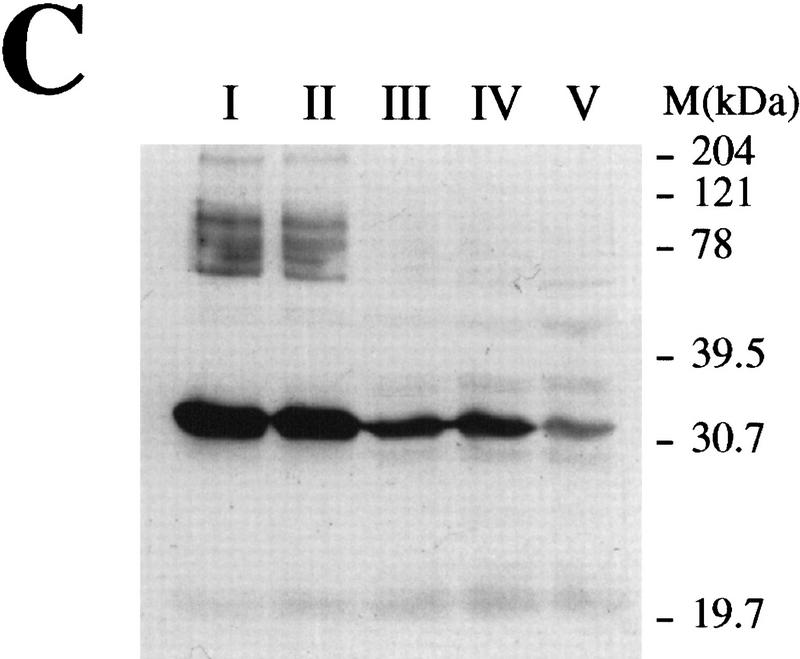
Distribution of Rad1 in different tissues and during spermatocyte development. (A) Western blots of whole cell extracts from various mouse tissues probed with anti-hRad1 affinity-purified antibody. HeLa cell nuclear extract (50 μg) is included on both Western blots. As a control, blots were also probed with anti-β actin antibody. The Rad1 band of ∼32 kD is indicated with an arrow. (B) Northern blot with various human tissues. The blot was probed separately with HRAD1 then with β-actin as a control. The major HRAD1 mRNA species is indicated with an arrow. (C) Western blot of rat testicular cells from 30-day-old rats separated by elutriation. All the lanes contain nuclear extracts normalized with respect to cell number (∼5 × 10 cells per lane). (Fraction I) Small cells, zygotene and early pachytene; (fraction II) cells in leptotene and zygotene some pachytene; (fraction III) mostly full pachytene; (fraction IV) late pachytene, and diplotene; (fraction V) postpachytene.
Rad1 levels are regulated during spermatogenesis and this protein localizes to chromosomes undergoing meiotic recombination
The discovery of high levels of HRAD1 mRNA in testis prompted us to test whether Rad1 protein levels change during spermatocyte development. For this purpose, developing rat spermatocytes were fractionated by size through the use of centrifugal elutriation (see Materials and Methods). This yielded five fractions containing cells at various stages of meiotic prophase I, which were then analyzed by Western blotting (Fig. 5C; see Fig. 6 for a schematic illustration of the stages of meiotic prophase I). Interestingly, hRad1 was found in relatively large amounts in the early stages of prophase I, corresponding mainly to late zygotene and early pachytene stage cells, whereas its levels declined in subsequent stages of prophase I, until it was almost undetectable in fraction V, which contained late pachytene and early diplotene stage cells.
Figure 6.
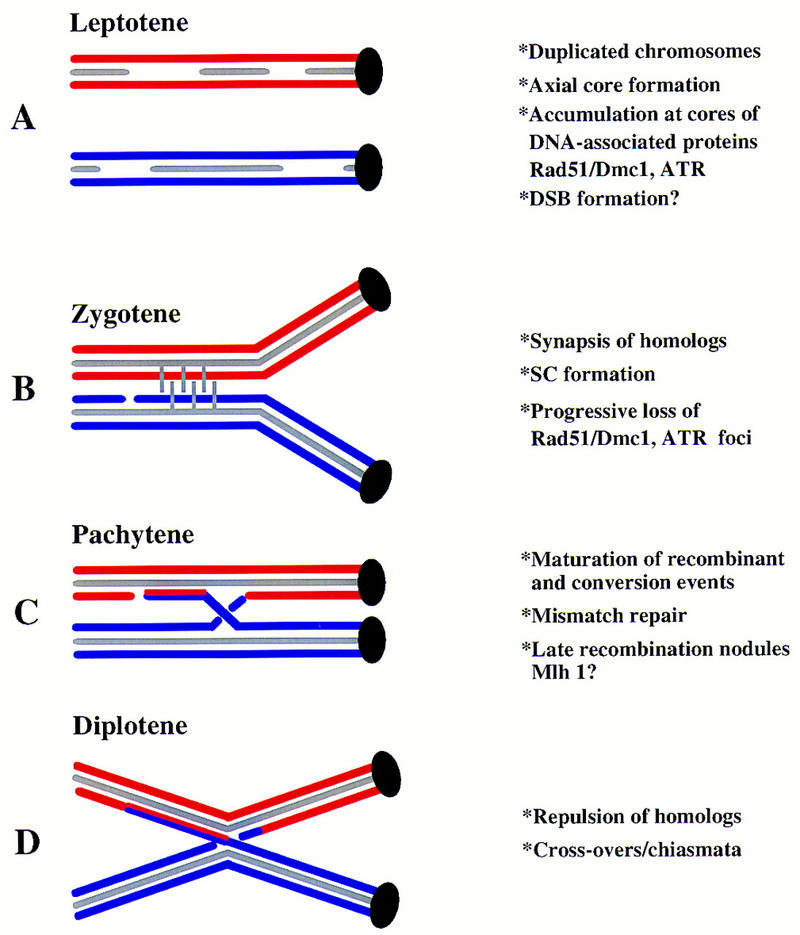
Diagrammatic summary of meiotic prophase stages and events. (A) At leptotene of meiotic prophase, S-phase is completed and chromosomal cores (gray) form in association with the pairs of sister chromatids (red and blue). Rad51/Dmc1 foci appear at the cores at this time. (B) At zygotene, the homologous chromosomes synapse and the cores align in parallel forming the synaptonemal complex (SC; indicated by vertical bars). The gap represents a DNA double-stranded break, which is thought to initiate meiotic recombination. At the onset of zygotene, the number of Rad51/Dmc1 foci reach their maximum and then start to decline. (C) Although the chromosomes are fully synapsed during the pachytene stage, the recombination processes are completed. The chromosomal crossover and heteroduplex DNA are indicted. (D) At the diplotene stage, the homologous chromosomes separate and the points of reciprocal exchanges are visible as chiasmata.
The above results suggest that Rad1 might perform a crucial role in meiotic progression, and are consistent with previous work implicating scRAD17 and scMEC1 in meiotic checkpoint control during the recombination events of meiotic prophase I (Lydall et al. 1996). In light of this, and because the two human homologs of scMec1p, ATR and ATM, were reported to be associated with meiotic chromosomes in certain stages of prophase I (Keegan et al. 1996), we tested whether mammalian Rad1 is also localized to prophase I chromosomes. Strikingly, immunofluorescence microscopy of rat and mouse spermatocytes labeled with the affinity-purified anti-hRad1 antiserum revealed bright foci along the axes of chromosomes from meiotic prophase nuclei (Fig. 7). This punctate pattern was specific for Rad1 because no staining was seen when the anti-Rad1 antibodies were preadsorbed with recombinant hRad1 protein, nor when various preimmune sera were used (data not shown). Double-labeling immunofluorescence microscopy with anti-Rad1 and an FITC-labeled secondary antibody, together with an antibody against a structural component of the axial element, chromosome core antigen (Cor1; Dobson et al. 1994) and a rhodamine-labeled secondary antibody, showed that the Rad1 foci are associated with chromosome cores or synaptonemal complexes (Fig. 7B,D,F and A,C,E, respectively).
Figure 7.
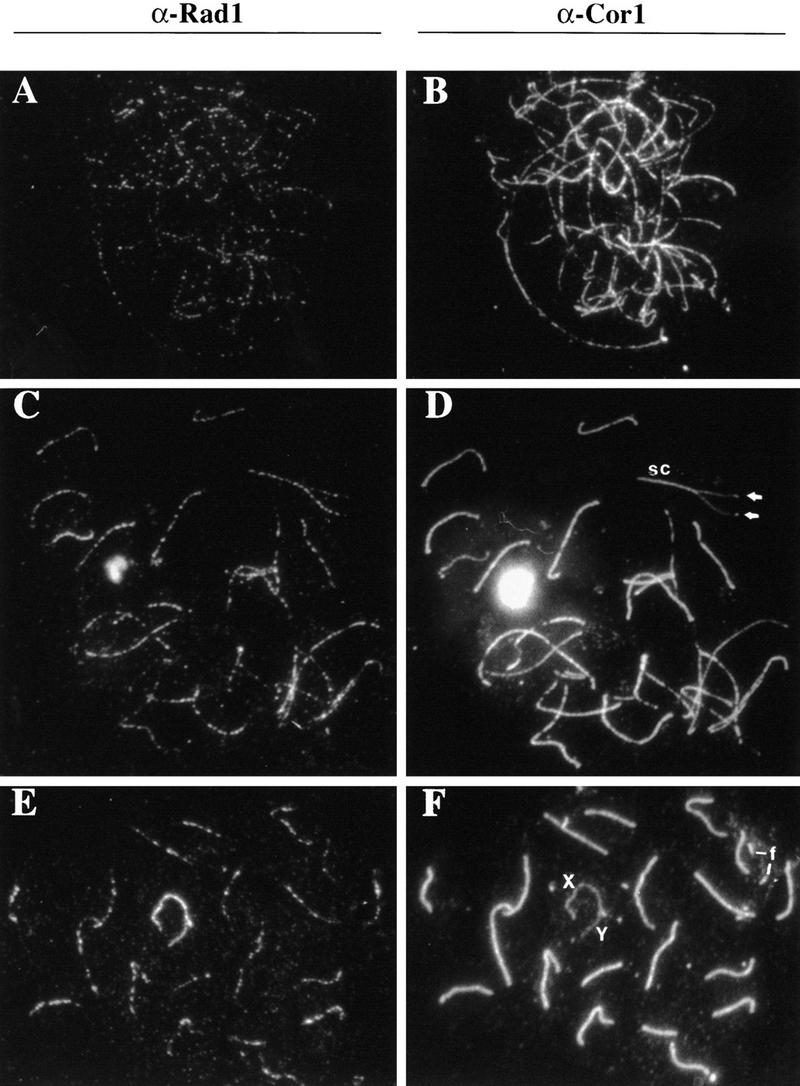
Immunofluorescence microscopy of meiotic chromosomes in different stages of meiosis. Mouse spermatocyte meiotic prophase nuclei were labeled with anti-hRad1 (A,C,E) or anti-Cor1 (B,D,F). (A,B) There are ∼390 hRad1 foci associated with the chromosome cores when synapsis is initiated. (C,D) During synapsis (zygotene), the number of Rad1 foci declines. There are ∼300 foci in this late zygotene nucleus and it is evident that the foci are associated with both unpaired chromosomes and chromosomes that are fully synapsed. (E,F) Extrachromosomal core fragments (f) do not have foci of any kind. There are ∼150 foci at this stage. In contrast, the X–Y chromosome pair remains heavily stained until diplotene.
Evidently, the Rad1 foci form on the cores before synapsis and remain there during the process in which pairs of homologous cores synapse to form the synaptonemal complex (Fig. 7A–D). The overall dynamics of mouse Rad1 localization in prophase I chromosomes are similar to those reported for Rad51 (Barlow et al. 1997; Moens et al. 1997), the mammalian homolog of bacterial recA that is involved in homologous recombination (Baumann et al. 1996). There are differences in detail, however; whereas the Rad51 foci appear as soon as the chromosome cores start to form at the onset of leptotene, the Rad1 foci become noticeable somewhat later when there is already extensive chromosome core formation (Fig. 7A,B). Also, there is a maximum of ∼250 Rad51 foci per nucleus at the end of leptotene and this number declines during zygotene, whereas there are up to 400 Rad1 foci per zygotene nucleus. The decline of Rad1 is also slower than that of Rad51, therefore an excess of Rad1 over Rad51 foci becomes pronounced by mid-pachytene. Furthermore, the X chromosome has more than the average number of foci containing both Rad51 and Rad1 but, whereas the Rad51 foci disappear from the X chromosome at mid-pachytene, the Rad1 antigen is abundant on the X and Y chromosomes at this time (Fig. 7E,F). Extrachromosomal accumulations of core protein are common in rodent nuclei and those fragments are not associated with Rad1 foci [Fig. 7E,F (f)]. Finally, for the X–Y chromosome pair, Rad51 foci are restricted to the X chromosome, whereas Rad1 foci appear on both the X and Y chromosomes (Fig. 7E).
To characterize further the distribution of Rad1 on meiotic chromosomes, we used the technique of immunoelectron microscopy (EM). Thus, the location of Rad1 was studied together with that of Dmc1, a mammalian meiosis-specific RecA homolog that is believed to play a specific role in meiotic recombination. Previous work has shown that Dmc1 and Rad51 often colocalize (P. Moens, unpubl.) and it has been proposed that they localize in the regions of the DNA where recombination events occur (Bishop 1994). Immunoelectron microscopy of rat spermatocytes labeled with anti-Rad1 and a secondary antibody conjugated to a 10-nm gold grain revealed that Rad1 is often located in clusters (Fig. 8), suggesting that the protein might be present in large complexes, as appears to be the case with Dmc1 (labeled with 5-nm gold grains; note that the largest 15-nm gold grains identify the centromeres; see legend to Fig. 8). At this level of resolution, Rad1 and Dmc1 usually do not colocalize (although they appear to by immunofluorescence microscopy; data not shown), suggesting that these proteins have discrete roles in meiosis.
Figure 8.
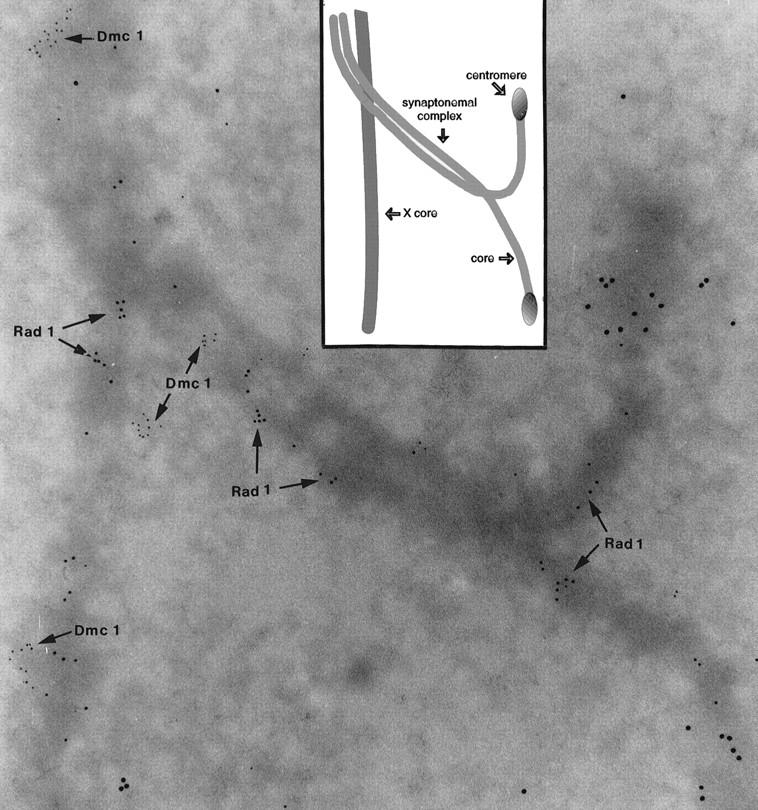
Immunogold localization of Rad1 and Dmc1 at zygotene. Rad1 is labeled with 10-nm, and Dmc1 with 5-nm gold particles. The centromeric ends (15-nm gold grains) are not yet synapsed, and the 10-nm grains on unpaired cores indicate the presence of Rad1 antigen in association with the cores. The SC and the X-chromosome core have both 10- and 5-nm gold clusters, but the two sizes of particle do not colocalize, suggesting distinct functions for Rad1 and Dmc1. A schematic representation of the EM image is shown (top, middle).
Discussion
S. pombe rad1+ and S. cerevisiae RAD17 are homologous genes that play important roles in bringing about cell cycle arrest after DNA damage. sprad1+ is also involved in triggering cell cycle arrest in response to stalled DNA replication forks. Furthermore, scRAD17 appears to play a similar role in the checkpoint processes that control progression through meiosis (Lydall et al. 1996). Certain other genes that function in these events have been identified both in fission and budding yeast, including sprad9+, and the homologous genes scMEC1 and sprad3+. The presence of related signaling systems in these two highly diverged yeasts suggests that the fundamentals of checkpoint control have been conserved highly throughout evolution. Consistent with this, the mammalian homolog of scMEC1/sprad3+, termed ATR, has been isolated, and both ATR and its close relative ATM have been shown to function in DNA damage-induced cell cycle checkpoint processes (Lavin and Shiloh 1997; Cliby et al. 1998). In addition, a human homolog of sprad9+ has been identified and its product, hRad9, has been demonstrated to complement partially the phenotype of a sprad9 mutant (Lieberman et al. 1996). These data suggest that other components of yeast DNA damage checkpoint pathways will be conserved in mammals. Indeed, as we report here, a structural and functional homolog of sprad1/scRAD17 exists in human and mouse.
hRad1 is a structural and functional homolog of scRad17p and spRad1
Sequence analyses reveal that the mammalian Rad1 homologs are related throughout their lengths to both scRad17p and spRad1; sequence identity is slightly greater to spRad1 than to scRad17p. Therefore, we focused on the potential functional overlap between hRad1 and its S. pombe counterpart. Strikingly, we found that HRAD1 significantly complements the sensitivity of the sprad1 mutant strain to UV irradiation. Furthermore, HRAD1 restores partially the cell cycle checkpoint defect of a rad1 mutant strain after UV irradiation. Thus, HRAD1 encodes a protein that can function in checkpoint processes. The complementation of the above phenotypes after UV irradiation, however, is in contrast to the weak complementation that is observed after γ-irradiation. Similarly, no detectable complementation of the S–M phase checkpoint defect after hydroxyurea treatment was observed. Therefore, although hRad1 appears to be able to substitute for spRad1 in functions associated with responses to UV, this is not the case for responses to ionizing radiation or hydroxyurea. A similar but reciprocal effect has been described for hRad9, which partially complements a sprad9-deficient strain after γ-irradiation but not after UV treatment (Lieberman et al. 1996). We can think of three possibilities to explain these observations. First, the mammalian proteins could only function partially in S. pombe, and the UV radiation response could have a lower threshold requirement for activity than the other pathways. Second, spRad1 or spRad9 might have other roles in DNA repair that are distinct from their roles in cell cycle arrest, as has been suggested previously for scRad17p (Lydall and Weinert 1995). Finally, spRad1 and spRad9 could possess multiple functional domains, only some of which are conserved sufficiently in the mammalian proteins. In support of this latter model, both scRad17p and spRad1 contain regions that are not present in the human and mouse proteins, and mutations in different regions of spRad1 have been shown previously to confer distinct phenotypes (Kanter-Smoler et al. 1995). Taken together, the available data indicate that hRad1 overlaps in function with spRad1 and suggest strongly that mammalian Rad1 proteins will prove to play crucial roles in DNA damage-induced cell cycle checkpoint control processes. Our finding that Rad1 levels are higher in proliferative tissues than in tissues where proliferation rates are low might, therefore, reflect the fact that higher levels of the DNA damage checkpoint components are necessary in tissues whose cells are dividing frequently.
Mammalian Rad1 localizes to meiotic chromosomes
We have found that Rad1 is localized to specific regions of the chromosomes of mammalian cells undergoing meiotic prophase I. Similar staining patterns were described previously for ATM and ATR (Keegan et al. 1996). Furthermore, hChk1 has been reported to be located on meiotic chromosomes and in addition has been proposed to function in checkpoint responses after DNA damage (Flaggs et al. 1997; Sanchez et al. 1997). These data, together with the fact that scMEC1, scRAD17, and scRAD24 are involved in both meiotic and mitotic checkpoint controls, suggest strong mechanistic similarities between the checkpoint that regulates meiotic progression and those that are triggered in the mitotic cell cycle by DNA damage or by stalled replication forks. An attractive model, therefore, is that these distinct checkpoint processes use a common set of core components. Although all the mammalian proteins listed above appear to be found on chromosomes undergoing meiotic exchanges, they have notably different spatial and temporal distribution patterns. Thus, Rad1 is located on the cores of both nonsynapsed and synapsed sister chromatids, whereas ATR and hChk1 have been found only on unsynapsed chromosomes in the early stages of prophase I (Keegan et al. 1996; Flaggs et al. 1997). Conversely, ATM has been reported to be located only on synapsed chromosomes (Keegan et al. 1996). These differences lead to a model in which different aspects of meiotic progression are sensed or signaled by distinct sets of checkpoint components. Significantly, Rad1 is the only one of these proteins so far reported to be present on both synapsed and nonsynapsed meiotic chromosomes, suggesting that this protein has a particularly central role in meiotic recombination or signaling meiotic progression. Consequently, an attractive model is that Rad1 or an associated component is a common target required for both ATM and ATR to assemble into the complex. Because spRad1 and spHus1, another DNA damage checkpoint protein, interact with each other, and a human homolog of spHus1 has been isolated (Kostrub et al. 1998; R. Freire and S.P. Jackson, unpubl.), it will be of great interest to study possible biochemical interactions between mammalian Rad1, Hus1, and the other proteins mentioned above, and to investigate by EM whether they colocalize on meiotic chromosomes.
Another noteworthy feature of the meiotic staining pattern of Rad1 is that it appears on meiotic chromosomes at stages of prophase I slightly after Rad51 foci appear but then remains associated with the chromatids after the Rad51 foci have dispersed. These characteristics would clearly fit with a model in which the Rad1-associated checkpoint apparatus assembles immediately after recombination begins, controlling the progress of recombination and keeping homologous chromosomes together until recombination is complete. The relatively low resolution of fluorescence microscopy obscures the relationship between chromosome core-associated RecA proteins Rad51 and Dmc1, and DNA damage-signaling proteins such as Rad1. Their relative locations can be assessed from EM, however, by the use of secondary antibodies conjugated to various sizes of colloidal gold particles. We observed that when Rad51 was labeled with 10-nm gold particles and Dmc1 with 5-nm gold particles, the two size of gold particles occurred together in the nodes (not shown). This is not the case for Rad1 and Dmc1. The 10-nm gold particles labeling Rad1 were mostly in groups separate from the groups of 5-nm gold particles that mark the Dmc1 antigen. Several of these groups would have appeared to “colocalize” if they had been viewed by immunofluorescence. The separate locations suggest discrete functions at meiotic prophase. Interestingly, the Rad51- and Dmc1-containing nodes are estimated to contain several hundred molecules and, by inference, it is likely that the Rad1 groups of 10-nm grains also represent a large number of Rad1 molecules.
In conclusion, the results that we present, together with those described elsewhere, provide strong support for the view that cell cycle checkpoint control events are conserved highly throughout eukaryotic evolution. Furthermore, they indicate that investigations of these events in yeast model systems will be very worthwhile for understanding checkpoint signaling in higher organisms. Key goals for the future will include a more complete biochemical characterization of key components, detailed analyses of the subcellular distribution of these components, and the generation of mice and cell lines that are deficient in them. In addition to revealing how checkpoint controls are executed, these studies will undoubtedly reveal further parallels between the pathways that operate in the meiotic and mitotic states of the cell cycle.
Materials and methods
Isolation of mammalian sprad1+ homologs and chromosomal localization of HRAD1
The human and mouse cDNAs were isolated as described in the Results section. Human cDNA was obtained from the plasmid library HPBALL (Simmons 1993) using the internal reverse and forward primers from the vector and the primers 5′-AAGTTCCCCACCTTGACTATCC-3′ and 5′-TTCCACAAAACATATTTGTCCATC-3′ that aligned with the cDNA of HRAD1. The different PCR-amplified products were cloned into pGEM-T (Promega) and sequenced. The whole human cDNA was amplified and cloned fused to the hexahistidine tag of the vector pQE-30 (Qiagen). The chromosomal localization was performed using a human–mouse and human–hamster hybrid cell line panel that each contained a single human chromosome (Human Genome Project; Sanger Centre, Cambridge, UK) and were used following the instructions provided. Two HRAD1 primers (5′-CTACTTTTGGAAATGCAGGAAGT-3′ and 5′-TCAAGACTCAGATTCAGGAACTTCA-3′) were designed that would amplify a region of 300 bp from the 3′ end of the human cDNA, but would not recognize the rodent homolog. When these were used with human genomic DNA as a template, the resulting product was a specific fragment of ∼550 bp suggesting that this region of the HRAD1 gene contains introns. Subsequently, this was verified by cloning and sequencing of the amplified DNA fragment. Importantly, this PCR product was not observed when mouse or hamster genomic DNA was used as a template, indicating that the primers used were indeed specific for the human gene. When genomic DNA from the various hybrid cell lines were used as a template, only that from cells bearing human chromosome 5 yielded a product. To define the location of HRAD1 on chromosome 5, a similar PCR approach was used with the Genebridge panel (Human Genome Project; Sanger Centre, Cambridge, UK).
S. pombe culture, plasmids, and manipulations
The S. pombe wild-type and rad1:ura4+ strains used in this study have already been described (Al-Khodairy and Carr 1992) and were cultured by standard techniques (Moreno et al. 1991). The plasmid used in these studies was expression vector pREP3X (Forsburg 1993). HRAD1 was cloned into the BamHI and SmaI sites of pREP3X (pREP3X/HRAD1) then verified by sequencing. In addition, the sprad1+ cDNA was amplified by RT–PCR and cloned into the XhoI and BamHI sites of pREP3X (pREP3X/sprad1). All the constructs and the empty vector were introduced into both rad1::ura4+ and WT strains by electroporation (Norbury and Moreno 1997). S. pombe viability studies were performed on cells grown in Edinburgh minimal medium (EMM; Moreno et al. 1991) with appropriate, recommended supplements at 30°C. Cells were cultured to saturation in liquid media, diluted and grown to early log phase. For γ-irradiation, the liquid culture was irradiated with the indicated doses using a 137Cs source at a dose rate of 0.18 kRad/min, then cells were plated at the appropriate density. For UV treatments, cells were first plated at the appropriate density and irradiated with the indicated doses using a Stratalinker 1800 (Stratagene). Plates were incubated for 4–6 days, until colonies were easily visible. For determination of septation, cells were grown to midlog phase in the same manner as for viability studies, then were washed in saline (0.9% NaCl), centrifuged, resuspended in saline at 5 × 106 cells/ml and irradiated at a dose of 50 J/m2. After irradiation, cells were centrifuged and inoculated into EMM at the same initial density. At the indicated time points, cells were collected, fixed with ethanol, washed once with 50 mm sodium citrate, and stained with 50 μg/ml of calcofluor white (Fluorescent brightener 28; SIGMA), containing 0.3 mg/ml p-phenylendiamine (Sigma) as an antifade. Sample were viewed under a fluorescence microscope and >300 cells/point were scored for the presence or absence of a septum.
Northern blot hybridization
Northern blots contained 2 μg of poly(A)+ RNA per lane and were obtained from Clontech. The complete HRAD1 cDNA was labeled with [α-32P]dATP using the Prime-a-gene labeling system (Promega). The blot was incubated and washed as recommended by the manufacturer.
Mammalian cell culture and transfections
HeLa and human osteosarcoma cells (U2OS) cells were routinely grown in DMEM supplemented with 10% fetal calf serum and grown at 37°C (7% CO2). U2OS cells were maintained in log-phase before being exposed to either 10 Grays of ionizing radiation or 50 J/m2 of UV. Cells were harvested at 1, 3, or 7 hr after irradiation. For expression in mammalian cells, HRAD1 cDNA was PCR amplified and cloned into the EcoRI and XbaI sites of PCI neo (Promega) including a Flag epitope tag at the 3′ end (PCI neo/hRad1–Flag). Subconfluent HeLa cells were transfected with the PCI neo/hRad1–Flag using lipofectamine plus reagent (GIBCO BRL) following the instructions of the manufacturer. After 36 hr, cells were collected and whole cell extracts were prepared.
Preparation of cell and tissue extract
To prepare U2OS and HeLa whole cell extract, cells were lysed in 300 μl of lysis buffer I [50 mm Tris (pH 8), 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, and complete protease inhibitors; Boehringer Mannheim]. The lysis mixture was incubated on ice for 20 min and cleared by centrifugation at 12,000g for 10 min at 4°C. HeLa nuclear extract were prepared as described previously (Jackson 1993). To prepare mouse tissue extracts, different tissues were washed in PBS several times and then disrupted in lysis buffer II [20 mm HEPES (pH 7.8), 450 mm NaCl, 25% glycerol, 50 mm NaF, 0.2 mm EDTA, and complete protease inhibitors] by 20 strokes in a tissue grinder. Then, extract were cleared by centrifugation at 12,000g for 10 min at 4°C. The elutriation protocol and the preparation of rat spermatocyte extracts were carried out as described previously (Heyting and Dietrich 1991).
Anti-hRad1 antibody production and purification
The HRAD1 coding sequence fused to the hexahistidine tag of the vector pQE-30 was expressed and purified according to the manufacturer’s instructions. Antibodies against the recombinant protein were raised in rabbits by standard procedures (Harlow and Lane 1988). Western immunoblot analyses were performed as described previously (Harlow and Lane 1988), and were developed by the Enhanced chemiluminescence reagent (Amersham) according to manufacturer’s instructions. The antitubulin antibody was purchased from Sigma, the anti-p53 antibody from Santa Cruz, and the anti-FLAG antibody from Kodak. Anti-Cor1 antibodies were generated in mice (12RB, 8L2) against a fusion protein generated in an E. coli strain containing an expression vector incorporating the gene for the 32-kD protein of the hamster meiotic chromosome core (Dobson et al. 1994). The Dmc1 antibody was generated in mouse (17RB) against an E. coli synthesized His-tagged fusion protein. The E. coli-expressed Dmc1 was Ni2+-NTA purified and then injected into mice. Later, the serum was collected and the cross-reacting Rad51 component removed by adsorption to immobilized Rad51 protein. The CREST anticentromere serum was obtained from a patient with the CREST syndrome (Moens et al. 1987). Recombinant hRad1 protein was attached to Sulfo-Link Coupling Gel (Pierce) according to manufacturer’s instructions and was used to immunoaffinity purify anti-hRad1 antibodies from crude rabbit serum as described previously (Lakin et al. 1996).
Immunocytology methods
Immunofluorescence studies in human cell lines were carried out as previously described using 4% paraformaldehyde as fixative (Görlich et al. 1995); anti-hRad1 affinity-purified antibody was used at a dilution 1:100. The protocol for detection of Rad1 antigen in mouse spermatocyte nuclei was that reported for hRad51 (Moens et al. 1997). Briefly, surface-spread (0.5% NaCl) testicular cells were attached to plastic-covered glass slides and were fixed in 2% (wt/vol) paraformaldehyde, washed in PBS and blocked with antibody dilution buffer (ADB; 10% goat serum, 3% BSA, 0.05% Triton X-100 in PBS). For EM, cells were treated briefly with 0.5 μg/ml DNase I in minimal essential medium (MEM). Slides were incubated with primary antibody overnight or for 2 hr at 37°C (anti-Cor1 was diluted 1:1000 in ADB; anti-hRad1 affinity-purified antiserum was diluted 1:10; anti-Dmc1 affinity purified antibody was diluted 1:1; and CREST serum was diluted 1:500). After washes, slides were incubated for 1 hr with a 1:500 dilution of secondary antibody (fluorochrome-conjugated goat anti-mouse, anti-rabbit, or anti-human). For EM, the primary sera were used at a lesser dilution and the secondary antibodies were diluted in ADB 1:50 goat anti-mouse conjugated to 5-nm gold grains, goat anti-rabbit conjugated to 10-nm gold grains, and goat anti-human with 15-nm gold grains. After washes and drying, coverslips were mounted with Prolong antifade (Molecular Probes, Eugene, OR). For EM, the plastic was floated off, nickel grids applied and later stained with 4% (wt/vol) osmium tetroxide.
Acknowledgments
We thank all members of the Jackson laboratory for their advice and support. In particular, we thank Fiona Lavin for conducting Flag–HRAD1 expression in HeLa cells, John Rouse for comments, and Carol Featherstone for invaluable input. We are grateful to D. Simmons for providing several cDNA plasmid libraries, G. Scott for providing mouse tissues, B. Spyropoulos for valuable help with figures, T. Morita, Osaka University, Japan, for providing the Dmc1 construct, A. Wynshaw-Boris for advice, Karim Labib for his help and advice with the S. pombe experiments, and to the Human Genome Project (Sanger Centre, Cambridge, UK) for providing the human genomic hybrid panel and the Genebridge panel. R.F. is a postdoctoral fellow supported first by a Federation of European Biochemical Societies fellowship and then by the European Commission. J.R.M. is supported by an European Molecular Biology Organization postdoctoral fellowship. P.B.M. and M.T. are supported by the National Research Council of Canada. This work was made possible by grants to S.P.J. from the Cancer Research Campaign and the A–T Medical Research Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The sequences reported in this paper have been deposited in the GenBank database under accession nos. AFO74717 and AFO74718.
Footnotes
E-MAIL spj13@mole.bio.cam.ac.uk; FAX (01223) 334089.
References
- Al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AL, Benson FE, West SC, Hultén MA. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 1997;16:5207–5215. doi: 10.1093/emboj/16.17.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, C., M. Liyanage, P.B. Moens, K. Nagahsima, K. Brown, S. Rottinghaus, S.P. Jackson, D. Tagle, T. Ried, and A. Wynshaw-Boris. 1998. ATM deficiency results in severe meiotic disruption as early as leptotenema in prophase I. Development (in press). [DOI] [PubMed]
- Baumann P, Benson FE, West SC. Human Rad51 promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra MF, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bohm M, Kirch H, Otto T, Rubben H, Wieland I. Deletion analysis at the DEL-27, APC and MTS1 loci in bladder cancer: LOH at the DEL-27 locus on 5p13-12 is a prognostic marker of tumor progression. Int J Cancer. 1997;74:291–295. doi: 10.1002/(sici)1097-0215(19970620)74:3<291::aid-ijc10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblast. Genes & Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: Occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Carr AM. Analysis of radiation-sensitive mutants of fission yeast. Methods Enzymol. 1997;283:471–494. doi: 10.1016/s0076-6879(97)83038-8. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Flaggs G, Plug AW, Dunks KM, Mundt KE, Ford JC, Quiggle MR, Taylor EM, Westphal CH, Ashley T, Hoekstra MF, Carr AM. ATM-dependent interactions of a mammalian chk1 homolog with meiotic chromosomes. Curr Biol. 1997;7:997–986. doi: 10.1016/s0960-9822(06)00417-9. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression sytems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartman E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Griffiths DJ, Barbet NC, McCready S, Lehmann AR, Carr AM. Fission yeast rad17: A homolog of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud’Homme JF, Dib C, Auffray C, Morissette J, Weissenbach J, Goodfellow PN. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, New York, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: A relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- Hassold TJ. Mismatch repair goes meiotic. Nature Genet. 1996;13:261–262. doi: 10.1038/ng0796-261. [DOI] [PubMed] [Google Scholar]
- Heyting C, Dietrich AJJ. Meiotic chromosome preparation and protein labeling. In: Hamkalo BH, Elgin SCR, editors. Methods in cell biology. New York, NY: Academic Press; 1991. pp. 177–202. [DOI] [PubMed] [Google Scholar]
- Hoekstra MF. Responses to DNA damage and regulation of cell cycle checkpoints by the ATM protein kinase family. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- Jackson SP. Identification and characterization of eukaryotic transcription factors. In: Hames BD, Higgins SJ, editors. Gene transcription: A practial approach. Oxford, UK: Oxford University Press; 1993. pp. 189–242. [Google Scholar]
- Jackson SP. Ataxia-telangiectasia at the cross-roads. Cur Biol. 1995;5:1210–1212. doi: 10.1016/s0960-9822(95)00238-7. [DOI] [PubMed] [Google Scholar]
- Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter-Smoler G, Knudsen KE, Jimenez G, Sunnerhagen P, Subramani S. Separation of phenotypes in mutant alleles of the Schizosaccharomyces pombe cell-cycle checkpoint gene rad1+. Mol Biol Cell. 1995;6:1793–1805. doi: 10.1091/mbc.6.12.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan KS, Holtzman DA, Plug AW, Christenson ER, Brainerd EE, Flaggs G, Bentley NJ, Taylor EM, Meyn MS, Moss SB, Carr AM, Ashley T, Hoekstra MF. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes & Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strands breaks are catalyzed by Spo11, a member of a widely converved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kelsell DP, Rooke L, Warne D, Bonzyk M, Cullin L, Cox S, West L, Povey S, Spurr NK. Development of a panel of monochromosomal somatic cell hybrids for rapid gene mapping. Ann Hum Genet. 1995;59:233–241. doi: 10.1111/j.1469-1809.1995.tb00743.x. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin ND, Weber P, Stankovic T, Rottinghaus ST, Taylor AM, Jackson SP. Analysis of the ATM protein in wild-type and Ataxia-Telangiectasia cells. Oncogene. 1996;13:2707–2716. [PubMed] [Google Scholar]
- Lavin MF, Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu Rev Inmunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- Lehmann AR. Molecular biology of DNA repair in the fission yeast Schizosaccharomyces pombe. Mutat Res. 1996;363:147–161. doi: 10.1016/0921-8777(96)00017-1. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lieberman HB, Hopkins KM, Nass M, Demetrick D, Davey S. A human homolog of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc Natl Acad Sci. 1996;93:13890–13895. doi: 10.1073/pnas.93.24.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KE, Sunnerhagen P, Subramani S. The Schizosaccharomyces pombe rad1 gene consists of three exons and the cDNA sequence is partially homologous to the Ustilago maydis REC1 cDNA. Gene. 1994;148:155–159. doi: 10.1016/0378-1119(94)90250-x. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Moens PB, Heyting C, Dietrich AJ, van Raamsdonk W, Chen Q. Synaptonemal complex antigen location and conservation. J Cell Biol. 1987;105:93–103. doi: 10.1083/jcb.105.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng HHQ, Spyropoulos B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetics of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Heiter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia-telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Norbury C, Moreno S. Cloning cell cycle regulatory genes by transcomplementation in yeast. Methods Enzymol. 1997;283:44–59. doi: 10.1016/s0076-6879(97)83006-6. [DOI] [PubMed] [Google Scholar]
- Onel K, Koff A, Bennett RL, Unrau P, Holloman WK. The REC1 gene of Ustilago maydis, which encodes a 3′ → 5′ exonuclease, couples DNA repair and completion of DNA synthesis to a mitotic checkpoint. Genetics. 1996;143:165–174. doi: 10.1093/genetics/143.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NGJ, Malcolm A, Taylor R, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI3-kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shapiro GI, Edwards CD, Ewen ME, Rollins BJ. p16INK4A participates in a G1 arrest checkpoint in response to DNA damage. Mol Cell Biol. 1998;18:378–387. doi: 10.1128/mcb.18.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- Siede W, Nusspaumer G, Portillo V, Rodríguez R, Friedberg EC. Cloning and characterization of RAD17, a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:1669–1675. doi: 10.1093/nar/24.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DL. Cloning cell surface molecules by transient expression in mammalian cells. In: Hartley D, editor. Cellular interactions and development. Oxford, UK: IRL Press; 1993. pp. 91–127. [Google Scholar]
- Storlazzi A, Xu L, Cao L, Kleckner N. Crossover and noncrossover recombination during meiosis: Timing and the pathway relationships. Proc Natl Acad Sci. 1995;92:8512–8516. doi: 10.1073/pnas.92.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen MP, Onel K, Holloman WK. The REC1 gene of Ustilago maydis involved in the cellular response to DNA damage encodes an exonuclease. J Biol Chem. 1994;269:747–754. [PubMed] [Google Scholar]
- Wieland I, Bohm M. Frequent allelic deletion at a novel locus on chromosome 5 in human lung cancer. Cancer Res. 1994;54:1772–1774. [PubMed] [Google Scholar]
- Wieland I, Bohm M, Arden KC, Ammermuller T, Bogatz S, Viars CS, Rajewsky MF. Allelic deletion mapping on chromosome 5 in human carcinomas. Oncogene. 1996;12:97–102. [PubMed] [Google Scholar]
- Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes & Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]