Abstract
Slow- and fast-twitch myofibers of adult skeletal muscles express unique sets of muscle-specific genes, and these distinctive programs of gene expression are controlled by variations in motor neuron activity. It is well established that, as a consequence of more frequent neural stimulation, slow fibers maintain higher levels of intracellular free calcium than fast fibers, but the mechanisms by which calcium may function as a messenger linking nerve activity to changes in gene expression in skeletal muscle have been unknown. Here, fiber-type-specific gene expression in skeletal muscles is shown to be controlled by a signaling pathway that involves calcineurin, a cyclosporin-sensitive, calcium-regulated serine/threonine phosphatase. Activation of calcineurin in skeletal myocytes selectively up-regulates slow-fiber-specific gene promoters. Conversely, inhibition of calcineurin activity by administration of cyclosporin A to intact animals promotes slow-to-fast fiber transformation. Transcriptional activation of slow-fiber-specific transcription appears to be mediated by a combinatorial mechanism involving proteins of the NFAT and MEF2 families. These results identify a molecular mechanism by which different patterns of motor nerve activity promote selective changes in gene expression to establish the specialized characteristics of slow and fast myofibers.
Keywords: Myosin, myoglobin, NFAT, MEF2, cyclosporin A, exercise
Subtypes of skeletal myofibers of adult vertebrates differ markedly with respect to contractile physiology, metabolic capabilities, ultrastructural morphology, and susceptibility to fatigue. The physiological and clinical importance of myofiber specialization has been recognized for several decades, and many studies have identified sets of specific contractile proteins and enzymes of intermediary metabolism, the selective expression of which establishes this physiological and biochemical diversity among skeletal myocytes (Saltin and Gollnick 1983; Booth and Baldwin 1996; Schiaffino and Reggiani 1996). Fiber-type-specific programs of gene expression can be detected at early stages of myogenic development in the embryo (DiMario et al. 1993; Ontell et al. 1993; Stockdale 1997), but remain plastic in adults, in whom they are subject to modification as a function of contractile load (e.g., exercise training), hormonal shifts, or systemic diseases (Holloszy and Coyle 1984; Massie et al. 1988; Ianuzzo et al. 1991; Sabbah et al. 1993; Williams and Neufer 1996).
A central role for motor nerve activity in determining skeletal muscle fiber composition was revealed bycross-innervation and electrical stimulation experiments, which demonstrated complete and reversible transformation of pre-existing myofibers by changing patterns of neuronal firing (Vrbova 1963; Williams et al. 1986; Pette and Vrbova 1992). Specifically, brief bursts of neural activity, interspersed between long periods of neuronal quiescence, promote the acquisition of fast-twitch, glycolytic fiber characteristics. Conversely, extended periods of tonic motor nerve activity stimulate a shift to the slow-twitch, oxidative myofiber phenotype. Neural stimulation provokes changes in the intracellular concentrations of several potential signaling molecules including calcium, cyclic AMP, and nitric oxide, as well as immediate-early gene products (c-fos) and molecular chaperones (hsp70) (Michel et al. 1994; Neufer et al. 1996; Williams and Neufer 1996), but specific signaling pathways and regulatory molecules that link motor nerve activity to fiber-typespecific gene expression have not been identified previously.
Basic, helix–loop–helix (bHLH) proteins of the MyoD family, MADS domain proteins of the MEF2 family, and several other varieties of transcription factors collaborate to establish myogenic cell lineages in the embryo, and are essential for muscle-specific gene transcription (Olson et al. 1995; Molkentin and Olson 1996; Black and Olson 1998), but no clear role for these proteins in fiber-type-specific gene expression has been established (Hughes et al. 1997). Thyroid hormone can exert profound effects on expression of genes encoding specific subtypes of myosin and other contractile proteins (Izumo and Mahdavi 1988), but thyroxin-mediated gene regulation does not account for myofiber diversity in the euthyroid state.
Calcineurin-dependent signaling mechanisms have been characterized extensively in the activation of cytokine gene expression in T and B lymphocytes responding to stimuli that elevate intracellular free calcium concentration ([Ca2+]i) (Rao et al. 1997). Binding of calcium to a calmodulin-calcineurin complex stimulates serine/threonine phosphatase activity of calcineurin, the major substrates of which are nuclear factor of activated t cells (NFAT) transcription factors. Dephosphorylation of NFATs by calcineurin promotes their translocation from the cytoplasm to the nucleus, where they bind a cognate nucleotide recognition sequence (Rao et al. 1997) and stimulate transcription of target genes that, in lymphocytes, include hematopoietic growth factors (e.g., GM-CSF) and inflammatory cytokines (e.g., IL-2). Recent studies demonstrate that calcineurin activity and the resulting nuclear translocation of NFAT are insensitive to transient, high-amplitude oscillations in [Ca2+]i that activate other calcium-dependent events (e.g., NF-κB or c-Jun amino-terminal kinase). Rather, the calcineurin-NFAT pathway responds preferentially to sustained, low-amplitude elevations of [Ca2+]i (Timmerman et al. 1996; Dolmetsch et al. 1997). This ability of a calcineurin-dependent signaling pathway to discriminate between different patterns in the amplitude and duration of changes in [Ca2+]i, in conjunction with previous data characterizing differences in intracellular calcium concentrations among specialized myofiber subtypes, provided the basis for our hypothesis that a calcineurin-dependent pathway influences fiber-type-specific gene expression.
Tonic motor nerve activity at 10–15 Hz is characteristic of slow-twitch fibers (Hennig and Lomo 1985) and results in a sustained elevation of [Ca2+]i within a concentration range between 100 and 300 nm (Chin and Allen 1996), a pattern predicted to activate calcineurin. In fast myofibers, resting [Ca2+]i is maintained at levels of only 50 nm (Westerblad and Allen 1991), and the high amplitude (∼1μm) calcium transients evoked by motor nerve activity are predicted to be of insufficient duration to evoke calcineurin-stimulated signaling. Chronic stimulation at 10 Hz of the motor nerve innervating fast myofibers results in sustained elevations of [Ca2+]i and promotes fast-to-slow fiber transformation (Williams et al. 1986; Sreter et al. 1987). Calcineurin and several NFAT isoforms are abundant in skeletal muscles (Hoey et al. 1995), though target genes that respond to this pathway in skeletal myocytes have not been identified previously, and a specific role for calcineurin in the control of myofiber specialization has not been proposed previously.
Here we demonstrate that fiber-type-specific gene expression in skeletal muscles is controlled by a signaling mechanism that involves calcineurin, a cyclosporin-sensitive, calcium-regulated serine/threonine phosphatase. This discovery has considerable explanatory power, in that it elucidates plausibly a complete signaling pathway linking motor nerve activity to selective changes in gene expression that establish diversity among myofibers.
Results
Selective activation of slow-fiber-specific promoters by forced expression of a constitutively active form of calcineurin
The myoglobin (Mb) and troponin I slow (TnIs) genes are expressed selectively in slow, oxidative skeletal muscle fibers (Levitt et al. 1995; Garry et al. 1996), whereas the muscle creatine kinase (MCK) gene is expressed most abundantly in the fast, glycolytic myofiber subtype (Yamashita and Yoshioka 1991). To test whether these genes might respond differently to a calcineurin-stimulated signaling pathway, we transfected skeletal myogenic cells with reporter genes linked to well-characterized control regions from these genes, along with an expression vector encoding a constitutively active (calcium-insensitive) form of calcineurin that retains sensitivity to inhibition by cyclosporin A (Manalan and Klee 1983; O’Keefe et al. 1992).
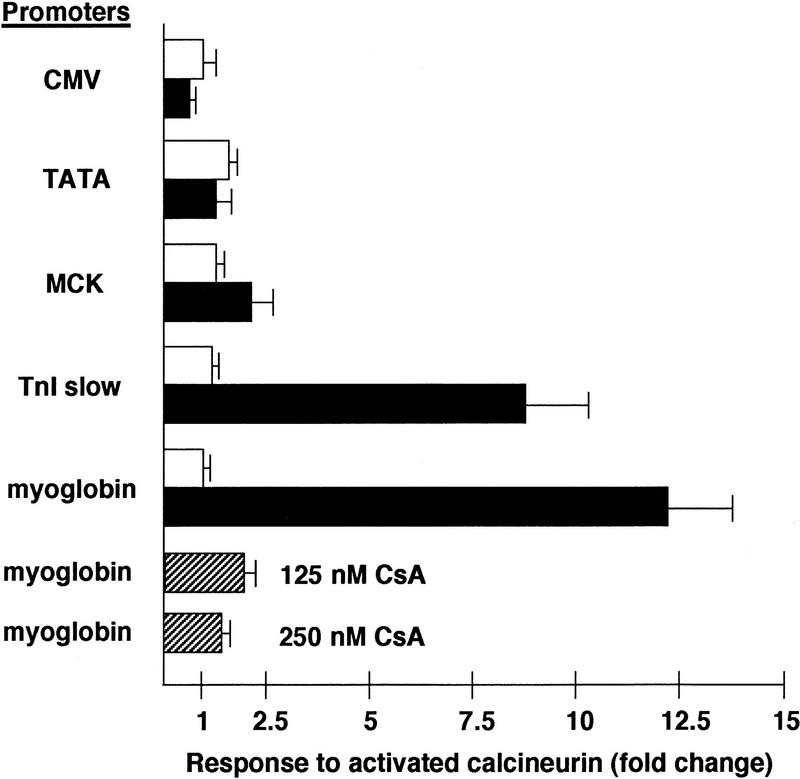
Transcriptional activity of the slow-fiber-specific myoglobin and TnIs promoters was stimulated in cultured skeletal myotubes (C2C12) by active calcineurin, as measured by expression of luciferase in cotransfection assays (Fig. 1). In contrast, activity of the fast-fiber-specific MCK promoter, or of other strong (CMV) or weak (minimal TATA element) promoters, was unaffected by activated calcineurin. The induction of the myoglobin promoter in the presence of the calcineurin expression plasmid was inhibited by cyclosporin A. This result indicates the specificity of the response, since the effect of cyclosporin A is to bind cyclophilin and form a protein complex that binds calcineurin and inhibits its protein phosphatase activity (Liu et al. 1991). The same relative potency of calcineurin-dependent transactivation (myoglobin and TnIs>>MCK, CMV, or TATA) was observed in Sol8 myotubes, a different myogenic cell line (data not shown). In contrast, forced expression of activated calcineurin had no effect on promoter activity in undifferentiated myoblasts (not shown) or in 3T3 fibroblasts (Fig. 1), demonstrating a requirement for muscle-specific factors in the calcineurin-stimulated pathway for transcriptional control of the myoglobin and TnIs promoters.
Figure 1.
Response of different promoters to forced expression of a constitutively active form of calcineurin (O’Keefe et al. 1992) in cultured C2C12 myotubes or NIH-3T3 fibroblasts. Promoter–reporter plasmids were constructed to link the indicated promoters: (CMV) cytomegalovirus; (TATA) a minimal promoter consisting of the TATA element from the human hsp70 gene; (MCK) a 4.8-kb 5′ flanking region from the murine muscle creatine kinase gene; (TnI slow) a 4.2-kb 5′ flanking region from the human slow-fiber-specific troponin I gene; (myoglobin) a 2-kb 5′ flanking region from the human myoglobin gene to a firefly luciferase reporter gene. The response to activated calcineurin was calculated as the fold change in luciferase activity induced by activated calcineurin above that measured following transfection of the empty vector alone, corrected for transfection efficiency (β-galactosidase activity). (Open bars) NIH-3T3 cells; (solid bars) C2C12 cells; (hatched bars) C2C12 + CsA. Cyclosporin A (CsA) was added to the culture medium at the indicated final concentrations. Histograms depict mean values (±s.e.) from 4–8 independent transfections in each cell background.
Calcineurin-stimulated transactivation of slow-fiber-specific promoters requires nucleotide sequence motifs characteristic of NFAT binding sites
The finding that the myoglobin and TnIs promoters can be transcriptionally regulated by a calcineurin-dependent mechanism suggested the participation of NFAT transcription factors in the signaling cascade. Examination of the complete nucleotide sequences of these functionally defined transcriptional control regions (2.0 and 4.2 kb, respectively) revealed two 8-bp elements within each that match the consensus-binding sequence for NFAT transcription factors (Rao et al. 1997). The response to activated calcineurin of the native promoter sequences was compared to that of mutated promoters in which these putative NFAT recognition elements were ablated by site-directed mutatagenesis.
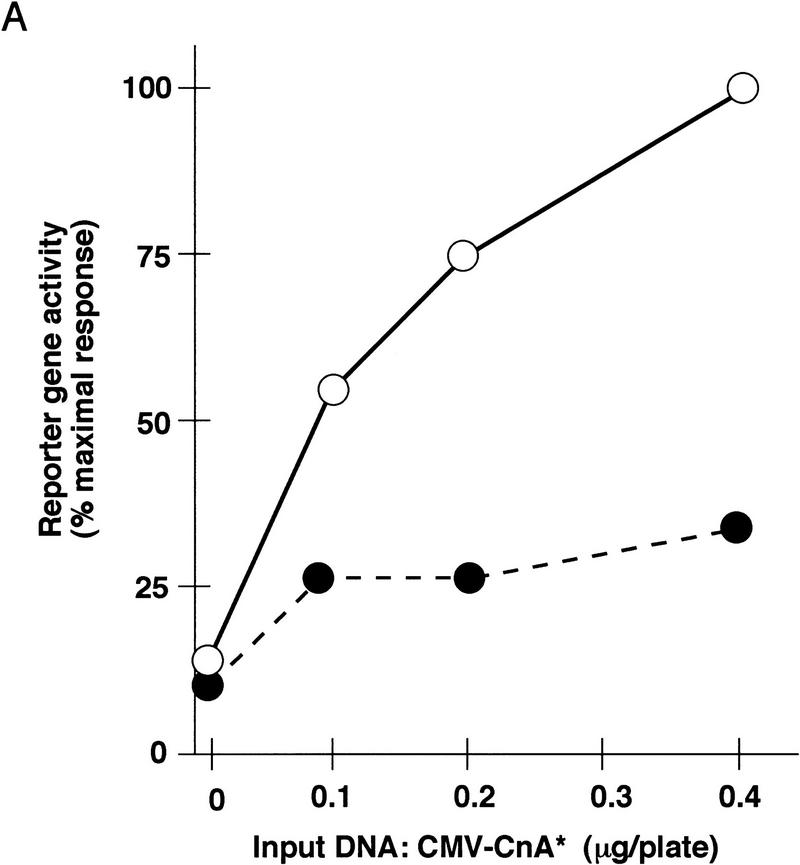
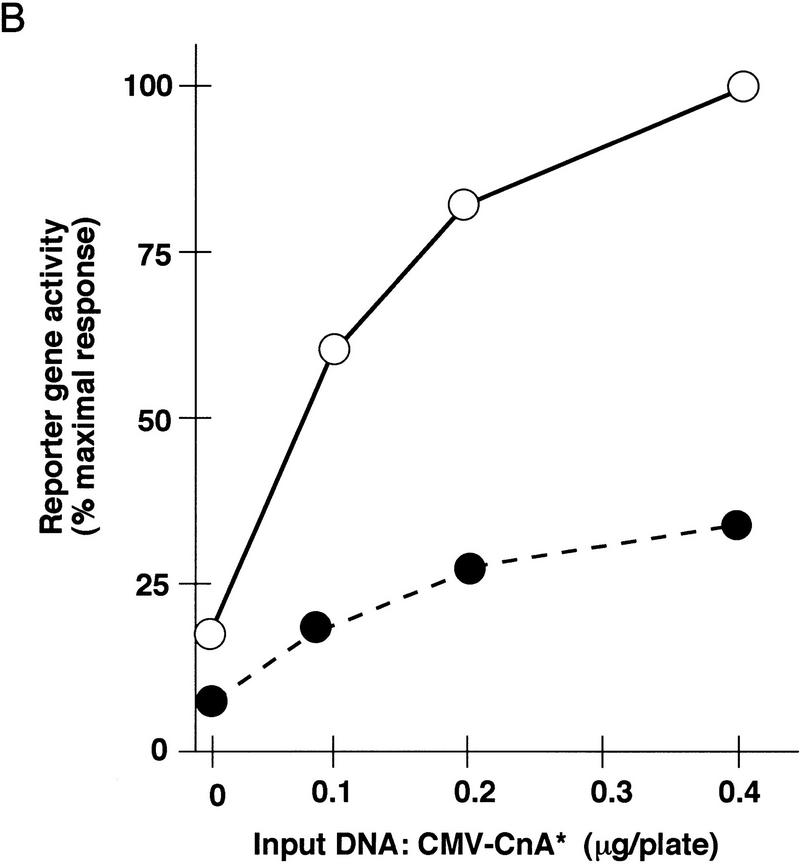
Disruption of putative NFAT recognition elements within both the myoglobin (Fig. 2A) and TnIs (Fig. 2B) promoters diminished the response to activated calcineurin, indicating that the transactivation mechanism is likely to involve DNA binding of NFAT proteins. Transduction of the calcineurin-directed signal to the native myoglobin and TnIs promoters exhibited a saturable dose-response relationship with respect to the activated calcineurin expression plasmid, and diminished reporter gene activation was evident across the entire dose range we examined. Some degree of calcineurin-dependent transactivation persisted after ablation of identifiable NFAT binding sites within these transcriptional control regions. Thus, either cryptic binding sites for NFAT proteins that cannot be recognized by inspection of the DNA sequence are present, or calcineurin-dependent signaling to these promoters can be driven without direct DNA binding of NFAT proteins.
Figure 2.
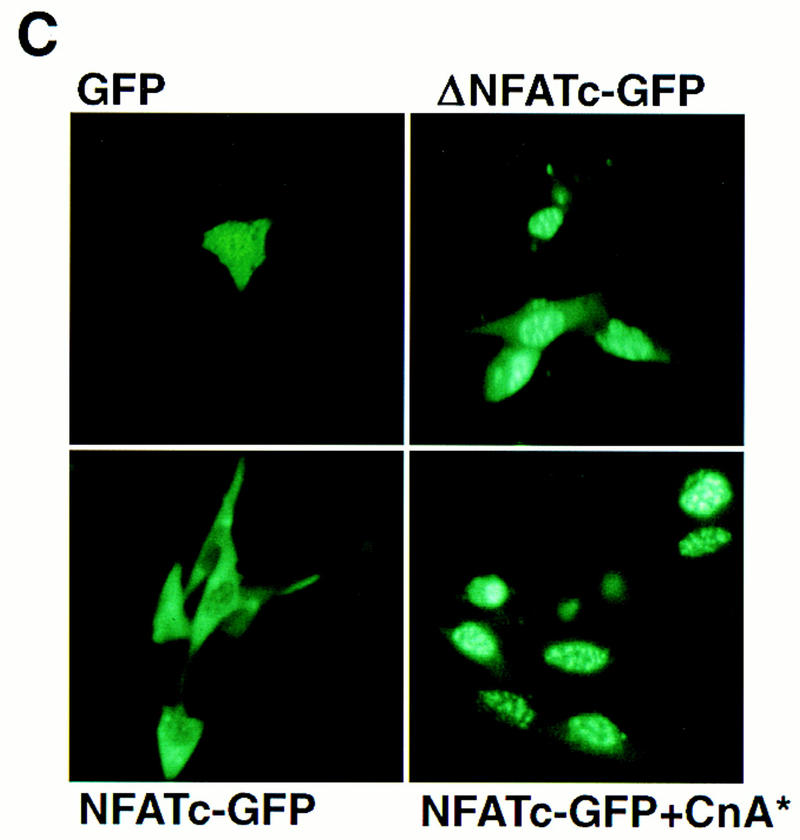
Role of NFAT proteins in calcineurin-dependent transactivation. Activity of wild-type and mutated myoglobin (A) or troponin I slow (B) gene promoters in differentiated C2C12 cells as a function of increasing doses of the calcineurin-expression plasmid. Consensus NFAT recognition motifs at the indicated positions relative to the transcriptional start sites (see Fig. 6) were altered (ΔNFAT) by site-directed mutagenesis, and transfections were performed as described in Figure 1. Data points represent mean values of luciferase activity, corrected for transfection efficiency (β-galactosidase activity), from duplicate transfections in a representative experiment, and expressed as a percentage of native promoter activity after transfection with the indicated amounts of activated calcineurin expression plasmid (CMV–CnA*). (A) (○) 2 kb of myoglobin; (•) ΔNFAT myoglobin (−690 and −232). (B) (○) 4.2 kb TnIs; (•) ΔNFAT TnIs (−743 and −683). (C) Activated calcineurin promotes nuclear translocation of NFAT proteins. C2C12 cells were transfected with plasmids expressing native GFP (upper left), a truncated variant of NFATc fused to GFP (ΔNFATc–GFP) that removes the amino-terminal regulatory domain controlled by calcineurin (upper right), or full-length NFATc fused to GFP (NFATc–GFP) in the absence (lower left) or presence (lower right) of activated calcineurin.
Nuclear localization of NFAT proteins in skeletal myocytes is under the control of calcineurin (Fig. 2C), as predicted from previously published results in lymphocytes (Timmerman et al. 1996). A fusion protein linking green fluorescent protein (GFP) to full-length NFATc (NFATc–GFP) is excluded from the nucleus in C2C12 cells under basal conditions, but undergoes nuclear translocation in the presence of activated calcineurin. As controls, an NFATc–GFP fusion protein lacking amino acids 1–318 of NFATc (ΔNFATc–GFP) is localized constitutively to the nucleus in the absence of activated calcineurin, whereas native GFP is distributed across both cytoplasmic and nuclear compartments. The amino-terminal segment of NFAT proteins (missing in ΔNFATc–GFP) includes the conserved SPRIEIT motif that constitutes the calcineurin targeting site (Aramburu et al. 1998).
Calcineurin-stimulated transactivation of slowfiber-specific promoters requires collaboration among multiple transcription factors
Muscle-specific transcription factors are required for calcineurin-dependent activation of the myoglobin and TnIs promoters, since no response was observed in a fibroblast cell background (Fig. 1). Previously, we defined two conserved upstream response elements within the myoglobin promoter, both of which are required for transcriptional activity in skeletal myotubes or cardiac myocytes (Devlin et al. 1989; Bassel-Duby et al. 1993; Grayson et al. 1995, 1998). These CCAC and A/T elements represent binding sites for Sp1 and MEF2 proteins, respectively, and function synergistically in muscle-specific gene regulation (Grayson et al. 1995, 1998). This prior work established a molecular basis for muscle-specific expression of myoglobin, but failed to account for selective expression of myoglobin in slow fiber types, since Sp1 and MEF2 proteins are equally abundant in slow and fast fibers (J. Grayson, R. Bassel-Duby, R.S. Williams, unpubl.).
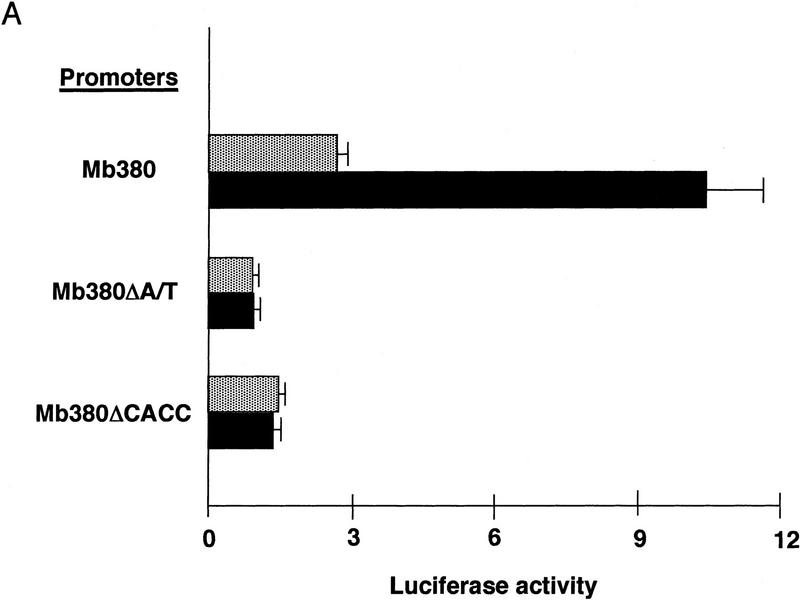
A myoglobin promoter segment truncated to nucleotides −373 to +7 (Mb380) was sufficient for muscle-specific expression in prior experiments (Devlin et al. 1989; Bassel-Duby et al. 1993; Grayson et al. 1995) and was responsive to calcineurin stimulation in our current studies (Fig. 3A). This region includes the CCAC and A/T motifs required for muscle-specific promoter activity, as well as a putative NFAT response element. Nucleotide substitutions within either the CCAC or A/T elements of Mb380 reduced basal transcription in differentiated myotubes, as observed previously, and abrogated the response to calcineurin (Fig. 3A). Thus, mutations that compromise binding of MEF2, Sp1, or other factors to the CCAC and A/T elements interdict the calcineurin-stimulated response, even when the NFAT consensus binding motif at −232 remains intact.
Figure 3.
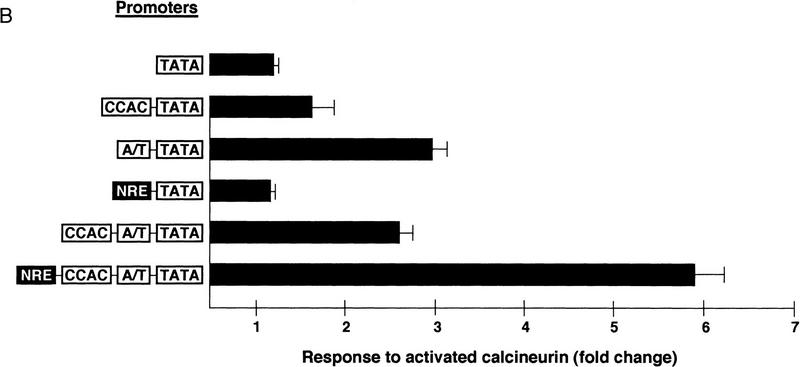
Upstream regulatory elements of the myoglobin gene participating in calcineurin-dependent transactivation. Data are presented as reporter–gene expression (mean ± s.e.m. of six independent transfections) normalized to activity of a cotranfected CMV–lacZ plasmid [luminometer units (×105)/well (1.9 × 105 cells)]. (A) Responses of native (Mb380) or mutated variants of a truncated segment (−373 to +7) of the human myoglobin gene promoter to activated calcineurin. Nucleotide substitutions were introduced into each of two upstream regulatory elements shown previously to be essential for muscle-specific promoter activity (Devlin et al. 1989; Bassel-Duby et al. 1993; Grayson et al. 1995, 1998). These mutated promoters (MbΔA/T and MbΔCCAC) are likewise defective for calcineurin-stimulated transactivation. (Stippled box) −CnA*; (solid bars) +CnA*. (B) Responses to activated calcineurin of synthetic promoters constructed with various combinations of multimerized oligonucleotide cassettes representing protein-binding motifs (CCAC) Sp1 binding site; (A/T) MEF2 binding site; (NRE) putative NFAT binding site; (TATA) TBP binding site and core promoter from the myoglobin promoter.
Functional interactions between transcription factors binding to motifs within the myoglobin promoter were examined further by linkage of various combinations of multimerized oligonucleotide cassettes representing cognate binding sites for MEF2, Sp1, and NFAT in promoter-reporter constructions. As assessed by cotransfection assays in C2C12 myotubes (Fig. 3B), forced expression of activated calcineurin only marginally enhanced transcription (less than twofold) of a construct bearing multiple copies of the CCAC motif. The response to calcineurin was somewhat more robust (threefold) if multimers of the A/T element were included within the synthetic promoters, either in the absence of heterologous protein binding sites, or when combined with multimerized CCAC sites. A reporter construction bearing multiple copies of the upstream (−690) NFAT response element (NRE) from the myoglobin promoter was minimally stimulated by activated calcineurin (less than twofold) in this cell background, but a construct combining NRE, A/T, and CCAC motifs was potently transactivated (sixfold). These results demonstrate that collaborative interactions among proteins binding to NRE, A/T, and CCAC elements from the myoglobin promoter are necessary for optimal transduction of the calcineurin-stimulated signal.
Administration of the calcineurin antagonist cyclosporin A to intact animals promotes slow-to-fast fiber transformation
To determine whether calcineurin-dependent activation of slow-fiber-specific promoters observed in cultured myotubes is pertinent to mature myofibers of intact animals, we assessed the proportion of fast versus slow fibers within soleus muscles of rats treated with cyclosporin A, a specific inhibitor of calcineurin (Liu et al. 1991; Clipstone and Crabtree 1992). The intraperitoneal administration of an immunosuppressant dose (5 mg/kg/day) of cyclosporin A for 6 weeks uniformly increased the proportion of fast fibers defined either by histochemical staining of myosin ATPase activity (Brooke and Kaiser 1970), or by specific immunohistochemical staining of fast skeletal myosin (Fig. 4A–D). In soleus muscles of 7 control animals, fast (type II) fibers represented 4%–24% (mean 14 ± 3%) of the total cell population, whereas 28%–37% (mean 31 ± 1%) of soleus fibers expressed fast myosin in 7 cyclosporin A-treated rats (P < 0.001) (Fig. 4E). This result is consistent with the hypothesis that physiological signals acting to establish and maintain the slow, oxidative myofiber phenotype in intact animals are transduced by a calcineurin-dependent pathway. Interdiction of the calcineurin-signaling pathway with cyclosporin A has reciprocal effects on expression of fast and slow myosin isoforms: Not only is slow myosin expression reduced (Fig. 4A,B), but fast myosin expression is enhanced (Fig. 4C,D). We infer, therefore, that calcineurin-dependent signaling both activates slow-fiber-specific genes and represses the fast fiber-specific program.
Figure 4.
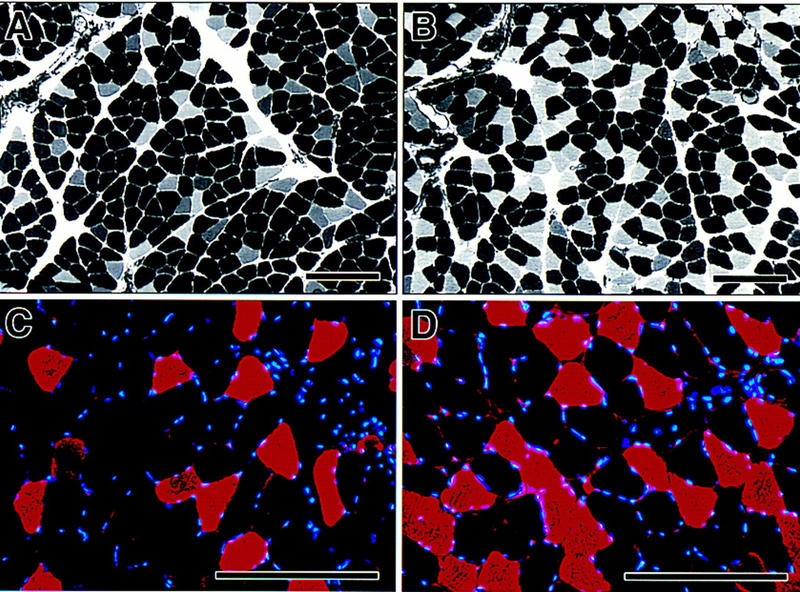
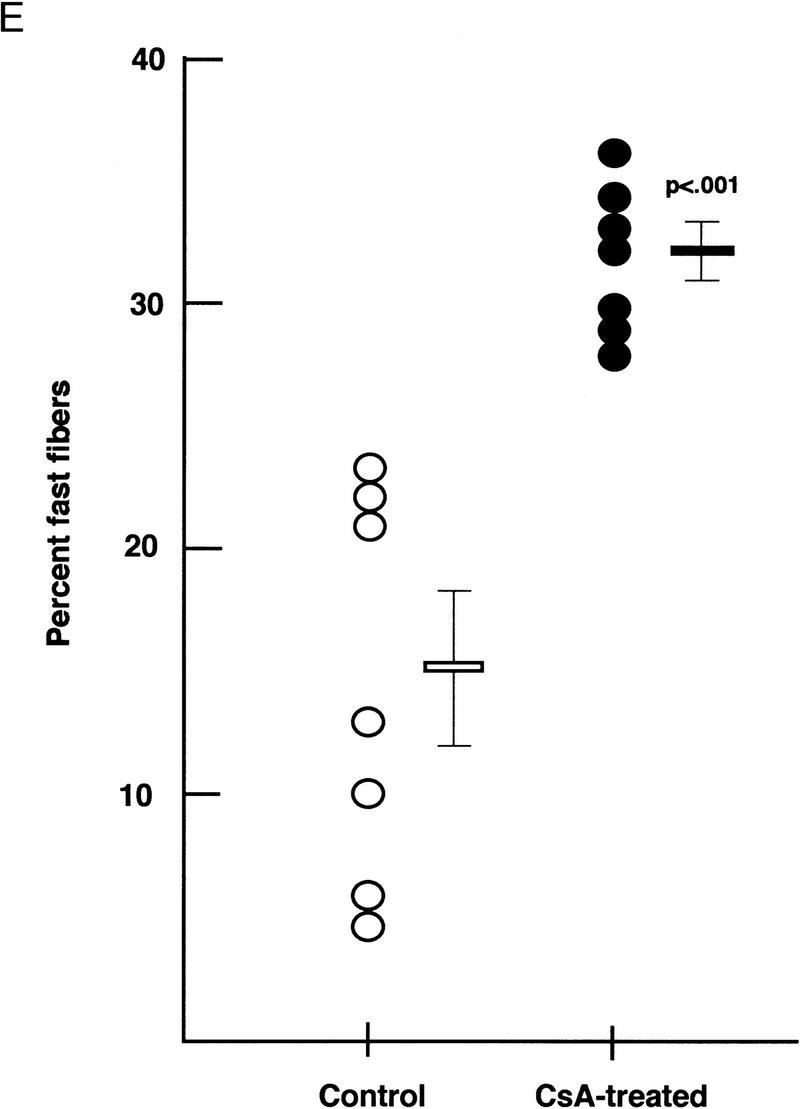
Fiber composition of soleus muscles from intact rats treated with cyclosporin A. Myosin ATPase activity determined by pH-dependent histochemistry distinguishes slow (darkly stained) and fast (unstained) fibers in sections of soleus muscle from vehicle-treated (A) and cyclosporin A-treated (B) rats. Immunohistochemistry using an antibody raised against fast myosin heavy chain identifies fibers expressing fast myosin (red) in sections of soleus muscle from vehicle-treated (C) and cyclosporin A-treated (D) rats. Nuclei are stained blue. (Bar, 200 μm). (E) Circles represent individual animals. (○) Vehicle treated; (•) cyclosporin A treated and mean values in each group (± s.e.) are shown as horizontal lines. The difference in group means was highly significant (P < 0.001 by unpaired Student’s t-test).
Discussion
A molecular basis for control of myofiber specialization by motor nerve activity
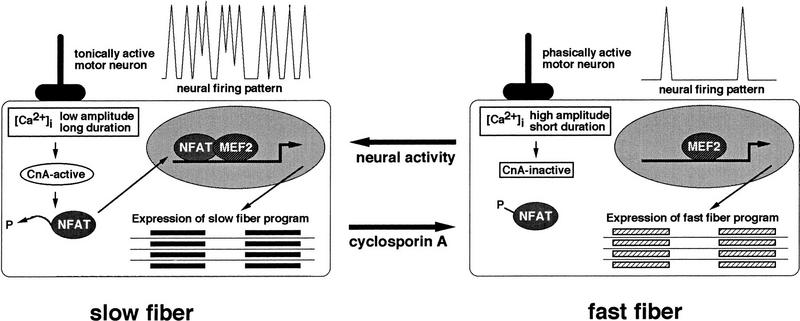
The results presented in this paper suggest a molecular model, not previously considered, to explain how motor-nerve activity controls programs of gene expression that define fast and slow subtypes of skeletal myofibers (Fig. 5). The model proposes that tonic motor nerve activity, characteristic of nerves innervating slow muscles, sustains [Ca2+]i at levels sufficient to activate the calcineurin–NFAT pathway. The protein phosphatase activity of calcineurin leads to dephosphorylation and nuclear localization of NFAT proteins. In the nucleus, NFAT proteins bind DNA in conjunction with other transcriptional regulators, including (but not limited to) MEF2, binding sites for which are clustered in promoter/enhancer regions controlling transcription of genes encoding proteins of the slow-fiber program. In fast fibers, high-amplitude calcium transients stimulated by infrequent, phasic firing of the motor nerve are of insufficient duration to maintain calcineurin in the active state, so NFAT proteins remain phosphorylated and are excluded from the nucleus. When NFAT proteins are unavailable for DNA binding and protein–protein interactions at target promoters, the slow-fiber-specific program is down-regulated, and genes encoding fast-fiber-specific proteins are transcribed.
Figure 5.
Model for a calcineurin-dependent pathway linking specific patterns of motor nerve activity to distinct programs of gene expression that establish phenotypic differences between slow and fast myofibers. MEF2 is shown to represent the requirement for collaboration between activated NFAT proteins and muscle-restricted transcription factors in slow-fiber-specific gene transcription, but other proteins (not shown) also are likely to participate.
This model accommodates the well-established associations between motor nerve activity and specialized fiber characteristics described in an extensive literature on muscle plasticity (Michel et al. 1994; Neufer et al. 1996; Williams and Neufer 1996). Fast-to-slow fiber transformation is evoked by increased motor nerve activity, stimulated by cross-innervation, electrical pacing, or exercise training. Slow-to-fast fiber transformation occurs as a consequence of decreased motor nerve activity, resulting from cross-innervation, certain disease states, hypogravity, or physical inactivity. The new and independent lines of evidence provided here support the contention that calcineurin-dependent signaling is an important mechanism central to these transformations.
Forced expression of constitutively active calcineurin selectively transactivates promoters from two genes that are expressed preferentially in slow versus fast skeletal myofibers. Thus, downstream effectors of a calcineurin-regulated signaling pathway are present and capable of transducing the signal in a muscle-cell background, and transcriptional regulatory elements capable of receiving the signal are present within genes representative of the slow-fiber program. Specific effector molecules appear to include NFAT proteins, as consensus NFAT binding motifs contained within slow-fiber-specific promoters participate in the response to activated calcineurin, and several NFAT isoforms are expressed in skeletal muscle (Hoey et al. 1995). Our data suggest, moreover, that DNA binding of NFAT proteins is not sufficient to transduce the calcineurin-generated signal in skeletal myocytes. Rather, NFAT transcription factors collaborate with MEF2 and other transcriptional regulatory proteins, the correct combination of which is present within differentiated myotubes, but absent from undifferentiated myoblasts or fibroblasts. Previous studies of calcineurin-stimulated transactivation of cytokine gene promoters in T cells, where AP-1 cooperates with NFAT in both DNA binding and transactivation (for review, see Rao et al. 1997), provide a precedent for synergistic combinatorial interactions between NFAT proteins and heterologous transcription factors.
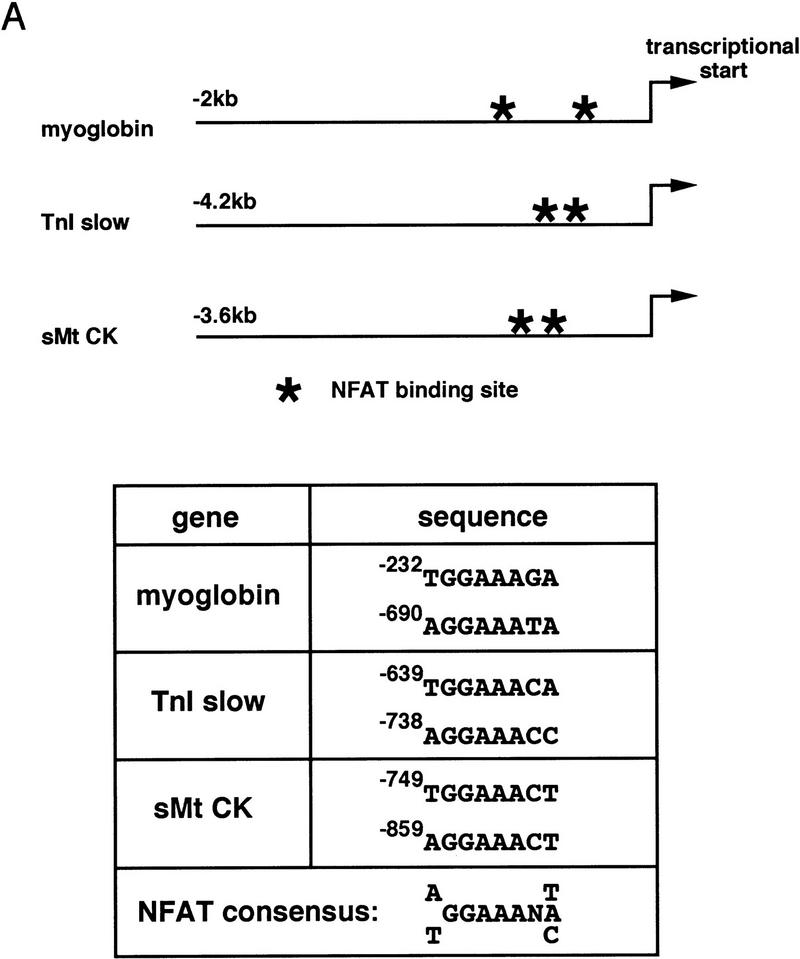
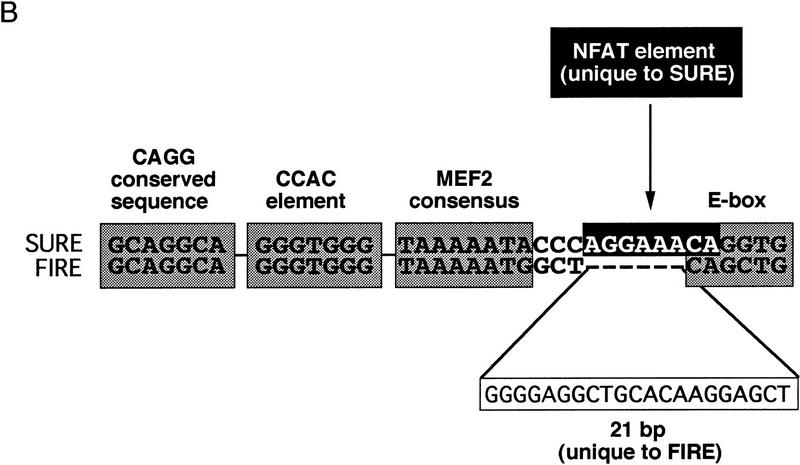
The organization of transcriptional control regions that confer fiber-type-specific expression (Fig. 6) is consistent with this viewpoint. Consensus NFAT binding sequences are conserved in the 5′ flanking region of myoglobin and TnIs genes from all vertebrate species in which promoter sequences are available, and in other slow-fiber-specific enhancers (Fig. 6A). Moreover, two studies that have mapped fiber-type-specific enhancers at the highest resolution provide additional support to our hypothesis that a calcineurin–NFAT signaling pathway is important to this mechanism. Buonanno and colleagues (Nakayama et al. 1996) identified a 128-bp element from the rat TnIs gene [slow upstream regulatory element (SURE)] that confers slow-fiber-specific transcription in transgenic mice, and a 144-bp element [fast intronic regulatory element (FIRE)] that directs fast-fiber-specific expression of a different isoform of troponin I (TnIf). These functionally distinctive SURE and FIRE elements contain similar or identical CAGG, CCAC, MEF2, and E box motifs (Fig. 6B), as found in many muscle-specific genes, so the basis for their reciprocal functions in specialized subtypes of myofibers has not been apparent. Examination of the SURE and FIRE elements in light of our new observations reveals a consensus NFAT recognition motif in the TnIs SURE element that is absent from the TnIf FIRE element. In the sarcomeric mitochondrial creatine kinase (sMtCK) gene, a 160-bp upstream element was shown to direct fiber-type-specific gene expression in transgenic mice (Qin et al. 1997). The sMtCK gene is expressed preferentially in slow, oxidative myofibers, in contrast to the MCK isoform (fast-fiber specific) that was studied in our experiments (Fig. 1). Like the myoglobin and TnIs gene enhancers, this sMtCK enhancer includes NFAT recognition motifs (Fig. 6A).
Figure 6.
NFAT consensus binding sequences are present within transcriptional control regions shown previously to direct transcription selectively in slow-oxidative myofibers (Parsons et al. 1993; Levitt et al. 1995; Qin et al. 1997). (A) Consensus NFAT binding motifs in myoglobin, troponin I slow (TnI slow), and sarcomeric mitochondrial creatine kinase (sMtCK) promoters. (B) Conserved sequence blocks (CAGG, CCAC, MEF2, and E box) common to a SURE from the rat troponin I slow gene and a FIRE from the quail troponin I fast gene (Nakayama et al. 1996). A predicted NFAT response element (darkly shaded) overlapping the E-box is a unique feature of the SURE element.
Finally, inhibition of calcineurin phosphatase activity in intact animals by administration of cyclosporin A leads to down-regulation of slow and induction of fast-fiber type-specific markers. This observation indicates that the conclusions drawn from experiments in cultured myotubes are likely to be pertinent to mature myofibers of adult animals.
Limitations of this model for fiber type determination
The model illustrated in Figure 5 reconciles our current findings with a substantial body of previous data concerning the regulation of skeletal muscle fiber type by motor nerve activity, the molecular basis for which was not apparent previously. However, for the sake of simplicity and clarity in the conduct and presentation of these initial experiments, we have considered only the distinction between fast and slow skeletal myofibers. Specialization among muscle cells is, of course, much more richly textured. Several additional subtypes of fast-twitch fibers can be defined on the basis of expression of different contractile protein isoforms (type IIa vs. IIb vs. IIx), distinctive metabolic properties [oxidative (IIa) vs. glycolytic (IIb)], or gross appearance [red (I and IIa) vs. white (IIb)]. Additional studies will be required to determine the degree to which calcium-regulated, calcineurin-dependent signaling participates in defining the complete spectrum of myofiber subtypes, and the manner in which this pathway interdigitates with other signaling processes.
This working model has other limitations that should be addressed in future studies. Several of the steps in the pathway are inferred by analogy to data acquired in nonmuscle-cell types and, therefore, will require direct confirmation in the muscle-cell background. For example, calcium-dependent regulation of calcineurin activity leading to DNA binding of NFAT proteins has not been assessed directly in skeletal muscles, but our model requires only that the fundamental principles established in T lymphocytes—selective activation by sustained but not transient elevations in [Ca2+]i (Timmerman et al. 1996; Dolmetsch et al. 1997)—can be extrapolated to predict the control of this pathway within a different cell type. Likewise, the conclusion that NFAT proteins transduce primarily the calcineurin-dependent signal to slow-fiber-specific promoters was suggested by analogy with calcineurin–NFAT signaling in other cell types, and has some experimental support from our current data, but additional studies will be necessary to provide rigorous proof of this concept. Our current data do not exclude the possibility that the calcineurin-stimulated signal is transduced to responsive target genes in skeletal muscle through mechanisms that are independent of NFAT proteins. Although no other transcription factors are known currently to be controlled directly by calcineurin, the activities of several muscle-specific transcription factors are regulated by phosphorylation events (Li et al. 1992) that conceivably could be modified by calcineurin. In addition, other muscle proteins known to be substrates of calcineurin [ryanodine and inositol 1,4,5-trisphosphate receptors (Cameron et al. 1995; Lam et al. 1995), and dystrophin (Walsh et al. 1995)] could participate plausibly in calcineurin-activated pathways that modulate transcription, without the involvement of NFAT proteins.
Finally, our current data cannot conclusively exclude the possibility that slow-to-fast fiber transformation induced by administration of cyclosporin A to intact animals is attributable to local or systemic drug effects that are unrelated to inhibition of calcineurin, or to inhibition of calcineurin in motor neurons or other cell types that communicate signaling information to skeletal myofibers. The experiments demonstrating calcineurin-dependent transactivation of slow-fiber-specific gene promoters in cultured myotubes support the notion that the effects of cyclosporin on skeletal myfibers of intact animals are cell autonomous, but additional studies will be required to verify this conclusion.
Calcineurin-dependent signaling in cardiac and skeletal muscles
Very recently, a calcineurin-dependent transcriptional pathway was shown to promote hypertrophic growth of the heart (Molkentin et al. 1998). In cardiac myocytes, this pathway was shown to involve collaborative interactions between activated NFAT proteins and GATA4, a cardiac-restricted transcription factor not present in skeletal muscle. Cardiomyocytes express several isoforms of MEF2, and many of the same genes that exhibit slow, oxidative fiber type-specific expression in skeletal muscle are transcriptionally active in cardiac myocytes [e.g., myoglobin or sMtCK (Parsons et al. 1993; Levitt et al. 1995; Qin et al. 1997)]. Thus, the functional interactions between NFAT and MEF2 proteins suggested by our current data in skeletal muscle are likely to be pertinent to the myocardium as well. Future studies will determine whether hypertrophic growth of skeletal muscles also can be stimulated by calcineurin-triggered signaling events, and should identify the molecular mechanisms by which overlapping, but distinctive, sets of genes are regulated by calcineurin in skeletal and cardiac myocytes.
Medical implications of calcineurin-regulated programs of gene expression in skeletal muscles
The discovery of a calcineurin-dependent pathway linked to specialization of myofiber subtypes reveals a potential for immunosuppressant drugs in current clinical use to modify skeletal muscle physiology and exercise performance in patients receiving such therapy. Our model is supported by a previous study in which pharmacologic blockade of calcineurin signaling in rats reduced the capacity for endurance exercise and diminished peak rates of oxidative phosphorylation in mitochondria isolated from skeletal muscles (Mercier et al. 1995). Clinical investigations to determine the degree to which changes in skeletal muscles account for unwanted side effects of immunosuppressant therapy are warranted.
In the long term, the discovery of a calcineurin-regulated pathway controlling fiber specialization in skeletal muscles could lead to novel strategies to enhance human health. For example, slow oxidative fibers are relatively resistant to the progressive myonecrosis that occurs with advancing age in individuals lacking dystrophin (Duchenne’s muscular dystrophy) (Webster et al. 1988). A strategy to promote fast-to-slow fiber transformation in these patients may reduce morbidity and prolong life until definitive gene therapy procedures can be developed. Patients with congestive heart failure, regardless of the primary cause, exhibit loss of slow, oxidative myofibers in their skeletal muscles (Massie et al. 1988; Sabbah et al. 1993), an abnormality that contributes to exercise intolerance in these individuals. A strategy to reverse this process could improve the quality of life of individuals suffering from heart failure, even if cardiac performance remains impaired (Sullivan et al. 1989). A decline in slow fibers is observed as a result of prolonged inactivity or hypogravity (Caiozzo et al. 1994), and the fiber composition of skeletal muscles influences insulin sensitivity (Kong et al. 1994) and lipoprotein metabolism (Tikkanen et al. 1996). Drugs or gene therapies capable of modifying calcineurin activity selectively in skeletal muscles plausibly could be used to increase the capacity for endurance exercise and to reduce risk for life-threatening diseases in human subjects.
Materials and methods
Cell culture and transfection conditions
NIH-3T3 cells or C2C12 myogenic cells were cultured, transfected with plasmid vectors, and assayed for luciferase and β-galactosidase, as described previously (Grayson et al. 1995, 1998). Each 35-mm dish of cells was cotransfected with promoter–reporter plasmid (0.5 μg), an expression plasmid that uses the CMV promoter to force expression of a constitutively active form of calcineurin (Manalan and Klee 1983; O’Keefe et al. 1992), or empty vector (pCI–NEO; 0.5 μg), along with a CMV–lacZ plasmid (0.5 μg) as an internal control for transfection efficiency. For dose-response experiments (Fig. 2), the total input DNA and the amount of promoter–reporter plasmid and CMV–lacZ was held constant, but the ratio of calcineurin expression vector to empty vector was varied.
Plasmid constructions
The expression plasmid used to stimulate calcineurin-regulated gene transcription was constructed by linking a CMV promoter carried in pCI–NEO (Promega) to a truncated variant of calcineurin A from which the carboxy-terminal region containing the autoinhibitory domain and a portion of the calmodulin-binding domain was deleted (O’Keefe et al. 1992). This form of calcineurin exhibits constitutive phosphatase activity, and is not subject to regulation by calcium-calmodulin in the manner of the native protein (O’Keefe et al. 1992). Promoter–reporter constructs were designed by linking the luciferase gene carried in pGL3 (Promega) to upstream promoter regions from the Mb, TnIs, and MCK genes, each of which have been shown previously to recapitulate the expression pattern of the respective endogenous genes when linked to a reporter gene and introduced into transgenic mice (Parsons et al. 1993; Levitt et al. 1995; Shield et al. 1996).
Other promoter–reporter plasmids used as controls (CMV–luciferase; TATA–luciferase; CMV–lacZ), or to identify upstream regulatory elements involved in transducing the signal derived from activated calcineurin (Mb380; MbΔA/T; MbΔCCAC; CCAC-TATA; A/T-TATA; CCAC-A/T-TATA) have been described in previous publications from this laboratory (Bassel-Duby et al. 1993; Grayson et al. 1995, 1998). Reporter constructions bearing five copies of the upstream NFAT response element from the myoglobin promoter (NRE-TATA and NRE-CCAC-A/T-TATA) were based on the oligonucleotide sequence 5′-AACCAGGAAATAGGATGCCCT-3′, and its complementary strand, representing nucleotide positions −694 to −674 in the human myoglobin promoter (underlined bases illustrate the NFAT consensus binding motif).
Putative NFAT binding sites within the myoglobin and troponin I slow promoters were disrupted using a PCR-based mutagenesis procedure, as described (Yang et al. 1997). The specific nucleotide sequence modifications included: myoglobin promoter (−690) AGGAAATA to GTCGACTA and (−232, reverse strand) TGGAAAGA to CTCGAGGA; TnI slow promoter (−738) AGGAAAC to AGCTAGC and (−639) TGGAAACA to ACTAGTCA.
Plasmids used to express NFAT–GFP fusion proteins were constructed in pEGFP–N1 (Clontech), using cDNA sequences encoding full-length (amino acids 1–716) or truncated (amino acids 319–716) NFATc (Northrop et al. 1994), modified at the carboxyl terminus for fusion in the correct reading frame to GFP. In the construct designed to express the truncated NFATc–GFP fusion protein (ΔNFATc–GFP), the native leucine residue at position 319 was converted to a methionine initiation codon. In both NFATc–GFP and ΔNFATc–GFP, the native stop codon was replaced with a 7 amino acid insertion preceeding the GFP coding sequence.
Fluorescence microscopy
An Olympus IMT-2 inverted fluorescence photomicroscope with FITC illumination and detection was used for evaluation and photography of C2C12 cells transfected with GFP expression plasmids. GFP fluorescence (excitation peak = 488 nm, emission peak = 507 nm) was photographed with Kodak Elite II 400 ASA slide film using an Olympus SC35 SLR camera back.
Histochemical analysis of fiber type in muscles from intact animals
Adult rats were treated with cyclosporin A (5 mg/kg) or vehicle administered by intraperitoneal injection daily for 6 weeks. Animal care was in accordance with institutional guidelines. Sections of soleus muscles from 7 animals in each group were stained histochemically for myosin ATPase activity at pH 4.54, as described (Brooke and Kaiser 1970). The proportion of fast and slow fibers was quantified by three observers who were blinded to the treatment status of the animals. Fibers expressing fast myosin were identified in 8 μm cryosections sections of the same muscles, postfixed in 4% paraformaldehype, by immunohistochemical analysis using a commercially available mouse monoclonal antibody (MY-32: Sigma, St. Louis, MO; 1:400) and LRSC goat anti-mouse IgG (Jackson Immunochemicals, West Grove, PA; 1:50). Nuclei were counterstained with Hoechst 33342 (Molecular Probes, Eugene, OR) at 0.6 μg/ml for 10 min.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants AR40849 and HL06296. Support for E.N.O. was provided by NIH grant HL49953. We are grateful to Robert C. Webb for technical contributions. Muscle tissues from cyclosporin A-treated and control rats were provided generously by Weiguo Zhang and Ron Victor.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL williams@ryburn.swmed.edu; FAX (214) 648-1450.
References
- Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan GG. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Grohe CM, Jessen ME, Parsons WJ, Richardson JA, Chao R, Grayson J, Ring WS, Williams RS. Sequence elements required for transcriptional activity of the human myoglobin promoter in intact myocardium. Circ Res. 1993;73:360–366. doi: 10.1161/01.res.73.2.360. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Booth FW, Baldwin KM. Muscle plasticity: Energy demand and supply processes. In: Rowell LB, Shepard JT, editors. The handbook of physiology: Integration of motor, circulatory, respiratory and metabolic control during exercise. Bethesda, MD: American Physiology Society; 1996. pp. 1075–1123. [Google Scholar]
- Brooke MH, Kaiser KK. Muscle fiber types: How many and what kind? Arch Neurol. 1970;23:369–379. doi: 10.1001/archneur.1970.00480280083010. [DOI] [PubMed] [Google Scholar]
- Caiozzo VJ, Baker MJ, Herrick RE, Tao M, Baldwin KM. Effect of spaceflight on skeletal muscle: Mechanical properties and myosin isoform content of a slow antigravity muscle. J Appl Physiol. 1994;76:1764–1773. doi: 10.1152/jappl.1994.76.4.1764. [DOI] [PubMed] [Google Scholar]
- Cameron AM, Steiner JP, Roskams AJ, Ali SM, Ronnett GV, Snyder SH. Calcineurin associated with the inositol 1,4,5-trisphosphate receptor-FKBP12 complex modulates Ca2+ flux. Cell. 1995;83:463–472. doi: 10.1016/0092-8674(95)90124-8. [DOI] [PubMed] [Google Scholar]
- Chin ER, Allen DG. Changes in intracellular free Ca2+ concentration during constant 10 Hz stimulation of mouse single skeletal muscle fibres. Physiologist. 1996;39:A–75. [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Devlin BH, Wefald FC, Kraus WE, Bernard TS, Williams RS. Identification of a muscle-specific enhancer within the 5′ flanking region of the human myoglobin gene. J Biol Chem. 1989;264:13896–13900. [PubMed] [Google Scholar]
- DiMario JX, Fernyak SE, Stockdale FE. Myoblasts transferred to the limbs of embryos are committed to specific fibre fates. Nature. 1993;362:165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Bassel-Duby RS, Richardson JA, Grayson J, Neufer PD, Williams RS. Postnatal development and plasticity of specialized muscle fiber characteristics in the hindlimb. Dev Genet. 1996;19:146–156. doi: 10.1002/(SICI)1520-6408(1996)19:2<146::AID-DVG6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Grayson, J., R. Bassel-Duby, and R.S. Williams. 1998. Transcriptional synergism in muscle-specific gene regulation: Physical and functional interactions between MEF-2 and Sp1. J. Cell. Biochem. (in press). [PubMed]
- Grayson J, Williams RS, Yu YT, Bassel-Duby R. Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol Cell Biol. 1995;15:1870–1878. doi: 10.1128/mcb.15.4.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig R, Lomo T. Firing patterns of motor units in normal rats. Nature. 1985;314:164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- Hoey T, Sun YL, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol. 1984;56:831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Koishi K, Rudnicki M, Maggs AM. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mech Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Ianuzzo CD, Hamilton N, Li B. Competitive control of myosin expression: hypertrophy vs. Hyperthyroidism. J Appl Physiol. 1991;70:2328–2830. doi: 10.1152/jappl.1991.70.5.2328. [DOI] [PubMed] [Google Scholar]
- Izumo S, Mahdavi V. Thyroid hormone receptor alpha isoforms generated by alternative splicing differentially activate myosin HC gene transcription. Nature. 1988;334:539–542. doi: 10.1038/334539a0. [DOI] [PubMed] [Google Scholar]
- Kong X, Manchester J, Salmons S, Lawrence JC. Glucose transporters in single skeletal muscle fibers. J Biol Chem. 1994;269:12963–12967. [PubMed] [Google Scholar]
- Lam E, Martin MM, Timerman AP, Sabers C, Fleischer S, Lukas T, Abraham RT, O’Keefe SJ, O’Neill EA, Wiederrecht GJ. A novel FK506 binding protein can mediate the immunosuppressive effects of FK506 and is associated with the cardiac ryanodine receptor. J Biol Chem. 1995;270:26511–26522. doi: 10.1074/jbc.270.44.26511. [DOI] [PubMed] [Google Scholar]
- Levitt LK, O’Mahoney JV, Brennan KJ, Joya JE, Zhu L, Wade R, Hardeman E. The human troponin I slow promoter directs slow fiber-specific expression in transgenic mice. DNA Cell Biol. 1995;14:599–607. doi: 10.1089/dna.1995.14.599. [DOI] [PubMed] [Google Scholar]
- Li L, Zhou J, James G, Heller-Harrison R, Czech MP, Olson EN. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell. 1992;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP–FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Manalan AS, Klee CB. Activation of calcineurin by limited proteolysis. Proc Natl Acad Sci. 1983;80:4291–4295. doi: 10.1073/pnas.80.14.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie BM, Conway M, Rajagopalan B, Yonge R, Frostick S, Ledingham J, Sleight P, Radda G. Skeletal muscle metabolism during exercise under ischemic condition in congestive heart failure: evidence for abnormalities unrelated to blood flow. Circulation. 1988;78:320–326. doi: 10.1161/01.cir.78.2.320. [DOI] [PubMed] [Google Scholar]
- Mercier JG, Hokanson JF, Brooks GA. Effects of cyclosporine A on skeletal muscle mitochondrial respiration and endurance time in rats. Am J Resp Crit Care Med. 1995;151:1532–1536. doi: 10.1164/ajrccm.151.5.7735611. [DOI] [PubMed] [Google Scholar]
- Michel JB, Ordway GA, Richardson JA, Williams RS. Biphasic induction of immediate early genes accompanies activity-dependent angiogenesis and myofiber remodeling of rabbit skeletal muscle. J Clin Invest. 1994;94:277–285. doi: 10.1172/JCI117318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Lu J, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:1–20. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements. Mol Cell Biol. 1996;16:2408–2417. doi: 10.1128/mcb.16.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufer PD, Ordway GA, Hand GA, Shelton JM, Richardson JA, Benjamin IJ, Williams RS. Continuous contractile activity induces fiber type specific expression of HSP70 in skeletal muscle. Am J Physiol. 1996;271:C1828–C1837. doi: 10.1152/ajpcell.1996.271.6.C1828. [DOI] [PubMed] [Google Scholar]
- Northrop JP, Ho SN, Chen L, Thomas DJ, Timmerman LA, Nolan GP, Admon A, Crabtree GR. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- O’Keefe SJ, Tamura J, Kincaid RL, Tocci MJ, O’Neill EA. FK-506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature. 1992;357:692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- Ontell MP, Sopper MM, Lyons G, Buckingham M, Ontell M. Modulation of contractile protein gene expression in fetal murine crural muscles:emergence of muscle diversity. Dev Dynm. 1993;198:203–213. doi: 10.1002/aja.1001980306. [DOI] [PubMed] [Google Scholar]
- Parsons WJ, Richardson JA, Graves KH, Williams RS, Moreadith RW. Gradients of transgene expression directed by the human myoglobin promoter in the developing mouse heart. Proc Natl Acad Sci. 1993;90:1726–1730. doi: 10.1073/pnas.90.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pette D, Vrbova G. Adaptation of mammalian skeletal muscle fibers to chronic electrical stimulation. Rev Physiol Biochem Pharmacol. 1992;120:115–202. doi: 10.1007/BFb0036123. [DOI] [PubMed] [Google Scholar]
- Qin W, Khuchua Z, Klein SC, Strauss AW. Elements regulating cardiomyocyte expression of the human sarcomeric mitochondrial creatine kinase gene in transgenic mice. J Biol Chem. 1997;272:25210–25216. doi: 10.1074/jbc.272.40.25210. [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Sabbah HN, Hansen-Smith F, Sharov VG, Kono T, Lesch M, Gengo PJ, Steffen RP, Levine TB, Goldstein S. Decreased proportion of type I myofibers in skeletal muscle of dogs with chronic heart failure. Circulation. 1993;87:1729–1737. doi: 10.1161/01.cir.87.5.1729. [DOI] [PubMed] [Google Scholar]
- Saltin B, Gollnick PD. Skeletal muscle adaptability: Significance for metabolism and performance. In: Peachey LDD, editor. Handbook of physiology: Skeletal muscle. Bethesda, MD: American Physiological Society; 1983. pp. 555–632. [Google Scholar]
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- Shield MA, Haugen HS, Clegg CH, Hauschka SD. E-box sites and a proximal regulatory region of the muscle creatine kinase gene differentially regulate expression in diverse skeletal muscles and cardiac muscle of transgenic mice. Mol Cell Biol. 1996;16:5058–5068. doi: 10.1128/mcb.16.9.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreter FA, Lopez JR, Alamo L, Mabuchi K, Gergely J. Changes in intracellular ionized Ca concentration associated with muscle fiber type transformation. Am J Physiol. 1987;253:C296–C300. doi: 10.1152/ajpcell.1987.253.2.C296. [DOI] [PubMed] [Google Scholar]
- Stockdale FE. Mechanisms of formation of muscle fiber types. Cell Struct Funct. 1997;22:37–43. doi: 10.1247/csf.22.37. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Higginbotham MB, Cobb FR. Exercise training in patients with chronic heart failure delays ventilatory anaerobic threshold and improves submaximal exercise performance. Circulation. 1989;79:324–329. doi: 10.1161/01.cir.79.2.324. [DOI] [PubMed] [Google Scholar]
- Tikkanen HO, Naveri H, Harkonen M. Skeletal muscle fiber distribution influences serum high-density lipoprotein cholesterol level. Atherosclerosis. 1996;120:1–5. doi: 10.1016/0021-9150(95)05652-1. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Clipstone NA, Ho SN, Northrop JP, Crabtree GR. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- Vrbova G. The effect of motoneurone activity on the speed of contraction of striated muscle. J Physiol (London) 1963;169:513–526. doi: 10.1113/jphysiol.1963.sp007276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP, Busaan JL, Fraser ED, Fu SY, Pato MD, Michalak M. Characterization of the recombinant C-terminal domain of dystrophin: Phosphorylation by calmodulin-dependent protein kinase II and dephosphorylation by type 2B protein phosphatase. Biochemistry. 1995;34:5561–5568. doi: 10.1021/bi00016a030. [DOI] [PubMed] [Google Scholar]
- Webster C, Silberstein L, Hays AP, Blau HM. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52:503–513. doi: 10.1016/0092-8674(88)90463-1. [DOI] [PubMed] [Google Scholar]
- Westerblad H, Allen DG. Changes in myoplasmic calcium concentration during fatigue in single mouse muscle fibers. J Gen Physiol. 1991;98:615–635. doi: 10.1085/jgp.98.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Neufer PD. Regulation of gene expression in skeletal muscle by contractile activity. In: Rowell LB, Shepard JT, editors. The handbook of physiology: Integration of motor, circulatory, respiratory and metabolic control during exercise. Bethesda, MD: American Physiology Society; 1996. pp. 1124–1150. [Google Scholar]
- Williams RS, Salmons S, Newsholme EA, Kaufman RE, Mellor J. Regulation of nuclear and mitochondrial gene expression by contractile activity in skeletal muscle. J Biol Chem. 1986;261:376–380. [PubMed] [Google Scholar]
- Yamashita K, Yoshioka T. Profiles of creatine kinase isoenzyme compositions in single muscle fibres of different types. J Muscle Res Cell Motil. 1991;12:37–44. doi: 10.1007/BF01781172. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bassel-Duby R, Williams RS. Transient expression of a winged-helix protein, MNF-β, during myogenesis. Mol Cell Biol. 1997;17:5236–5243. doi: 10.1128/mcb.17.9.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]