Abstract
Mesodermal development is a multistep process in which cells become increasingly specialized to form specific tissue types. In Drosophila and mammals, proper segregation and patterning of the mesoderm involves the bHLH factor Twist. We investigated the activity of a Twist-related factor, CeTwist, during Caenorhabditis elegans mesoderm development. Embryonic mesoderm in C. elegans derives from a number of distinct founder cells that are specified during the early lineages; in contrast, a single blast cell (M) is responsible for all nongonadal mesoderm formation during postembryonic development. Using immunofluorescence and reporter fusions, we determined the activity pattern of the gene encoding CeTwist. No activity was observed during specification of mesodermal lineages in the early embryo; instead, the gene was active within the M lineage and in a number of mesodermal cells with nonstriated muscle fates. A role for CeTwist in postembryonic mesodermal cell fate specification was indicated by ectopic expression and genetic interference assays. These experiments showed that CeTwist was responsible for activating two target genes normally expressed in specific subsets of nonstriated muscles derived from the M lineage. In vitro and in vivo assays suggested that CeTwist cooperates with the C. elegans E/Daughterless homolog in directly activating these targets. The two target genes that we have studied, ceh-24 and egl-15, encode an NK-2 class homeodomain and an FGF receptor (FGFR) homolog, respectively. Twist activates FGFR and NK-homeodomain target genes during mesodermal patterning of Drosophila and similar target interactions have been proposed to modulate mesenchymal growth during closure of the vertebrate skull. These results suggest the possibility that a conserved pathway may be used for diverse functions in mesodermal specification.
Keywords: CeTwist, C. elegans, ceh-24, egl-15, mab-5, CeE/DA, NdE box
Caenorhabidtis elegans has an intricate and reproducible pattern of mesodermal cell types, including striated muscles, nonstriated muscles, and nonmuscle cells (Waterston 1988; Moerman and Fire 1997). The striated muscles are the most numerous: these attach to the outer body wall and contract longitudinally to allow movement of the animal (Waterston 1988). Nonstriated muscles are a more diverse group of single sarcomere cells that include pharyngeal muscles (essential for ingestion and grinding of food) intestine-associated muscles (defecation), uterine and vulval muscles (egg laying), and male-tail associated muscles (mating behavior). Nonmuscle mesodermal cells include six coelomocytes: oblong mesodermal cells that inhabit the body cavity and derive from a mesodermal lineage but show no characteristics of muscle (Sulston and Horvitz 1977; Sulston et al. 1983). Although the function of the coelomocytes is unclear, their ability to take up a variety of molecules from the coelomic fluid suggests a scavenger function akin to that of macrophages (Chitwood and Chitwood 1974; Fire et al. 1998a).
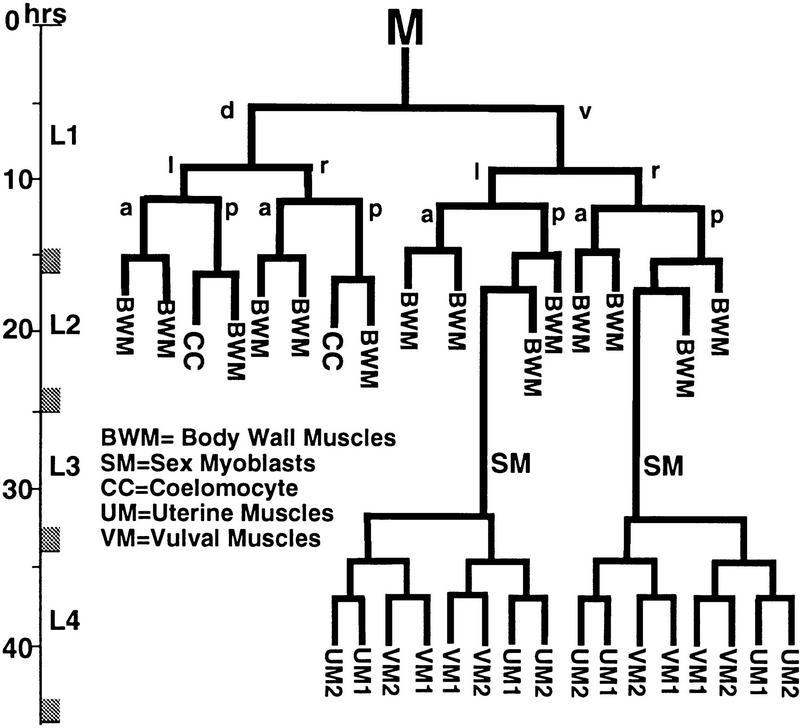
Different aspects of the mesodermal pattern in C. elegans are specified during embryonic and postembryonic development. Mesodermal cells produced during embryogenesis include the majority of body wall muscles (81), the four intestine-associated muscles, and four coelomocytes (Sulston et al. 1983). These cells are all retained throughout postembryonic development, with each cell growing in size to accommodate the dramatic growth of the animal. During postembryonic development a single mesodermal blast cell (M) divides to produce a small number of additional mesodermal cells (Sulston and Horvitz 1977). In hermaphrodites, M divisions in early larval development yield 14 striated body wall muscles, two sex myoblasts (SMs), and two coelomocytes (Sulston and Horvitz 1977; see Fig. 1). The SMs, which are born in the posterior of L2 animals, migrate toward the anterior of the animal during later development to eventually flank the center of the developing gonad (Sulston and Horvitz 1977). There they divide to produce the 16 sex muscles (8 vulval and 8 uterine muscles) used in hermaphrodite egg laying.
Figure 1.
Postembryonic mesodermal lineage M in C. elegans. Times indicated are post-hatching at 25°C (lineage redrawn from Sulston and Horvitz 1979).
Studies of mesodermal development in vertebrate systems and Drosophila have been remarkably effective in identifying molecules and interactions that lead to cell-type specification and to overall tissue patterning. The vertebrate and Drosophila systems offer only a limited knowledge of the mesodermal cell lineage and are thus less amenable to an understanding of mesodermal pattern formation at single-cell resolution. The availability of a full cell lineage for C. elegans raises the hope that we might be able to understand the formation of a relatively simple mesodermal pattern on a single-cell mechanistic level. Realization of this goal will require that we identify many or all of the components involved in specification of the mesodermal pattern in C. elegans and characterize their precise expression patterns, their roles in cell-type specification, and their functional interactions with other components of the specification machinery.
In this report, we describe a gene activation pathway that plays a critical role in generating patterns of nonstriated muscles during the postembryonic phase of C. elegans mesoderm development. This pathway is deduced from an analysis of promoter and enhancer function during the specification of cell type, and is shown to involve four transcription factors: the Antennapedia class homeodomain factor MAB-5 (Kenyon 1986; Costa et al. 1988), a novel C. elegans homolog of the Twist family, the C. elegans E/Daughterless (E/DA) homolog (Krause et al. 1997), and a member of the NK-2 homeodomain family (Harfe et al. 1998).
Results
Fine structure analysis of a nonstriated muscle enhancer
Regulation of the ceh-24 promoter provided us a means to study transcriptional activation in mesodermal cells with nonstriated muscle character (Harfe and Fire 1998). Functional analysis of the upstream region had identified a 22-bp fragment that was sufficient (when concatamerized) for enhancer activity in M-derived vulval, uterine, and intestine-associated muscles (Harfe and Fire 1998). This 22-bp sequence contains two E boxes; we have named these motifs NdE boxes for the NdeI restriction enzyme that recognizes this sequence (CATATG; Harfe and Fire 1998). Further characterization of the 22-bp enhancer element was carried out by use of a detailed point mutational analysis (Fig. 2). This analysis revealed the following: (1) The first base pairs of two NdE-box motifs were necessary for activity (Fig. 2A, 1m); (2) Spacing was important for activity. Insertion of 21 bp between NdE boxes abolished activity (Fig. 2A,11m); (3) Activity was abolished by mutations that changed several base pairs flanking the NdE boxes (Fig. 2A, 3m, 4M); (4) Mutation of both NdE boxes to a consensus MyoD-binding site, CAGCTG, eliminated enhancer activity (Fig. 2A, 10m).
Figure 2.
Sequence requirements for in vivo activation by the NdE-box motif. (A) Wild-type and mutated forms of the ceh-24 NdE-box vulval muscle enhancer were concatamerized and assayed for activation of a truncated pes-10 promoter fragment. Activity was measured using a lacZ or GFP reporter construct. The pes-10 promoter used in this assay is not intrinsically active, but can be activated in a variety of tissues by use of additional enhancer sequences for activation (A. Fire, S. Xu, and G. Seydoux, pers. comm.). NdE boxes are underlined. Mutations are indicated with lowercase letters. All constructs were injected into wild-type hermaphrodite animals and were assayed in at least two independent transgenic lines. A wild-type ceh-24 NdE box enhancer assayed in front of the deleted pes-10 promoter was capable of activating reporter transcription only when more than two NdE boxes were present. ceh-24 NdE box enhancer mutations 1m, 2m, 4m, 5m, 8m, 11m, and all wild-type ceh-24 NdE box enhancers were assayed with both lacZ and GFP reporter genes. All others were assayed with lacZ. A construct containing nine copies of a wild-type ceh-24 NdE box enhancer, in addition to vulval, intestinal, uterine, and anal depressor muscle expression had additional expression in one cell in the head. This cell has been tentatively identified as the head mesodermal cell. Expression in this cell has not been observed with any other construct. (B) Summary of NdE box mutations. The two NdE boxes are underlined. Base pairs that were required for reporter expression are boxed. Base pairs highlighted in gray resulted in loss of expression only when mutated concurrently. Separate mutation of these base pairs had no evident effect on reporter expression (Fig. 2A, 5m, 6m, 7m, and 8m).
Figure 3.
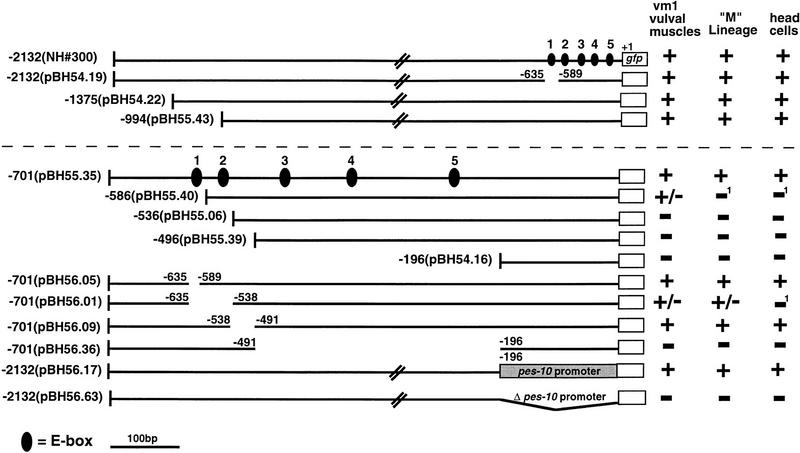
Cis-acting elements controlling egl-15 promoter activity. Deletion constructs derived from an egl-15::gfp translational fusion are diagrammed. All constructs were tested for activity in at least two independent lines. Larvae, embryos, and adults were examined for expression of GFP in early M lineage descendants, head neurons, and vm1 vulval muscles, respectively. (+) GFP expression levels similar to that of the full-length egl-15 promoter construct (NH#300). (−) no observed reporter expression; (+/−) expression that was only marginally detectable. Each construct contained the egl-15 ATG codon (+1) and was fused in-frame with GFP. NH#300 was a gift of C. Branda and M. Stern (Yale University, New Haven, CT). Construct pBH56.17 contained the pes-10 promoter in place of the first 196 bp of the egl-15 promoter. The five NdE-like E boxes are numbered 1 (−602: taaCATATGcac), 2 (−572: ctcCAGATGtga), 3 (−471: taaCATCTGgac); 4 (−371: caaCATATGcgt), and 5 (−239: ttaCATATGgtg). 1Reporter expression in the vm1 vulval muscles of constructs pBH55.40 and pBH56.01 was greatly reduced relative to the full length egl-15::gfp fusion. A similar decrease in M lineage activity (which is much less evident in the parent construct) would likely result in a complete failure to detect the reporter.
Figure 4.
bHLH comparisons among Twist family members and related factors. Amino acids identical to CeTwist are indicated by dashes. Sources for bHLH domain sequence are: Dtwist (Drosophila, Thisse et al. 1988); Mtwist; (Mus, Wolf et al. 1991); Htwist (Human, Howard et al. 1997; Ghouzzi et al. 1997); Xetwist (Xenopus, Hopwood et al. 1989). Dermo-1 (Mus, Li et al. 1995), and dHand (Mus, Hollenberg et al. 1995; Srivastava et al. 1995). No close homolog of CeTwist exists in the C. elegans genome database (80% completion; 5/98). On the basis of BLAST similarity searches, the distantly-related CeMyoD (Krause et al. 1990) may be the nearest homolog of CeTwist in C. elegans.
Figure 10.
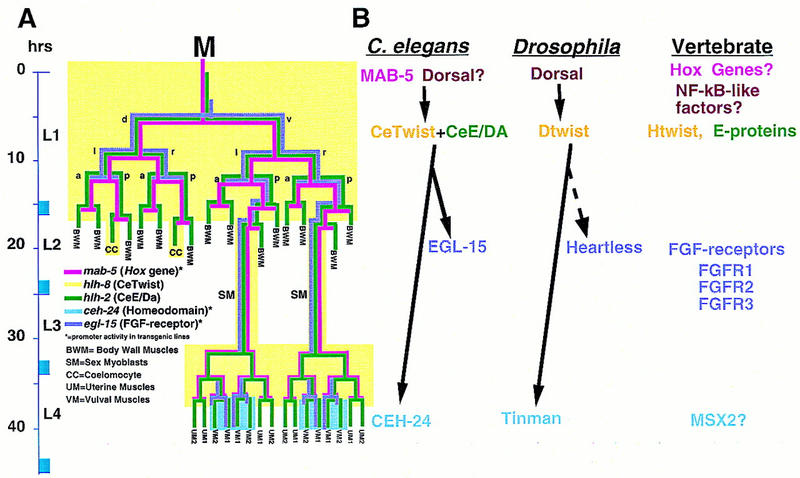
Aspects of a mesodermal patterning pathway are conserved. (A) A proposed pathway of genes active in the M lineage. Expression patterns for mab-5 and hlh-2 are derived from antibody staining (S. Salser and C. Kenyon, unpubl.; Krause et al. 1997, this work). mab-5 expression may extend to later stages in the SM lineages. Expression patterns for ceh-24 and egl-15 are predicted from reporter constructs (Harfe and Fire 1998; C. Branda and M. Stern, unpubl.). The hlh-8 pattern is inferred from both types of data (see text). The proposed pathway, inferred from genetic and interference experiments as described in the text, includes a requirement for MAB-5 in the early activation of hlh-8 and a role for the resulting CeTwist–CeE/DA heterodimer in the activation of ceh-24 and egl-15. Also as expected from this model (data not shown), MAB-5 was required for early (but not late) M lineage activity of egl-15 reporter constructs. (B) Comparative aspects of a Twist-dependent mesodermal diversification pathway. The vertical location of each protein name corresponds to the proposed time of action. In Drosophila, Dorsal can directly activate Dtwist by binding NF-κB/Dorsal sites upstream of Dtwist (Thisse et al. 1991). Dtwist can activate heartless (FGFR) expression (Shishido et al. 1997), although it is not known if this interaction is direct (dashed arrow). hlh-8 requires a consensus NF-κB/Dorsal site for promoter activity. No dorsal gene has been identified in C. elegans. Dominant mutations in human Twist (Htwist), FGFRs, and the homeodomain MSX2 can result in craniosynostosis (Jabs et al. 1993; Muenke et al. 1994; Meyers et al. 1995; Przylepa et al. 1996; Ghouzzi et al. 1997; Howard et al. 1997).
Requirements for activity of the egl-15 promoter in the M lineage
egl-15 encodes a member of the FGF receptor (FGFR) family that is required for the proper migration of the SMs (Stern and Horvitz 1991; DeVore et al. 1995). The egl-15 promoter is active in many early M lineage descendants and later in the four vm1 vulval muscles (C. Branda and M. Stern, unpubl.). Although this promoter activity pattern is distinct from that of ceh-24, the activity of each in the later M lineage suggested the possibility of a common factor specifying M lineage activity of the two genes.
An egl-15 promoter fragment of 701 bp was sufficient to drive reporter expression in the M lineage (Fig. 3, pBH55.35). This fragment contained five matches to the E-box consensus. Three were precise NdE boxes (CATATG), whereas the other two differed from this consensus by a single base pair (Fig. 3). Deletion analysis suggested critical roles for the E-box motif and for additional elements in egl-15 promoter activity: Specifically, M-lineage activity was eliminated by promoter truncations that removed the first two NdE-like boxes (Fig. 3, pBH55.06 and pBH55.39), and by a deletion that removed the three proximal NdE-like boxes (Fig. 3, pBH56.36).
C. elegans contains a member of the twist family
A sequence similar to the NdE box has been reported in Drosophila to be capable of binding the bHLH protein Twist (Ip et al. 1992; Lee et al. 1997; Yin et al. 1997). The analysis of the ceh-24 vulval muscle enhancer and the egl-15 promoter indicated a possible role for twist bHLH family members in specification of nonstriated muscle. In a search of the C. elegans genome database (currently at 80% completion), we identified one candidate twist homolog. The identified sequence contained a bHLH region that was more similar to the bHLH region of the twist family than to any known C. elegans bHLH gene (Fig. 4). In keeping with standard nomenclature (Krause et al. 1997), this gene has been named hlh-8 (helix–loop–helix). The protein product will be referred to as CeTwist. Within the bHLH domain, CeTwist shared 59%–63% identity to published Twist family members in other species. Outside of the bHLH domain, there was no obvious homology between CeTwist and other Twist family members.
CeTwist is present in postembryonic mesodermal cells
To analyze the spatial pattern of hlh-8 activity, we employed a series of hlh-8 reporter fusions with GFP- and lacZ-coding regions and a set of polyclonal antisera prepared against full-length CeTwist protein.
We observed a consistent pattern of expression with a variety of reporter fusions containing extensive sequences surrounding hlh-8 (Fig. 5). This pattern will be referred to as the hlh-8 promoter activity pattern. During embryogenesis, the most striking focus of hlh-8 promoter activity was in the M blast cell (Fig. 5A,B). This activity was first observed in threefold stage embryos, after M had finished its posterior migration. Expression of hlh-8::gfp chimeras was also evident after hatching in a set of rectum-associated cells that appeared to be defecation-associated muscles (this required the full-length hlh-8 genomic fragment; see Fig. 8A, below). During larval development, hlh-8 reporter constructs were active in M and all undifferentiated descendants of M (Fig. 5A–J). This activity was lost as cells differentiated into body wall or sex muscles.
Figure 5.
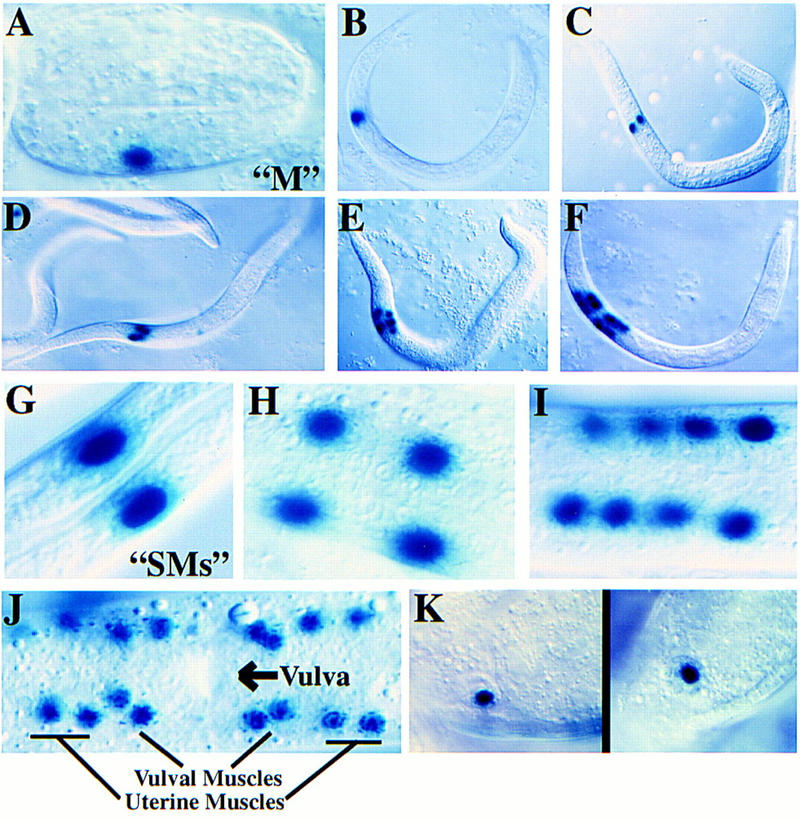
Activity pattern of hlh-8 reporter fusions. (A–K). Pattern of expression from an integrated hlh-8::lacZ reporter construct (pBH47.74; see Fig. 8). An identical expression pattern was observed with GFP as a reporter (data not shown). (A) Threefold embryo with M labeled; (B) L1 animal with M labeled; (C) M daughters; (D) M granddaughters; (E) eight descendants of the M blast cell; (F) eighteen descendants of the M blast cell; (G) SMs; (H–I) Reporter expression in sex myoblast descendants; (J) Ventral view of L4 larva with 16 sex muscles (8 vulval and 8 uterine) arranged around the vulva; (K) An hlh-8::lacZ fusion with additional 5′ sequences (pBH47.72, see Fig. 8) showed reporter activity in coelomocytes. The reporter activity in these cells was nuclear, and was seen with both hlh-8::lacZ and hlh-8::gfp fusion constructs. These two properties confirm that reporter expression was intrinsic to these cells (coelomocytes can also accumulate GFP secreted by other cells, but this process produces vesicular GFP and is not observed with lacZ; Fire et al. 1998b).
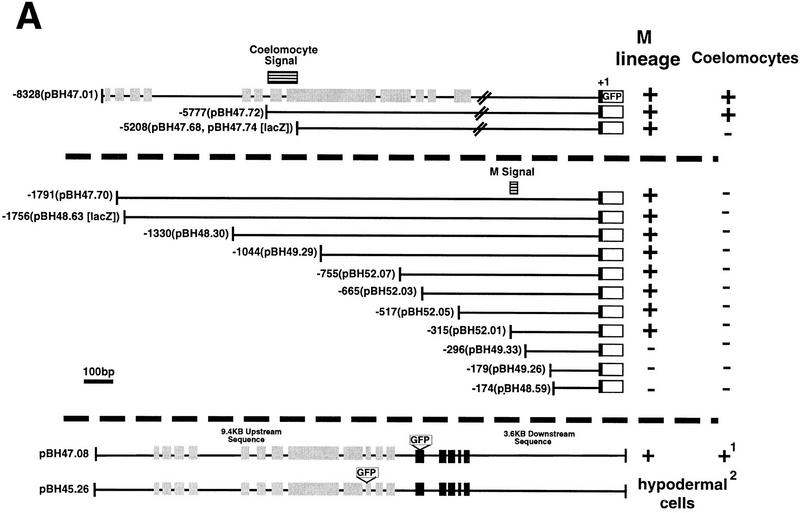
Figure 8.
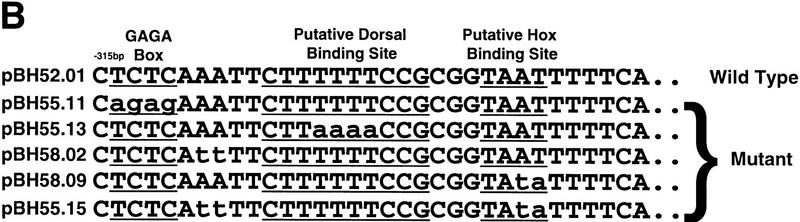
Cis-acting signals controlling hlh-8. All constructs were assayed in at least two independent transgenic lines. Reporter activity was scored in all M lineage cells and in all six coelomocytes (four embryonic and two postembryonic). (+) Reporter-positive cells, (−) reporter-negative cells. In − constructs reporter expression was absent in the entire M lineage or all coelomocytes. All translational fusion constructs contained the first nine amino acids of hlh-8 (black boxes) fused in frame to GFP. (A) Identification of hlh-8 cis-acting sequences. Gray boxes represent the entire predicted coding region of an uncharacterized putative gene upstream of hlh-8. Black boxes and white boxes represent hlh-8 and GFP coding regions, respectively. The location of DNA elements required for expression in coelomocytes and the M lineage are indicated (horizontal lined boxes). Two NdE box elements were found, at −128 (acgCATATGttg) and −48 (tttCATATGttt). 1Antibody staining indicated CeTwist protein accumulated in defecation-associated muscles. This activity was evident in tagged constructs with the complete CeTwist coding region (pPD47.08), but not in any of the hlh-8–reporter fusions. 2Insertion of GFP into an ORF (gray boxes) of the predicted gene upstream of hlh-8 produced reporter activity in hypodermal cells (pBH45.26; data not shown). (B) Point mutation analysis of a 21-bp segment essential for M-lineage activation of the hlh-8 promoter. All constructs contained 315 bp of DNA 5′ of the start of hlh-8 transcription and the first nine amino acids of hlh-8 fused in frame to GFP. A construct containing 315 bp of wild-type sequence was active in the M lineage (pBH52.01; Fig. 8A). Deletion of 21 bp of 5′ hlh-8 promoter sequence resulted in an inactive construct (pBH49.33; Fig. 8A). The GAGA motif, putative NF-κB site, and putative HOX-binding site are underlined. Mutationsare indicated with lowercase letters. All mutations tested abolished reporter activity.
The hlh-8 promoter remained active in the two M-derived coelomocytes throughout adult life. Starting in the L2 larval stage, activity was also seen in all four non-M-derived embryonic coelomocytes (Fig. 5K). These cells are born during embryogenesis (Sulston et al. 1983). Activity was detected in embryonic coelomocytes after the birth of both postembryonic M-derived coelomocytes. Additional hlh-8 reporter expression was observed in a group of head cells. These were likely to be neuronal, on the basis of position and morphology; their precise identities were not determined.
Immunofluorescence staining with antisera to CeTwist indicated an expression pattern similar to that seen with hlh-8 reporter fusions. CeTwist was first detected at the L1 stage in defecation-associated muscles and in a small number of neuron-like cells in the head. In later larvae, CeTwist was seen in SMs and in their descendants. As with the reporter fusions, no activity of CeTwist was observed in differentiated body wall or sex muscles. Although the antibody and reporter patterns coincided in a number of key aspects, several differences were evident. Expression in M and its descendants prior to the SM stage was detected with reporter fusions but not with antibody. Expression in mature coelomocytes was likewise only detected in reporter fusions. These differences could reflect the limited sensitivity of the antibody (see Materials and Methods) and/or negative control elements that were missing or inactive in the reporter transgenes.
The ceh-24 NdE-box enhancer contains a Twist-responsive control element
The expression of CeTwist precedes ceh-24 promoter activity in the developing vulval muscles during the L4 to adult molt. To test the possibility that CeTwist could be participating, either directly or indirectly, in the activation of the ceh-24 promoter, we used expression vectors to engineer expression of CeTwist in a variety of different C. elegans tissue types in vivo (see Materials and Methods). The resulting animals were assayed for ectopic activity of the ceh-24 22-bp NdE box enhancer. These experiments were carried out in a genetic background containing an integrated GFP reporter construct driven by the ceh-24 22-bp NdE box enhancer. GFP expression was then monitored in the presence of transgenes designed to misexpress CeTwist in diverse tissue types. We found that CeTwist was capable of inducing modest GFP expression from the integrated ceh-24 22-bp NdE box enhancer in most C. elegans tissues (Fig. 6).
Figure 6.
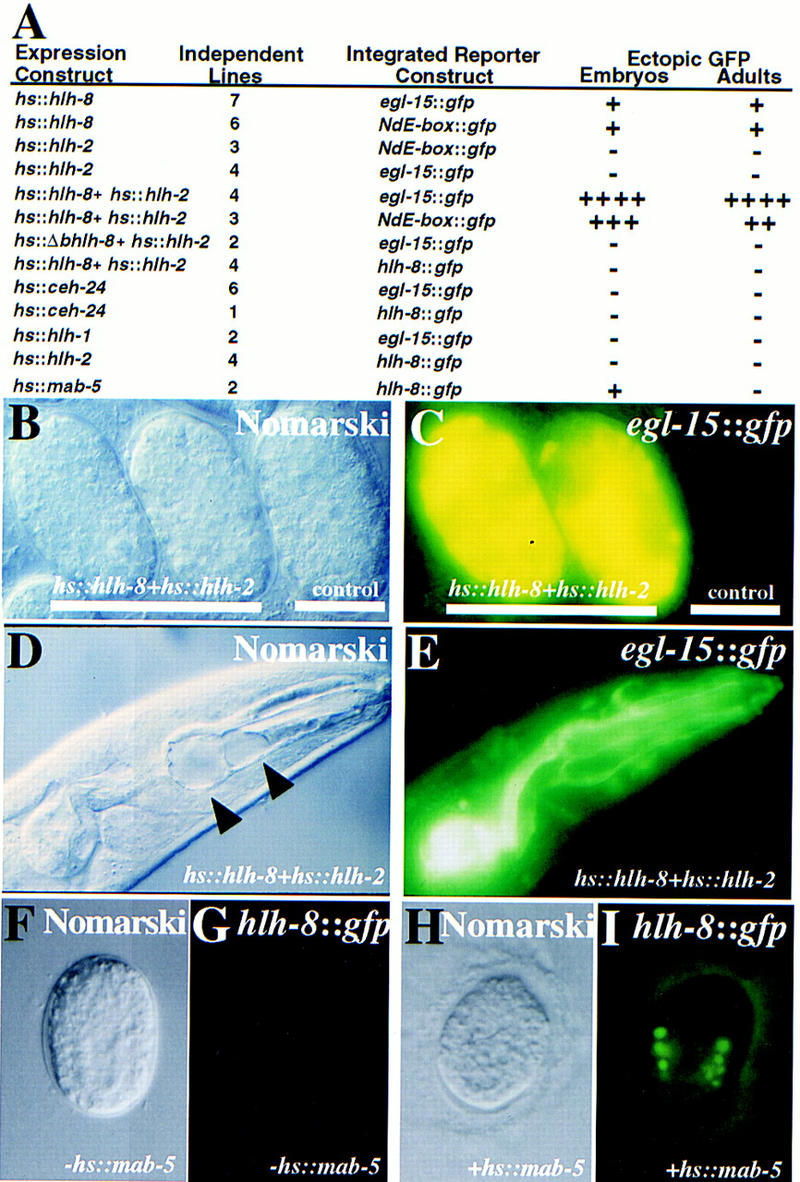
Consequences of ectopic CeTwist and CeE/DA expression. Ectopic heat shock expression constructs were injected into animals already containing an integrated GFP reporter construct (see Materials and Methods). Heritable extrachromosomal arrays of each ectopic expression construct were produced before phenotypic effects were assayed. Genes listed correspond to the following proteins: (hlh-8) CeTwist; (hlh-2) CeE/DA; (hlh-1) CeMyoD; (bhlh-8) CeTwist missing its basic domain and helix I. (A) Ectopic protein was produced from heat shock constructs by placement of stable transgenic lines at 37°C for 2 hr. Heat-shocked animals were allowed to recover at room temperature for >2 hr before being assayed for expression of the integrated GFP reporter. (+) Modest levels of GFP expression. Each additional + indicates a dramatic increase in GFP expression. Animals negative for ectopic reporter expression are indicated by a −. In heat-shocked animals with at least one +, reporter expression was seen in most cells of embryos and adults. (B,C) Example of hsp16-2::hlh-8+hsp16-2::hlh-2 embryos examined 4 hr after the initial heat shock. (B) Nomarski photograph. (C) CeTwist–CeE/DA driven ectopic expression from an egl-15::gfp integrated reporter construct. No expression was seen in embryos without the hs::hlh-8 construct. (D,E) Example of hsp16-2::hlh-8+hsp16-2::hlh-2 adult animals 24 hr after the initial heat shock. (D) Nomarski photograph. (E) CeTwist–CeE/DA driven ectopic expression from an egl-15::gfp integrated reporter construct. Large vacuoles were seen in the pharynx (arrowheads). (F–I) MAB-5-driven ectopic expression from an hlh-8::gfp reporter construct. An hsp16-1::mab-5 fusion construct (Salser et al. 1992) was introduced into a strains with an integrated hlh-8::gfp reporter. (F,G) Embryo in the absence of heat shock; (H,I) embro following heat shock.
Synergistic effects of CeTwist and CeE/DA
Transcription factors of the bHLH family are thought to function as dimers. These factors can homodimerize and in many cases heterodimerize with other members of the bHLH family (Murre et al. 1989; Lassar et al. 1991). One component of heterodimeric complexes in vertebrates is often a member of the ubiquitous family of bHLH factors called E proteins (Murre et al. 1989). In Drosophila, the Daughterless protein is the only known E family homolog (Cronmiller et al. 1988). Mouse and Drosophila Twist have been shown in vitro to heterodimerize with E/DA (Spicer et al. 1996). C. elegans contains a single known E/DA homolog, CeE/DA (Krause et al. 1997). CeE/DA is expressed in many tissues, including all undifferentiated M lineage cells (Krause et al. 1997; M. Krause, unpubl.). This suggested that CeE/DA and CeTwist might functionally heterodimerize in vivo.
To determine whether CeTwist and CeE/DA were capable of functioning together, we coexpressed the two proteins ectopically in C. elegans (see Materials and Methods). This resulted in a dramatic increase in ceh-24 NdE box enhancer activity, relative to induction by CeTwist alone (Fig. 6). CeE/DA was not capable of inducing transcription from the 22-bp ceh-24 NdE box enhancer element when expressed alone (Fig. 6A). Ectopic expression of either the C. elegans bHLH MyoD protein (CeMyoD; Krause et al. 1990) or an NK-2 class homeodomain (CEH-24; Harfe and Fire 1998) was not sufficient for activation of the ceh-24 NdE box enhancer (Fig. 6A).
Activation of the egl-15 promoter by CeTwist + CeE/DA
To determine whether CeTwist was also involved in activation of egl-15, we ectopically expressed CeTwist either alone or together with CeE/DA in a strain containing an integrated egl-15::gfp fusion (see Materials and Methods). The egl-15 promoter was modestly activated by CeTwist alone, with a dramatic stimulation in the presence of both CeTwist and CeE/DA (Fig. 6). Expression of CeE/DA alone was not sufficient to activate the egl-15 promoter (Fig. 6A). A functional CeTwist protein was required to drive reporter expression from the egl-15 promoter: CeTwist protein lacking the basic DNA-binding domain and helix I (when expressed with CeE/DA) could not activate the egl-15 promoter in vivo (Fig. 6A).
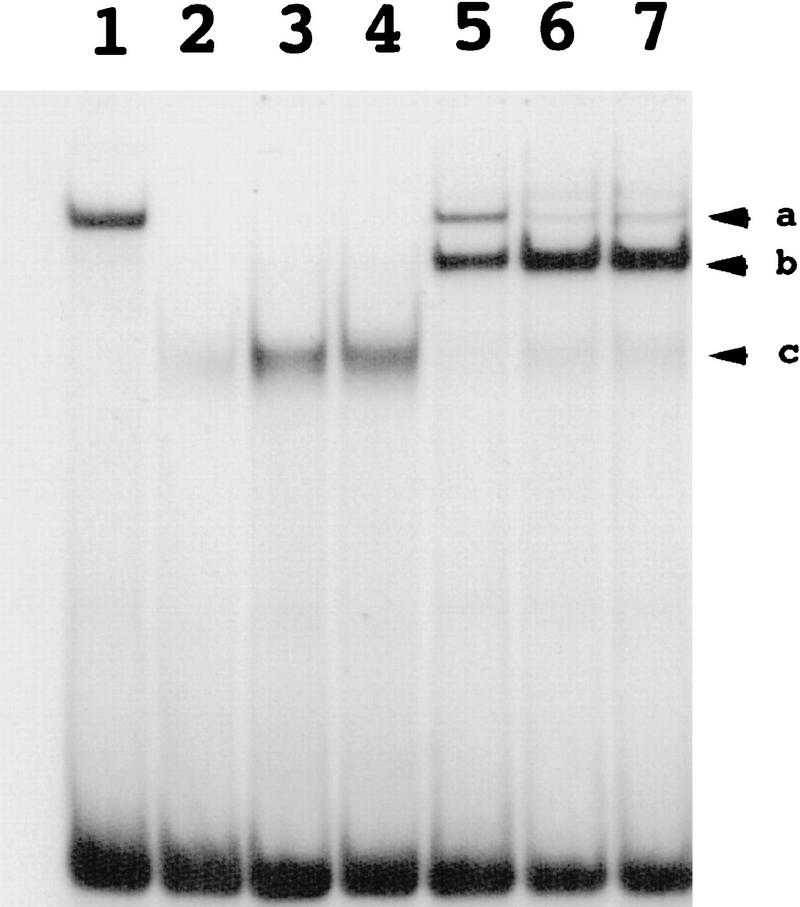
CeTwist and CeE/DA preferentially form heterodimers on E boxes in vitro
In vitro gel shift assays supported the hypothesis that CeTwist and CeE/DA work in concert to activate transcription. The capability for heterodimerization by the two proteins was first assayed with a canonical E box (CAGGTG). Homodimers of either protein were able to bind to this target (Fig. 7). When the two proteins were mixed, we observed a preferential formation of a heterodimeric complex with intermediate mobility (Fig. 7). Heterodimerization was also evident with the NdE-box sequence (CATATG). In this case, we observed binding of isolated CeTwist but no binding of isolated CeE/DA; mixing of the two proteins produced a novel shifted band as expected from heterodimer formation (data not shown). A mutant NdE-box gATATG, which lacked the first base of the NdE-box consensus motif, was not capable of binding either CeTwist or CeTwist–CeE/DA heterodimers. This mutated NdE box was not functional in vivo (Fig. 2A).
Figure 7.
In vitro DNA-binding activities of CeTwist and CeE/DA. CeTwist and CeE/DA preferentially bound E boxes as heterodimers in vitro. CeE/DA-fusion protein was obtained as described previously (Krause et al. 1997). Equimolar starting concentrations of CeE/DA and CeTwist fusion proteins were arbitrarily defined as 1×. (Lane 1) CeE/DA (1×); (lanes 2–4) CeTwist at 1×, 2×, and 3×, respectively; (Lanes 5–7) CeE/DA at 1× and CeTwist at 1×, 2×, and 3×, respectively. Arrowheads: (a) CeE/DA homodimer; (b) CeTwist–CeE/DA heterodimer; (c) CeTwist homodimer. Gel shifts for this figure were performed with a probe that contained the E-box sequence CAGGTG. Equivalent binding results were obtained with a consensus NdE box (CATATG) with the exception that little or no binding was seen with the CeE/DA homodimer, while no binding by either CeTwist or CeTwist–CeE/DA was seen with a mutated E box (gATATG; data not shown).
Consequences of ectopic CeTwist expression
Ectopic expression of CeTwist via an inducible heat shock promoter resulted in a variety of phenotypic consequences: In addition to activation of the ceh-24 NdE-box element and the egl-15 promoter (Fig. 6), heat shocked adult animals were both uncoordinated and egg-laying defective (data not shown). Transgenic adults that expressed a CeTwist protein that was missing its basic domain and helix I were essentially wild type.
The broad spatial activity of the heat shock promoter made it difficult to analyze CeTwist effects on individual tissues. We therefore examined effects of driving CeTwist expression from a variety of promoters that express in more limited sets of cells. First, we expressed CeTwist from the body muscle specific myo-3 promoter (the myo-3 promoter is active in differentiated body wall, defecation, and sex muscles; Okkema et al. 1993). This resulted in ectopic activity of both the egl-15 promoter and ceh-24 NdE box enhancer in body wall muscles (data not shown). These animals were somewhat sluggish but able to move, indicating that some muscle function was retained in myo-3::hlh-8 affected animals. Body wall muscle cells did not appear morphologically altered when visualized with polarized light or Nomarski optics. The majority of myo-3::hlh-8 animals were also egg-laying defective, contained extra vulval muscles, and had fewer M-derived coelomocytes. We were surprised by the apparent ability of myo-3::hlh-8 to drive transformations from coelomocytes to SMs; this effect may indicate a low level of myo-3::hlh-8 activity in nondifferentiated cells of the M lineage.
We next expressed CeTwist from the pharyngeal muscle-specific myo-2 promoter (Okkema et al. 1993). We were unable to recover stable transgenic lines containing this construct. F1 myo-2::hlh-8 animals were found to ectopically activate the egl-15 promoter in a variable subset of pharyngeal muscles. These pharyngeal muscles appeared deformed, with many cells containing large vacuoles.
Postmitotic expression of CeTwist in mechanosensory cells (using the mec-3 promoter; Way et al. 1991) did not result in a visible phenotype: The animals exhibited normal mechanosensory behavior. No activation of the egl-15 promoter was seen in these cells.
Consequences of reduced CeTwist expression
As a means to determine the effects of decreased CeTwist, we have used RNA-mediated genetic interference (Guo et al. 1993; Fire et al. 1998b). RNA injection in C. elegans has been shown to be an effective tool for interfering with the function of endogenous genes. In recent work, it has become clear that this interference is particularly potent and specific with double-stranded RNA (dsRNA) corresponding to the gene of interest (Fire et al. 1998b).
We first examined effects on progeny animals after injection of dsRNA for hlh-8 into adult animals. A strain carrying GFP driven by the ceh-24 NdE-box element was used as a recipient. Adults from this strain injected with dsRNA for hlh-8 produced broods with 40% of progeny lacking ceh-24 NdE box activity in the anal depressor muscle. As a control, injection of an unrelated dsRNA (ceh-24) into this strain had no effect on NdE-box driven GFP expression in these cells. These results were consistent with the hypothesis that CeTwist is required for the activation of the NdE-box element in the anal depressor muscle.
Experiments in which RNA is injected into a parent animal are limited in that interference effects are not generally observed in sex muscles in the next generation (the RNA may be diluted out by the time that these muscles form; Fire et al. 1998b). One useful property of RNA-mediated interference is the ability of injected dsRNA to enter mesodermal cells from the body cavity. This allows a single injection of dsRNA to produce interference in many cells of the mesoderm of the injected animal (Fire et al. 1998b). To circumvent the apparent dilution of RNA in the generation following injection, we carried out hlh-8 dsRNA injections into L1 larvae and examined the injected animals as adults (C. Mello, pers. comm.; S. Xu and A. Fire, unpubl.). In these experiments, we observed an egg-laying defective phenotype in 21% of injected animals (n = 48); in each case, the egg-laying defect was associated with a loss or decrease in the number of cells expressing the NdE box-driven reporter in the vulval/uterine region. Similar effects of hlh-8 dsRNA were observed with an egl-15::gfp reporter (data not shown).
The hlh-8 promoter is activated by distinct cis-acting elements
To identify sequences responsible for hlh-8 promoter activity, we performed a unidirectional deletion analysis of the hlh-8 promoter (Fig. 8A). The reporter genes used in these experiments were lacZ and GFP. Identical results were obtained with each reporter. We tested each construct for expression in both the M lineage and coelomocytes.
The hlh-8 deletion analysis identified two cis-acting elements: a promoter-proximal element and a promoter-distal element (Fig. 8A). The 569-bp promoter-distal element was responsible for reporter activity in all six mature coelomocytes. The 315-bp promoter-proximal element was required for hlh-8 promoter activity in undifferentiated cells of the M lineage.
Deletion of 21 bp from the 5′ end of the promoter-proximal element resulted in a complete loss of activity (Fig. 8A, pBH49.33). Within this 21-bp region, the promoter-proximal M lineage element contained a single putative NF-κB/Dorsal-binding site and a GAGA motif (Biggin and Tjian 1988; Thisse et al. 1991). Targeted mutations in either site abolished activity of the 315-bp minimal promoter in the entire M lineage (Fig. 8B, pBH55.11 and pBH55.13). Interestingly, alteration of a DNA sequence 3′ of the NF-κB/GAGA motif region also resulted in inactivation of the hlh-8 promoter (Fig. 8B, pBH58.09). The sequences in this region showed high homology to binding sites for the Antennapedia class of homeodomains (Damante et al. 1994).
The 315-bp promoter-proximal element also contained two putative CeTwist-binding sites (Fig. 8). In vivo ectopic CeTwist + CeE/DA coexpression experiments revealed no evidence of hlh-8 autoregulation (Fig. 6A).
Early CeTwist expression requires the homeotic selector protein MAB-5
The C. elegans Hox cluster genes mediate many anterior–posterior cell-fate decisions, affecting both mesodermal and ectodermal lineages (Kenyon 1986; Chisolm 1991; Clark et al. 1993; Wang et al. 1993). The presence of binding sites similar to Antennapedia class homeodomain sites in critical regions of the hlh-8 promoter prompted us to introduce an integrated hlh-8::gfp reporter transgene into strains containing available Hox cluster mutations.
MAB-5 was a good initial candidate for an activator of hlh-8. MAB-5 protein is prominent in the M mesoblast and in descendants of M for at least four cell divisions; MAB-5 expression is maintained in the SMs and their undifferentiated progeny while expression trails off in M-lineage cells that differentiate as striated muscle (Cowing and Kenyon 1996; S. Salser and C. Kenyon, unpubl.). This expression pattern within the M lineage is similar to that inferred for CeTwist. We found that embryos homozygous for a mab-5 null mutation (e1239; Kenyon 1986) lacked hlh-8 promoter activity in the mesoblast M. This requirement continued during the period immediately after hatching: L1 and L2 stage mab-5(e1239) larvae showed little or no hlh-8 promoter activity in M descendants. Surprisingly, the activity of hlh-8 in later M lineages did not require mab-5: SMs and their progeny in mab-5(e1239) animals expressed the hlh-8::gfp reporter at levels comparable to wild type.
It would have been surprising if MAB-5 were the sole component responsible for the hlh-8 activity pattern. In particular, the presence of MAB-5 in ectodermal structures (in which hlh-8 is apparently not active) suggested the existence of additional regulatory mechanism(s) with a restrictive role in hlh-8 regulation. Additional evidence for such mechanisms came from experiments with an inducible hs::mab-5 construct that can produce MAB-5 protein in virtually all tissues (Stringham et al. 1992; Salser and Kenyon 1992). We found that broad misexpression of MAB-5 led to ectopic expression of the hlh-8::gfp reporter, but only in a limited pattern of mesodermal precursor cells in the embryo (Fig. 6I). No activation of hlh-8 was observed in nonmesodermal embryonic tissue or in any region of adult or larval animals. These results suggest a combinatorial mechanism by which hlh-8 is activated in the presence of both MAB-5 activity (mid-posterior positional identity) and a yet-to-be-defined mesoderm-specific activation factor(s).
A unique role for mab-5 in the early activation of hlh-8 was suggested by analysis of the neighboring HOX factors, egl-5 or lin-39. No defects in the level or pattern of hlh-8 activity were seen in egl-5 or lin-39 mutants. To address the possibility of redundant roles for egl-5 and mab-5 in activation of hlh-8, mab-5(e1239)egl-5(n945) double mutant animals were analyzed; the double mutant behaved identically to mab-5(e1239).
Requirements for MAB-5 in M lineage patterning
Given the expression of mab-5 in the early M lineage and the possible activator–target relationship between MAB-5 and hlh-8, we anticipated that MAB-5 might play a role in patterning within the lineage. It should be stressed that we expect MAB-5 to activate a number of targets in the course of M lineage specification; hence the M-lineage phenotypes of mab-5 mutants will likely reflect numerous contributions in addition to the early activation of hlh-8. The initial description of the mab-5 null phenotype reported a number of alterations in the cellular products derived from the M lineage (Kenyon 1986). To examine more closely the nature of the MAB-5-dependent events, we followed the M lineage in detail in a number of mutant animals (Fig. 9).
Figure 9.
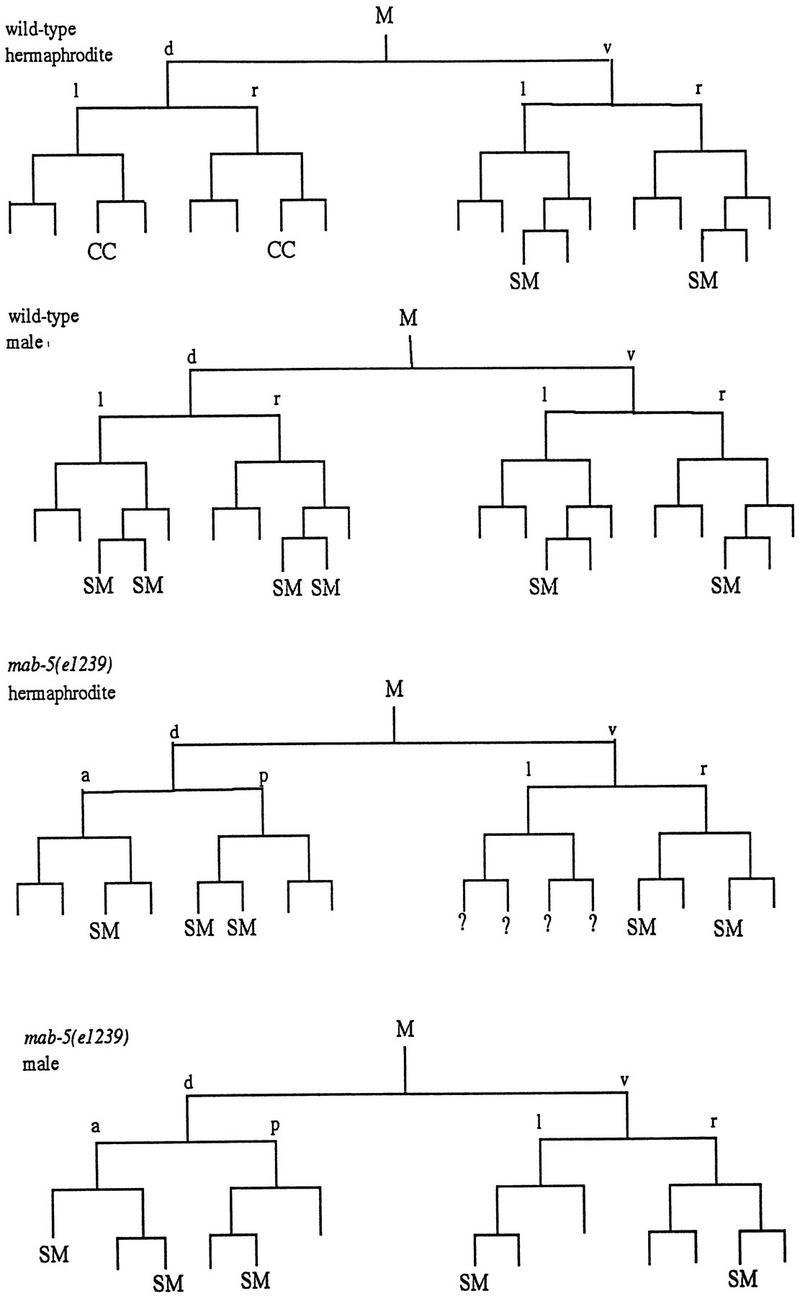
Requirements for MAB-5 in M lineage patterning. Divisions and cell fates in the M lineage are shown for wild-type (Sulston and Horvitz 1979) and sample animals of genotype mab-5(e1239); him-5(e1490) (SM) Sex myoblast; (CC) Coelomocyte; (?) not followed; Cells that apparently adopted a body wall muscle fate are unmarked. To characterize the extent of lineage variability and identify consistent features in the mutant, lineages were followed from the L1 to L3 stage in three hermaphrodites and two males. General features of the mutant lineages are summarized as follows: at hatching, M had migrated toward the posterior (as in wild type) in all mutant animals but was often situated dorsal to wild type. The first division of M was generally normal (although M divided along the anterior–posterior axis in one animal). In the second division, an abnormality was seen in the cleavage plane of Md: this cell frequently divided along the anterior–posterior instead of left–right body axis (n = 5/9 divisions observed in hermaphrodites and 2/3 divisions observed in males). The most striking and consistent phenotypes seen in mab-5(−) hermaphrodites were the loss of the two M-derived coelomocytes (100% of spot-checked animals, with n > 200) and the production of cells that appeared to be supernumary SMs. Like wild-type SMs, these supernumerary myoblasts became enlarged and divided during the L3 stage. The putative SMs arose in multiple, variable positions after M had undergone four divisions (generally without the additional round of cell division that normally occurs in the Mvl/rpa position). The following behavior was seen in the two lineaged hermaphrodites not shown in the figure: The first generated one dorsal SM (Md.ppa) and one ventral SM (Mv.rpa) on the right and two SMs (origins unknown) on the left. All of these cells migrated toward the gonadal anchor cell, enlarged, and underwent multiple cell divisions during the L3. In a second animal in which te Md/vl lineages were followed to mid-L2, the M.vlpa cell became an SM (which migrated toward the gonad) and there were no apparent dorsal SMs. In the additional male lineage, M.drpa and M.vrpa both became apparent SMs that migrated toward the gonad, enlarged, and divided during the L3. To augment this analysis, the early M cell division pattern and the pattern of SMs were observed in larger numbers of animals by spot-checking cell number and position (by Nomarski microscopy) in appropriately staged larvae. Among 27 left or right sides observed in L3 hermaphrodites, 1 side had zero SMs, 7 sides had one SM, 11 sides had two SMs, 6 sides had three SMs, 1 side had four SMs, and 1 side had six SMs. Among 4 left or right sides spot-checked in males, 1 had one SM, 2 had two SMs, and 1 side had four SMs. Generally the resulting cells could be seen in the ventral quadrant, having migrated to flank the center of the gonad; in a minority of cases, the cells apparently migrated to dorsal midbody region (presumably dorsally generated cells that migrated anteriorly but not ventrally) or, less frequently, in more posterior ventral or dorsal positions (presumably SM cells that failed to migrate). The male SMs normally migrate posteriorly, indicating that mab-5 controls the direction of migration of these cells in males. M-derived male mating structures are generally absent in mab-5(0) mutants (see Kenyon 1986).
First, we saw frequent defects in the initial division patterns in the M lineage. The most frequent defect was in the second division, in which the dorsal and ventral M daughters normally divide left–right, to produce a single mesoblast in each quadrant of the animal. In the mab-5 mutant, we saw frequent division of the dorsal daughter in the anterior–posterior direction, yielding animals in which M derivatives were missing from one of the four body quadrants.
A second defect occurred later in the lineage and concerned the M-derived coelomocytes: These coelomocytes were not observed in the mab-5 mutant, although the animals were still capable of producing the embryonic coelomocytes.
Finally, the majority of mab-5 hermaphrodites produced supernumary cells that looked and behaved like SMs. These transformations suggest that MAB-5 targets play a key role in the specification of fates within the M lineage.
Discussion
A common cis-acting element participates in activation of two differentially expressed mesodermal genes
To identify components that specify gene expression in the postembryonic mesoderm, we studied the regulation of two genes expressed in the mesoderm: ceh-24, which encodes an NK-2 class homeodomain factor activated during vulval muscle differentiation, and egl-15, which encodes an FGFR family member expressed broadly within the M lineage. For both genes, our analysis led to identification of a control element with a core sequence CAtaTG. Searches of known transcription factor-binding targets suggested to us that this sequence could act as a binding site for members of the Twist family of bHLH regulatory factors.
Involvement of a C. elegans Twist family member in activation of gene expression in postembryonic mesoderm
hlh-8 is the closest known homolog in C. elegans of the Drosophila and vertebrate twist genes. Several properties of hlh-8 support the hypothesis that it is involved directly in activation of gene expression in the postembryonic mesoderm. (1) The hlh-8 promoter is active in all of the undifferentiated cells of the postembryonic mesodermal lineage derived from the M blast cell. Activity in the M lineage begins late in embryogenesis, preceding expression of at least two putative target genes (ceh-24 and egl-15); (2) In vitro, the hlh-8 product is capable of binding to the NdE box, an element from the ceh-24 and egl-15 promoters that we have shown to be sufficient for M-lineage expression; (3) Animals in which CeTwist activity has been reduced in early development by RNA-mediated interference exhibit a decrease in activity of at least two putative CeTwist targets (the ceh-24 NdE box and the egl-15 promoter); (4) Ectopic expression of hlh-8 in naive cells is sufficient to activate mesoderm-specific control elements within the ceh-24 and egl-15 promoters; (5) Although CeTwist is sufficient to bind and carry out activation as a homodimer, maximal activity (in vitro and in vivo) depends on the presence of the more generally expressed bHLH factor CeE/DA (the C. elegans homolog of the E12 family and Daughterless); CeE/DA is present throughout the M lineage (Krause et al. 1997; M. Krause, unpubl.).
Analysis of sequences involved in hlh-8 regulation suggested that a protein of the HOX family might be an additional upstream component in this regulatory pathway. Our results suggest that the homeotic selector factor MAB-5 plays an essential role in the early activation of hlh-8. MAB-5 activation cannot, however, account for the entire hlh-8 activity pattern, and we propose that additional mesoderm-specific components (perhaps interacting with an upstream NF-κb/Dorsal control element) cooperate with MAB-5 in the initial activation of hlh-8.
Divergent and conserved aspects of Twist activity in mesodermal specification
Developmental roles for Twist have evidently varied rather dramatically in evolution. During Drosophila gastrulation, Twist is expressed in all mesodermal precursors. Drosophila mutants lacking Twist fail to complete gastrulation and lack all mesodermal tissues. Subsequent expression occurs in visceral and heart precursors, with Twist required for formation of those structures. Although vertebrate Twist factors are similar to Drosophila Twist in their predominantly mesodermal expression, developmental activation is later (well beyond gastrulation) and less extensive (only a subset of mesodermal precursors) as compared to Drosophila. Chan and Behringer (1995) show that specification and differentiation of mesodermal cell types can occur in a mouse mutant lacking Twist, but that numerous defects in mesodermal patterning are observed. Our data suggest the situation in C. elegans to be closer to the vertebrate case, with CeTwist activity mediating a defined subset of mesodermal cell-fate decisions, rather than the entirety of mesodermal fate specification.
Although the developmental and anatomical aspects of Twist activity differ among the insect, vertebrate, and nematode systems, there are numerous surprising features indicating a conservation of biochemical function and perhaps conservation of the identity of certain targets (Fig. 10). Our data indicate an interaction of CeTwist with CATATG consensus sequences in promoters for at least two genes, the NK-class homeodomain ceh-24 and the FGFR-like gene egl-15. Among the known direct targets for Drosophila Twist are homologous sequence motifs in the NK-class homeobox gene tinman (Lee et al. 1997; Yin et al. 1997). Drosophila Twist is also required for activation of the FGFR gene DFR1/heartless (Shishido et al. 1997). In contrast to what is observed in C. elegans, Drosophila does not appear to require the E/DA homolog (Daughterless) for full Twist activity (Yin et al. 1997).
Although specific targets for Twist regulation are not defined in vertebrate systems, an intriguing set of observations from human genetics raises the possibility of conservation in at least one set of targets. This analogy is based on a set of specific human genetic syndromes that include as a prominent feature the premature closure of the skull (craniosynostosis). These syndromes have been shown to result from dominant mutations in Twist, three FGFR genes (FGFR1, FGFR2, FGFR3), and the NK-related homeobox gene MSX2 (Jabs et al. 1993; Muenke et al. 1994; Meyers et al. 1995; Przylepa et al. 1996; Ghouzzi et al. 1997; Howard et al. 1997). Disruption of twist in mice has also been shown to cause a haploinsufficient craniofacial abnormality similar to that seen in humans (Ghouzzi et al. 1997). The connection between NK and NK-like homeodomains is not a convincing argument for target conservation: The NK and NK-related homeodomain classes are rather extensive families in which ceh-24, tinman, and MSX2 are by no means the natural orthologs. The Twist–FGF connection, by contrast, makes a much stronger case for analogy. Although vertebrate Twist is likely to have a large number of targets, the remarkable similarity in human heterozygote phenotypes between Twist and FGFR mutations has been taken as a strong suggestion of a regulator–target relationship.
Materials and methods
Construction of plasmids and transgenic lines used in ectopic expression experiments
cDNAs encoding a gene of interest were cloned into expression constructs containing either inducible or tissue-specific promoters (Mello and Fire 1995). The hsp::mab-5 construct was from Salser and Kenyon (1992). Other heat shock (hs)-inducible constructs were as follows: hs::hlh-8, pBH48.8 and pBH48.13; hs::hlh-2, pKM1035 and pKM1034; hs::ceh-24, pBH23.4 and pBH23.19; hs::hlh-1, pPD50.63 and pPD50.66; and hs::Δbhlh-8, pBH56.45 and pBH56.51.
For each coding region, the first construct of each pair uses the hsp16-2 promoter, while the second uses hsp16-41. These promoters are active in most somatic tissue types (Stringham et al. 1992). The hs::Δbhlh-8 constructs lack the putative DNA binding (basic) region and helix I of hlh-8. These constructs were created by deletion of bases 61–145 from the hlh-8 cDNA (GenBank accession no. AF037063).
GFP reporter strains used were as described below. Plasmids included in each transgene are underlined. All transgenes were integrated as described previously (Mello and Fire 1995), and all arrays containing the plasmid pMH86 were integrated in a dpy-20(e1282) genetic background: NH2447[ayIs2(IV)], egl-15::gfp (NH#300+pMH86), shows expression in adult vm1 vulval muscles (C. Branda and M. Stern, in prep; Harfe et al. 1998). PD4666[ayIs6(X)] and PD4667[ayIs7(IV)], hlh-8::gfp (pBH47.70+pMH86), serves as a marker for M and undifferentiated cells in the M lineage. PD4656[ccIs4656(IV)], NdE-box::gfp (pBH34.21+pMH86), shows expression in adult vulval muscles, uterine muscles, intestinal muscles, and anal depressor muscle (Harfe and Fire 1998). PD4521[ccIs4521(II)], myo-3::gfp (pSAK-2+pSAK-4+pMH86), shows expression in all body wall and vulval muscles (Fire et al. 1998b).
Generation of Twist protein and antibodies
Expression vector pRSETA was used to express CeTwist protein in Escherichia coli BL21 (lys-S) cells as a His-tagged fusion protein. CeTwist–His protein was purified with a Ni affinity column in denaturing conditions (8 m urea, 500 mm NaCl, 20 mm NaP04 at pH 4.0). The eluted protein was pure and was used directly in gel-shift assays. For polyclonal antibody production in mice, purified CeTwist-His protein was dialyzed to remove urea and lyophilized before injection. Polyclonal antisera were used to localize CeTwist protein in synchronized populations of animals, as described previously (Krause et al. 1997). Immunolocalization of CeTwist was a relatively low-sensitivity procedure: Expression in positive cells was just above the limit of detection with fluorescent secondary antibody. Thus a failure to stain with the antibody could represent a severalfold decrease in level or complete loss. This is in contrast to the GFP fusions, which were readily detected under standard conditions (and could be detected even if reduced substantially).
Gel-shift assays
Gel-shift assays with a canonical E box were performed essentially as described in Krause et al. (1997), using the probe GCAATCGAAACAACAGGTGTTCGCCCGTTGATC. The E box is underlined. The sequences of NdE box elements used in gel-shift assays are shown in Figure 2.
The assay for CeTwist–CeE/DA binding in vitro may be somewhat less stringent than requirements for activation in vivo. Three of the mutants in Figure 2 (3m, 10m, and 11m) were bound by CeTwist–CeE/DA in gel-shift assays but were not activated in vivo, either in wild-type sex muscles or by ectopic CeE/DA–CeTwist (Fig. 2; data not shown).
Acknowledgments
We thank C. Branda and M. Stern for integrating the hlh-8::gfp construct, providing strain NH2447, and for information about the egl-15 expression pattern, S. Salser for providing information about the mab-5 expression pattern, and E. Jabs, M. Frasch, C.M. Fan, E. Hedgecock, J. Maloof, C. Mello, S. Xu, A. Corsi, and S. Kostas for help and suggestions. This work was supported by the NIH (grants R01GM37706 to A.F., R01GM37053 to C.K., T32GM07231 to B.H., F32HD08331 to J.L., and Z01DK36117 to M.K.) C.K. acknowledges the MRC Laboratory of Molecular Biology for hospitality during the early phases of the lineage project. Some strains used in this work were obtained from S. Clark, N. An, and H.R. Horvitz, and from the Caenorhabditis Genetics Center (supported by a grant from the NIH Center for Research Resources).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL fire@mail1.ciwemb.edu; FAX (410) 243-6311.
References
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. Twist is required in head mesenchyme for cranial tube morphogenesis. Genes & Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- Chitwood BG, Chitwood MB. Introduction to nematology. Baltimore, Maryland: University Park Press; 1974. [Google Scholar]
- Clark S, Chisholm AD, Horvitz HR. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- Costa M, Weir M, Coulson A, Sulston J, Kenyon C. Posterior pattern formation in C. elegans involves position-specific expression of a gene containing a homeobox. Cell. 1988;55:747–756. doi: 10.1016/0092-8674(88)90131-6. [DOI] [PubMed] [Google Scholar]
- Cowing DW, Kenyon C. Expression of the homeotic gene mab-5 during Caenorhabditis elegans embryogenesis. Development. 1992;116:481–490. doi: 10.1242/dev.116.2.481. [DOI] [PubMed] [Google Scholar]
- Cronmiller C, Schedl P, Cline TW. Molecular characterization of daughterless, a Drosophila sex determination gene with multiple roles in development. Genes & Dev. 1988;2:1666–1676. doi: 10.1101/gad.2.12a.1666. [DOI] [PubMed] [Google Scholar]
- Damante G, Fabbro D, Pellizzari L, Civitareale D, Guazzi S, Polycarpou-Schwartz M, Cauci S, Quadrifoglio F, Formisano S, Di Lauro R. Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore DL, Horvitz HR, Stern MJ. An FGF Receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell. 1995;83:611–620. doi: 10.1016/0092-8674(95)90101-9. [DOI] [PubMed] [Google Scholar]
- Fire A, Ahnn J, Kelly W, Harfe B, Kostas S, Hsieh J, Hsu M, Xu S. GFP applications in C. elegans. In: Chalfie M, Kain S, editors. GFP: Green fluorescent protein strategies and applications. New York: John Wiley and Sons; 1998a. . (In press.) [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas S, Driver S, Mello C. Potent and specific genetic interference by double stranded RNA in C. elegans. Nature. 1998b;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nature Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Fire A. Muscle and nerve-specific regulation of a novel NK-2 class homeodomain factor in C. elegans. Development. 1998;125:421. doi: 10.1242/dev.125.3.421. [DOI] [PubMed] [Google Scholar]
- Harfe, B.D., C. Branda, M. Krause, M. Stern, and A. Fire. 1998. MyoD and the specification of muscle and non-muscle fates in postembryonic development of the C. elegans mesoderm. Development (In press). [DOI] [PubMed]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood ND, Plunk A, Gurdon JB. Xenopus twist is expressed in response to induction in the mesoderm and neural crest. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, De Luna R, Garcia Delgado C, Gonzalez-Ramon M, Kline A, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nature Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes & Dev. 1992;6:1518–1529. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB, Snead ML, Maxson R. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- Kenyon C. A gene involved in the development of the posterior body region of C. elegans. Cell. 1986;46:477–487. doi: 10.1016/0092-8674(86)90668-9. [DOI] [PubMed] [Google Scholar]
- Krause M, Fire A, White Harrison S, Priess J, Weintraub H. CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell. 1990;63:907–919. doi: 10.1016/0092-8674(90)90494-y. [DOI] [PubMed] [Google Scholar]
- Krause M, Park M, Zhang J, Yuan J, Harfe B, Zu S, Greenwald I, Cole M, Paterson B, Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–2189. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47- like proteins in vivo. Cell. 1991;66:301–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lee MY, Park T, Schulz RA, Kim Y. Twist-mediated activation of the NK-4 homeodomain gene in the visceral mesoderm of Drosophila requires two distinct clusters of E-box regulatory elements. J Biol Chem. 1997;28:17531–17541. doi: 10.1074/jbc.272.28.17531. [DOI] [PubMed] [Google Scholar]
- Li L, Cserjesi P, Olson E. Dermo-1: A novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- Mello CC, Fire A. DNA transformation. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern biological analysis of an organism. New York, NY: Academic Press; 1995. pp. 452–482. [Google Scholar]
- Meyers GA, Day D, Goldberg R, Daentl D, Przylepa KA, Abrams LJ, Graham J, Feingold M, Moeschler J, Rawnsley E, Scott AF, Jabs EW. FGFR2 exon IIIa and IIIc mutations in Crouzon, Jackson-Weiss, and Pfeiffer syndromes: Evidence for missense changes, insertions and a deletion due to alternative RNA splicing. Am J Hum Genet. 1996;58:491–498. [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Fire A. Muscle: Structure, function, and development. In: Riddle DL, Blumenthal T, Meyer BJ, Preiss JR, editors. Caenorhabditis elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S, Winter RM. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nature Genet. 1994;8:269–274. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in C. elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przylepa KA, Paznekas W, Zhang M, Golabi M, Bias W, Bamshad MJ, Carey JC, Hall BD, Stevenson R, Orlow S, Cohen MM, Jabs EW. Fibroblast growth factor receptor 2 mutations in Beare-Stevenson cutis gyrata syndrome. Nature Genet. 1996;13:492–494. doi: 10.1038/ng0896-492. [DOI] [PubMed] [Google Scholar]
- Salser SJ, Kenyon C. Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature. 1992;355:255–258. doi: 10.1038/355255a0. [DOI] [PubMed] [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNA. Development. 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and mef-2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Stern MJ, Horvitz HR. A normally attractive cell interaction is repulsive in two C. elegans mesodermal cell migration mutants. Development. 1991;113:797–803. doi: 10.1242/dev.113.3.797. [DOI] [PubMed] [Google Scholar]
- Stringham EG, Dixon DK, Jones D, Candido EPM. Temporal and spatial expression patterns of the small heat-shock (HSP16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomas JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thisse B, Stoetzel C, Gorostiza-Thisse C, Perrin-Schmitt F. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of the early Drosophila embryos. EMBO J. 1988;7:2175–2183. doi: 10.1002/j.1460-2075.1988.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- Wang BB, Muller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- Waterston RH. Muscle. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 281–335. [Google Scholar]
- Way JC, Wang L, Run JQ, Wang A. The mec-3 gene contains cis-acting elements mediating positive and negative regulation in cells produced by asymmetric cell division in Caenorhabditis elegans. Genes & Dev. 1991;5:2199–2211. doi: 10.1101/gad.5.12a.2199. [DOI] [PubMed] [Google Scholar]
- Wolf C, Thisse C, Stoetzel C, Thisse B, Gerlinger P, Schmitt-Perrin F. The M-twist gene of Mus is expressed in subsets of mesodermal cells and is closely related to the Xenopus X-twi and Drosophila twist genes. Dev Biol. 1991;143:363–373. doi: 10.1016/0012-1606(91)90086-i. [DOI] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]