Abstract
The mechanisms responsible for translational silencing of certain mRNAs in growing oocytes, and for their awakening during meiotic maturation, are not completely elucidated. We show that binding of a ∼80-kD protein to a UA-rich element in the 3′ UTR of tissue-type plasminogen activator mRNA, a mouse oocyte mRNA that is translated during meiotic maturation, silences the mRNA in primary oocytes. Translation can be triggered by injecting a competitor transcript that displaces this silencing factor, without elongation of a pre-existing short poly(A) tail, the presence of which is mandatory. During meiotic maturation, cytoplasmic polyadenylation is necessary to maintain a poly(A) tail, but the determining event for translational activation appears to be the modification or displacement of the silencing factor.
Keywords: mouse, oocytes, tPA mRNA, 3′ UTR, poly(A), translation
The progression of gametes through the last steps of meiosis and the initial stages of embryonic development requires multiple changes in gene expression. These changes do not rely on modifications of gene transcription, which is at best very limited in mature gametes and early zygotes. The timely synthesis of developmentally important products involves the regulated translation of mRNAs that have accumulated earlier during gametogenesis. Translational control is thus a critical feature of meiotic maturation and early development, and the elucidation of the molecular mechanisms responsible for this control may also be relevant to a better understanding of the modulation of gene expression in somatic cells.
In most animal species, fully grown primary oocytes contain a pool of mRNAs that are stored, stable and untranslated, in the cytoplasm. The translational activation of these dormant mRNAs occurs during meiotic maturation and after fertilization. In contrast, other mRNAs that are translated before meiotic maturation are silenced when meiosis resumes. Studies on oocytes from a variety of species have suggested possible mechanisms for such changes in translation (Curtis et al. 1995; Richter 1995; Vassalli and Stutz 1995; Wickens et al. 1995; Seydoux 1996; Osborne and Richter 1997; Stebbins-Boaz and Richter 1997). The observed changes in the translational activity of many maternal mRNAs are associated with changes in the length of their poly(A) tails: In general, an increase in translation is associated with poly(A) tail elongation, whereas silencing is correlated with poly(A) tail shortening or removal.
The awakening of many dormant oocyte mRNAs appears to be controlled by sequences in their 3′ UTR. These cis-acting determinants, which are involved in both poly(A) tail elongation and translational activation, include the AAUAAA hexanucleotide that is also recognized by the cleavage and polyadenylation machinery responsible for processing of nuclear transcripts, and a more variable U-rich sequence termed the cytoplasmic polyadenylation element (CPE) (Richter 1995; Osborne and Richter 1997). Proteins that interact with CPEs and/or the AAUAAA hexanucleotide have been identified in Xenopus and mouse oocytes (McGrew and Richter 1990; Bilger et al. 1994; Hake and Richter 1994; Simon and Richter 1994; Ballantyne et al. 1995; Gebauer and Richter 1995, 1996; Stebbins-Boaz et al. 1996; Wu et al. 1997). Although the determinants involved in cytoplasmic polyadenylation are progressively being identified, precisely how this modification affects translation of previously dormant mRNAs remains elusive.
Another unresolved issue relates to the mechanism of translational silencing of oocyte mRNAs. This occurs under two distinct sets of circumstances: A number of mRNAs translated during oocyte growth and in fully grown primary oocytes are silenced during meiotic maturation (Bachvarova et al. 1985; Paynton et al. 1988; Fox and Wickens 1990; Varnum and Wormington 1990), whereas other mRNAs destined to be translated during meiotic maturation or after fertilization are rendered dormant around the time of their transcription in the growing oocyte (Huarte et al. 1992; Robbie et al. 1995). The shortening of the poly(A) tail of oocyte mRNAs is invariably coupled with inhibition of their translation. In growing mouse primary oocytes, newly transcribed tissue-type plasminogen activator (tPA) mRNA is initially endowed with the usual long poly(A) tail characteristic of nuclear transcripts, and it subsequently undergoes poly(A) tail shortening and translational silencing (Huarte et al. 1992). Until resumption of meiosis, this mRNA is then stored in the cytoplasm of primary oocytes in a dormant form that is characterized by the presence of an unusually short poly(A) tail of only 40–60 nucleotides. When meiotic maturation occurs, tPA mRNA undergoes elongation of it’s poly(A) tail and becomes translationally active. Interestingly, the same 3′ UTR UA-rich region is required for both deadenylation and silencing and, together with the canonical AAUAAA signal, for readenylation and translation of tPA mRNA. Because both deadenylation and readenylation involve overlapping sequences, this region has been termed the adenylation control element (ACE) (Huarte et al. 1992).
Although it is clear that changes in the translational status of oocyte mRNAs are correlated with changes in the length of their poly(A) tail, the dormancy of certain maternal transcripts in fully grown primary oocytes is intriguing. Several studies have shown that, in general, mRNAs with a long poly(A) tail are translated with greater efficiency than those lacking poly(A) (Jacobson 1995), but this effect appears to be most pronounced between 5 and 32 adenylate residues. Recently, an interaction between poly(A) binding protein (PABP) and yeast translation initiation factors (eIF-4E or eIF-4G) has been shown; it could account for the influence of the poly(A) tail on translation efficiency (Sachs et al. 1997). The length of the poly(A) tail that remains on dormant oocyte mRNAs (∼40–50 adenosine residues) should, however, be sufficient for binding of at least one PABP, and, hence, for interaction with initiation factors. Thus, maternal mRNA dormancy may not be accounted for by the presence of a relatively short poly(A) tail, even for those mRNAs that undergo partial deadenylation in growing oocytes and CPE-dependent readenylation and translation following resumption of meiosis.
The regulation of translation of certain mRNAs has been shown to be specifically a consequence of protein/RNA interactions involving the 3′ UTR of the transcript, independent of changes in the poly(A) tail length (Standart and Jackson 1994; Decker and Parker 1995; Murata and Wharton 1995). In Xenopus oocytes, for instance, sequence determinants in the 3′ UTR of dormant FGF receptor-1 mRNA inhibit translation of an injected reporter transcript in immature oocytes, a repression that is released at meiosis without apparent changes in polyadenylation of this transcript (Robbie et al. 1995). In extracts of clam oocytes, ablation of the 3′ UTRs of dormant ribonucleotide reductase and cyclin A mRNAs (so called masked mRNAs) prevents the binding of an 82-kD protein and leads to their premature translational activation (unmasking) (Standart et al. 1990). Similar mechanisms could also be involved in the transient silencing of certain ACE-containing mouse oocyte mRNAs.
Recently, we have shown that sequences in the 3′ UTR of dormant tPA mRNA in primary mouse oocytes are in a masked configuration (Stutz et al. 1997). Injection of antisense oligodeoxynucleotides (as-ODNs) complementary to different parts of the endogenous transcript revealed two regions that are protected from hybridization, that is, the ACE and the AAUAAA hexanucleotide. Early on resumption of meiosis, part of the ACE and the AAUAAA region become accessible to as-ODN hybridization; this unmasking, which occurs concomitantly with the very first stages of poly(A) tail elongation and well before translation can be detected, may represent the release from a silencing mechanism responsible for dormancy of tPA mRNA in primary oocytes.
In view of these results, we reasoned that the ACE may function as a cis-acting silencing determinant in primary oocytes, and that it might be possible to perturb such a dormancy mechanism by competitive displacement of a putative trans-acting translational repressor. We have tested this model in a system that explores the translational control of an endogenous mRNA in the context of the live cell. Our results show that masking mechanisms and poly(A) tail metabolism, previously considered to be alternative means of modulating protein synthesis during meiotic maturation, cooperate to control the translation of a maternal mRNA in mammalian oocytes.
Results
Activation of tPA mRNA translation in meiotically arrested primary oocytes
In growing oocytes, newly transcribed polyadenylated tPA mRNA undergoes partial deadenylation and, concomitantly, becomes translationally silent (Huarte et al. 1992); both processes require the presence of the ACE in the 3′ UTR (Fig. 1A). Silencing could be a consequence of the unusually short poly(A) tail; alternatively, deadenylation and silencing may be unrelated. Because the ACE and the AAUAAA regions are protected from as-ODN hybridization in primary oocytes (Stutz et al. 1997), indicating that they are in a masked configuration, competitive displacement of putative masking factor(s) might affect the fate of the mRNA. Unlabeled synthetic transcripts (Fig. 1B) corresponding to the ACE [positions 2401–2442 in tPA cDNA (Rickles et al. 1988)] or the AAUAAA (tPA-d, positions 2467–2504) regions or to an upstream sequence in the 3′ UTR [tPA-c, positions 2280–2320, a region that is not protected from as-ODN hybridization (Stutz et al. 1997)], were injected in fully grown primary oocytes. The oocytes were cultured for 6 hr in presence of dB-cAMP, to prevent resumption of meiosis, and translation of tPA mRNA was assessed by analyzing tPA enzymatic activity in oocyte extracts (Fig. 2A). No tPA was detected in noninjected primary oocytes (lanes 1–3), nor in primary oocytes injected with transcripts tPA-c (lanes 10–15) or tPA-d (not shown), or with a poly(U) homopolymer transcript (not shown). In contrast, ACE-injected primary oocytes were induced to synthesize the protein, and the effect was related to the dose of competitor transcript (lanes 4–9); the amount of tPA was comparable with that in noninjected maturing oocytes cultured for the same length of time (lanes 16–18).
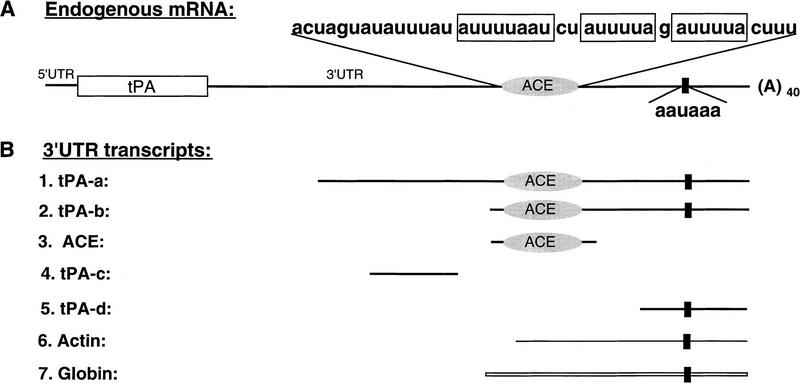
Figure 1.
Schematic representation (not drawn to scale) of endogenous tPA mRNA and of different 3′-UTR RNA transcripts. (A) Endogenous tPA mRNA. (Open rectangle) Coding region; (solid lines) 5′ and 3′ UTRs; (oval symbol) ACE region, the sequence of which is indicated above (three UA-rich sequences are boxed). (Solid, vertical rectangle) Polyadenylation signal (AAUAAA). The poly(A) tail of ∼40 adenosine residues present on tPA mRNA in primary oocytes is indicated [(A)40]. (B) 3′ UTR transcripts. Two different 3′-UTR transcripts containing both the ACE region and the polyadenylation signal are named tPA-a (479 nucleotides) and tPA-b (115 nucleotides); ACE (83 nucleotides), tPA-c (83 nucleotides) and tPA-d (80 nucleotides) correspond respectively to the ACE region detailed in A (plus vector-derived 5′ and 3′ sequences) and to two regions flanking the ACE; tPA-d encompasses the polyadenylation signal. Actin and Globin represent the 3′ terminal fragments of β-actin (133-nucleotide) and β-globin (165-nucleotide) mRNAs, respectively.
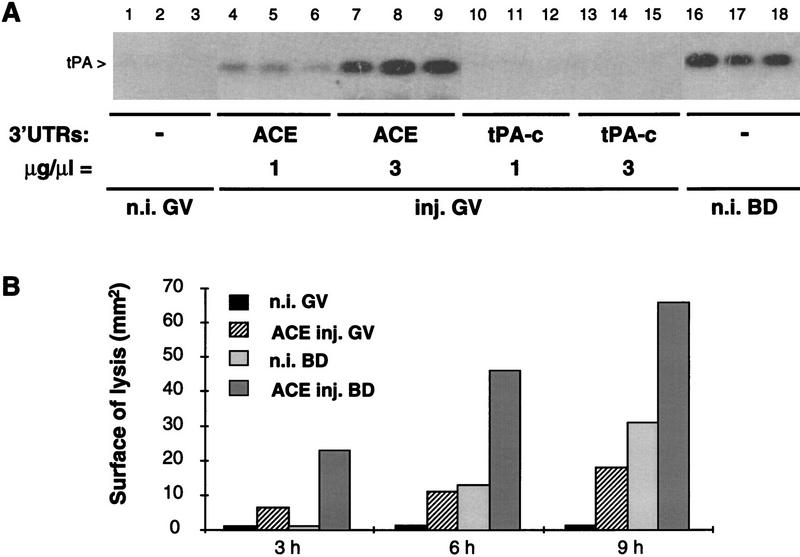
Figure 2.
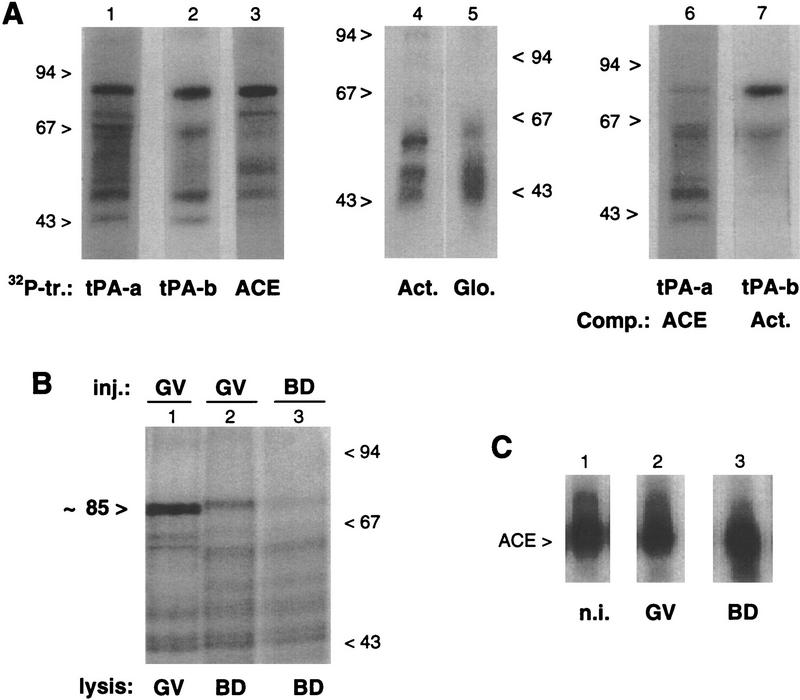
Translational awakening of tPA mRNA in oocytes. (A) Induction of tPA synthesis in primary oocytes. The indicated 3′-UTR transcripts were injected in primary oocytes (GV) at 1 or 3 μg/μl (lanes 4–15); transcript injected at 1 μg/μl, are present in oocytes at a ∼104-fold molar excess over endogenous tPA mRNA. After 6 hr of culture in the presence of dB-cAMP, arrested primary oocytes were lysed in groups of five and assayed in triplicate by zymography (Huarte et al. 1985). Groups of five control noninjected arrested primary oocytes (n.i. GV, lanes 1–3) or maturing oocytes lysed 6 hr after GVBD (n.i. BD, lanes 16–18) were analyzed in parallel. The zymogram was incubated for 40 hr at 37°C. (B) Time course of tPA accumulation in arrested primary and in maturing oocytes. Noninjected primary oocytes and primary oocytes injected with the ACE transcript at 2 μg/μl were incubated in the presence or absence of dB-cAMP. After the indicated periods of time, arrested primary oocytes (GV) or maturing oocytes (BD) were lysed in groups of five, and the samples were assayed in duplicate on a zymogram that was incubated for 40 hr at 37°C; tPA activity was quantitated by measuring the surface of the proteolytic zones [n.i., noninjected; inj., injected (Stutz et al. 1997)].
Injection of the ACE competitor transcript did not affect the pattern of protein synthesis in primary oocytes metabolically labeled with [35S]methionine, as determined by SDS-PAGE and autoradiography (not shown). Thus, the observed effect on tPA synthesis appears specific, in that it is neither related to a general increase in protein synthesis nor accompanied by changes in the rate of synthesis of other proteins detectable by metabolic labeling. In fully grown primary oocytes, the nucleus is arrested at the prophase of the first meiotic division, and transcription is almost completely absent. However, to verify that the effect of the injected ACE transcript is caused by translational recruitment of dormant tPA mRNA, and not by translation of newly transcribed mRNA, primary oocytes were injected with the ACE transcript and incubated for 6 hr in medium containing α-amanitin (10 μg/ml). The presence of this inhibitor of transcription did not prevent tPA synthesis in ACE-injected oocytes (not shown).
We conclude that injection of a synthetic transcript corresponding to the ACE region of tPA mRNA induces translational awakening of this mRNA in arrested primary oocytes. This suggests that the injected transcript relieves the mRNA from an active silencing process, presumably by competing for a trans-acting translational repressor.
Effect of the ACE competitor transcript on the translational activation of tPA mRNA in meiotically arrested and maturing oocytes
During meiotic maturation, tPA is first detectable 4 hr after germinal vesicle breakdown (GVBD) (Huarte et al. 1985). Primary oocytes were injected with ACE transcript, incubated for increasing periods of time under conditions that prevent or allow maturation, and analyzed for tPA protein by zymography (Fig. 2B). In primary oocytes (GV) that had been injected with ACE, tPA was detectable 3 hr after injection, whereas, as expected, the enzyme was not detectable in noninjected maturing oocytes (BD) 3 hr after GVBD. Accumulation of tPA was comparable in primary oocytes 6 hr after ACE injection and in noninjected maturing oocytes 6 hr after GVBD; at later times, tPA accumulated more rapidly in maturing oocytes. Thus, the initial stages of tPA mRNA translational activation occur more rapidly in ACE-injected primary oocytes than in maturing oocytes.
To determine the effect of ACE transcript injection on tPA production by maturing oocytes, injected primary oocytes were cultured under conditions allowing resumption of meiosis. This resulted in the rapid accumulation of tPA, to levels significantly higher than in noninjected maturing oocytes (Fig. 2B). These observations suggest that the putative competitive unmasking of the cis-acting silencing region of tPA mRNA both induces awakening of the mRNA in arrested primary oocytes and primes the mRNA for rapid translational activation during meiotic maturation.
Effects of the ACE competitor transcript on the poly(A) metabolism of exogenous and endogenous tPA RNAs
Translational regulation of tPA mRNA is accompanied by changes in its poly(A) tail: Silencing occurs concomitantly with partial deadenylation, whereas readenylation appears necessary for translation during meiotic maturation. To explore possible effects of ACE transcript injection on this poly(A) tail metabolism, we coinjected a 32P-labeled partial tPA transcript (tPA-a, Fig. 1B), which mimics the regulation of the endogenous tPA mRNA (Huarte et al. 1992). When injected alone, in vitro polyadenylated tPA-a [tPA-a (A+)] undergoes partial deadenylation in primary oocytes (Fig. 3A, lane 2; Huarte et al. 1992); coinjection (at a 50-fold molar excess) of the ACE transcript (lane 3), but not that of a control transcript (tPA-c) (lane 4), markedly reduced the extent of deadenylation achieved 6 hr after injection. In contrast, coinjection of the ACE transcript with 32P-labeled polyadenylated actin 3′ UTR (Huarte et al. 1992) did not influence the fate of its poly(A) tail (not shown). Lower amounts of the ACE transcript (25- and 10-fold molar excess) also decreased deadenylation of tPA-a (A+), albeit to a lower degree (not shown). The default deadenylation of the 32P-labeled polyadenylated 3′ UTR actin transcript in maturing oocytes (Huarte et al. 1992) was not affected by coinjection of the ACE transcript (not shown), thus excluding a nonspecific effect on deadenylation in general. These results suggest that the ACE competitor can sequestrate a trans-acting factor involved in the sequence-specific deadenylation of tPA mRNA in growing primary oocytes.
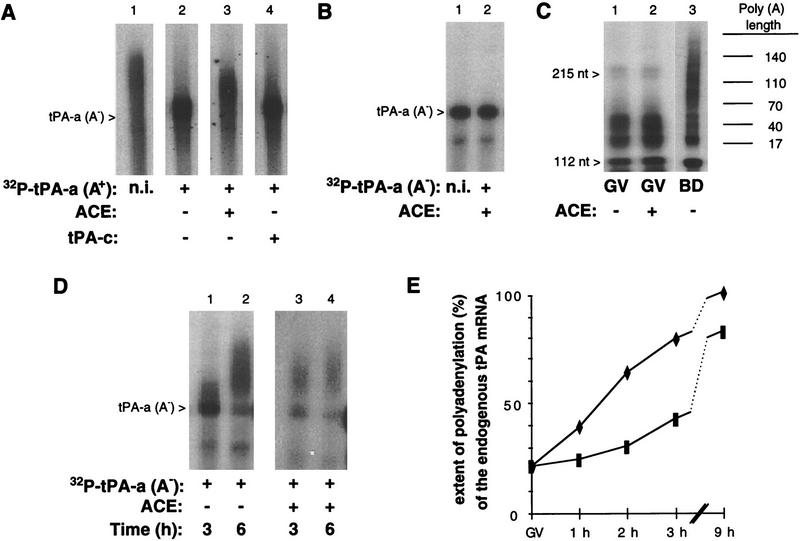
Figure 3.
Poly(A) tail metabolism in ACE-injected oocytes. (A) Effect of the ACE competitor transcript on a polyadenylated tPA 3′ UTR fragment in arrested primary oocytes. The 32P-labeled tPA-a transcript was polyadenylated in vitro to generate a poly(A) tail of ∼150–300 adenosines residues [32P-labeled tPA-a (A+), lane 1] and injected in primary oocytes, alone (lane 2) or together with either the ACE (lane 3) or the tPA-c (lane 4) transcripts at a 50-fold molar excess. The injected transcript migrates as a smear as a consequence of the heterogeneity of poly(A) length resulting from the in vitro polyadenylation reaction. Groups of 10 injected oocytes were cultured for 8 hr in the presence of dB-cAMP and lysed. The recovered 32P-labeled transcripts were analyzed on a 6% acrylamide–urea gel by autoradiography. The open arrowhead indicates the position of migration of a nonpolyadenylated tPA-a [tPA-a(A−)] transcript. (B) Effect of the ACE competitor transcript on a nonadenylated tPA 3′-UTR fragment in arrested primary oocytes. Primary oocytes coinjected with the 32P-labeled tPA-a(A−) fragment and the ACE transcript were lysed after 6 hr of culture in the presence of dB-cAMP (lane 2). The RNA was analyzed as for A. The noninjected 32P-labeled tPA-a(A−) transcript is shown in lane 1 (n.i.). (C) Effect of the ACE competitor transcript on the poly(A) status of endogenous tPA mRNA in arrested primary oocytes. Noninjected primary oocytes (lane 1) and primary oocytes injected with the ACE transcript at 2 μg/μl (lane 2) were incubated for 6 hr in the presence of dB-cAMP. Total RNA was extracted and tPA mRNA was analyzed by a RT–PCR-poly(A) test. Noninjected maturing oocytes were lysed 6 hr after GVBD were analyzed in parallel. Poly(A) tail lengths (in nucleotides) were determined by comparison with a sequencing ladder loaded on the same gel. Each lane corresponds to approximately one oocyte. (D) Effect of the ACE competitor transcript on a nonadenylated tPA 3′-UTR fragment in maturing oocytes. Primary oocytes injected with the 32P-labeled tPA-a(A−) fragment alone (lanes 1,2) or together with the ACE transcript (lanes 3,4) were cultured in the absence of dB-cAMP, and lysed 3 or 6 hr after GVBD. The recovered 32P-labeled transcripts were analyzed as for A. The migration of the 32P-labeled tPA-a(A−) transcript is indicated by the open arrowhead. (E) Effect of the ACE competitor transcript on the poly(A) status of endogenous tPA mRNA in maturing oocytes. Primary oocytes injected with the ACE transcript at 2 μg/μl (♦) or with an equal volume of KCl (▪) were cultured in the absence of dB-cAMP and collected before or 1, 2, 3, and 9 hr after GVBD. Total RNA was extracted and tPA mRNA was analyzed with the RT–PCR–poly(A) test. Poly(A) tail lengths were determined by comparison with a sequencing ladder loaded on the same gel. The extent of polyadenylation at the different time points, expressed as a percentage of the polyadenylation achieved 9 hr after GVBD in ACE-injected oocytes, was computed after determining the poly(A) tail length of the longest abundant amplification product in each sample.
To determine whether the translation of tPA mRNA in ACE-injected primary oocytes is accompanied by its polyadenylation, we first coinjected a 32P-labeled poly(A−) tPA-a [tPA-a (A−)] fragment together with the ACE transcript. After 6 hr incubation in presence of dB-cAMP, the migration of the recovered radiolabeled fragment (Fig. 3B, lane 2) was identical to that of the nonadenylated molecule (lane 1). A possible effect of the ACE competitor on the poly(A) status of endogenous tPA mRNA was analyzed by use of a RT–PCR poly(A) test (Salles and Strickland 1995) on total RNA prepared from primary noninjected and ACE-injected oocytes. The amplification products obtained with RNA from control noninjected and ACE-injected primary oocytes indicated that, under both conditions, the length of the poly(A) tail on tPA mRNA was similar, that is, ∼50 residues (Fig. 3C, lanes 1,2). In contrast, 6 hr after GVBD, tPA mRNA in maturing oocytes had undergone a substantial elongation of its poly(A) tail (lane 3). Thus, unlike what happens in noninjected oocytes during meiotic maturation, the translational activation of tPA mRNA induced by the ACE competitor in arrested primary oocytes is not accompanied by a readenylation of its poly(A) tail.
Similar experiments were performed on maturing oocytes, in which injection of the ACE transcript causes early translational activation of tPA mRNA (see above). Coinjection of the 32P-labeled tPA-a (A−) fragment with the ACE transcript resulted in an acceleration of polyadenylation, which was almost complete 3 hr after GVBD (Fig. 3D, lane 3), whereas it was only partial in oocytes injected with 32P-labeled tPA-a (A−) alone (lane 1). Polyadenylation of endogenous tPA mRNA was measured by the RT–PCR poly(A) test (Fig. 3E). Here again, injection of the ACE competitor resulted in a precocious elongation of the poly(A) tail of tPA mRNA that was already conspicuous 1 hr after GVBD. Competitive displacement of a putative silencing factor in primary oocytes thus accelerates both tPA mRNA polyadenylation and translational activation when the oocytes are allowed to undergo meiotic maturation.
Translational awakening of a dormant mRNA requires the presence of a poly(A) tail
The experiments summarized above show that in ACE-injected meiotically arrested oocytes, translation of endogenous tPA mRNA can be achieved without readenylation of the partially deadenylated transcript by the ACE competitor. To investigate the role of the poly(A) tail in this awakening, we prepared chimeric transcripts (Fig. 4A) containing the 5′ UTR and the coding region of urokinase type plasminogen activator (uPA) mRNA, and the 3′ UTR of either actin (Ch-1) or tPA (Ch-2) mRNAs. The activity of uPA can be revealed by use of the same zymographic assay that reveals tPA. The 3′ UTR of actin does not contain ACE determinants involved in silencing or activation, so that actin is constitutively synthesized in primary oocytes (Bachvarova et al. 1989; Paynton and Bachvarova 1994).
Figure 4.

Translation of injected chimeric and endogenous tPA mRNAs. (A) Schematic representation of the endogenous tPA mRNA and injected chimeric mRNA constructs. (Open rectangles) Coding regions of tPA and uPA mRNAs. Chimeric mRNA-2 and -1 constructs comprise the 5′ UTR, coding region, and part of the 3′ UTR of uPA mRNA and the 3′ end of the 3′ UTR of either tPA (Ch-2) or β-actin (Ch-1) mRNAs. (Shaded ovals and rectangles) ACE and AAUAAA sequences, respectively. (A)40 represents the ∼40-nucleotide poly(A) tail present on the endogenous tPA mRNA in primary oocytes. (A)n symbolizes the 3′ end of the constructs: (A)0, no poly(A) tail; (A)30, poly(A) tail of 30 nucleotides. (B) Translation of injected chimeric mRNAs. Primary oocytes were injected with Ch-2-(A)0 (lanes 1–4) or Ch2-(A)30 (lanes 5–8) mRNAs (200 ng/μl), either alone (lanes 1,2,5,6) or together with ACE transcript (2 μg/μl) (lanes 3,4,7,8), or with Ch-1-(A)0 (lanes 9,10) or Ch1-(A)30 (lanes 11,12) mRNAs (200 ng/μl). After 6 hr of culture in the presence of dB-cAMP, primary oocytes (GV) were lysed in duplicate groups of five (lanes 1–12). (Lanes 13–14) Oocytes injected with Ch-1-(A)30 mRNA, incubated in the absence of dB-cAMP and lysed in groups of five maturing oocytes (BD) 6 hr after GVBD. Synthesis of tPA and uPA was assessed by zymography. (C) Translation of endogenous tPA mRNA. The injected ACE transcript and the as-ODN 2504 (5′-AAAGTGTGAAAAATACCTCTG) are represented above and below the endogenous tPA mRNA, respectively. (Top) Primary oocytes were injected with the ACE transcript (2 μg/μl) either alone (lanes 3,4) or with as-ODN 2504 (1 μg/μl, lanes 5,6). After 6 hr of culture in the presence of dB-cAMP, groups of five primary oocytes (GV) were lysed and tPA synthesis was assessed by zymography. (Lanes 1,2) Groups of five noninjected primary oocytes cultured in parallel; (lanes 7,8) groups of five oocytes coinjected with the ACE transcript and as-ODN 2504, cultured in the absence of dB-cAMP, and lysed 6 hr after GVBD (BD).
As reported previously (Huarte et al. 1992), neither Ch-1 nor Ch-2 are translated in primary oocytes (Fig. 4B, lanes 1,2 and 9,10). Addition of a short poly(A) tail (30 A’s) to Ch-1 is sufficient to promote its translation in primary oocytes (lanes 11,12), whereas a similar short poly(A) tail does not allow translation of Ch-2 (lanes 5,6). Ch-2 (A)30 thus mimics the behavior of endogenous tPA mRNA, because it is silent in primary oocytes and translated during maturation (lanes 13,14). These observations indicate that a short poly(A) tail is necessary, but not sufficient, for translation in primary oocytes, and hence, that a distinct silencing mechanism operates on Ch-2.
Because injection of the ACE competitor relieves the silencing mechanism responsible for tPA mRNA dormancy, we coinjected the ACE transcript with Ch-2. The ACE transcript did not elicit translation of Ch-2 (A)0 but, as expected, did trigger translation of endogenous tPA mRNA (lanes 3,4). In contrast, Ch-2 bearing a short poly(A) tail was awakened in the presence of ACE, just like endogenous tPA mRNA (lanes 7,8).
We conclude that the translational activation of dormant Ch-2 can be achieved by injection of the ACE competitor, but that the presence of a short poly(A) tail is indispensable for translation. To verify whether the short poly(A) tail remaining on dormant tPA mRNA in primary oocytes is mandatory, we used as-ODN amputation (Stutz et al. 1997) to remove the poly(A) tail of the endogenous transcript. As-ODN 2504, complementary to the 3′ end of tPA mRNA, downstream of the AAUAAA sequence, promotes amputation of the poly(A) tail in primary oocytes, without preventing tPA mRNA translational activation during meiotic maturation, because the required regulatory regions (ACE and AAUAAA) are not removed (Stutz et al. 1997). The ACE competitor failed to induce translation of amputated (i.e., deadenylated) tPA mRNA (Fig. 4C, lanes 5,6), whereas, as expected, it did awaken the native mRNA (lanes 3,4), and the amputated mRNA was translated in maturing oocytes (lanes 7,8).
The partial deadenylation that accompanies silencing of tPA mRNA in growing oocytes is thus not sufficient to induce dormancy; interaction with a putative ACE-binding silencing factor appears critical. Conversely, competitive removal of this factor is sufficient to trigger translation of the mRNA in arrested primary oocytes, provided that a poly(A) tail is present; the poly(A) tail acts as a cis-acting determinant indispensable, but not sufficient, for translation.
Role of cytoplasmic readenylation during meiotic maturation
The translational activation of tPA mRNA in ACE-injected primary oocytes does not require elongation of its preexisting poly(A) tail. What is the role of the extensive readenylation of tPA mRNA in maturing oocytes? To explore this issue, we compared the translation of chimeric mRNAs (Ch-2, see Fig. 4A) comprising a wild-type AAUAAA polyadenylation signal or a mutant nonfunctional AAGAAA sequence. In the absence of a poly(A) tail, the Ch-2 transcript was translated in maturing oocytes, as evidenced by the synthesis of uPA, provided it had a wild-type polyadenylation signal (Fig. 5, lanes 3,4); as expected, the AAGAAA mutant transcript was not translated (lanes 11,12), because it cannot acquire the indispensable poly(A) tail during maturation. In contrast, both the wild-type (lanes 7,8) and the mutant (lanes 15,16) transcripts were translated during meiotic maturation when they were injected with a 40 nucleotide poly(A) tail at their 3′ end. All the injected transcripts were efficiently silenced in primary oocytes (lanes 1,2,5,6,9,10,13,14,17,18), and translated with comparable efficiency in vitro in a reticulocyte lysate (not shown).
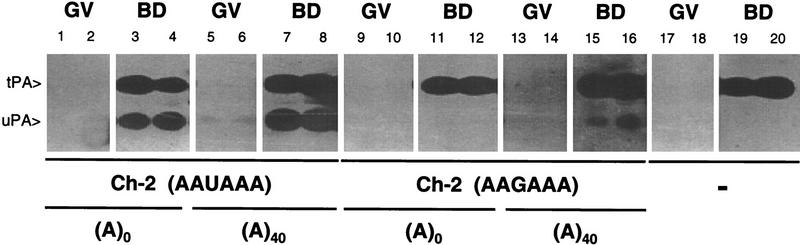
Figure 5.
Role of cytoplasmic readenylation during meiotic maturation. Primary oocytes were injected with the following transcripts (150 ng/μl): Ch-2 (AAUAAA) without [(A)0, lanes 1–4] or with [(A)40, lanes 5–8] a 40-nucleotide poly(A) tail, and a mutant form of Ch-2 lacking a functional polyadenylation signal, Ch-2 (AAGAAA) without [(A)0, lanes 9–12] or with [(A)40, lanes 13–16] a 40-nucleotide poly(A) tail. Injected oocytes were cultured in the presence (GV) or absence of dB-cAMP (BD) and lysed in groups of five after ∼10 hr. Noninjected control oocytes (−) (lanes 17–20) were cultured and lysed in parallel. Synthesis of tPA and uPA were assessed by zymography.
Thus, whereas translation requires the presence of a poly(A) tail, it need not be longer than that present on tPA mRNA in primary oocytes (∼50 nucleotide), and translational activation can take place even if polyadenylation itself is blocked, as is the case with the AAGAAA mutant. This indicates that cytoplasmic readenylation of tPA mRNA is not necessary for its translational awakening after GVBD. However, the amount of uPA in oocytes injected with the polyadenylated mutant Ch-2 transcript (Fig. 5, lanes 15,16) is clearly less than that in oocytes injected with the wild-type transcript (lanes 3,4,7,8). We have shown previously that an AAGAAA mutant form of the tPA-a transcript (see Fig. 1B), polyadenylated in vitro prior to injection, is extensively deadenylated in maturing oocytes, presumably by the default deadenylation pathway (Huarte et al. 1992). This deadenylation of the mutant Ch-2 transcript, that cannot be readenylated because it lacks a functional polyadenylation signal, probably accounts for the lower level of uPA accumulation.
Identification of an ACE-binding protein
The striking effect of the ACE transcript on tPA mRNA translation in primary oocytes suggests that it competes with the endogenous mRNA for binding of a silencing factor. To reveal macromolecules with which the ACE transcript may interact, we exposed meiotically arrested oocytes injected with different 32P-labeled transcripts to UV light. Oocyte lysates were digested with RNase T1, and the spectrum of proteins that had become isotopically tagged by label transfer was revealed by SDS-PAGE and autoradiography. A conspicuous radiolabeled complex of 85 kD was observed (Fig. 6A) following the injection of different tPA mRNA 3′ UTR fragments, that is, the 479-nucleotide tPA-a transcript (lane 1) and the 115-nucleotide tPA-b transcript (lane 2), both of which contain the ACE and AAUAAA regulatory regions, and the 83-nucleotide ACE competitor transcript itself (lane 3), which lacks the AAUAAA region. In contrast, no radiolabeled species of Mr greater than ∼60 kD was observed in samples from oocytes injected with actin or globin 3′ UTRs (lanes 4,5), or with the 80-nucleotide 3′-terminal fragment from tPA mRNA (tPA-d) (Fig. 1B, not shown). Because the 85-kD complex was observed with the tPA-a, tPA-b, and ACE transcripts, but not with the 3′ UTR of other mRNAs, the proposed RNA–protein interaction involves the ACE itself, and does not appear to require other structures present in tPA-a such as three upstream CPE-like sequences and the AAUAAA polyadenylation signal.
Figure 6.
In vivo UV-induced cross-linking of 32P-labeled 3′ UTR transcripts. (A) Primary oocytes were injected with different 32P-labeled transcripts (tr.) from the 3′ UTRs of tPA (tPA-a, tPA-b, and ACE, lanes 1–3), β-actin (Act., lane 4) and β-globin (Glo., lane 5) mRNAs. Alternatively, primary oocytes were coinjected with 32P-labeled tPA-a and the ACE competitor (Comp.) fragment (at a 50-fold molar excess) (lane 6), or with 32P-labeled tPA-b and the β-actin 3′ UTR fragment (at a 20-fold molar excess) (lane 7). Following 5 hr of culture in the presence of dB-cAMP, the oocytes were exposed to UV light and lysed. The samples were digested with RNase T1 and analyzed by SDS-PAGE and autoradiography. The migration of molecular-mass markers is indicated in kD. (B) In maturing oocytes, primary oocytes were injected with the 32P-labeled ACE transcript and cultured for 4 hr in medium with (lane 1) or without (lane 2) dB-cAMP, before being exposed to UV light. Maturing oocytes cultured without dB-cAMP were injected with the 32P-labeled ACE transcript 2 hr after GVBD and exposed to UV light 2 hr later (lane 3). Following lysis of the oocytes, the samples were processed and analyzed as for A. (C) ACE transcript in injected oocytes. Primary oocytes injected with the 32P-labeled ACE transcript were cultured for 6 hr in the presence (lane 2) or absence (lane 3) of dB-cAMP. The 32P-labeled transcripts recovered from arrested primary oocytes (GV, lane 2) or maturing oocytes (BD, lane 3) were analyzed on a 6% acrylamide–urea gel. In lane 1, the noninjected (n.i.) 32P-labeled ACE transcript was analyzed in parallel.
Competition with cold ligand confirmed that the 85-kD complex involves the ACE: coinjection of unlabeled ACE competitor with the labeled tPA-a fragment resulted in a marked decrease in the intensity of the 85-kD band (Fig. 6A, cf. lanes 1 and 6), whereas this band remained conspicuous when an unlabeled actin 3′ UTR transcript was injected together with the labeled tPA-b fragment (Fig. 6A, cf. lanes 2 and 7). Addition of the protease trypsin to extracts containing the 85-kD complex resulted in its degradation (not shown), suggesting that it is formed by UV-induced covalent cross-linking of the ACE with an oocyte protein.
Together with our previous results showing that the ACE region of tPA mRNA is protected from as-ODN hybridization in primary oocytes (Stutz et al. 1997), these observations suggest that an ∼80-kD protein specifically interacts with this cis-acting regulatory element in the dormant mRNA. Cross-linking of 32P-labeled transcripts injected in 50 μm diameter growing primary oocytes revealed that this protein is already present when the tPA gene is transcribed (not shown). Thus, this protein may be the silencing factor responsible for inducing and maintaining dormancy of newly transcribed tPA mRNA. If this is the case, interaction of this protein with its target region may be modified when the oocytes enter meiotic maturation and translation of the mRNA is activated. Thus, we compared the pattern of radiolabeled RNA–protein complexes obtained in arrested primary and in maturing oocytes by UV-induced cross-linking of the injected 32P-labeled ACE transcript (Fig. 6B). When injected oocytes were allowed to enter meiotic maturation before cross-linking, the migration of the 85-kD complex was slightly slower than in arrested primary oocytes, and its abundance was significantly reduced (cf. lanes 1 and 2). Similarly, when oocytes were injected immediately after GVBD, only small amounts of the slower-migrating complex were observed (lane 3). Oocytes injected in parallel with the 32P-labeled ACE transcript were lysed 6 hr later without cross-linking, and the integrity of the radiolabeled RNA fragment was verified (Fig. 6C). No significant alteration was observed in primary (GV, lane 2) or in maturing (BD, lane 3) oocytes; in particular, and as expected because it lacks the AAUAAA signal, the ACE transcript did not undergo polyadenylation in maturing oocytes. These observations suggest that a structural modification occurs in the ACE-binding protein when the oocytes undergo meiotic maturation, before tPA mRNA translation can be detected (i.e., ∼4 hr after GVBD), and that this modification accompanies a change in its binding to its target sequence. They are in accord with the hypothesis that the ACE-binding protein is a silencing factor that is released from the cis-acting regulatory region of tPA mRNA to allow its translation.
Discussion
The deadenylation and silencing of tPA mRNA in growing and fully grown primary oocytes and its readenylation and translational activation following resumption of meiosis depend on the presence of specific 3′ UTR sequences. The ACE region of the 3′ UTR is protected from as-ODNs in primary mouse oocytes, suggesting that it is engaged in an intra- or intermolecular interaction. This interaction could be directly involved in the silencing of tPA mRNA. In support of this hypothesis, part of the ACE region becomes accessible to as-ODNs early during meiotic maturation, before tPA synthesis can be detected (Stutz et al. 1997). Masking and unmasking may thus be part of the translational control mechanism of tPA mRNA.
The identification of RNA-masking translational repressors most often relies on experiments using cell-free extracts (Standart et al. 1990; Ostareck-Lederer et al. 1994). Experiments that explore translational repression of an endogenous mRNA in intact cells represent a useful alternative. Injection of an ACE-containing transcript in the cytoplasm of arrested primary oocytes resulted in the specific and very rapid translational awakening of tPA mRNA, in the absence of other changes characteristic of meiotic maturation, such as GVBD. Furthermore, ACE-injected oocytes allowed to undergo meiotic maturation produced more tPA than noninjected maturing oocytes. Injection of the ACE transcript presumably displaces a masking factor bound to the ACE region of tPA mRNA, and this appears sufficient to trigger translation. Arrested primary oocytes thus contain all the machinery required for translation of a dormant mRNA, and masking is responsible for the dormancy of tPA mRNA.
The silencing of tPA mRNA in growing primary oocytes is accompanied by its partial deadenylation. The ACE-binding translational repressor could thus target a deadenylase to tPA mRNA, or itself have deadenylase activity. In Xenopus oocytes, a nuclear deadenylase released after GVBD is implicated in the default deadenylation of mRNAs lacking CPEs (Varnum et al. 1992; Wormington et al. 1996). In mouse oocytes, such a default deadenylation was not prevented by the ACE competitor, suggesting that the default process is different from that involved in the specific deadenylation of tPA mRNA. In accord with this view, deadenylation of tPA mRNA in primary oocytes leaves a short poly(A) tail of ∼40 residues, whereas default deadenylation in maturing oocytes appears complete. The relationship between the cytoplasmic deadenylation of ACE-containing transcripts and their translational silencing in primary oocytes is not clear. A polyadenylated ACE-containing reporter transcript is only transiently translated in primary oocytes, being silenced at a time when deadenylation is still ongoing (Huarte et al. 1992). Thus, deadenylation may not be a prerequisite for silencing.
The translational activation of tPA mRNA in ACE-injected arrested primary oocytes occurs without elongation of its poly(A) tail. Hence, cytoplasmic readenylation is not required, and factor(s) responsible for polyadenylation in maturing oocytes may be lacking or inactive in primary oocytes (Paynton and Bachvarova 1994). However, the presence of a short poly(A) tail is mandatory for translation of endogenous tPA mRNA, as it is for translation of injected reporter mRNAs. Interestingly, the poly(A) stretch that remains at the 3′ end of partially deadenylated tPA mRNA in primary oocytes is of a size sufficient for binding of PABP, which stimulates the formation of translation initiation complexes (Munroe and Jacobson 1990). We do not know whether competitive displacement of the ACE-binding repressor is sufficient for translation, or whether it triggers a modification of tPA mRNA other than poly(A) tail elongation. In this context, the causal relationship between polyadenylation, cap ribose methylation, and translational activation of a dormant mRNA in Xenopus oocytes may be relevant (Kuge and Richter 1995). In maturing Xenopus oocytes, methylation is secondary to polyadenylation; if cap ribose methylation does occur in primary mouse oocytes, it is unlikely to be related to polyadenylation because the awakening of tPA mRNA is not accompanied by elongation of its poly(A) tail.
To search for trans-acting effector(s) of tPA mRNA silencing in primary oocytes, we injected radiolabeled 3′ UTR fragments and exposed the live oocytes to UV light. An 85-kD RNA–protein complex was obtained following RNase T1 digestion of cross-linked extracts from oocytes injected with different ACE-containing transcripts. Given the size of the largest RNase T1 fragment of the ACE transcript (25 nucleotides, i.e., ∼8 kD), the ACE-binding protein should have a molecular mass of ∼77 kD. Proteins with high affinity for U-rich 3′ UTR sequences have been described from a variety of sources (McGrew and Richter 1990; Gillis and Malter 1991; Fox et al. 1992; Simon and Richter 1994; Decker and Parker 1995; Simon et al. 1996; Wu et al. 1997). CPEB, responsible for polyadenylation of a maternal transcript in maturing Xenopus oocytes, has been purified (Hake and Richter 1994) and a cDNA encoding its murine homolog (mCPEB) has been characterized (Gebauer and Richter 1996). The size of mCPEB (62 kD), and the fact that it is responsible for events occuring after GVBD, rather than in arrested primary oocytes, distinguish it from the ACE-binding protein. Interestingly, in Spisula solidissima oocytes, a 3′ UTR-binding 82-kD protein represses translation of the mRNAs encoding cyclin A and the small subunit of ribonucleotide reductase (Standart et al. 1990; Walker et al. 1996); this protein, reported to be 67% identical to Xenopus CPEB in the carboxy-terminal region (Gebauer et al. 1994; Gebauer and Richter 1996), thus has a function and a size similar to the ACE-binding protein.
On entry in meiotic maturation, shortly after GVBD, the ∼80-kD ACE-binding protein appears to undergo a modification that results in a slower migration of the RNA–protein complex; the recovered complex also decreases in abundance. Although direct evidence for this is lacking, a post-translational modification of the protein may decrease its affinity for its target sequence, thus leading to unmasking of the ACE region of tPA mRNA. This unmasking, like that achieved by injection of the competitor ACE transcript, frees the mRNA for translation. Our previous demonstration that the ACE region becomes partially accessible to as-ODN hybridization shortly after GVBD (Stutz et al. 1997) is in accord with this model; also in support of the notion that the ACE sequence is not involved in RNA–protein interactions after GVBD is the observation that the ACE competitor by itself does not prevent polyadenylation or translation of tPA mRNA during meiotic maturation.
Different post-translational modifications could account for a change in electrophoretic migration of the ACE–protein complex during meiotic maturation. For instance, ubiquitination would mark the protein for degradation (Hochstrasser 1996), a change in its redox state could influence its interaction with the mRNA (Hentze et al. 1989; Gillis and Malter 1991; Malter and Hong 1991). However, phosphorylation of the protein, possibly by a cell cycle kinase, appears particularly likely: Several precedents illustrate the importance of phosphorylation in modulating the activity of RNA-binding proteins controlling cytoplasmic polyadenylation and/or translation (Paris et al. 1991; Kwon and Hecht 1993; Hake and Richter 1994; Walker et al. 1996). Furthermore, translation of tPA mRNA can be induced, together with GVBD, in incompetent mouse oocytes by the injection of mRNAs encoding cdc2 and cyclin B1, resulting in the induction of MPF activity (de Vantery et al. 1997).
The translational awakening of tPA mRNA, together with the modification and probable decrease in affinity of the ACE-binding protein when meiosis resumes, suggest a model in which masking and unmasking are critical for translational silencing and later awakening of this mRNA in oocytes. What then is the role of the extensive readenylation of tPA mRNA during meiotic maturation? Although readenylation is not necessary for transient translational awakening, the presence of a short poly(A) tail is required. Thus, cytoplasmic readenylation of tPA mRNA is not required for translational activation per se, but it is essential to maintain a poly(A) tail on the mRNA in the face of a default deadenylation process affecting many other oocyte mRNAs. This view is in accord with the concept that the poly(A) tail length of CPE-containing mRNAs is determined by an equilibrium between two competing reactions: polyadenosine addition and removal (Wickens 1992). Recent observations on the respective contributions of unmasking and polyadenylation in the translational control of FGF receptor mRNA in Xenopus laevis oocytes are also fully consistent with our findings: These two modifications can be uncoupled, and unmasking appears critical to trigger translation (Culp and Musci 1998).
The respective roles of masking, unmasking, and readenylation in modulating the expression of a maternal mRNA should be viewed in the more general perspective of translational control in other cell types (Decker and Parker 1995; Spirin 1995). In male gametes, mRNA silencing by binding of regulatory proteins to 3′ UTRs is well documented (Kwon and Hecht 1991; Goodwin et al. 1993; Fajardo et al. 1994; Schafer et al. 1995), and translational activation does not appear to require polyadenylation (Kleene 1989). In Drosophila embryos, nanos and pumilio promote posterior morphology by triggering specific deadenylation and translational repression of the maternal hunchback mRNA (Wreden et al. 1997). In somatic cells also, there is growing evidence for an important role of specific 3′ UTR sequences in silencing certain mRNAs (Kruys et al. 1989; Gueydan et al. 1996; Kern et al. 1996; Ostareck et al. 1997; Ranganathan et al. 1997) in addition to their function in controlling mRNA stability and degradation (Jacobson 1996); here again, cytoplasmic readenylation does not appear to play a part in reactivating translation. We propose that what may be particular to oocytes is neither the use of translational control as one way to modulate gene expression, nor the critical role of 3′ UTR masking and unmasking to regulate protein synthesis, but a particular need to maintain a poly(A) tail on mRNAs that are translated, whereas others are deadenylated and degraded. In somatic cells, under conditions that lead to activation of a previously silent mRNA, the mRNA will be translated, deadenylated, and degraded, and new transcripts can be generated that will not be silenced. In oocytes and early embryos, unmasking allows translation and cytoplasmic readenylation is necessary to maintain a poly(A) tail, and thereby a translatable mRNA, because new transcripts cannot be synthesized.
This hypothesis predicts that the cellular machinery required for cytoplasmic readenylation could be, at least in part, specific to oocytes. Whereas there is no evidence for an oocyte-specific form of CPSF, the presence of multiple forms of poly(A) polymerase in Xenopus oocytes (Ballantyne et al. 1995; Gebauer and Richter 1995) suggests that cytoplasmic readenylation might require a specific form of the enzyme. Similarly, CPEB appears to be restricted mostly to the gonads and, in the ovary, is detectable only in oocytes (Gebauer and Richter 1996). In contrast, trans-acting elements such as the ∼80-kD protein that binds to the ACE in tPA mRNA might operate to control translation of selected mRNAs in somatic cells also.
Translational awakening of oocyte tPA mRNA can be achieved in two ways. The modification and probable detachment of a masking protein from its 3′ UTR target sequence appears as the physiological mechanism responsible for tPA synthesis during meiotic maturation. Experimentally, however, competition by an injected transcript corresponding to the cis-acting ACE sequence also results in translation of the mRNA. This suggests that untranslated RNA transcripts, or untranslated regions of mRNAs, have the potential of relieving translational repression operating on a subset of mRNAs. It is reminiscent of the striking biological effects on cell growth and differentiation that have been attributed to 3′ UTRs of mRNAs encoding muscle structural proteins (Rastinejad and Blau 1993; Rastinejad et al. 1993; Ranganathan et al. 1995, 1997), and ribonucleotide reductase (Amara et al. 1995; Fan et al. 1996). Thus, translational regulation may involve multiple mechanisms whereby the masking effect of trans-acting repressors can be overcome, to yield the rapid and specific induction of protein synthesis.
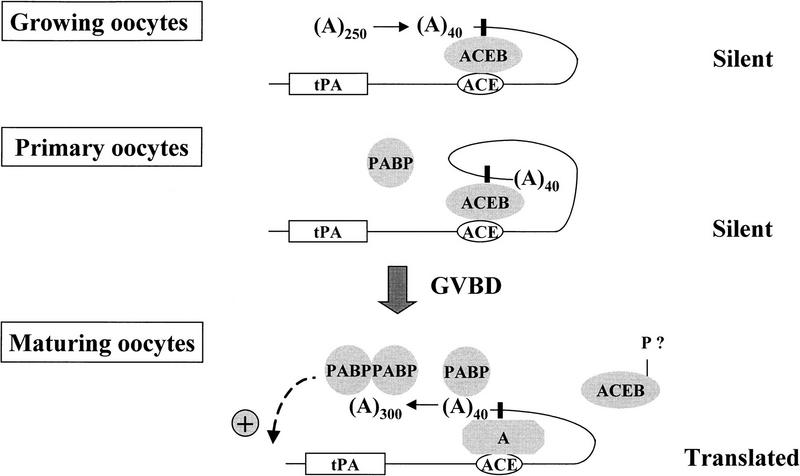
In conclusion, our results show how masking and unmasking mechanisms and poly(A) tail metabolism cooperate to control translation of a transiently dormant mRNA in mouse oocytes. They suggest the following model (Fig. 7): In growing primary oocytes, the newly-transcribed polyadenylated (∼250 A’s) mRNA is exported from the nucleus to the cytoplasm, in which a specific cis-acting silencing determinant (ACE) interacts with a trans-acting translational repressor (ACEB). This interaction also induces a partial deadenylation, which leaves only a short poly(A) tail (∼40 A’s) on the dormant mRNA. In fully grown oocytes, the repressor is required to maintain dormancy. This could happen by preventing the PABP from binding to the short poly(A) tail and thereby from contributing to translation initiation. Early after resumption of meiosis, the repressor is modified (e.g., phosphorylated) and released from the mRNA. PABP can then bind to the poly(A) tail, thus allowing translational activation of the mRNA. Concomitantly, readenylation, itself dependent on specific structural elements in the 3′ UTR, that is, the ACE and the polyadenylation signal, occurs at a time when many other oocyte mRNAs are rapidly deadenylated. This allows the persistance of a poly(A) tail, which is indispensable for translation. In this view, readenylation is necessary to maintain, possibly to stimulate, translation, but it is not directly involved in the awakening of the dormant mRNA. This underlines the critical role played by masking and unmasking mechanisms in the translational control of a mammalian maternal mRNA.
Figure 7.
Model for tPA mRNA translational control. (ACEB) ACE-binding protein; (P) putative post-translational modification (phosphorylation?); (PABP) poly(A) binding protein; (A) factors necessary for cytoplasmic polyadenylation. The solid, vertical rectangle represents the AAUAAA cleavage and polyadenylation signal.
Materials and methods
Oocyte collection, injection, and culture
Procedures for oocyte collection from Swiss albino mice, injection, and culture have been described previously (Huarte et al. 1985, 1987; Strickland et al. 1988). For injection, primary oocytes were incubated in DMEM containing 5% fetal calf serum, 25 μg/ml sodium pyruvate, and 2.5 mg/ml polyvinylpyrrolidone (PVP, Pharmacia); a volume of ∼10 pl was injected in the cytoplasm of the oocytes. Oocytes were cultured either in the presence of dB-cAMP (100 μg/μl), to prevent resumption of meiosis, or in its absence to allow meiotic maturation. Growing primary oocytes of ∼50 μm diameter were prepared from 10-day-old females according to Eppig (1977).
Construction of plasmids and in vitro transcription
The cytoplasmic rabbit β-globin 3′ UTR fragment was transcribed with SP6 polymerase from a plasmid [A(3), (Vassalli et al. 1989)]; this generates a transcript of 165 nucleotides.
A 140-nucleotide cDNA fragment (positions 1753–1892) of mouse cytoplasmic β-actin 3′ UTR (Tokunaga et al. 1986) was subcloned in pGEM2 (Promega Corp., Madison, WI). This plasmid was linearized with ApaLI and transcribed with T3 RNA polymerase to generate a transcript of 133 nucleotides.
The tPA mRNA 3′ UTR transcripts, tPA-a, tPA-b, and tPA-d (of 479, 115, and 80 nucleotides, respectively) were prepared from a template generated by PCR amplification of plasmids containing the 455, 104, and 37 3′-terminal nucleotides of mouse tPA cDNA [transcripts 1, 5, and 6 in Huarte et al. (1992)], with a 5′ primer (SP6 primer, 5′-GGCTTGTACATATTGTCGTTA) corresponding to plasmid sequences upstream of the SP6 promoter and a 3′ primer complementary to the last 21 nucleotide of tPA cDNA (tPA primer, 5′-AAAGTGTGAAAAATACCTCTG). Two 51-bp synthetic ApaI–XhoI double-stranded ODNs, containing either the ACE region of tPA cDNA (ACE, positions 2401–2442) or a different tPA 3′ UTR-sequence (tPA-c positions, 2275–2314), were subcloned into a digested ApaI–XhoI plasmid coding for the Ch-1 transcript. ACE-containing ODN (5′-CACTAGTATATTTATATTTTAATCTATTTTAGATTTTACTTTC and its complementary sequence, 3′-CCGGGTGATCATATAAATATAAAATTAGATAAAATCTAAAATGAAAGAGCT) were hybridized before subcloning. tPA-c-containing ODN (5′-CCTGTACTCCACACTCCTCAACTCTTGGGACATATCCACTGAC and its complementary sequence 3′-CCGGGGACATGAGGTGTGAGGAGTTGAGAACCCTGTATAGGTGACTGAGCT) were also hybridized before subcloning. The ACE or tPA-c transcripts (both 83 nucleotides long) were obtained with T3 polymerase, after linearizing the plasmids with XbaI.
Chimeric mRNA-1 and mRNA-2 (Ch-1 and Ch-2) transcripts were prepared from plasmids as described previously (Huarte et al. 1992). Two XbaI–XbaI fragments corresponding either to the Ch-1 or to the Ch-2 DNA inserts were subcloned into a pSP65 poly(A)30 vector. Chimeric mRNA-1 (A)30 [Ch-1 (A)30] and mRNA-2 (A)30 [Ch-2 (A)30] transcripts were prepared with SP6 polymerase from templates generated by PCR amplifications using the 5′ SP6 primer and a 3′ primer complementary at its 5′ end to the poly(A)30 tract and at its 3′ end to 3 nucleotides of the multiple cloning site of the pSP65 poly(A)30 vector [5′-(T)30 AGC]. Chimeric transcripts mRNA-2 mutated in the poly(A) signal [Ch-2-AAGAAA (A)40 or (A)0] and their wild-type controls [Ch-2-AAUAAA (A)40 or (A)0] were all generated from PCR amplification. Ch-2-AAGAAA (A)40 and (A)0 were amplified with the 5′ SP6 primer and a 3′ primer complementary to the last 36 nucleotides of tPA cDNA except for the C/A substitution in the poly(A) signal with or without a 40 T residue tail [5′-(T)40 or (T)0 AAAGTGTGAAAAATACCTCTGAATTTCTTATTTAAG]. The 3′ primer used for the Ch-2 (A)0 is complementary to the same last 36 nucleotide without the G substitution in the poly(A) signal.
All transcripts were capped and prepared as described previously (Huarte et al. 1987). The transcripts used either in the cross-linking experiment or analyzed directly on acrylamide urea gels are labeled with [32P]UTP at 4 μCi/μl and a total concentration of UTP of 50 μM and injected at ∼5 × 106 cpm/μl. Competitor and chimeric transcripts used in competition displacement or in translation assays were prepared using [32P]UTP at 0.5 μCi/μl and a total concentration of UTP of 500 μm and were injected at 2 μg/μl or ∼150–200 ng/μl, respectively. When indicated, the transcripts were polyadenylated in vitro as described previously (Vassalli et al. 1989). The purified RNAs were dissolved in 150 mm KCl before injection.
In vivo UV-induced cross-linking and gel electrophoresis of RNA–protein complexes
Oocytes injected with 32P-labeled transcripts (5 × 106 cpm/μl) were placed on ice and exposed for 7 min to UV radiation in a Stratalinker device (1 μJ/cm2). Following lysis in 20 μl of 10 mm Tris-Cl at pH 7.4, 1 mm EDTA, 1 mm DTT, 0.25% Triton X-100, 1 μg/μl yeast tRNA, and 1 U/μl RNasin, RNase T1 was added (0.5 U/μl) and the samples (20–30 oocytes per sample) were digested for 30 min at 37°C. An equal volume of double-strength reducing sample buffer was added, and the samples were subjected to a 10% SDS-PAGE (Laemmli 1970) and autoradiography.
ODN synthesis and purification
ODNs were synthesized on an Applied Biosystems model 380A DNA synthesizer with the phosphoramidite method. All ODNs were purified by extraction with n-butanol (Sawadogo and Van Dyke 1991), extracted once with phenol/chloroform and precipitated with ethanol. The purified ODNs were dissolved in 150 mm KCl at 1 μg/μl (1 A260 = 33 μg/ml).
RNA analysis and translation assays
RNA extractions were performed as described previously (Huarte et al. 1987). Size analysis was performed by electrophoresis in 6% polyacrylamide-urea gels and autoradiography. For translation assays, cultured oocytes were collected in groups of five, lysed in 0.25% Triton X-100 and 1 μg/μl bovine serum albumin, and assayed by zymography (Huarte et al. 1985). For biosynthetic labeling of proteins, primary oocytes were cultured in modified Biggers medium (Paleos and Powers 1981) for 6 hr in the presence of 200 μCi of Tran35S-label (1.079 Ci/mmole, ICN Biomedicals, Inc) per milliliter. Oocyte extracts were subjected to a 10% SDS-PAGE (Laemmli 1970) and autoradiography.
Poly(A) test
The length of the poly(A) tail of tPA mRNA was measured by the poly(A) test (PAT) (Salles and Strickland 1995). Total RNA was annealed with p(dT)12–18 in the presence of T4 DNA ligase; an oligo(dT17)–anchor [5′-CGAATTCTCGAGGATCCGTCGAC(T)17] was then added to the reaction. Reverse transcription was followed by PCR amplification with a tPA 5′-specific oligonucleotide: 5′-CCACACTCCTCAACTCTTGGGAC and with an adaptor oligonucleotide: 5′-CGAATTCTCGAGGATCCGTCGAC; the reaction was labeled with 5 μCi of [32P]dATP (3000 Ci/mmole, Amersham). The amplification products were electrophoresed on 6% denaturing polyacrylamide–urea gels.
Acknowledgments
We thank Dominique Belin for helpful advice throughout this work. This work was supported by grants from the Fonds National Suisse de la Recherche Scientifique.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Andre.stutz@medecine.unige.ch; FAX 41-22 70 25 260.
References
- Amara FM, Chen FY, Wright JA. Defining a novel cis element in the 3′-untranslated region of mammalian ribonucleotide reductase component R2 mRNA: Role in transforming growth factor-beta 1 induced mRNA stabilization. Nucleic Acids Res. 1995;23:1461–1467. doi: 10.1093/nar/23.9.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvarova R, De Leon V, Johnson A, Kaplan G, Paynton BV. Changes in total RNA, polyadenylated RNA, and actin mRNA during meiotic maturation of mouse oocytes. Dev Biol. 1985;108:325–331. doi: 10.1016/0012-1606(85)90036-3. [DOI] [PubMed] [Google Scholar]
- Bachvarova R, Cohen EM, De Leon V, Tokunaga K, Sakiyama S, Paynton BV. Amounts and modulation of actin mRNAs in mouse oocytes and embryos. Development. 1989;106:561–565. doi: 10.1242/dev.106.3.561. [DOI] [PubMed] [Google Scholar]
- Ballantyne S, Bilger A, Astrom J, Virtanen A, Wickens M. Poly(A) polymerases in the nucleus and cytoplasm of frog oocytes: Dynamic changes during oocyte maturation and early development. RNA. 1995;1:64–78. [PMC free article] [PubMed] [Google Scholar]
- Bilger A, Fox CA, Wahle E, Wickens M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes & Dev. 1994;8:1106–1116. doi: 10.1101/gad.8.9.1106. [DOI] [PubMed] [Google Scholar]
- Culp PA, Musci TJ. Translational activation and cytoplasmic polyadenylation of FGF receptor-1 are independently regulated during Xenopus oocyte maturation. Dev Biol. 1998;193:63–76. doi: 10.1006/dbio.1997.8785. [DOI] [PubMed] [Google Scholar]
- Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- de Vantery C, Stutz A, Vassalli JD, Schorderet-Slatkine S. Acquisition of meiotic competence in growing mouse oocytes is controlled at both translational and posttranslational levels. Dev Biol. 1997;187:43–54. doi: 10.1006/dbio.1997.8599. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Analysis of mouse oogenesis in vitro: Oocyte isolation and the utilization of exogenous energy sources by growing oocytes. J Exp Zool. 1977;198:375–382. doi: 10.1002/jez.1401980311. [DOI] [PubMed] [Google Scholar]
- Fajardo MA, Butner KA, Lee K, Braun RE. Germ cell-specific proteins interact with the 3′ untranslated regions of Prm-1 and Prm-2 mRNA. Dev Biol. 1994;166:643–653. doi: 10.1006/dbio.1994.1344. [DOI] [PubMed] [Google Scholar]
- Fan H, Villegas C, Huang A, Wright JA. Suppression of malignancy by the 3′ untranslated regions of ribonucleotide reductase R1 and R2 messenger RNAs. Cancer Res. 1996;56:4366–4369. [PubMed] [Google Scholar]
- Fox CA, Wickens MP. Poly(A) removal during oocyte maturation: A default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes & Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- Fox CA, Sheets MD, Wahle E, Wickens M. Polyadenylation of maternal mRNA during oocyte maturation: Poly(A) addition in vitro requires a regulated RNA binding activity and a poly(A) polymerase. EMBO J. 1992;11:5021–5032. doi: 10.1002/j.1460-2075.1992.tb05609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Richter JD. Cloning and characterization of a Xenopus poly(A) polymerase. Mol Cell Biol. 1995;15:1422–1430. doi: 10.1128/mcb.15.3.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Mouse cytoplasmic polyadenylylation element binding protein: An evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc Natl Acad Sci. 1996;93:14602–14607. doi: 10.1073/pnas.93.25.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Xu W, Cooper GM, Richter JD. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis P, Malter JS. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991;266:3172–3177. [PubMed] [Google Scholar]
- Goodwin EB, Okkema PG, Evans TC, Kimble J. Translational regulation of tra-2 by its 3′ untranslated region controls sexual identity in C. elegans. Cell. 1993;75:329–339. doi: 10.1016/0092-8674(93)80074-o. [DOI] [PubMed] [Google Scholar]
- Gueydan C, Houzet L, Marchant A, Sels A, Huez G, Kruys V. Engagement of tumor necrosis factor mRNA by an endotoxin-inducible cytoplasmic protein. Mol Med. 1996;2:479–488. [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Rouault TA, Harford JB, Klausner RD. Oxidation-reduction and the molecular mechanism of a regulatory RNA–protein interaction. Science. 1989;244:357–359. doi: 10.1126/science.2711187. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–825. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- Huarte J, Belin D, Vassalli J-D. Plasminogen activator in mouse and rat oocytes: Induction during meiotic maturation. Cell. 1985;43:551–558. doi: 10.1016/0092-8674(85)90184-9. [DOI] [PubMed] [Google Scholar]
- Huarte J, Belin D, Vassalli A, Strickland S, Vassalli J-D. Meiotic maturation of mouse oocytes triggers the translation and polyadenylation of dormant tissue-type plasminogen activator mRNA. Genes & Dev. 1987;1:1201–1211. doi: 10.1101/gad.1.10.1201. [DOI] [PubMed] [Google Scholar]
- Huarte J, Stutz A, O’Connell ML, Gubler P, Belin D, Darrow AL, Strickland S, Vassalli J-D. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- Jacobson A. Poly(A) metabolism and translation: The closed-loop model. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 451–480. [Google Scholar]
- ————— Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Kern PA, Ranganathan G, Yukht A, Ong JM, Davis RC. Translational regulation of lipoprotein lipase by thyroid hormone is via a cytoplasmic repressor that interacts with the 3′ untranslated region. J Lipid Res. 1996;37:2332–2340. [PubMed] [Google Scholar]
- Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989;106:367–373. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- Kuge H, Richter JD. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: Implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Hecht NB. Cytoplasmic protein binding to highly conserved sequences in the 3′ untranslated region of mouse protamine 2 mRNA, a translationally regulated transcript of male germ cells. Proc Natl Acad Sci. 1991;88:3584–3598. doi: 10.1073/pnas.88.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Binding of a phosphoprotein to the 3′ untranslated region of the mouse protamine 2 mRNA temporally represses its translation. Mol Cell Biol. 1993;13:6547–6557. doi: 10.1128/mcb.13.10.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Malter JS, Hong Y. A redox switch and phosphorylation are involved in the post-translational up-regulation of the adenosine-uridine binding factor by phorbol ester and ionophore. J Biol Chem. 1991;266:3167–3171. [PubMed] [Google Scholar]
- McGrew LL, Richter JD. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: Characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990;9:3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Osborne HB, Richter JD. Translational control by polyadenylation during early development. Prog Mol Subcell Biol. 1997;18:173–198. doi: 10.1007/978-3-642-60471-3_8. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleos GA, Powers RD. The effect of calcium on the first meiotic division of the mammalian oocyte. J Exp Zool. 1981;217:409–416. doi: 10.1002/jez.1402170312. [DOI] [PubMed] [Google Scholar]
- Paris J, Swenson K, Piwnica Worms H, Richter JD. Maturation-specific polyadenylation: In vitro activation by p34cdc2 and phosphorylation of a 58-kD CPE-binding protein. Genes & Dev. 1991;5:1697–1708. doi: 10.1101/gad.5.9.1697. [DOI] [PubMed] [Google Scholar]
- Paynton BV, Bachvarova R. Polyadenylation and deadenylation of maternal mRNAs during oocyte growth and maturation in the mouse. Mol Reprod Dev. 1994;37:172–180. doi: 10.1002/mrd.1080370208. [DOI] [PubMed] [Google Scholar]
- Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- Ranganathan G, Ong JM, Yukht A, Saghizadeh M, Simsolo RB, Pauer A, Kern PA. Tissue-specific expression of human lipoprotein lipase. Effect of the 3′-untranslated region on translation. J Biol Chem. 1995;270:7149–7155. doi: 10.1074/jbc.270.13.7149. [DOI] [PubMed] [Google Scholar]
- Ranganathan G, Vu D, Kern PA. Translational regulation of lipoprotein lipase by epinephrine involves a trans-acting binding protein interacting with the 3′ untranslated region. J Biol Chem. 1997;272:2515–2519. doi: 10.1074/jbc.272.4.2515. [DOI] [PubMed] [Google Scholar]
- Rastinejad F, Blau HM. Genetic complementation reveals a novel regulatory role for 3′ untranslated regions in growth and differentiation. Cell. 1993;72:903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- Rastinejad F, Conboy MJ, Rando TA, Blau HM. Tumor suppression by RNA from the 3′ untranslated region of alpha-tropomyosin. Cell. 1993;75:1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- Richter JD. Dynamics of poly(A) addition and removal during development. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 481–503. [Google Scholar]
- Rickles RJ, Darrow AL, Strickland S. Molecular cloning of complementary DNA to mouse tissue plasminogen activator mRNA and its expression during F9 teratocarcinoma cell differentiation. J Biol Chem. 1988;263:1563–1569. [PubMed] [Google Scholar]
- Robbie EP, Peterson M, Amaya E, Musci TJ. Temporal regulation of the Xenopus FGF receptor in development: A translation inhibitory element in the 3′ untranslated region. Development. 1995;121:1775–1785. doi: 10.1242/dev.121.6.1775. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: Translation initiation in eucaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Salles FJ, Strickland S. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl. 1995;4:317–321. doi: 10.1101/gr.4.6.317. [DOI] [PubMed] [Google Scholar]
- Sawadogo M, Van Dyke MW. A rapid method for the purification of deprotected oligodeoxynucleotides. Nucleic Acids Res. 1991;19:674. doi: 10.1093/nar/19.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer M, Nayernia K, Engel W, Schafer U. Translational control in spermatogenesis. Dev Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- Seydoux G. Mechanisms of translational control in early development. Curr Opin Genet Dev. 1996;6:555–561. doi: 10.1016/s0959-437x(96)80083-9. [DOI] [PubMed] [Google Scholar]
- Simon R, Richter JD. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol Cell Biol. 1994;14:7867–7875. doi: 10.1128/mcb.14.12.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Wu L, Richter JD. Cytoplasmic polyadenylation of activin receptor mRNA and the control of pattern formation in Xenopus development. Dev Biol. 1996;179:239–250. doi: 10.1006/dbio.1996.0254. [DOI] [PubMed] [Google Scholar]
- Spirin AS. Masked and translatable messenger ribonucleoproteins in higher eukaryotes. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 319–334. [Google Scholar]
- Standart N, Jackson RJ. Regulation of translation by specific protein/mRNA interactions. Biochimie. 1994;76:867–879. doi: 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- Standart N, Dale M, Stewart E, Hunt T. Maternal mRNA from clam oocytes can be specifically unmasked in vitro by antisense RNA complementary to the 3′-untranslated region. Genes & Dev. 1990;4:2157–2168. doi: 10.1101/gad.4.12a.2157. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Richter JD. Translational control during early development. Crit Rev Eukaryot Gene Expression. 1997;7:73–94. doi: 10.1615/critreveukargeneexpr.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocytes maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- Strickland S, Huarte J, Belin D, Vassalli A, Rickles RJ, Vassalli J-D. Antisense RNA directed against the 3′ noncoding region prevents dormant mRNA activation in mouse oocytes. Science. 1988;241:680–684. doi: 10.1126/science.2456615. [DOI] [PubMed] [Google Scholar]
- Stutz A, Huarte J, Gubler P, Conne B, Belin D, Vassalli JD. In vivo antisense oligodeoxynucleotide mapping reveals masked regulatory elements in an mRNA dormant in mouse oocytes. Mol Cell Biol. 1997;17:1759–1767. doi: 10.1128/mcb.17.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K, Taniguchi H, Yoda K, Shimizu M, Sakiyama S. Nucleotide sequence of a full-length cDNA for mouse cytoskeletal beta-actin mRNA. Nucleic Acids Res. 1986;14:2829. doi: 10.1093/nar/14.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum SM, Wormington WM. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: A default mechanism for translational control. Genes & Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]
- Varnum SM, Hurney CA, Wormington WM. Maturation-specific deadenylation in Xenopus oocytes requires nuclear and cytoplasmic factors. Dev Biol. 1992;153:283–290. doi: 10.1016/0012-1606(92)90113-u. [DOI] [PubMed] [Google Scholar]
- Vassalli J-D, Stutz A. Translational control. Awakening dormant mRNAs. Curr Biol. 1995;5:476–479. doi: 10.1016/s0960-9822(95)00095-9. [DOI] [PubMed] [Google Scholar]
- Vassalli J-D, Huarte J, Belin D, Gubler P, Vassalli A, O’Connell ML, Parton LA, Rickles RJ, Strickland S. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes & Dev. 1989;3:2163–2171. doi: 10.1101/gad.3.12b.2163. [DOI] [PubMed] [Google Scholar]
- Walker J, Dale M, Standart N. Unmasking mRNA in clam oocytes: Role of phosphorylation of a 3′ UTR masking element-binding protein at fertilization. Dev Biol. 1996;173:292–305. doi: 10.1006/dbio.1996.0024. [DOI] [PubMed] [Google Scholar]
- Wickens M. Forward, backward, how much, when: Mechanisms of poly(A) addition and removal and their role in early development. Semin Dev Biol. 1992;3:399–412. [Google Scholar]
- Wickens M, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 411–450. [Google Scholar]
- Wormington M, Searfoss AM, Hurney CA. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Wu L, Good PJ, Richter JD. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]