Abstract
The developmental ontogeny of the vascular system (consisting of xylem, phloem and [pro]cambium) is poorly understood despite its central role in plant physiology. We show that in the Arabidopsis root meristem, xylem cell lineages are specified early, whereas phloem and procambium are established through a set of asymmetric cell divisions. These divisions require the WOODEN LEG (WOL) gene. The WOL gene encodes a novel two-component signal transducer with an unusual tandem arrangement of two receiver domains. It is expressed specifically in the vasculature from the early stages of embryogenesis on, consistent with a role as a sensor for vascular morphogenesis.
Keywords: WOODEN LEG, procambium, asymmetric division, DhkA, embryogenesis
The plant vascular system is responsible for transporting water, nutrients, and photosynthates between plant organs. It also undergoes developmental adaptations, such as wood formation, which involve specific proliferation of the vascular tissue. Therefore, the pattern of cell divisions is an important determinant of the cellular organization of this tissue (Esau 1977). Vascular tissue is first established during embryogenesis as an undifferentiated procambial tissue in the innermost domain of the plant embryo enclosed by the epidermal and ground tissue layers (Esau 1977; Steeves and Sussex 1989). After differentiation of the phloem and xylem strands within this domain, cell proliferation originates primarily from the initial cells of the procambial tissue immediately proximal to the mitotically quiescent regions of the terminal meristems (Esau 1977; Steeves and Sussex 1989; Scheres et al. 1994). Later in development, a lateral meristem (the cambium) is formed as the undifferentiated cells begin to divide in the procambial tissue between the phloem and xylem strands. There is a high degree of diversity of the cell-division patterns within the vascular tissue in plants, especially with regard to the formation and activity of the cambium. As these patterns are species specific, it is conceivable that the control of cell proliferation within the vascular tissue is largely under genetic regulation.
Several factors have been implicated in the regulation of cell proliferation of the vascular tissue. On the basis of mutation analyses, signal transduction pathways related to auxin (Carland and McHale 1996; Oyama 1997; Hardtke and Berleth 1998; Steinmann et al. 1999; Hobbie et al. 2000; Koizumi et al. 2000) and brassinosteroid (Jang et al. 2000; Schrick et al. 2000) phytohormones are involved. Physiological and genetic experiments have also indicated a role for other phytohormones (such as gibberellins, cytokinins, and ethylene; Aloni 1987; Eriksson et al. 2000), for sucrose (Warren Wilson 1978), and for physical pressure (Brown 1964). Furthermore, a few genetic loci have been identified that are essential for normal cell proliferation but function in an as yet uncharacterized molecular context (Scheres et al. 1995; Carland et al. 1999).
We report here the genetic and molecular characterization of the WOL locus of Arabidopsis. We show that the morphogenesis of vascular tissue in the root and hypocotyl region is established through a set of asymmetric cell divisions requiring WOL, a putative two-component histidine kinase with a receptor domain similar to that of the DhkA of Dictyostelium discoideum. The deduced domain structure of WOL suggests it represents a novel class among the two hybrid molecules characterized to date from plants, some other eukaryotes and bacteria. Further analysis of the WOL locus should permit the eventual characterization of the upstream and downstream components of a signal transduction pathway likely to regulate vascular cell proliferation and differentiation.
Results
WOL is essential for vascular asymmetric cell divisions
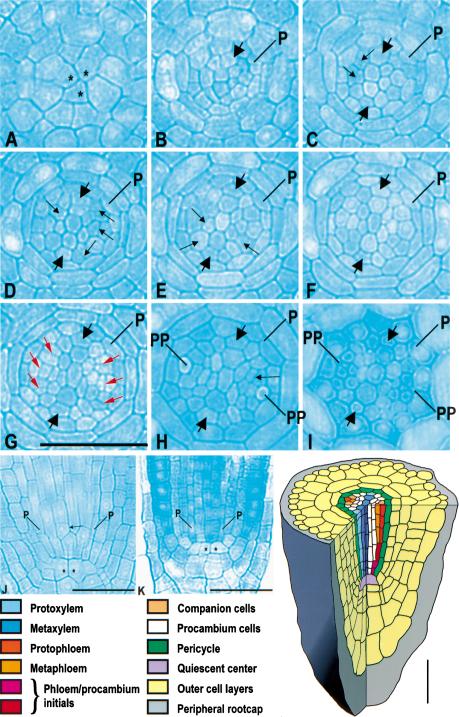
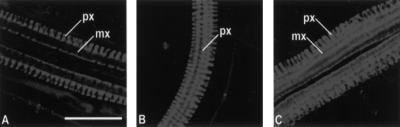
The WOL locus was defined by a recessive mutation that results in reduced cell number and exclusive xylem differentiation within the vascular tissue (Scheres et al. 1995). This reduced cell number is first observed during embryogenesis, when a cell division process fails to take place in the root and lower hypocotyl region soon after the torpedo stage. Subsequently, this pattern is elaborated by the procambial initials during primary root development (Scheres et al. 1995). To identify the WOL-dependent divisions that result in the patterning of xylem and phloem of the root, we determined the vascular cell lineage relationships in the primary root of wild type and wol (Fig.1; Table 1). In serial sections of the primary root meristem (Fig. 1), we observed that xylem cell lineages form an axis composed of four to five cell files very close to the underlying quiescent center (Fig. 1C, schematic). Two domains of initials (two to five cells each; Fig. 1B,C) that give rise to the phloem cell lineages and to the undifferentiated procambial cell lineages through asymmetric cell divisions (Fig. 1C–G; schematic) flank this axis. The number and exact pattern of these procambial divisions show some variability between individual seedlings, which is in contrast to the invariant pattern of cell lineages in the endodermis and outer layers (Scheres et al. 1994). These divisions are asymmetric in the sense that they give rise to multiple cell lineages with different fates (Horvitz and Herskowitz 1992). The histologically early phloem differentiation as compared to xylem differentiation (Fig. 1H,I) is consistent with previous descriptions in other species (Esau 1977).
Figure 1.
Cell lineages in the vascular bundle of primary root in Arabidopsis. (A–I) A cross-section series of the wild-type primary root (3-μm-thick sections). (A) At the level of the quiescent center (QC). (B) 3 μm, (C) 6 μm, (D) 9 μm, (E) 12 μm, (F) 15 μm, (G) 27 μm, (H) 69 μm, and (I) ∼120 μm above the QC. (J,K) Longitudinal sections of the wild-type and wol primary root, respectively. Cells of the QC are labeled with asterisks. Newly formed cell walls after the cell divisions in the procambium in C–G are indicated with small black arrows. The specific set of newly formed cell walls associated with phloem development (G) is indicated in red. Prospective protoxylem cells are indicated with thick arrows. (p) Pericycle; (pp) sieve elements of the protophloem. Scale bar, 30 μm. Schematic: cell lineages of the vascular bundle. The colors refer to the cell lineages only; the differentiation status of the cells is not known. The first and second maturing sieve elements are indicated as protophloem and metaphloem, respectively.
Table 1.
Cell numbers in the primary root of Arabidopsis
|
|
Initial cells
|
Differentiated cells
|
|---|---|---|
| wt | 11 ± 1.49 | 31 ± 1.71 |
| wol | 8 ± 2.23 | 9 ± 2.07 |
Number of initial cells were counted in sections made directly above the quiescent center, numbers of differentiated cells were made ∼200 μm above the quiescent center. Mean cell numbers and ± SD are indicated. (wt n = 8; wol n = 6).
In wol, the periclinal cell divisions described above are absent (Fig. 1J–K, schematic; Fig. 3C, below) with the exception of a few divisions that slightly increase cell number in the vascular cylinder (Table 1). Furthermore, the number of vascular initials in wol is slightly lower than in wild type, indicating that some of the embryonic divisions required to form the initials do not occur. Taken together, WOL activity is associated with a set of formative divisions that first take place during the late stages of embryogenesis (Scheres et al. 1995) and that continue to propagate the pattern in the root meristem.
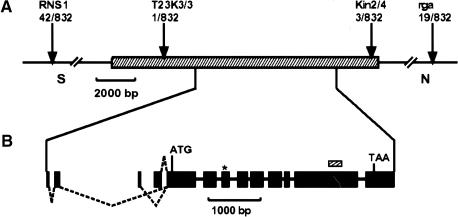
Figure 3.
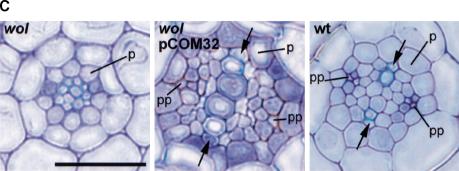
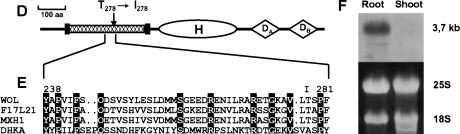
Molecular cloning of the WOL locus. (A) Fine mapping. RNS1 and rga are preexisting CAPS markers in top of chromosome 2. T23K3/3 and Kin2/4 are the closest markers (designed in this study) for the wol mutation in the BAC T23K3. The number of recombination events between markers and the wol locus is indicated below the markers. Hatched bar represents the 13.8-kb MscI fragment of the T23K3 BAC clone fragment (cloned in pCOM32), which complemented the wol mutation. (B) Structure of the WOL gene. Exons are indicated as solid bars. Three combinations of dotted lines show the alternative splicing variations of the WOL gene (deposited in GenBank). All result in the identical longest open reading frame with an ATG in position 13572–13574 of the BAC T23K3. The wol mutation is located in the third exon (asterisk), where it is converting T278 (ACT) to I278 (ATT). The small hatched bar represents the 256-bp probe used in hybridization analyses. (C) Complementation of the wol mutation. Left to right: Cross sections of wol, wol transformed with the pCOM32 construct, and wild-type primary roots. (Arrows) Protoxylem in wild-type and rescued root; (p) pericycle; (pp) sieve elements of protophloem. Scale bar, 30 μm. (D) Predicted domain structure of WOL. Extracellular receptor domain (hatched bar) is located near the N terminus between the two transmembrane regions (vertical solid bars). C-terminal domain consists of a histidine kinase domain (H) and two receiver domains (DA and DB). (E) Amino acid sequence alignment representing a region surrounding the wol mutation site in the putative receptor domain of three WOL-like proteins (WOL, F17L21.11, MXH1.16) and Dictyostelium discoideum DHKA. Identical amino acids are indicated by black boxes. The altered amino acid residue in wol (I278) is shown above the alignment. (F) Northern blot of total RNA from wild-type roots and shoots. Ethidium bromide–stained ribosomal RNA is shown as a loading control.
WOL is required for vascular morphogenesis
The role of WOL in phloem specification was analyzed previously in the context of a double mutant of wol and fass, a mutation resulting in supernumerary cell layers. In the wol × fass double mutant, there was an increase in the number of vascular cell layers with phloem markers observed in the vascular cylinder, indicating that WOL is not essential for phloem development. A model was proposed in which xylem specification temporally precedes phloem development. Because the wol mutant has a reduced number of vascular precursor cells, all the available cells are specified as xylem (Scheres et al. 1995). More recently, it was shown that xylem organization is also altered in wol (Cano-Delgado et al. 2000). The xylem axis in wild type consists of two types of cells: protoxylem, the two outermost cells with predominantly annular (ring-like) cell wall thickenings, which differentiates early, and metaxylem, the central cells with reticulate (more continuous) wall thickenings, which differentiates later (Fig. 2A). In contrast, the vascular cylinder of the wol primary root consists solely of protoxylem (Fig. 2B; Cano-Delgado et al. 2000). Therefore, we investigated the status of xylem in the wol × fass background and found both protoxylem and metaxylem to be present (Fig. 2C). Thus, WOL does not appear to be necessary for metaxylem differentiation but has a more indirect influence on xylem differentiation by controlling the number of cells in the vascular cylinder.
Figure 2.
WOL is not necessary for metaxylem development. (A) wt: Metaxylem is gradually differentiating in an axis flanked by two files of protoxylem cells. (B) wol: The entire vascular bundle is differentiated as protoxylem. (C) wol × fass: Protoxylem and metaxylem organization analogous to that in wt. The confocal images were taken from whole mounts of seedlings stained with fuchsin. Scale bar, 30 μm.
WOL is a member of a novel family of two component hybrid molecules
To determine the molecular nature of the WOL gene product, we identified the WOL gene through positional cloning (Fig. 3A). The wol locus was mapped to an 11-kb region between two CAPS markers (cleaved amplified polymorphic sequences) in the BAC (bacterial artificial chromosome) clone T23K3 (Fig. 3A). The annotation (Lin et al. 1999) predicts only one gene (T23K3.2, a putative two component histidine kinase) in this region. The region between the CAPS markers was completely sequenced in wol, and only one point mutation was found, converting T278 to I278 in the deduced amino acid sequence. To confirm the identity of the two component hybrid molecule as WOL, we complemented the mutation with a 13.8-kb fragment of genomic DNA (pCOM32; Fig. 3A). Homozygous wol plants containing the transgene had indeterminate root growth (not shown), normal cell number, and a wild-type pattern of xylem, phloem, and procambial cells (Fig. 3C).
To identify the WOL coding region, we sequenced the longest (T20648) of several ESTs homologous to the WOL locus. Subsequently, the cDNA was extended using RT–PCR and 5′RACE–PCR techniques. By cloning the major amplification products resulting from the 5′RACE, three different cDNA species corresponding to three different transcription start sites and alternative splicing schemes could be detected (Fig. 3B). All result in an identical longest ORF of 1057 amino acid residues, indicating that the alternative splicing does not have an impact on the amino acid sequence of the gene product.
The predicted WOL protein (Schultz et al. 1998) shows extensive similarity to two-component hybrid molecules, suggesting that it functions as a signal transducer (Fig. 3D). It has a short N-terminal cytoplasmic domain (residues 1–105) followed by an extracellular receptor (R) domain (residues 127–400) flanked by two transmembrane regions and the C-terminal phosphorelay domain system with a histidine kinase domain (H; residues 449–737) and two receiver domains (Da and Db; residues 762–893 and 922–1044, respectively). In the Arabidopsis database, there are two other WOL-like genes (MXH1.16 and F17L21.11) with a similar deduced protein domain structure (Fig. 3E), indicating that WOL belongs to a small protein family. The deduced sequence of the WOL protein is 54% identical to MXH1.16 and 57% identical to F17L21.11. Furthermore, a WOL-like sequence has been reported in the genome of Hordeum vulgare. Among the characterized two-component receptor molecules, WOL-like proteins are unique in having two putative D domains. The closest homolog of WOL-like genes is DhkA (Wang et al. 1996, 1999) of the slime mold Dictyostelium discoideum (Fig. 3E). WOL and DhkA are 24% identical in the R, 44% in the H, and 39% in the Db domains, respectively. However, the Da domain (with the absolutely conserved residues D768, D825, and K882 characteristic to the receiver domains) is absent in DhkA. The functional role of the phosphorelay domains remains to be determined. The mutation in wol results in a nonconservative change of an amino acid residue of the putative receptor domain that is conserved within the WOL protein family (Fig. 3E). This indicates that the fully recessive wol mutation may reduce the ability of the molecule to bind a ligand or transmit the information of ligand binding to the C-terminal part of the molecule.
WOL expression is specific for the vascular cylinder
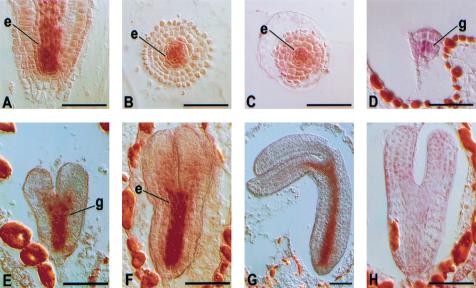
Organ specificity of WOL expression was determined using a gene specific probe (Fig. 3B). Southern blot analysis indicated that hybridization was specific for the WOL gene under the conditions used. In the RNA blot analysis, we detected a single major band of ∼3.7 kb that was more abundant in root than shoot (Fig. 3F). To determine the specificity of WOL expression at the cellular level, we performed in situ localization of WOL mRNA on embryonic and root sections (Fig. 4). In the wild-type primary root, WOL is expressed in the vascular cylinder and pericycle (Fig. 4A,B). The expression pattern of WOL is established during the early stages of embryogenesis. At the globular stage of embryogenesis, WOL mRNA is detected in the four innermost cells, which are the precursors of the vascular tissue (procambium; Fig. 4D). During the heart, torpedo, and nearly mature stages of embryogenesis (Fig. 4E–G), expression is apparent in the procambium of the cotyledon shoulders, prospective hypocotyl, and embryonic root. WOL expression in the wol mutant primary root can be detected in the vascular cells, indicating that WOL is not necessary for its own expression (Fig. 4C).
Figure 4.
Localization of WOL mRNA during embryonic and primary root development by in situ hybridization. (A–G) antisense probe. (A) Longitudinal and (B) cross sections of the wild-type primary root. (C) Cross section of wol primary root; (D) globular stage; (E) late heart stage; (F) torpedo stage; (G) bent-cotyledon stage of the wild-type embryo. (H) Torpedo-stage embryo hybridized with a sense WOL probe. Hybridizations of other sections with sense RNA probes did not reveal signals. Scale bar, 50 μm. The endodermal (e) and the innermost ground tissue (g) layer next to the WOL expression domain are indicated.
Discussion
WOL expression both spatially and temporally coincides with the divisions of the procambial cells of the embryonic and primary root that are defective in the wol mutant. Therefore, we postulate that the WOL gene product is a receptor molecule that controls the asymmetric cell divisions of the vascular initials through a specific signal transduction pathway involving a phosphotransfer reaction characteristic of the two component receptors. In the root tip, WOL expression can be detected both in the region of asymmetric cell divisions and in the zone of differentiation just above it (Fig. 4B). The persistent WOL expression after the asymmetric cell divisions is reminiscent of the expression pattern of the SCARECROW gene, which controls periclinal cell divisions during ground tissue development (Di Laurenzio et al. 1996). The persistent expression may be a safeguard mechanism or play an as yet undiscovered role in differentiation. WOL expression during pericycle development may also be related to its role in regulating asymmetric cell divisions, as the pericycle is ontogenetically part of the vascular cylinder (Scheres et al. 1994, 1995) and WOL expression can be detected before the separation of the pericycle cell lineages (Fig. 4D). Otherwise, the pericycle appears anatomically normal in wol. Furthermore, molecular markers diagnostic for pericycle (J0121, J0272; see Materials and Methods) are present in the wol pericycle (M. Bonke, unpubl.). It is possible that redundancy exists in the regulation of the pericycle-forming cell divisions or that the missense mutation studied here does not affect this particular division. Whether WOL or its homologs have a more general role in regulating cell divisions during vascular development will have to wait for the identification of null alleles for these loci.
The exclusive differentiation of protoxylem in wol precludes the possibility of determining if WOL is required for divisions of the cambium found between xylem and phloem. The pattern of the WOL-dependent procambial cell divisions around the developing xylem provides an intriguing parallel to the pattern of the cambial cell divisions between phloem and xylem, which are very prominent in wood and storage root development (Esau 1977). It is therefore possible that at least some aspects of developmental regulation are common to the procambial and cambial cell divisions and that WOL-like genes contribute to the diversity of vascular patterns found in plants. Support for this hypothesis comes from our recent identification of WOL-like genes active in the cambial zone of the trunk of the silver birch (Betula pendula) tree (L. Kauppinen, M. Riikonen, unpubl.).
Further analysis of the WOL-related signal transduction pathway should lead to the eventual characterization of the upstream and downstream components required for vascular cell proliferation and differentiation. The wol phenotype appears to be distinct from the known mutants related to phytohormone responses. Moreover, the WOL expression pattern does not favor any of the currently characterized pathways involved in the proliferation of the vascular system, as many hormonal and nutritional substances are transported in the root vascular cylinder. Because the wol mutation does not affect WOL expression, it is probable that the WOL gene itself is not under the control of the WOL-related signal transduction pathway. The role of phytohormones and other factors regulating the WOL gene or the signal transduction pathway remains to be studied.
The WOL domain organization and receptor domain structure place it in a distinct subclass among previously identified plant two-component receptors. The other receptors have been shown to function in ethylene or cytokinin signal transduction or in osmosensing (Chang et al. 1993; Kakimoto 1996; Urao et al. 1999, 2000). The tandem arrangement of two putative receiver domains has been previously reported for an uncharacterized gene slr0322 of Synechochystis (Mizuno et al. 1996). However, this gene is not very homologous to the WOL-like genes and may not be very closely related functionally. Among the two component molecules of bacteria, plants, and some other eukaryotes characterized to date, the putative receptor domain, the histidine kinase domain, and the latter one of the two receiver domains (Db) of WOL are most similar to DhkA of Dictyostelium discoideum. DhkA is essential for cellular organization during fruiting body development (Mizuno et al. 1996), and its ligand may be a peptide (Wang et al. 1999). It is therefore possible that a homologous receptor–ligand interaction is specifying development both in slime molds and plants.
Materials and methods
Materials
GenBank accession numbers for the genetic material reported here are: AC007069 (T23K3 BAC); AA586219, AI992824, T20648 (ESTs representing the WOL gene); AJ278528, AJ278529, AJ278530 (three different splice variants representing WOL); U42597 (DhkA); AC004557 (BAC containing F17L21.11); AB011485 (BAC containing MXH1.16); and AJ234550 (WOL-like genomic fragment in H. vulgare). Enhancer trap lines J0121 and J0272 are described at http://www.plantsci.cam.ac.uk/Haseloff/CATALOGUES/JLines/index.html and were obtained through the Nottingham Arabidopsis Stock Centre (NASC).
Histological techniques
For serial sectioning seedlings were fixed according to Scheres et al. (1994) and embedded using Leica Historesin. Sections (2–3 μm) were made on a Leica RM2165 rotary microtome, using a Leica microtome knife. Sections were stained in a 0.05% toluidine blue solution in water and photographed on an Olympus Provis microscope using a Sensicam 12-bit cooled camera (PCO, ccd imaging). Images were processed with Adobe Photoshop 4.0.1.
To single out the double mutants from the cross described in Scheres et al. (1995), we analyzed the fass-looking plants of the F3 segregating in the wol background. Wol × fass double mutant seedlings were cleared with acidified methanol (10 mL of methanol, 2 mL of concentrated HCl (37%), 38 mL of H2O) and incubated at 55°–57°C for 15 min. Then the acidified methanol was replaced with basic solution (7% NaOH in 60% ethanol) and incubated for 15 min at room temperature. Rehydrating the seedlings took place in several steps, each time replacing the incubation solution. First in 40%, then 20%, and finally in 10% ethanol. The seedlings were stained for 5 min in 0.01% basic fuchsin solution, destained in 70% ethanol, and rehydrated to 10% ethanol solution. An equal amount of 50% glycerol was added and then incubated another 30 min. Seedlings were mounted in 50% glycerol. CLSM images were taken on an Axiovert 135M confocal microscope with an argon ion laser (568 + 488-nm emission). Image processing consisted of Kalman filtering during image acquisition (Bio-Rad software). Optical sections were projected together to form a composite image of the vascular bundle.
Molecular techniques
WOL locus was cloned using chromosome walking techniques. CAPS (Konieczny and Ausubel 1993) markers T23K3/3 and Kin2/4 (and six other markers, data not shown) between preexisting markers RNS1 and rga were created by detecting polymorphism between Landsberg and Columbia ecotypes. Complementation vector pCOM32 was created by cloning a 13.8-kb Msc1 fragment into pBIN19 derivative pRD400 (Datla et al. 1992). The construct was transformed into Agrobacterium tumefaciens strain C58C1 (pGV2260; essentially as in Bevan 1984). wol plants were transformed with pCOM32 (Clough and Bent 1998). Transgenic seedlings (T1) with long roots were selected by resistance to kanamycin. Presence of the transgene in these plants was confirmed by PCR. Segregation of the transgene in T2 generation was confirmed.
cDNA cloning was performed with gene-specific primers by using the RobusT RT–PCR Kit (Finnzymes Oy) according to the manufacturer's instructions. To determine the 5′-UTR of the WOL mRNA, the 5′ RACE system for rapid amplifications of cDNA ends version 2.0 (Life Technologies) was used and also executed according to the manufacturer's instructions.
Total RNA was isolated from 17-d-old Arabidopsis thaliana ecotype Columbia root and shoot (Carpenter and Simon 1998). Northern blot analysis was carried out with 15 μg of total RNA. The membrane was hybridized with a 32P-labeled gene-specific WOL probe (a 256-bp PCR fragment [Fig. 3B] representing nucleotides 10143–10398 of the BAC clone T23K3). Genomic DNA preparation (Doyle and Doyle 1990) was performed from 17-d-old seedlings. Southern blot analysis was performed with 15 μg of DNA and the 32P-labeled 256-bp PCR fragment (described above) of WOL or a 1067 bp cDNA fragment corresponding to nucleotides 10055–11204 of WOL genomic sequence as a probe (Maniatis et al. 1982). Both probes were specific for the WOL gene.
In situ hybridization analysis was performed essentially as described by Di Laurenzio et al. (1996) except that tissue samples were cleared in xylene and embedded in Histoplast (Shandon). Proteinase K was used at 10 μg/mL. Seedlings that were 5 d old were used. WOL gene-specific sense and antisense probes (a 256-bp PCR fragment or a 1067-bp cDNA fragment as in Northern blot analysis) were labeled using DIG RNA Labeling Kit (Boehringer) according to manufacturer's protocol. A probe concentration of 50 ng/mL/kb was used in the hybridization. Sections (7 μm thick) were mounted in 50% glycerol or Immu-Mount (Shandon). Images were taken using differential interference contrast (DIC) settings.
Acknowledgments
We want to thank J. Lim and M. Kivimäki for technical assistance; V. Willemsen for help with histological techniques; G. Schindelman for help with fine mapping techniques; T.H. Teeri and K. Aspegren for help in designing the agrotransformation strategy; O. Levälampi for help with agrotransformation; J. Jung for help with in situ hybridization; J. Hiltunen and H. Vihinen for help with microscopical techniques; B. Scheres for the fass × wol F2 seeds, discussions, and helpful comments on the manuscript; L. Di Laurenzio for creation of the mapping line, help in the rough mapping, discussions, and helpful comments on the manuscript; M. Bevan and A. Cano-Delgado for communicating unpublished results; V. Albert, M.-T. Hauser, K. Törmäkangas, and M. Saarma for discussions; and an anonymous reviewer for helpful comments on manuscript. This work was supported by the Academy of Finland and TEKES. Work in the Benfey lab on radial patterning is supported by a grant from the NIH (GM43778).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yhelariu@operoni.helsinki.fi; FAX 358-9-19159366.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.189200.
References
- Aloni R. Differentiation of vascular tissues. Annu Rev Plant Physiol. 1987;38:179–204. [Google Scholar]
- Bevan MW. Binary Agrobacterium vectors for plant transformation. Nucleic Acid Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CL. The influence of external pressure on the differentiation of cells and tissues cultured in vitro. In: Zimmerman M, editor. The formation of wood in forest trees. New York: Academic Press; 1964. pp. 389–404. [Google Scholar]
- Cano-Delgado AI, Metzlaff K, Bevan MW. The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development. 2000;127:3395–3405. doi: 10.1242/dev.127.15.3395. [DOI] [PubMed] [Google Scholar]
- Carland FM, McHale NA. LOP1: A gene involved in auxin transport and vascular patterning in Arabidopsis. Development. 1996;122:1811–1819. doi: 10.1242/dev.122.6.1811. [DOI] [PubMed] [Google Scholar]
- Carland FM, Berg BL, FitzGerald JN, Jinamornphongs S, Nelson T, Keith B. Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell. 1999;11:2123–2137. doi: 10.1105/tpc.11.11.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter CD, Simon AE. Methods in molecular biology: Arabidopsis protocols, Vol. 82. Totowa, NJ: Humana Press; 1998. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Datla RS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W. Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene. 1992;122:383–384. doi: 10.1016/0378-1119(92)90232-e. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Eriksson ME, Israelsson M, Olsson O, Moritz T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol. 2000;7:784–788. doi: 10.1038/77355. [DOI] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. 2nd ed. New York: Wiley; 1977. [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Lui N Y, Bandyopadhyay A, Estelle M. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development. 2000;127:23–32. doi: 10.1242/dev.127.1.23. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Jang J-C, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes & Dev. 2000;14:1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Koizumi K, Sugiyama M, Fukuda H. A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular network: Calling the auxin signal flow canalization hypothesis into question. Development. 2000;127:3197–3204. doi: 10.1242/dev.127.15.3197. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Mizuno T, Kaneko T, Tabata S. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 1996;3:407–414. doi: 10.1093/dnares/3.6.407. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;120:2475–2487. [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser M-T, Janmaat K, Weisbeek P, Benfey PN. Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development. 1995;121:53–62. [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes & Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. Cambridge: Cambridge University Press; 1989. [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Gälweiler L, Palme K, Jürgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki I, Shinozaki I. Two-component systems in plant signal transduction. Trends Plant Sci. 2000;5:67–74. doi: 10.1016/s1360-1385(99)01542-3. [DOI] [PubMed] [Google Scholar]
- Wang N, Shaulsky G, Escalante R, Loomis WF. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 1996;15:3890–3898. [PMC free article] [PubMed] [Google Scholar]
- Wang N, Soderbom F, Anjard C, Shaulsky G, Loomis WF. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol Cell Biol. 1999;7:4750–4756. doi: 10.1128/mcb.19.7.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren Wilson J. The position of regenerating cambia: Auxin/sucrose ratio and the gradient induction hypothesis. Proc R Soc Lond Ser B. 1978;203:153–176. doi: 10.1098/rspb.1978.0098. [DOI] [PubMed] [Google Scholar]