Abstract
Context
The Wnt signaling pathways promote cell growth and are best known for their role in embryogenesis and cancer. Several lines of evidence suggest these pathways might also be involved in bipolar disorder (BP).
Objective
We tested for the association of candidate genes in the Wnt signaling pathways with disease susceptibility in a family-based BP study
Design
227 tagSNPs from 34 genes were successfully genotyped. Initial results led us to focus on the gene PPARD, in which we genotyped an additional 13 SNPs for follow-up.
Setting
Nine academic medical centers in the United States.
Participants
554 BP offspring and their parents from 317 families.
Main Outcome Measures
We tested for family-based association using FBAT and HBAT. Exploratory analyses testing for interactions of PPARD SNPs with clinical covariates and with other Wnt genes were conducted with GENASSOC.
Results
In the initial analysis, the most significantly associated SNP was rs2267665 in PPARD (nominal p=0.0003). This remained significant at p=0.05 by permutation after accounting for all SNPs tested. Additional genotyping in PPARD yielded four SNPs in one haplotype block that were significantly associated with BP at p<0.01, the most significant being rs9462082 (p=0.0001). Exploratory analyses revealed significant evidence (p<0.01) for interactions of rs9462082 with poor functioning on the Global Assessment Scale (OR = 3.36, 95% CI = 1.85–6.08), and with SNPs in WNT2B (rs3790606, OR = 2.56, 95% CI = 1.67–4.00) and WNT7A (rs4685048, OR = 1.79, 95% CI 1.23–2.63).
Conclusions
We found evidence for association of BP with PPARD, a gene in the Wnt signaling pathway. The consistency of this result with one from the Wellcome Trust Case-Control Consortium encourages further study. If the finding can be confirmed in additional samples, it may illuminate a new avenue for understanding the pathogenesis of severe BP and developing more effective treatments.
Introduction
Wnt proteins are a family of molecules that locally activate cell signaling pathways, which regulate cell fate and play an important role in development [for a review see (Nusse, 2005)1]. Aberrations in these pathways have been implicated in a number of chronic diseases, such as cancer. There are several lines of evidence to suggest these pathways may also be involved in the etiology of bipolar disorder (BP) [for a review see (Gould & Manji, 2002)2].
First, Wnt signaling pathways influence neuroplasticity, cell survival, and adult neurogenesis, and recent studies have suggested that BP may involve impairments in these functions. For example, Wnt7a was found to be critical to axon and growth cone remodeling in the cerebellum3, mice with inactivated Wnt1 genes failed to develop large portions of their brain4, and Wnt3 increased neurogenesis in adult rat hippocampus5.
Second, mood stabilizing drugs and antipsychotic medications used to treat BP are known to affect Wnt signaling pathways, particularly through glycogen synthase kinase 3β (GSK3β), a key enzyme in these pathways. Lithium inhibits this enzyme leading to activation of Wnt signaling6, and haploinsufficiency of GSK3β mimics lithium effects in a mouse model7. Clozapine8 and haloperidol9 have also been found to inhibit GSK3β, and one study showed that valproic acid similarly activates Wnt signaling10. Further, another study showed that dopamine increases GSK3β activity and that this increase is reversed by dopamine D2 receptor blockade and by lithium11.
Third, it has been shown that monozygotic twins who are discordant for BP have differential expression of genes in Wnt signaling pathways12. Among 292 genes found to be differentially expressed, eight were in the Wnt pathway. One of these was TCF7, a transcription factor activated by β-catenin. This supports a role for the Wnt pathway in BP pathophysiology. At the level of etiology, this study directly suggests a role for environmental and epigenetic factors given that monozygotic twins are genetically identical, though it also indirectly suggests that variations in genes that alter Wnt signaling could be etiologic factors in BP.
Finally, in an association study of BP with 22 genes on chromosome 2213, evidence for association was found with HMG2L1, a gene which influences Wnt signaling by interacting with NLK, a negative regulator of TCF714.
Motivated by these considerations, we sought to systematically test whether variation in candidate genes of the Wnt signaling pathways was associated with susceptibility for BP in a family-based study.
Methods
Study Sample
The families in the current study were ascertained through one of three projects: the NIMH Genetics Initiative Bipolar Disorder Consortium involving nine different sites across the United States (including Johns Hopkins University, Indiana University, Washington University in St. Louis, University of California, San Diego, University of Iowa, University of Pennsylvania, University of Chicago, Rush-Presbyterian Medical Center, University of California, Irvine, and the NIMH Intramural Program); a separate collaboration involving the University of Chicago, Johns Hopkins University, and the NIMH Intramural Program (here referred to as CHIP); or the Clinical Neurogenetics (CNG) collection. In all three samples, the families were recruited opportunistically through probands with bipolar I disorder (BPI) and one or more relatives affected with a mood disorder. The NIMH Genetics Initiative families were assessed with the Diagnostic Interview for Genetic Studies (DIGS)15, and diagnoses were assigned using best estimate procedures based on DSM-III-R or DSM-IV criteria. The CHIP and CNG families were assessed with the Schedule for Affective Disorders and Schizophrenia-Lifetime Version (SADS), and diagnoses were made using similar best estimate procedures based on Research Diagnostic Criteria. We have previously reported high diagnostic reliability for major mood episodes and diagnoses using both the SADS (kappas 0.72–1.0) (Simpson et al, 2002) and the DIGS (kappas 0.78–0.96) (Nurnberger, Jr. et al, 1994). We have also found, in a small sample of 33 subjects originally diagnosed with BPI by RDC criteria after assessment with the SADS, that on re-interview an average of 10 years later using the DIGS, that 31 of them were re-diagnosed with BPI and two with BPII, both by RDC and DSM-IV criteria (unpublished data). We used all independent quads (two parents and two affected offspring) and trios (two parents and one affected offspring) from these families with DNA available for genotyping. This yielded 1,118 subjects in 237 quads and 80 trios from 317 families. Among the affected offspring, there were 491 with BPI, 39 with bipolar II disorder (BPII), and 24 with schizoaffective manic disorder or schizoaffective disorder, bipolar type (SAM or SABP). Of these, 60.5% were female, the average age of onset was 18, and the average age at interview was 41. Ninety-six percent of subjects had been treated. Of the 634 parents in our samples, 613 (97%) were of European ancestry (defined as ≥ 90% European) as assessed by a STRUCTURE analysis in which genotype data from 250 SNPs in our samples were analyzed along with data from HapMap CEPH, Yoruban, Chinese, and Japanese samples. This broke down by study sample as follows: NIMH: 458/470 (97% with European ancestry); CHIP: 147/154 (95%); CNG: 10/10 (100%). The 21 other parents came from 14 families and included five with African ancestry (defined as ≥ 90% African), two with Chinese ancestry (defined ≥ 90% Chinese) and 14 with mixed ancestry.. All subjects provided informed consent at the institutions where they were seen, and all protocols were approved by the local IRBs.
Genotyping
We genotyped, in these subjects, 34 candidate genes from the Wnt signaling pathways (see Table 1 for details). We compiled a list of genes from the tables and diagrams reflecting various aspects of Wnt signaling, including Wnt ligands, receptors, interactors, and targets, that were present on the Wnt Homepage (http://www.stanford.edu/~rnusse/wntwindow.html) as of June 2005. We localized 131 of these genes to chromosomal locations using the University of California, Santa Cruz Genome Browser. We selected candidates from these based on whether they were: a) expressed in the brain; and b) located in a chromosomal region that had been implicated by previous linkage studies of bipolar disorder and/or schizophrenia. Evidence for linkage included a significant result from one of two meta-analyses16,17, genome-wide significance in at least one family study, or consistent replication of a “suggestive” finding18 across more than one independent family study. Two Wnt signaling genes were included although they did not meet all of the above criteria. These were GSK3β because of its well-established inhibition by lithium6 and NLK because it interacts with HMG2L1, which was previously associated with psychotic bipolar disorder13,14.
Table 1.
Candidate genes involved in Wnt signaling pathways
| Chr | Gene | Location | Exons | SNPs | Size (kb)a | 1 SNP/N kb | % Captureb | Avg D′ | Avg r2 | Blocksc | Functiond |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | WNT2B | 1p13 | 5 | 10 | 73.87 | 7.39 | 0.65 | 0.42 | 0.08 | 4 | Wnt protein |
| 1 | BCL9 | 1q21 | 10 | 8 | 104.83 | 13.10 | 0.34 | 0.55 | 0.15 | 4 | bind β-catenin, promote transcription |
| 1 | BGLAP | 1q25-q31 | 4 | 6 | 38.84 | 6.47 | 0.78 | 0.62 | 0.20 | 2 | TG - cell adhesion |
| 1 | WNT9A | 1q42 | 4 | 2 | 46.90 | 23.45 | 0.33 | 0.34 | 0.09 | 2 | Wnt protein |
| 1 | WNT3A | 1q42 | 4 | 5 | 74.21 | 14.84 | 0.89 | 0.95 | 0.30 | 1 | Wnt protein |
| 1 | RHOU | 1q42.11-q42.3 | 3 | 3 | 31.54 | 10.51 | 0.65 | 0.99 | 0.47 | 1 | mediates Wnt1 signaling |
| 3 | WNT7A | 3p25 | 4 | 9 | 81.54 | 9.06 | 0.43 | 0.51 | 0.12 | 5 | Wnt protein |
| 3 | CTNNB1 | 3p21 | 15 | 2 | 61.00 | 30.50 | 0.94 | 0.99 | 0.25 | 1 | β-catenin |
| 3 | GSK3B | 3q13.3 | 11 | 6 | 286.97 | 47.83 | 0.56 | 0.91 | 0.21 | 2 | GSK3-β receptor |
| 4 | CTBP1 | 4p16 | 9 | 5 | 57.68 | 11.54 | 0.60 | 0.82 | 0.22 | 2 | phosphorylation |
| 5 | NEUROG1 | 5q23-q31 | 1 | 1 | 21.66 | 21.66 | 0.57 | N/A | N/A | 1 | TG - neuronal differentiation |
| 5 | WNT8A | 5q31 | 8 | 5 | 27.43 | 5.49 | 1.00 | 1.00 | 0.22 | 1 | Wnt protein |
| 5 | PTTG1 | 5q35.1 | 6 | 6 | 26.88 | 4.48 | 0.58 | 0.70 | 0.19 | 3 | TG - chromosome stability, DNA repair |
| 6 | PPARD | 6p21.2-p21.1 | 8 | 2 | 105.62 | 52.81 | 0.41 | 1.00 | 0.59 | 1 | TG - neurodevelopment |
| 6 | VEGFA | 6p12 | 8 | 7 | 36.27 | 5.18 | 0.58 | 0.53 | 0.12 | 3 | TG - binds neurophilin |
| 6 | WISP3 | 6q21 | 5 | 5 | 35.61 | 7.12 | 0.96 | 0.97 | 0.19 | 1 | TG - skeletal growth |
| 6 | GJA1 | 6q21-q23.2 | 2 | 4 | 34.13 | 8.53 | 0.46 | 0.57 | 0.16 | 3 | TG - transmembrane channels |
| 8 | FZD3 | 8p21 | 8 | 5 | 90.19 | 18.04 | 0.96 | 0.57 | 0.12 | 3 | frizzled receptor |
| 8 | ENPP2 | 8q24.1 | 25 | 9 | 101.79 | 11.31 | 0.68 | 0.62 | 0.20 | 4 | TG - lysophosphatidic acid |
| 8 | MYC | 8q24.12-q24.13 | 3 | 3 | 25.36 | 8.45 | 0.36 | 0.55 | 0.16 | 2 | TG - transcription of growth gene |
| 8 | WISP1 | 8q24.1-q24.3 | 5 | 18 | 58.26 | 3.24 | 0.64 | 0.41 | 0.11 | 8 | TG - cell proliferation |
| 10 | DKK1 | 10q11.2 | 4 | 3 | 23.38 | 7.79 | 0.75 | 0.87 | 0.17 | 1 | inhibits Wnt signaling; feedback TG |
| 10 | NODAL | 10q22.1 | 3 | 4 | 29.35 | 7.34 | 0.79 | 0.80 | 0.36 | 2 | TG – forms mesoderm, axial organization |
| 11 | MMP7 | 11q21-q22 | 6 | 5 | 30.24 | 6.05 | 0.44 | 0.62 | 0.08 | 3 | TG - matrix metalloproteinase |
| 11 | MMP3 | 11q22.3 | 10 | 5 | 27.81 | 5.56 | 0.76 | 0.88 | 0.30 | 1 | TG - matrix metalloproteinase |
| 12 | WNT10B | 12q13 | 5 | 2 | 26.42 | 13.21 | 0.50 | 1.00 | 0.76 | 1 | Wnt protein |
| 12 | IGF1 | 12q22-q23 | 4 | 9 | 104.65 | 11.63 | 0.40 | 0.65 | 0.16 | 4 | TG - mediates growth hormone |
| 12 | FZD10 | 12q24.33 | 1 | 5 | 23.25 | 4.65 | 0.29 | 0.70 | 0.16 | 3 | frizzled receptor |
| 13 | EFNB2 | 13q33 | 5 | 13 | 65.24 | 5.02 | 0.67 | 0.49 | 0.10 | 3 | TG - mediates developmental events |
| 16 | AXIN1 | 16p13.3 | 10 | 10 | 85.02 | 8.50 | 0.40 | 0.63 | 0.20 | 7 | tumor suppressor |
| 17 | NLK | 17q11.2 | 11 | 5 | 173.72 | 34.74 | 0.68 | 0.96 | 0.25 | 1 | chromosome separation; feedback TG |
| 18 | SMAD4 | 18q21.1 | 13 | 2 | 69.53 | 34.77 | 0.89 | 1.00 | 0.52 | 1 | mediates TGFβ1 |
| 18 | TCF4 | 18q21.1 | 20 | 42 | 386.30 | 9.20 | 0.82 | 0.42 | 0.14 | 10 | transcription factor 4 |
| 22 | CBY1 | 22q12 | 5 | 1 | 37.20 | 37.20 | 0.90 | N/A | N/A | 1 | inhibits Wnt signaling |
Size reflects the size of the most inclusive RefSeq gene definition ± 10kb; For BGLAP, one SNP lay outside of the most inclusive RefSeq gene definition ± 10kb and the distance to this SNP was added to the total size
Percent of known common variation in the gene ± 10 kb according to HapMap Phase II captured by the SNPs typed in the gene ± 10 kb
Number of LD blocks in the gene ± 10 kb based on the typed SNPs and defined by a solid spine D′ ≥ 0.8
TG = target gene
We selected for genotyping non-synonymous coding SNPs identified from NCBI (http://www.ncbi.nlm.nih.gov) as well as a set of tag SNPs chosen to capture the known common genetic variation (minor allele frequency [MAF] > 0.1) across each gene ±10 kb at an r2 ≥ 0.8. The tag SNPs were selected using LDSelect19 with HapMap Phase I data (http://www.hapmap.org) which was available at the time of the design of the study. These SNPs were genotyped by the Illumina Integrated BeadArray System at Illumina. A total of 265 SNPs were originally selected for genotyping. Of these, 34 SNPs could not be genotyped by Illumina. Another 6 replacement SNPs were identified, resulting in 237 successfully genotyped SNPs. Of these, 10 SNPs were excluded because they were either monomorphic (n=2) or not in Hardy-Weinberg equilibrium (estimated among the founders) at p<0.05 (n=8), leaving 227 SNPs for the initial analyses.
For follow-up, we sought to do denser genotyping in our most significant gene, PPARD (see below). We used the web-based tool QuickSNP20 to select an additional 18 tag SNPs from HapMap Phase II data that captured the known common variation in the gene with a MAF > 0.05 at an r2 > 0.9. We used the TaqMan-5′ nuclease assay to genotype these SNPs. Two of the assays failed and three SNPs were excluded because of missing data > 5.0%, leaving 13 additional SNPs for analyses.
Statistical Analyses
We carried out two main analyses. First, we conducted a single-locus analysis in which we individually tested each SNP for association with BP using FBAT. FBAT is a flexible program for testing allelic associations with family data. Under certain conditions, it reduces to the commonly used transmission/disequilibrium test (TDT). However, FBAT is more general and allows tests of association that are robust to population confounds in the case where parental data is missing and/or other offspring are included in the analysis21. We used the biallelic mode and examined the additive model for counting alleles. We specified the option to calculate the variance empirically in order to provide valid tests of association in the presence of linkage22, because each of the candidate genes were selected based on prior evidence of linkage. We used permutations to estimate the significance of our best finding after accounting for all the tests carried out with FBAT. After randomly shuffling the haplotypes across each of the genes that was transmitted to the offspring, we re-calculated the FBAT tests for each SNP and counted the number of replicates out of 10,000 in which there was a more significant finding than what we observed.
Second, we conducted a multi-locus analysis in which we tested haplotypes of adjacent SNPs for association with BP using HBAT23. It has been shown that in some situations (such as when r2=1 between the risk variant and a particular multi-SNP haplotype) haplotypes may provide more information for association than corresponding single-locus tests24. HBAT is an elaboration of FBAT that allows for family-based association tests of haplotypes, even when phase is ambiguous. We used a sliding window approach to test haplotypes of 2, 3 and 4 adjacent SNPs. Haplotypes were only allowed to cross intergenic regions when <1 kb separated SNPs in the two genes. This happened three times for the following gene pairs: WNT9A-WNT3A, WNT2A-RHOU, MMP7-MMP3. Significance values were obtained for tests of each specific haplotype.
In exploratory analyses of our most significant findings in the PPARD gene, we separately tested for sub-group differences in the evidence of association by certain clinical features or for interactions with SNPs from other genes in the Wnt signaling pathways. For both analyses, we used the method implemented by the program genassoc in STATA 9.0 (http://www-gene.cimr.cam.ac.uk/clayton/software/stata/genassoc/) which allows for tests of interactions. Here, the family dataset is converted into cases and matched pseudocontrols and analyzed for genotypic associations with a SNP of interest using conditional logistic regression models. These models may then be expanded to incorporate interactions between the SNP and other covariates. The significance of the interactions is determined by comparing models with and without the interaction terms using likelihood ratio tests (LRT). For the analysis looking at clinical features, we assumed the cases and pseudocontrols were matched on the feature under investigation. This is artificial because the pseudocontrols do not have clinical features, but it allows us to test for sub-group differences in the association with a SNP among the cases by including interaction terms between the SNP and a covariate for the feature. The different features tested in this way, selected to capture various aspects of severity, included the following: number of depressive episodes, number of manic episodes, age at onset (dichotomized at 21 or younger), ever attempted suicide, ever hospitalized, history of psychosis, history of mood incongruent psychosis, poor functioning in the past month (as assessed by the Global Assessment Scale [GAS], which is a measure of general functioning ranging from 0–100 with a score lower than the mean of 68 representing poor functioning). For the analyses looking at two-way interactions between PPARD and the other genes examined in the Wnt signaling pathway, we individually tested interaction terms for the best SNPs in PPARD with the single best SNP from each of the other genes. In all models for both analyses, we used a dominant coding for the SNPs being tested.
Results
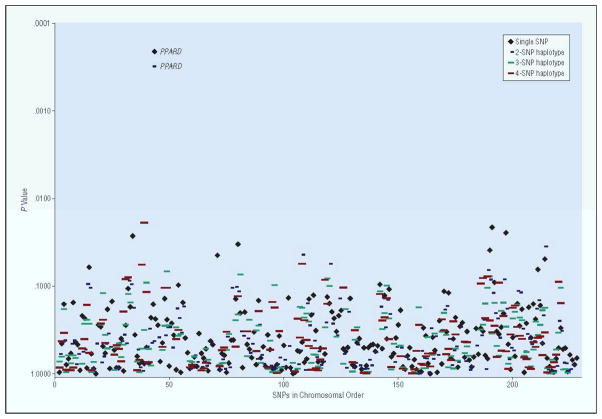
The results of the single SNP and 2, 3, and 4 SNP haplotype analyses are shown in Figure 1. The most significant single SNP was rs2267665 in PPARD (nominal p=0.0003). The association with this SNP was significant at p=0.05 by permutation after accounting for all the tests carried out with the SNPs. The most significant haplotype association was with a two SNP haplotype that included rs2267665 (p=0.0009). No other single SNP or haplotype was associated with BP at the nominal p<0.01.
Figure 1.
Association of 227 SNPs in Wnt signaling pathway genes with bipolar disorder. P-values for allelic single SNP analyses (black diamonds) and haplotype analyses (blue, green and red lines) on the log scale. Results with nominal p<0.01 are labeled with the gene name.
To corroborate the finding with PPARD, we consulted the publicly available results from the Wellcome Trust Case-Control Consortium (WTCCC) Genome-Wide Association Study (GWAS) with BP. The SNP rs2267665 was not genotyped in this study, but a SNP (rs9470015) that was in perfect linkage disequilibrium with it (r2=1.0) was, and this SNP was associated with BP at p=0.02 in the analysis in which all available controls were used to maximize the power of the sample. In both the SNP from our study and from the WTCCC GWAS, the common allele was over-represented in BP.
Encouraged by these findings, we sought to genotype a denser panel of SNPs in PPARD in order to provide better coverage of the gene and refine the boundaries of the apparent association. The PPARD gene, which is located on chromosome 6p21.2-p21.1, has 8 exons and spans approximately 85.6 kb (http://www.ncbi.nlm.nih.gov/RefSeq/). In the initial genotyping effort, we successfully genotyped only two intronic SNPs (including rs2267665 described above) that captured approximately 44% of the known variation in the gene according to the HapMap phase II data with a MAF>0.05 at an r2>0.80. With the successful genotyping of 13 additional tagSNPs we increased the coverage to capture 86% of the known variation with a MAF>0.01 at an r2>0.90.
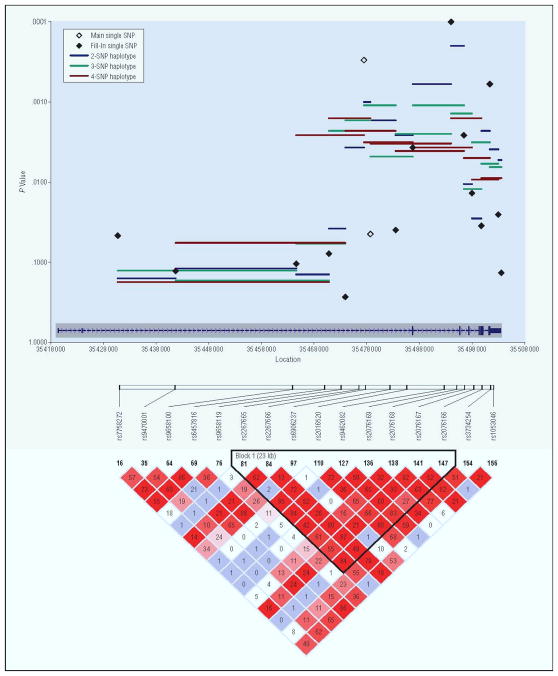
The results of the single SNP and 2, 3, and 4 SNP haplotype analyses with all 15 SNPs in PPARD are graphically depicted in Figure 2 and the details shown in Table 2. Of the 13 new tagSNPs, four were significantly associated with BP at p<0.01. The most significant of these (rs9462082) was associated at p=0.0001. The most significant haplotype was a two SNP haplotype that included rs9462082 (p=0.0003). All of the SNPs associated at p<0.01 resided in a single haplotype block that spanned exons 3 to 7 of the gene. The two most significant SNPs overall rs2267665 and rs9462082 were highly correlated at r2=0.84.
Figure 2.
Specific association results for PPARD. Plotted are empiric p-values for single-SNP TDT tests (black diamonds) and 2- 3- and 4-SNP haplotype windows (blue, green and red solid lines). Diamonds representing the SNP results for the 2 SNPs in the main analysis are open and diamonds representing the SNP results for the 13 fill in SNP are closed. The exon structure of the gene is included at the bottom of the graph in blue. The LD plot is shown below the graph with r2 values in the diamonds and the corresponding LD blocks42.
Table 2.
Details of association results with all genotyped SNPs in PPARD
| Allelicb | Haplotypicc | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Location | Function | Alleles | MAFa | Informative Families | S | E(S) | Var(S) | p | 2 SNP p | 3 SNP p | 4 SNP p |
| rs7744392 | 35430741 | intron | A/G | 0.048 | 31 | 31 | 22.5 | 18.25 | 0.0466 | 0.1588 | 0.1271 | 0.1770 |
| rs9470001 | 35441719 | intron | G/C | 0.079 | 44 | 50 | 41.5 | 31.25 | 0.1284 | 0.1195 | 0.1689 | 0.0010 |
| rs9658100 | 35464618 | intron | T/G | 0.08 | 44 | 52 | 42.5 | 34.25 | 0.1045 | 0.1421 | 0.0011 | 0.0008 |
| rs6457816 | 35470826 | intron | T/C | 0.085 | 51 | 61 | 50.0 | 39.00 | 0.0782 | 0.0008 | 0.0004 | 0.0006 |
| rs2267665 | 35477469 | intron | G/A | 0.155 | 94 | 58 | 89.5 | 75.25 | 0.0003 | 0.0010 | 0.0012 | 0.0036 |
| rs2267666 | 35478706 | intron | T/A | 0.237 | 110 | 105 | 124.5 | 94.25 | 0.0446 | 0.0016 | 0.0052 | 0.0035 |
| rs6906237 | 35483504 | intron | C/A | 0.073 | 44 | 52 | 40.5 | 31.25 | 0.0397 | 0.0029 | 0.0026 | 0.0041 |
| rs2016520 | 35486756 | 5′ UTR | A/G | 0.184 | 95 | 72 | 98.0 | 80.00 | 0.0037 | 0.0005 | 0.0012 | 0.0041 |
| rs9462082 | 35494019 | intron | G/A | 0.143 | 86 | 48 | 79.5 | 67.25 | 0.0001 | 0.0003 | 0.0012 | 0.0020 |
| rs2076169 | 35496457 | intron | T/C | 0.095 | 64 | 40 | 62.0 | 53.50 | 0.0026 | 0.0106 | 0.0132 | 0.0049 |
| rs2076168 | 35497977 | intron | A/C/T | 0.201 | 98 | 81 | 102.5 | 76.25 | 0.0138 | 0.0268 | 0.0032 | 0.0096 |
| rs2076167 | 35499765 | synonymous | A/G | 0.223 | 106 | 93 | 112.5 | 85.75 | 0.0352 | 0.0021 | 0.0062 | 0.0089 |
| rs2076166 | 35501382 | intron | C/T | 0.137 | 88 | 50 | 78.0 | 67.00 | 0.0006 | 0.0040 | 0.0064 | |
| rs3734254 | 35502988 | 3′ UTR | T/C | 0.187 | 100 | 88 | 108.5 | 84.25 | 0.0255 | 0.0051 | ||
| rs1053046 | 35503556 | 3′ UTR | G/A | 0.06 | 38 | 42 | 34.5 | 25.25 | 0.1356 | |||
MAF = Minor Allele Frequency
Allelic results from FBAT; statistics for the minor allele shown
Haplotypic results from HBAT; Results presented for 2-, 3-, and 4-SNP sliding window haplotypes. The p-value for each haplotype is listed in the row for the first SNP in the haplotype.
In an exploratory analysis, we tested whether the association with the two best SNPs in the PPARD gene varied depending upon certain covariates related to the clinical presentation of BP. Of the different clinical covariates examined, only poor functioning as measured by the Global Assessment Scale (GAS) showed a significant interaction (LRT p<0.01) with these two SNPs. The SNP rs9462082 was significantly associated with BP among all subjects with an odds ratio (OR) of 1.46 (95% CI = 1.16, 1.85) for those who were homozygous for the common allele compared to others. However, among subjects with poor functioning (GAS < 68), this risk increased to an OR of 3.36 (95% CI = 1.85, 6.08). There was no apparent association of this SNP with BP among those who were high functioning (OR=1.28, 95% CI = 0.85, 1.93). The results were nearly identical for rs2267665.
In further exploratory analyses, we tested for two-way interactions between the two best SNPs in PPARD and the best SNP from each of the other Wnt signaling pathway genes. Evidence for significant interactions (LRT p<0.01) was detected with WNT2B and WNT7A. In particular, rs9462082 was significantly associated with BP only among those who were carriers of the rare allele at rs3790606 (MAF=0.31) in WNT2B (OR = 2.56, 95% CI = 1.67, 4.00), but not among those who were homozygous for the common allele (OR = 1.16, 95% CI = 0.79, 1.72). The results were nearly identical for rs2267665. Similarly, rs9462082 was significantly associated with BP only among those who were carriers of the rare allele at rs4685048 (MAF=0.49) in WNT7A (OR = 1.79, 95% CI = 1.23, 2.63), but not among those who were homozygous for the common allele (OR = 0.75, 95% CI = 0.42, 1.32).
Discussion
Several converging lines of evidence suggest that Wnt signaling pathways, which play an important role in a number of cellular functions, may also contribute to the etiology of BP. We sought to test whether variation in genes of the Wnt signaling pathways is associated with susceptibility to BP using a family-based design. We observed an association with a SNP in the PPARD gene that remained significant after correcting for the multiple tests carried out. Interestingly, another SNP that was in perfect LD with this SNP was genotyped in the WTCCC GWAS and was also nominally associated with BP. Further genotyping in our own sample suggested the association was delimited within a single haplotype block that spanned exons 3 to 7 of the gene. Moreover, the association appeared to be strongest among BP subjects with the poorest functioning and among those carrying a rare allele at rs3790606 in WNT2B or at rs4685048 in WNT7A. These findings merit further investigation in order to identify the putative causal variant(s) within this region of the gene and to confirm their effects on poor functioning BP in particular.
The PPARD gene is located on chromosome 6p21. Chromosome 6p has not directly been implicated by linkage studies of BP, although one study found evidence of interaction between loci on 6q and 6p25. By contrast, this region has been implicated in schizophrenia by a number of linkage studies. A rigorous meta-analysis of 20 different genome-wide linkage scans26 identified two broad loci on 6p, one stretching from 6pter-6p22.3 and another from 6p22.3-21.1, that were genome-wide significant and among the top 10 signals across the entire genome. This region contains several interesting candidate genes, including one (DTNBP1) that has been associated with both schizophrenia and psychotic BP27,28. PPARD itself has not previously been reported to be associated with either BP or schizophrenia.
The PPARD gene encodes a member of the peroxisome proliferator-activated receptor (PPAR) family. The PPARs are nuclear hormone receptors that mediate a wide variety of cellular and biochemical processes, including peroxisomal functioning, lipid oxidation, lipid synthesis, cell proliferation and inflammation [for a review see (Burger & Moller)29]. They act by dimerizing with the retinoid X receptor and upon binding with various ligands serve as transcription factors for a number of different target genes that have peroxisome proliferation response elements (PPREs) within their promoters. PPARD is expressed in a wide range of tissues, but most notably the brain, adipose and skin30. Studies have shown that in the murine brain it is expressed at particularly high levels in the entorhinal cortex, hypothalamus and hippocampus, as well as the corpus callosum and neostriatum29. Interestingly, PPARD appears to be expressed at its highest levels in embryonic brain, suggesting it may help regulate differentiation of cells during neurodevelopment30. Consistent with this, several studies have shown that PPARD agonists augment differentiation and myelogenesis of cultured murine oligodendrocytes31,32, and PPARD null mice have diminished myelination levels of the corpus callosum and other neurodevelopmental abnormalities31. These findings suggest that if PPARD influences susceptibility to BP, it could be through neurodevelopmental processes. Further, agonists of PPARD were neuroprotective in rat models of stroke and of neurodegenerative disease33.
The increased evidence of association for PPARD among those with poor functioning is consistent with a potential role for Wnt dysfunction in severe bipolar disorder. The GAS score captures both social-occupational and interpersonal functioning in the month prior to the interview. We have previously shown that social functioning was the most highly familial feature of BP, among 40 variables tested, and that loss of employment was also familial34. We have previously hypothesized35 that psychotic features in BP represent clinical manifestations of etiologic overlap between BP and schizophrenia. We found no evidence to support that hypothesis in these data. However, functional impairment represents another potential clinical indicator of etiologic overlap, given that the most impaired BP patients are, by this metric, the most similar to schizophrenia patients, who are typically chronically impaired.
Several prior studies have implicated Wnt signaling genes in BP as well as in schizophrenia. For schizophrenia, the frizzled 3 gene (FZD3), which encodes a receptor for Wnt ligands, has been associated in three different samples36–38, and Proitsi et al.39, employing a Wnt pathway approach similar to ours, found evidence for association with DKK4. We did not find any support for the former gene, and did not study the latter. For BP, one study reported a positive association with GSK3B40, but we did not observe a similar finding with this gene in our sample. There are three potential explanations for our failure to find signals in FZD3 and GSK3B. First, these genes may not be related to BP etiology. Second, these genes may be involved in BP, but we may have failed to assay the relevant SNPs. This possibility is particularly relevant for GSK3B for which we assayed only 56% of known common variation. Third, we could have assayed SNPs that are etiologically relevant to BP (or are in high LD with relevant SNPs), but our sample may have had insufficient power to detect the potentially very modest effect sizes the risk alleles confer (see below). More recently, a GWAS using pooled samples of BP subjects41, some of which were also used in the current study, reported replicated associations with several genes involved with the Wnt signaling pathway, including NXN, A2BP1 and DFNB31.. Unfortunately, we did not study these genes. The Baum et al. (2007) study did not detect significant associations with PPARD, WNT7A, or WNT2B. There are several potential reasons for this. Despite some overlap in the BP subjects included in the two studies, there were substantial differences in sample composition which may have introduced heterogeneity. Additionally, Baum et al.41 used a pooling approach for genotyping which likely reduced their power to detect genetic variants with modest effect sizes.
We also found evidence for two-way interactions of PPARD with WNT2B and WNT7A. These Wnt ligand genes are at the “upstream” end of the Wnt signaling pathways while PPARD is on the “downstream” end of the beta-catenin Wnt pathway. There are three Wnt signaling pathways: the beta-catenin pathway (the canonical pathway), the planar cell polarity pathway and the Wnt/Ca2+ pathway; PPARD is a target gene in the beta-catenin pathway, which is the one that has been implicated in the mechanism of action of bipolar disorder medications. The current findings may suggest that multiple “hits” along the Wnt beta-catenin pathway are needed in order to substantially influence BP susceptibility. We note, however, that the findings of statistical interactions are exploratory and further confirmation is required.
The current study has several limitations that merit attention. First, the sample size may not have been sufficient to detect associations with loci of smaller effects on BP. We estimated that this sample had 80% power to detect association with a locus exerting a genotypic relative risk (GRR) of approximately 1.6, assuming an additive model, a disease prevalence of 1%, and a conservative α = 0.00022 that is Bonferoni adjusted for the 227 SNPs tested). However, BP appears to be a very genetically complex disease, and it is possible the GRRs of BP susceptibility genes may be smaller than 1.6. Second, when we designed the experiment the HapMap Phase II data was not yet available, and as a result our selection of tag SNPs provided incomplete coverage of the currently know common variation in the candidate genes under study. Consequently, we may have missed some of the relevant associations in these candidate genes. Third, we only studied 34 of at least 131 known Wnt-related genes, chosen largely because of their location in BP and/or schizophrenia linkage regions. Because of the limited robustness of linkage studies, we may have omitted genes with an etiologic role in BP. This problem should soon be overcome as the results of several large whole genome association studies in BP and schizophrenia will soon be available. Combining these datasets, as is planned, will allow for comprehensive study of all Wnt-related genes. The study also has several important strengths. Most notably, we used a family-based design with data available on both parents. Thus, we were able to extract the maximal information from this sample for association testing, and the findings are robust to potential confounding by population stratification.
In summary, we found evidence for association with a gene that is a downstream target of Wnt signaling, PPARD. If the finding of association can be confirmed in additional samples, it may illuminate a new avenue for understanding the pathogenesis of severe BP and developing more effective treatments.
Acknowledgments
This work was supported by grants from the NIMH (R01 MH-042243 [JBP], R01 MH-061613 [ESG] and K01 MH-072866 [PPZ]), the National Alliance for Research on Schizophrenia and Depression, and the Stanley Medical Research Institute, and the NIMH Intramural Research Program (FJM). PB was supported by a gift from the Alex Brown Foundation. VLW and JBP were also supported by Margaret Price Investigatorships. Some DNA samples were prepared and distributed by The Rutgers University Cell and DNA Repository under a contract from the NIMH.
We are grateful to B. Schweizer, Y. Huo, K. Miao, and B. Craighead for their contributions. We are also grateful to the many interviewers and diagnosticians who contributed to this project, and to the families who devoted their time and effort to the study.
The BP Phenome Group consists of F. J. McMahon, J. Steele, J. Pearl, L. Kassem, V. Lopez from the Genetic Basis of Mood and Anxiety Disorders Unit, Mood and Anxiety Program, National Institute of Mental Health, National Institutes of Health, Bethesda, MD; J. Potash, D. MacKinnon, E. Miller, J. Toolan from the Department of Psychiatry, Johns Hopkins School of Medicine, Baltimore, MD; P. Zandi from the Department of Mental Health, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD; T. Schulze from the Division of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Ruprecht-Karls-University of Heidelberg, Mannheim, Germany; S. Simpson from the Department of Psychiatry, University of Colorado at Denver, Denver, CO.
The NIMH Genetics Initiative Bipolar Disorder Consortium who contributed to this paper consists of William Byerley, William Coryell, J. Raymond DePaulo, John Kelsoe, Francis McMahon, John Nurnberger, and John Rice
Some of the data and biomaterials were collected in four projects that participated in the NIMH BP Genetics Initiative from 1991–98. The Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, U01 MH46282, J. Nurnberger, M.D., Ph.D., M. Miller, M.D., and E. Bowman, M.D.; Washington University, St. Louis, MO, U01 MH46280, T. Reich, M.D., A. Goate, Ph.D., and J. Rice, Ph.D.; Johns Hopkins University, Baltimore, MD U01 MH46274, J. R. DePaulo, Jr., M.D., S. Simpson, M.D., MPH, and C. Stine, Ph.D.; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, E. Gershon, M.D., D. Kazuba, B.A., and E. Maxwell, M.S.W. Other data and biomaterials were collected in ten NIMH BP Genetics Initiative projects from 1999–2003. The Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, J. Nurnberger, M.D., Ph.D., M. J. Miller, M.D., E. S. Bowman, M.D., N. L. Rau, M.D., P. R. Moe, M.D., N. Samavedy, M.D., R. El-Mallakh, M.D. (at University of Louisville), H. Manji, M.D., D.A. Glitz, M.D. (at Wayne State University), E.T. Meyer, M.S., C. Smiley, R.N., T. Foroud, Ph.D., L. Flury, M.S., D. M. Dick, Ph.D., H. Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, J. Rice, Ph.D., T. Reich, M.D., A. Goate, Ph.D., L. Bierut, M.D.; Johns Hopkins University, Baltimore, MD, R01 MH59533, M. McInnis M.D., J. R. DePaulo, Jr., M.D., D. F. MacKinnon, M.D., F. M. Mondimore, M.D., J. B. Potash, M.D., P. P. Zandi, Ph.D., D. Avramopoulos, Ph.D. and J. Payne, M.D.; University of Pennsylvania, PA, R01 MH59553, W. Berrettini M.D., Ph.D.; University of California at Irvine, CA, R01 MH60068, W. Byerley M.D., and M. Vawter M.D.; University of Iowa, IA, R01 MH059548, W. Coryell M.D., and R. Crowe M.D.; University of Chicago, IL, R01 MH59535, E. Gershon, M.D., J. Badner Ph.D., F. McMahon M.D., C. Liu Ph.D., A. Sanders M.D., M. Caserta, S. Dinwiddie M.D., T. Nguyen, D. Harakal; University of California at San Diego, CA, R01 MH59567, J. Kelsoe, M.D., R. McKinney, B.A.; Rush University, IL, R01 MH059556, W. Scheftner M.D., H. M. Kravitz, D.O., M.P.H., D. Marta, B.S., A. Vaughn-Brown, MSN, RN, and L. Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, F. McMahon, M.D., L. Kassem, PsyD, S. Detera-Wadleigh, Ph.D., L. Austin, Ph.D., D. L. Murphy, M.D.
References
- 1.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15(1):28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 2.Gould TD, Manji HK. The Wnt signaling pathway in bipolar disorder. Neuroscientist. 2002;8(5):497–511. doi: 10.1177/107385802237176. [DOI] [PubMed] [Google Scholar]
- 3.Hall AC, Lucas FR, Salinas PC. Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell. 2000;100(5):525–35. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 4.McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62(6):1073–85. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 5.Lie DC, Colamarino SA, Song HJ, Desire L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437(7063):1370–5. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 6.Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93(16):8455–9. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien WT, Harper AD, Jove F, Woodgett JR, Maretto S, Piccolo S, Klein PS. Glycogen synthase kinase-3beta haploinsufficiency mimics the behavioral and molecular effects of lithium. J Neurosci. 2004;24(30):6791–8. doi: 10.1523/JNEUROSCI.4753-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang UG, Seo MS, Roh MS, Kim Y, Yoon SC, Kim YS. The effects of clozapine on the GSK-3-mediated signaling pathway. FEBS Lett. 2004;560(1–3):115–9. doi: 10.1016/S0014-5793(04)00082-1. [DOI] [PubMed] [Google Scholar]
- 9.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36(2):131–7. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Huang LD, Jiang YM, Manji HK. The mood-stabilizing agent valproate inhibits the activity of glycogen synthase kinase-3. J Neurochem. 1999;72(3):1327–30. doi: 10.1046/j.1471-4159.2000.0721327.x. [DOI] [PubMed] [Google Scholar]
- 11.Beaulieu JM, Sotnikova TD, Yao WD, Kockeritz L, Woodgett JR, Gainetdinov RR, Caron MG. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc Natl Acad Sci U S A. 2004;101(14):5099–104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matigian N, Windus L, Smith H, Filippich C, Pantelis C, McGrath J, Mowry B, Hayward N. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol Psychiatry. 2007;12(9):815–25. doi: 10.1038/sj.mp.4001998. [DOI] [PubMed] [Google Scholar]
- 13.Potash JB, Buervenich S, Cox NJ, Zandi PP, Akula N, Steele J, Rathe JA, Avramopoulos D, tera-Wadleigh SD, Gershon ES, Depaulo JR, Jr, Feinberg AP, McMahon FJ. Gene-based SNP mapping of a psychotic bipolar affective disorder linkage region on 22q12. 3: association with HMG2L1 and TOM1. Am J Med Genet B Neuropsychiatr Genet. 2008;147(1):59–67. doi: 10.1002/ajmg.b.30574. [DOI] [PubMed] [Google Scholar]
- 14.Yamada M, Ohkawara B, Ichimura N, Hyodo-Miura J, Urushiyama S, Shirakabe K, Shibuya H. Negative regulation of Wnt signalling by HMG2L1, a novel NLK-binding protein. Genes Cells. 2003;8(8):677–84. doi: 10.1046/j.1365-2443.2003.00666.x. [DOI] [PubMed] [Google Scholar]
- 15.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51(11):849–59. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 16.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7(4):405–11. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 17.Segurado R, tera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI, Jr, Craddock N, DePaulo JR, Baron M, Gershon ES, Ekholm J, Cichon S, Turecki G, Claes S, Kelsoe JR, Schofield PR, Badenhop RF, Morissette J, Coon H, Blackwood D, McInnes LA, Foroud T, Edenberg HJ, Reich T, Rice JP, Goate A, McInnis MG, McMahon FJ, Badner JA, Goldin LR, Bennett P, Willour VL, Zandi PP, Liu J, Gilliam C, Juo SH, Berrettini WH, Yoshikawa T, Peltonen L, Lonnqvist J, Nothen MM, Schumacher J, Windemuth C, Rietschel M, Propping P, Maier W, Alda M, Grof P, Rouleau GA, Del-Favero J, Van BC, Mendlewicz J, Adolfsson R, Spence MA, Luebbert H, Adams LJ, Donald JA, Mitchell PB, Barden N, Shink E, Byerley W, Muir W, Visscher PM, Macgregor S, Gurling H, Kalsi G, McQuillin A, Escamilla MA, Reus VI, Leon P, Freimer NB, Ewald H, Kruse TA, Mors O, Radhakrishna U, Blouin JL, Antonarakis SE, Akarsu N. Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: Bipolar disorder. Am J Hum Genet. 2003;73(1):49–62. doi: 10.1086/376547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 19.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grover D, Woodfield AS, Verma R, Zandi PP, Levinson DF, Potash JB. QuickSNP: an automated web server for selection of tagSNPs. Nucleic Acids Res. 2007;35(Web Server issue):W115–W120. doi: 10.1093/nar/gkm329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7(5):385–94. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 22.Lake SL, Blacker D, Laird NM. Family-based tests of association in the presence of linkage. Am J Hum Genet. 2000;67(6):1515–25. doi: 10.1086/316895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26(1):61–9. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 24.Clayton D, Chapman J, Cooper J. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27(4):415–28. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- 25.Schulze TG, Buervenich S, Badner JA, Steele CJ, tera-Wadleigh SD, Dick D, Foroud T, Cox NJ, MacKinnon DF, Potash JB, Berrettini WH, Byerley W, Coryell W, Depaulo JR, Jr, Gershon ES, Kelsoe JR, McInnis MG, Murphy DL, Reich T, Scheftner W, Nurnberger JI, Jr, McMahon FJ. Loci on chromosomes 6q and 6p interact to increase susceptibility to bipolar affective disorder in the national institute of mental health genetics initiative pedigrees. Biol Psychiatry. 2004;56(1):18–23. doi: 10.1016/j.biopsych.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O’Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O’Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raybould R, Green EK, Macgregor S, Gordon-Smith K, Heron J, Hyde S, Caesar S, Nikolov I, Williams N, Jones L, O’Donovan MC, Owen MJ, Jones I, Kirov G, Craddock N. Bipolar disorder and polymorphisms in the dysbindin gene (DTNBP1) Biol Psychiatry. 2005;57(7):696–701. doi: 10.1016/j.biopsych.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS. Genetic variation in the 6p22. 3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet. 2002;71(2):337–48. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–35. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 30.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137(1):354–66. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 31.Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20(14):5119–28. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saluja I, Granneman JG, Skoff RP. PPAR delta agonists stimulate oligodendrocyte differentiation in tissue culture. Glia. 2001;33(3):191–204. [PubMed] [Google Scholar]
- 33.Iwashita A, Muramatsu Y, Yamazaki T, Muramoto M, Kita Y, Yamazaki S, Mihara K, Moriguchi A, Matsuoka N. Neuroprotective efficacy of the peroxisome proliferator-activated receptor delta-selective agonists in vitro and in vivo. J Pharmacol Exp Ther. 2007;320(3):1087–96. doi: 10.1124/jpet.106.115758. [DOI] [PubMed] [Google Scholar]
- 34.Schulze TG, Hedeker D, Zandi P, Rietschel M, McMahon FJ. What is familial about familial bipolar disorder? Resemblance among relatives across a broad spectrum of phenotypic characteristics. Arch Gen Psychiatry. 2006;63(12):1368–76. doi: 10.1001/archpsyc.63.12.1368. [DOI] [PubMed] [Google Scholar]
- 35.Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, Depaulo JR, Jr, McInnis MG. The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. Am J Psychiatry. 2001;158(8):1258–64. doi: 10.1176/appi.ajp.158.8.1258. [DOI] [PubMed] [Google Scholar]
- 36.Katsu T, Ujike H, Nakano T, Tanaka Y, Nomura A, Nakata K, Takaki M, Sakai A, Uchida N, Imamura T, Kuroda S. The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci Lett. 2003;353(1):53–6. doi: 10.1016/j.neulet.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Si T, Ling Y, Ruan Y, Han Y, Wang X, Zhang H, Kong Q, Li X, Liu C, Zhang D, Zhou M, Yu Y, Liu S, Shu L, Ma D, Wei J, Zhang D. Association study of the human FZD3 locus with schizophrenia. Biol Psychiatry. 2003;54(11):1298–301. doi: 10.1016/s0006-3223(03)00291-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Yu X, Yuan Y, Ling Y, Ruan Y, Si T, Lu T, Wu S, Gong X, Zhu Z, Yang J, Wang F, Zhang D. Positive association of the human frizzled 3 (FZD3) gene haplotype with schizophrenia in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2004;129(1):16–9. doi: 10.1002/ajmg.b.30076. [DOI] [PubMed] [Google Scholar]
- 39.Proitsi P, Li T, Hamilton G, Di FM, Collier D, Killick R, Chen R, Sham P, Murray R, Powell J, Lovestone S. Positional pathway screen of wnt signaling genes in schizophrenia: association with DKK4. Biol Psychiatry. 2008;63(1):13–6. doi: 10.1016/j.biopsych.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Benedetti F, Bernasconi A, Lorenzi C, Pontiggia A, Serretti A, Colombo C, Smeraldi E. A single nucleotide polymorphism in glycogen synthase kinase 3-beta promoter gene influences onset of illness in patients affected by bipolar disorder. Neurosci Lett. 2004;355(1–2):37–40. doi: 10.1016/j.neulet.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nothen MM, Georgi A, Schumacher J, Schwarz M, Abou JR, Hofels S, Propping P, Satagopan J, tera-Wadleigh SD, Hardy J, McMahon FJ. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13(2):197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]