Abstract
The type 1 insulin-like growth factor receptor (IGF1R) and its ligands (IGF1 and IGF2) have been implicated in a variety of physiological processes and in diseases such as cancer. In addition to IGF1R, IGF2 also activates the insulin receptor (IR) isoform A and therefore antibodies against IGF2 can inhibit cell proliferation mediated by the signaling through both IGF1R and IR triggered by IGF2. We identified a new human monoclonal antibody (mAb), m708.2, which bound to IGF1 and IGF2 but not to insulin. m708.2 potently inhibited signal transduction mediated by the interaction of IGF1 or IGF2 with the IGF1R and IGF2 with the IR. It also inhibited the growth of the breast cancer cell line MCF-7. An affinity-matured derivative of m708.2, m708.5, bound to IGF1 with equilibrium dissociation constant, KD = 200 pM and to IGF2 with KD = 60 pM. m708.5 inhibited signal transduction mediated by IGF1 and IGF2 and cancer cell growth more potently than m708.2. These results suggest that m708.5 could have potential as a candidate therapeutic for cancers driven by the IGF1,2 interactions with IGF1R and IR.
Keywords: mAb, IGF1, IGF2, picomolar affinity, phage display, yeast display
Introduction
The type 1 insulin-like growth factor receptor (IGF1R) and its ligands (IGF1 and IGF2) have been implicated in a variety of physiologic processes and in pathologic conditions such as cancer [1–4]. Although the role of the IGF system in cancer has been recognized many years ago, it is not until recent years that the system’s components have been targeted and shown to affect cell transformation, proliferation, survival, motility, and migration in tissue cultures and in mouse models of cancers [5–7]. The IGF1R shares a similar tetrameric α2β2 structure with insulin receptor (IR). The IGF-mediated signaling is initiated by the binding of IGF1, 2 to their receptor IGF1R, which induces conformational changes in the transmembrane domain of the β subunit. The conformational changes further lead to trans-phosphorylation of the kinase domain in the two β chains [8]. The phosphorylated and fully activated IGF1R recruits adaptor proteins, such as insulin receptor substrate (IRS) 1, IRS2 and Src-Homology Collagen (SHC) [9]. The phosphorylation of IRS1 regulates the activity of phosphoinositide 3-kinase and protein kinase B (also known as Akt) and triggers the activation of some transcription factors which control the expression of many genes that are important for cell proliferation and growth [3]. Numerous studies demonstrate that IGF1R is expressed in a broad panel of tumors, suggesting that inhibition of IGF1R signaling may have both proapoptotic and antiproliferative consequences [10]. Thus, it seems that the modulation of the activity of the IGF system could add to the arsenal of anticancer therapeutic approaches [11]. Different strategies aimed at targeting the IGF system have been investigated, including small molecule tyrosine kinase inhibitors, antisense technologies [12], monoclonal antibodies (mAbs) targeting the IGF1R [10, 13–14], IGF1 [15] and IGF2 [11, 16]. They inhibit the kinase activity or disrupt the interaction of IGF1R with its ligands IGF1, 2, and down-regulate the levels of IGF-1R.
MAbs have been highly successful as cancer therapeutics. Great progress in clinical and preclinical stages has been made in developing antibodies aimed at targeting IGF system components [17]. During the past several years, a number of mAbs against the IGF1R have been identified with cancer cell growth inhibitory activity [10, 13, 18–19]. However, the IGF1R is not very significantly overexpressed in tumors and the mAbs could bind to normal cells expressing the receptor with possible toxic effects. Antibodies against the ligands IGF1 and IGF2 that inhibit the interaction with IGF1R may not have these undesirable effects. Most anti-IGF1R antibodies do not inhibit IGF1R/IR hybrid receptor. IGF2-mediated signaling through the hybrid receptor could escape the blockage by IGF1R antibodies. In addition, another consideration is that binding to the ligands, which is not cell-associated but is diffusible, may not require significant penetration into solid tumors, thus avoiding problems associated with penetration [11]. A rat monoclonal antibody KM1468, which is cross-reactive to human IGF1 and IGF2, could suppress the development of new bone tumors, and the progression of established tumors [20–21]. KM1468 inhibits tumor growth and increases survival in a mouse model of hepatitic metastasis induced by human colorectal cancer cell line, HT29 [21]. More recently, several human mAbs against IGF2 have been reported [11, 16] and an IGF1,2 cross-reactive mAb is also described [22]. These results suggest that binding to IGF1 and IGF2 can markedly affect tumor growth and that mAbs against the two ligands are promising candidate therapeutics.
Here, we describe the identification and characterization of a human mAb (m708.5) which bound with high affinity to IGF1 (KD = 200 pM) and IGF2 (KD = 60 pM). It potently inhibited signal transduction mediated by the IGF1R interaction with IGF1 and IGF2, and the interaction of the IR with IGF2, resulting in the inhibition of phosphorylation of IGF1R, IR and cancer cell growth. These results suggest that m708.5 may have potential as a candidate therapeutic.
Materials and Methods
Panning of a Large Phage Displayed Fab Library
Recombinant human IGF1 was used to pan a human naive Fab phage library containing 1010 unique clones as previously described [23].
Mutagenesis by Error-prone PCR
Error-prone PCR of the entire scFv gene was performed using Stratagene GeneMorph® II Random Mutagenesis Kit according to the instructions of the manufacturer. Briefly, PCR was done in a 50-µL reaction containing 1 × Mutazyme II reaction buffer, 0.5 µM each of primers ERRORF (5’ GATATATCCATGGCCCAGGCGGCC 3’) and ERRORR (5’ ACCACTAGTTGGGCCGGCCTG 3’), 0.2 mM (each) dNTPs, 1 ng of DNA template, 2 µM 8-oxo-deoxyguanosine triphosphate, 2 µM 2'-deoxy-p-nucleoside-5'-triphosphate, and 2.5 U of Mutazyme II DNA polymerase. The reaction mixtures were denatured at 95 °C for 2 min, cycled 35 times at 95 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min, and finally extended at 72 °C for 10 min. The PCR products were gel-purified and amplified in four 100-µL PCR reactions containing 1× Accuprime PCR reaction mix (Invitrogen), 1 µM of primers YDRDF (5’ CTTCGCTGTTTTTCAATATTTTCTGTTATTGCTTCAGTTTTGGCCCAGGCGGCC 3’) and YDRDR (5’ GAGCCGCCACCCTCAGAACCGCCACCCTCAGAGCCACCACTAGTTGGGCCGGCCTG 3’), 120 ng of error-prone PCR product, and 2.5 U of Accuprime pfx DNA polymerase (Invitrogen). The reactions were thermally cycled using the same conditions except that 30 cycles were used. Amplified insert was gel-purified and concentrated using vacuum concentrator.
Construction of Yeast Displayed Mutant Libraries
The yeast display vector used for library construction, pYD7, was modified from pCTCON2 [24]. Two SfiI restriction sites of pYD7 before and after the scFv matched the cloning sites of the phage display vector. This allowed fragments to be conveniently shuttled between the yeast vector and phage display vector. Furthermore, to better present displayed antibodies on yeast surface, scFvs were fused at the N-terminal of the yeast protein agglutinin (aga2p).
The pYD7 vector was digested with SfiI and gel-purified. Multiple aliquots of 12 µg of mutagenized scFv DNA and 4 µg of plasmid DNA were concentrated into 20–30 µL of water. The pYD7-scFv libraries were then electroporated into competent yeast cells EBY100 prepared as described previously [25]. Briefly, 10 ml of EBY100 yeast cells in YPD media (10 g/l yeast nitrogen base, 20 g/l peptone and 20 g/l glucose) were grown at 30°C with shaking at 250rpm overnight. The culture was inoculated into 100 ml of fresh YPD and allowed to grow to OD600 of 1.6 before collecting by centrifugation. The cell pellet was washed twice with 50 ml of cold water, and once with 50 ml cold electroporation buffer (1 M sorbitol/1 mM CaCl2). Cells were conditioned in 20 ml of 0.1 M LiAc/10 mM DTT by incubation at 30°C with shaking at 250rpm for 30 min. Cells were washed one more time with 50 ml of electroporation buffer, then resuspended in the same buffer to reach 1 ml volume. Each electroporation mixture included 400 µl of yeast cell suspension, 4 µg linearized vector, and 12 µg insert DNA. Cells were electroporated at 2.5 kV and 25 µF in BioRad GenePulser cuvettes (0.2 cm electrode gap). After electroporation, cells were resuspended in 20 ml of 1:1 (v:v) mix of 1 M sorbitol: YPD media and incubated at 30°C for 1 h. Finally, the cells were collected and amplified in SDCAA media (20 g/l glucose, 6.7 g/l yeast nitrogen base without amino acids, 5.4 g/l Na2HPO4, 8.6 g/l NaH2PO4.H2O and 5 g/l casamino acids) at 30°C with shaking at 250rpm for 24–48 h.
Selection of Binders from the Yeast Libraries
Typically, amplified yeast library was transferred to SG/RCAA media (20 g/l galactose, 20 g/l Raffinose, 1 g/l glucose, 6.7 g/l yeast nitrogen base without amino acids, 5.4 g/l Na2HPO4, 8.6 g/l NaH2PO4.H2O and 5 g/l casamino acids) for induction at 20°C for 16–18 h in culture density and volume appropriate for the size of the library.
The methodology for generating and isolating higher affinity mutants was as described in references [24, 26–27]. Antigen concentrations are chosen based on the equilibrium dissociation constant (KD) of the parental antibody. The antigen incubation volume must be large enough to allow at least tenfolds excess of the antigen over amounts of scFv displayed on yeast (assuming 5×104 scFv/yeast cell [24]). Antigen incubation times were chosen to ensure that the reaction reached at least 90% of equilibrium calculated as described in reference [28]. Before FACS sorting, induced yeast library (1×109) was incubated with 10 µg-IGF1-conjugated magnetic beads for 1 h at room temperature in PBSA buffer (0.1% BSA in PBS), followed by separation with a magnetic stand. The isolated yeast cells were washed 3 times with PBSA buffer, and added into 10 ml of SDCAA media for amplification overnight in a 30°C shaker with 250 rpm. The amplified yeast cells were induced in SG/RCAA media for 18 h at 20°C with 250 rpm shaking. For the first FACS selection of the 1st round, approximately 1 × 108 yeast cells were pelleted, washed twice with PBSA buffer and resuspended in 1 ml PBSA buffer with 3 nM biotinylated IGF1 and 2 µg/ml of mouse anti-c-myc antibody (Invitrogen). After incubation, yeast cells were washed 3 times and then resuspended in 1 ml PBSA buffer. Both 1:50 diluted R-phycoerythrin conjugated streptavidin (Invitrogen) and Alexa Fluor 488 conjugated goat anti-mouse IgG antibody (Invitrogen) were added to yeast cells. Cell were incubated at 4 °C for 30 min, and washed 3 times with PBSA buffer, and then resuspended in PBSA buffer for sorting. Sorting gates were determined to select only the population with higher antigen binding signals. Collected cells were grown overnight in SDCAA media at 30 °C and induced in SG/RCAA for the next sorting. For the next two FACS selections, approximately 1–2×107 yeast cells were used for staining with 1 nM and 0.3 nM biotinylated IGF-1, respectively.
Two and three FACS selections were performed in 2nd and 3rd round. Yeast cells were pelleted, washed in PBSA buffer, resuspended in PBSA buffer with biotinylated IGF1 (ranging from 3 nM to 0.1 nM) and mouse anti-c-myc antibody, and incubated on ice. Cells were then washed with PBSA twice and resuspended in 1 ml PBSA with R-phycoerythrin conjugated streptavidin and Alexa Fluor 488 conjugated goat anti-mouse antibody.
After the 1st and 2nd round, yeast plasmids were isolated using Zymoprep Yeast Plasmid Miniprep II Kit (Zymo Research) according to the manufacturer's instructions and used as templates for error-prone PCR to amplify gene repertoire for the construction of the 2nd and 3rd mutant libraries. Cells from the last selection of the 3rd round were spread on SDCAA plates. Monoclonal yeast cells were characterized and isolated antibodies with improved affinity were sequenced.
Conversion to IgG1, and expression of Fab, scFv and IgG1
We used the same procedure as previously described [29]. Fab and scFv were expressed and purified as previously described [30–31].
ELISA Binding Assay
Antigens (50 ng) per well were coated on 96-well ELISA plates overnight at 4°C. For phage ELISA, ~1×1010 pfu phages were incubated with antigen for 1 h. Bound phages were detected with anti-M13-HRP mAb (1:3000, GE Healthcare). For Fab and IgG1 ELISA, Fabs and IgG1s with different dilutions were incubated with antigens for 1 h. Bound Fabs were detected with anti-FLAG-HRP mAb (1:3,000; Sigma). Bound IgG1s were detected with anti-human Fc-HRP mAb (1:2,000; Invitrogen). The 2,2’-azino-bis-(3-ethylbenzthiazoline-6-sulfonic acid) substrate (Sigma) was added and the reaction was read at 450 nm.
Affinity Measurement of Yeast-displayed scFvs
The equilibrium dissociation constant was determined essentially as previously described [32].
Affinity Determination by Surface Plasmon Resonance
Interactions between various isolated antibodies and IGF1 and IGF2 were analyzed by surface plasmon resonance technology using a Biacore X100 instrument (GE healthcare). IGF1 or IGF2 was covalently immobilized onto a sensor chip (CM5) using carbodiimide coupling chemistry. A control reference surface was prepared for nonspecific binding and refractive index changes. For analysis of the kinetics of interactions, varying concentrations of antibodies were injected at flow rate of 30 µl/min using running buffer containing 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% Surfactant P-20 (pH 7.4). The association and dissociation phase data were fitted simultaneously to a 1:1 model by using BIAevaluation 3.2. All the experiments were done at 25°C.
Competition Assay
scFv or IgG1 m708.5 (10 nM) was incubated with 5 nM biotinylated human IGF1 and 1 nM human biotinylated IGF2 at room temperature for 20 min. The mixtures were added to 5×105 MCF-7 cells in 50 µl PBSA buffer and incubated for 30 min on ice. After washing once, cells were incubated with a 1:50 dilution of R-phycoerythrin conjugated streptavidin for 30 min on ice, then washed again and resuspended in 0.5 ml PBSA buffer. Analysis was performed using a BD Bioscience FACScalibur. Irrelevant anti-gp41 MPER scFv-3A2a (unpublished data) and anti-Nipah/Hendra viruses IgG1-m102.4 [33] were used as negative controls.
Phosphorylation Assay
Measurement of IGF1R and IR posphorylation was performed as previously described [11] except that the treatment medium was made by adding 1 nM human IGF1 or 5 nM human IGF2.
Cell Growth Assay
Cell growth and inhibition were measured as previously described [11] except that IGF1 (2.5 nM) was also used in addition to IGF2 (2.5 nM).
Results
Identification of a high-affinity IGF1,2 cross-reactive human mAb, m708.2
To develop human mAbs against human IGF1, we used a large size (~ 1010 independent clones) naive human Fab phage-displayed library. A recombinant human IGF1 was conjugated to magnetic beads and used as the antigen for pannings. After three rounds of panning, more than 200 random individual phage clones were screened by phage ELISA against IGF1. Clones that exhibited significant binding to IGF1 were sequenced. Finally, three clones with unique sequences were found. They were expressed in bacteria as soluble Fabs, purified, and tested for binding activity in ELISA. Two Fabs, designated m705 and m706, showed binding specifically to IGF1 only, while one Fab, m708, bound to both IGF1 and IGF2 in ELISA (supplementary Figure 1). m708 was selected for affinity maturation by light chain shuffling.
Two rounds of panning of the light chain shuffled library (containing 2×108 independent clones) against IGF1 conjugated magnetic bead were performed and 200 clones from the second round of panning were screened by phage ELISA. Among them, Fab m708.2 showed markedly higher binding to both IGF1 and IGF2 than parental m708 version (data not shown) and was selected for further improvement.
Construction of m708.2 mutant libraries displayed on yeast and selection of a pM affinity binder, m708.5
To further increase the affinity of m708.2, it was converted to a scFv and randomly mutagenized by error-prone PCR. We used yeast display for further maturation because it allows fine discrimination between mutants by flow cytometry and our attempts to increase affinity by phage display were not successful. Therefore, the mutant library was displayed on yeast cells by homologous recombination with a vector containing a C-terminal Aga2 protein and c-myc tag. To obtain large amounts of DNA insert and avoid improper incorporation at the site of homologous recombination, purified DNA obtained from the first PCR reaction was re-amplified in a second PCR reaction. Thus, 30–40% of the transformed cells displayed scFvs as verified by flow cytometry (data not shown).
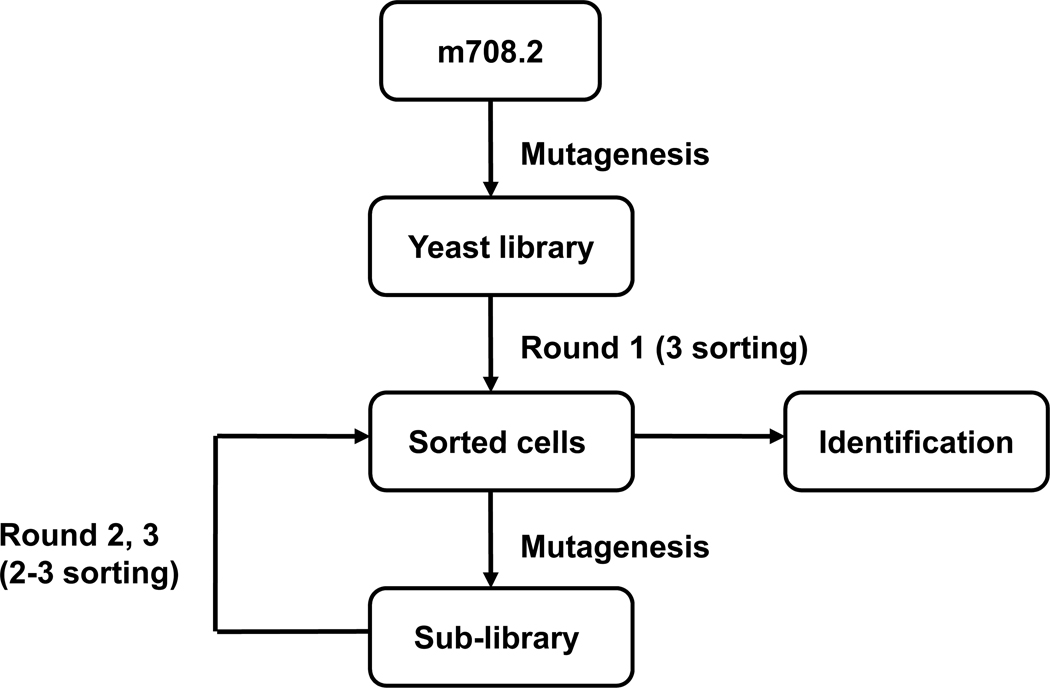
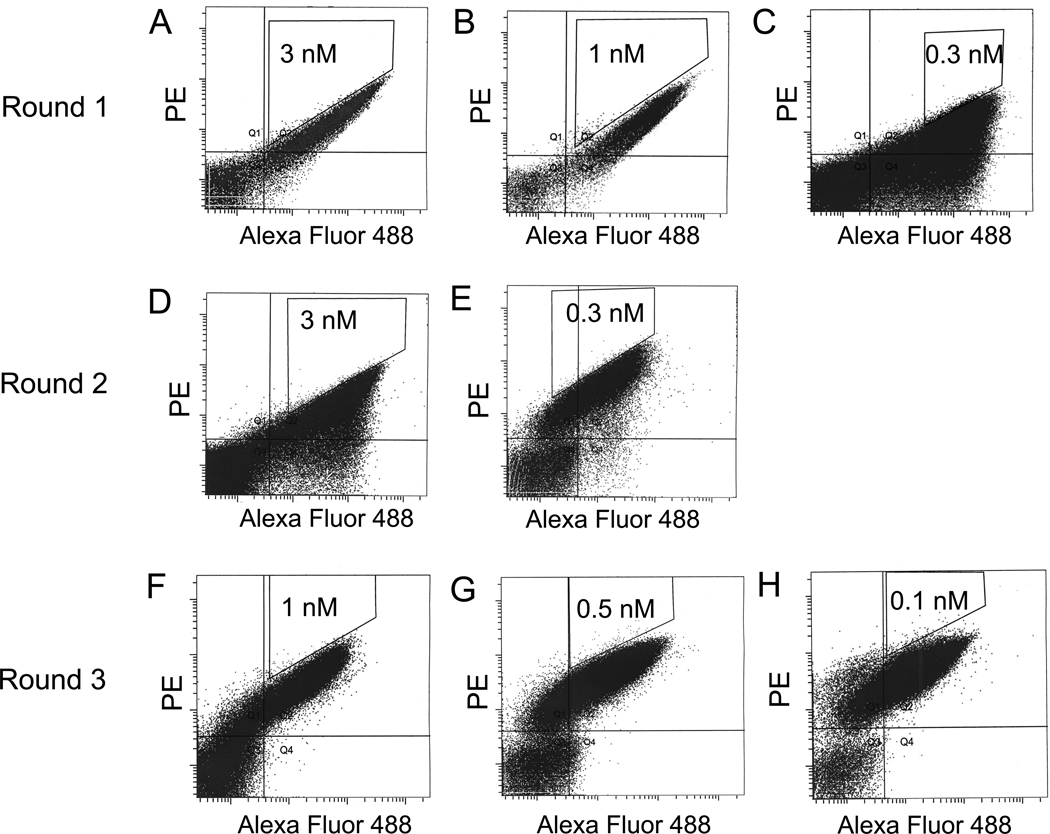
Mutant yeast library of relatively large (up to 109) size was generated and subjected first to one round of selection by using IGF1 conjugated magnetic beads according to the method described in references [34–35]. This allowed elimination of yeast cells that ddi not express antibodies or bound weakly to IGF1. The library was then sorted several times by FACS for binding to IGF1. The sorted scFvs were mutated by error-prone PCR of the entire gene to yield a new sub-library. The process of sorting and mutagenesis was then cyclically repeated (Figure 1). The highest affinity clones from the final round of maturation were identified (Figure 2) and their sequences were analyzed.
Figure 1. Affinity maturation scheme.
Figure 2. FACS for affinity maturation.
Yeast libraries were labeled with mouse anti-c-myc antibody followed by goat anti-mouse dye as well as biotinylated IGF1 followed by streptavidin-dye. (A, B, C) During three FACS selections of round 1, yeast cells were stained with concentrations of biotinylated IGF1 at 3 nM, 1nM and 0.3 nM, respectively. (D, E) During two FACS selections of round 2, yeast cells were stained with concentrations of biotinylated IGF1 at 3 nM, and 0.3 nM, respectively. (F, G, H) During three FACS selections of round 3, yeast cells were stained with concentrations of biotinylated IGF1 at 1 nM, 0.5nM and 0.1 nM. The 0.1–0.3% cells were selected from sort gates.
One dominant clone, m708.5, was identified by sequence analysis. Compared with m708.2, m708.5 had two amino acid substitutions in the heavy chain CDR2 (H2), two amino acid substitutions in H3 and nine amino acid substitutions in the framework region (Table 1). There were four substitutions in the framework regions of the light chain. These substitutions resulted in a remarkable improvement of affinity but still retaining cross-reactivity as measured by two different methods. The first method was based on yeast-displayed scFvs that were incubated with varying concentrations of biotinylated IGF1 or IGF2. The affinity of yeast-displayed m708.5 scFv for IGF1 was increased 39-fold compared to that of m708.2 (1×10−10 M vs 3.9×10−9 M), whereas its affinity for IGF2 was increased 27-fold (4.1×10−11 M vs 1.1×10−9 M) (Table 2). Similar values were obtained for isolated soluble scFv m708.5 as measured with surface Plasmon resonance (supplementary Figure 2): 2×10−10 M for IGF1 and 6.1×10−11 M for IGF2, compared to scFv m708.2 (2.2×10−9 M for IGF1 and 1.8×10−9 M for IGF2) (Table 2). Therefore, the cross-reactive antibody m708.2 was successfully matured to a scFv, m708.5, with pM affinity for both IGF1 and IGF2.
Table 1.
Sequences of the m708.2 and m708.5 heavy chain CDRs.
| Clone | H1a | H2b | H3c |
|---|---|---|---|
| m708.2 | G G T F S S Y A | I I P I LG I A | A R G P R G Y S Y N F D Y |
| m708.5 | - - - - - - - - | - - - T - - -V | - G - - - - - - - - - - N |
Identical residues are denoted by -.
H1 is CDR1 of heavy chain;
H2 is CDR2 of heavy chain;
H3 is CDR3 of heavy chain.
Table 2.
Binding rate constants and affinities of scFv m708.2,5 as measured by FACS and Biacore.
| Antibody | Antigen | FACSa | Biacoreb | ||
|---|---|---|---|---|---|
| KD (M) | kon (M−1s−1) | koff (s−1) | KD(M) | ||
| m708.2 scFv | IGF1 | 3.9×10−9 | 1.1×106 | 2.4×10−3 | 2.2×10−9 |
| IGF2 | 1.1×10−9 | 2.3×105 | 4.0×10−4 | 1.8×10−9 | |
| m708.5 scFv | IGF1 | 1.0×10−10 | 1.4×106 | 2.8×10−4 | 2.0×10−10 |
| IGF2 | 4.1×10−11 | 4.1×106 | 2.5×10−6 | 6.1×10−11 | |
Values for yeast-displayed scFvs.
Values for purified soluble scFvs.
Avidity of IgG1 m708.5
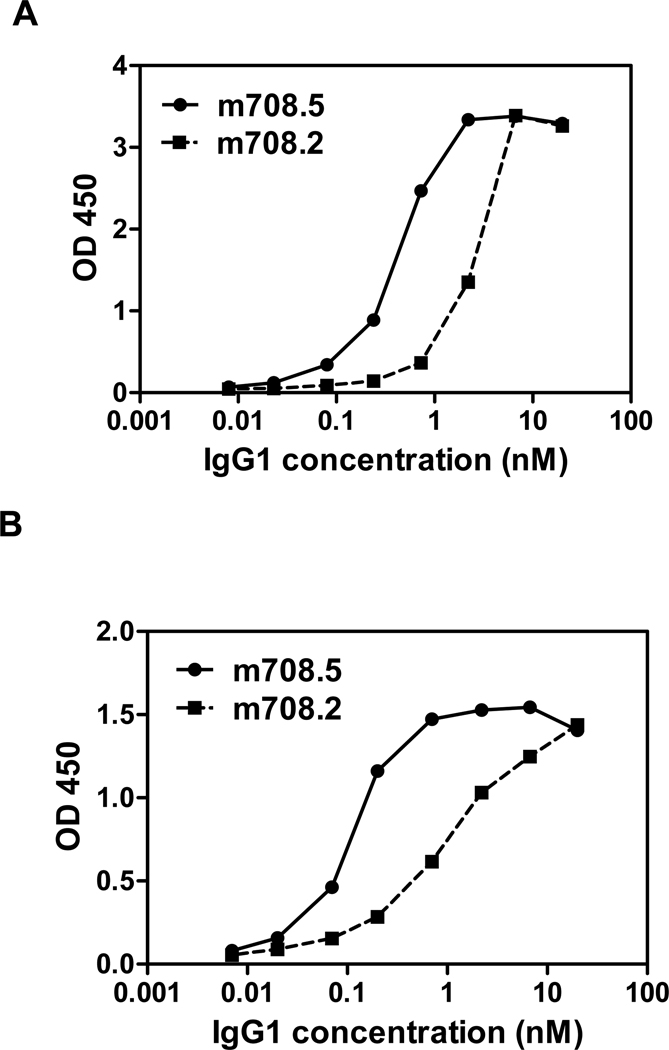
The scFv m708.5 was converted to an IgG1 format and its avidity measured with surface Plasmon resonance. IgG1 m708.5 showed increased binding for both IGF1 and IGF2 compared to scFv m708.5. The effective KD of m708.5 for IGF1 and IGF2 was 1000-fold and 60-fold higher than KD for scFv, respectively (data not shown). The measured KD (<10−12 M) is below the sensitivity of the Biacore instrument (106 M−1S−1 for kon and 10−6 S−1 for koff) and therefore may not be reliable. IgG1 m708.5 was also much more effective in binding to both IGF1 and IGF2 than m708.2 as measured by ELISA (Figure 3). Thus, m708.5 exhibited very high avidity in IgG1 format resulting in much stronger binding than in scFv format.
Figure 3. Binding of IgG1 m708.2 and m708.5 to IGF1,2.
IgG1 m708.2 and m708.5 were serially diluted and added to wells coated with IGF1 (A) or IGF2 (B). Bound IgG1 was detected with an HRP conjugated anti-human Fc antibody and measured as optical densities (O.D.) at 450 nm. The data are representatives of three separate experiments performed in duplicates with variations not exceeding 10%.
m708.5 inhibits binding of IGF1,2 to IGF1R and IR and their phosphorylation
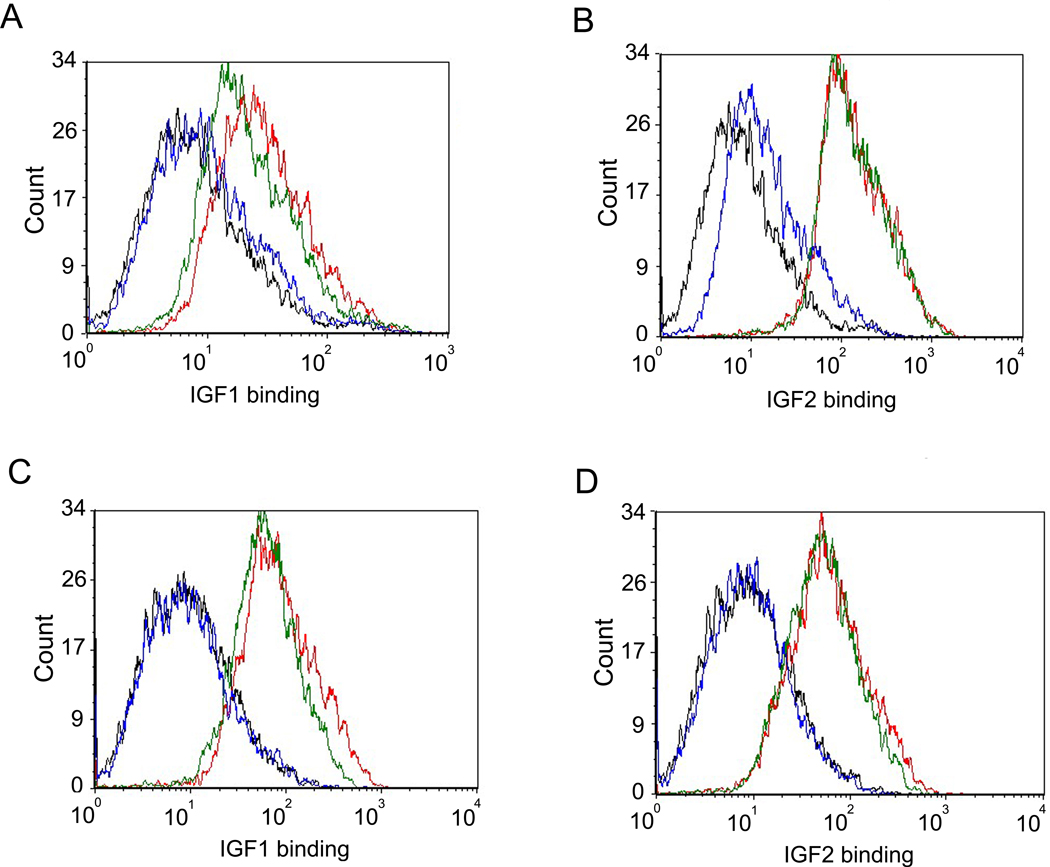
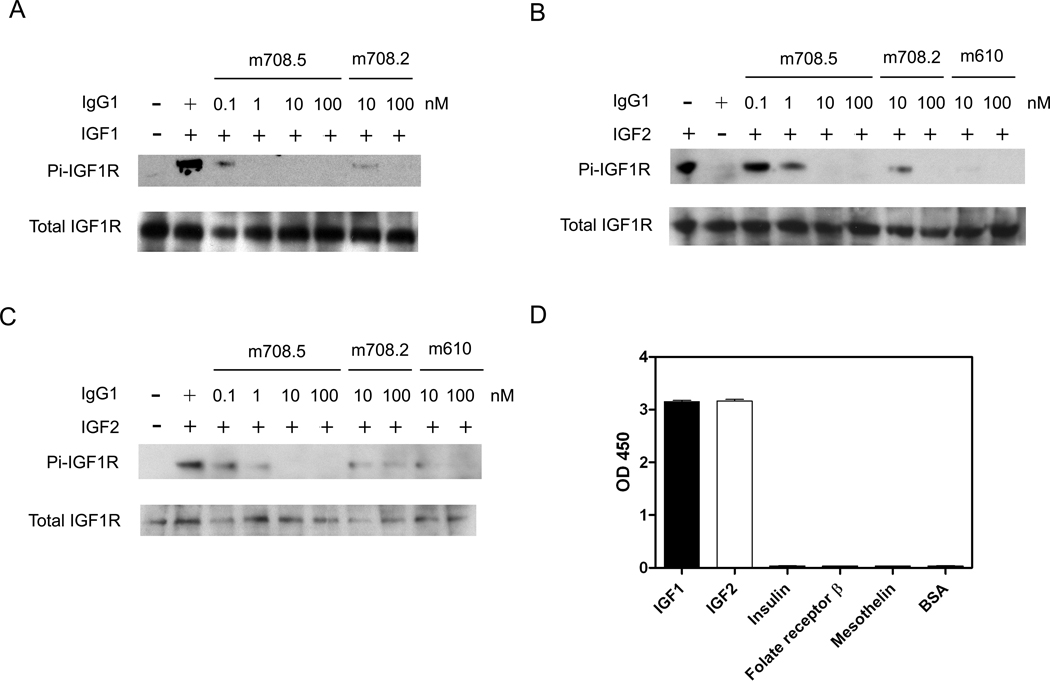
IGFs bind and activate IGF1R and the signals transduced by this pathway lead to cell proliferation. To find whether m708.5 inhibits binding of IGF1 and IGF2 to IGF1R and IR, we used the breast cancer cell line MCF-7, which have been shown to express both receptors. m708.5 in scFv and IgG1 completely blocked IGF1 and IGF2 binding to their receptors on MCF-7 cells (Figure 4). Control scFv and IgG that did not recognize IGFs had no effect. The antibody also inhibited IGF1-induced (1 nM) or IGF2-induced (5 nM) phosphorylation. Immunoblottings (Figure 5A) showed that from 1 nM to 100 nM, m708.5 completely inhibited the IGF1-induced phosphorylation of IGF1R. Similar inhibitory activity was also observed with m708.2, which however only partly blocked IGF1 induced phosphorylation at 10 nM. In addition to IGF1, both m708.2 and m708.5, as well as m610 were capable of inhibiting IGF2-induced phosphorylation of IGF1R (Figure 5B). However, m708.5 was superior to m708.2 (Figure 5B) and competed with m610. The antibodies also inhibited IGF2-mediated phosphorylation of the IR induced by 5 nM of IGF2 (Figure 5C). The inhibition was independent on human insulin which m708.5 did not bind (Figure 5D). Therefore, m708.5 could effectively inhibit ligand-induced phosphorylation of the two receptors without inhibiting insulin-IR interaction.
Figure 4. Inhibition of IGF1 and IGF2 binding to MCF-7 cells by m708.5.
(A–B) scFv m708.5 scFv and control scFv 3A2a scFv were pre-incubated with biotinylated IGF1 (A) and biotinylated IGF2 (B) for 20 min at room temperature. Then mixtures were incubated with MCF-7 cells for 30 min on ice. (C–D) IgG1 m708.5 and control IgG1 m102.4 were pre-incubated with biotinylated IGF1 (C) and biotinylated IGF2 (D) for 20 min at room temperature. Then mixtures were incubated with MCF-7 cells for 30 min on ice. After the staining of R-phycoerythrin conjugated streptavidin for 30 min on ice, cells were detected by flow cytometry. In all the graphs, black lines are for cells without any antibody labeling. Blue lines are for tested antibodies, and green lines are for the control antibodies. Red lines show binding with IGF1 or IGF2 alone. Data shown are representatives of three separate experiments performed in duplicates with similar results.
Figure 5. Inhibition of IGF1R and IR phosphorylation by m708.2,5 and absence of m708.5 binding to human insulin.
(A–B) MCF-7 cells were starved in serum free medium for 5 h first, followed by addition of treatment medium with 1 nM IGF1 (A) or 5 nM IGF2 (B) with indicated concentrations of IgG1s. Thirty minutes later cells were chilled and lysed. IGF1R was immunoprecipitated and the phosphorylated IGF1R was detected with a phosphor-tyrosine specific antibody. The total amount of IGF1R was detected by the same polyclonal antibody used for the immunoprecipitation. (C) MCF-7 cells were starved and treated with 5 nM IGF2 with indicated concentrations of IgG1s. The phosphorylated IR was detected with a phospho-tyrosine specific antibody. The total amount of IR was detected. (D) IgG1 m708.5 (10 nM) was added to wells coated with IGF1, IGF2, human insulin and irrelevant antigens. Bound IgG1 was detected with an HRP conjugated anti-human Fc antibody and measured as O.D. at 450 nm. Data shown are representatives of three separate experiments performed in duplicates with similar results.
Inhibition of MCF-7 cell growth by m708.5
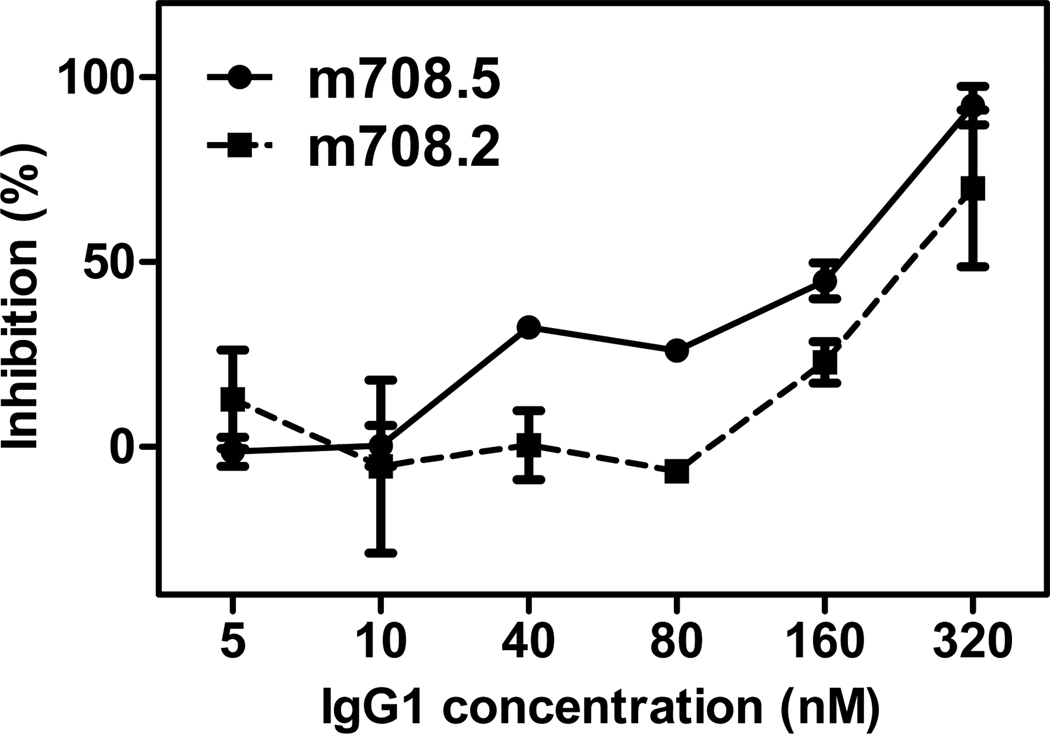
In order to investigate whether the blocking of IGF binding by m708.5 is potent enough to inhibit the proliferation of cells, we tested the activity of m708.5 in a cell growth assay using MCF-7 cells. After 3 days of treatment, the cell growth was almost completely inhibited at 320 nM of m708.5 (Figure 6). At concentrations higher than 40 nM m708.5 treatment resulted in more than 40% cell growth inhibition. In contrast, m708.2 could only inhibit cancer cells at concentrations higher than 80 nM and 40% inhibition was achieved above 160 nM. It is noteworthy that MCF-7 cells produce significant amounts of IGF2-concentrations of up to 35 nM were observed after 3 days [11]. These data showed that the potency of m708.5 was remarkably improved compared with m708.2 and it effectively inhibited tumor cell proliferation in vitro.
Figure 6. Growth inhibition of MCF-7 cells by m708.2 and m708.5.
MCF-7 cells were incubated in complete medium overnight. Different concentrations of IgG1 m708.2 and m708.5 were pre-incubated with added IGF1 (2.5 nM) and IGF2 (2.5 nM) for 15 min. The media of MCF-7 cells were replaced by the mixture of IgG1 and ligands immediately. Cells were allowed to grow for 3 days, and MTS substrate was added to detect viable cells. The reaction was monitored by absorbance at 450 nM. Positive control was cells in serum free medium with IGF ligands. Blank control was cells in serum free medium without any IGFs. Shown are data with mean ± SEM calculated from three separate experiments.
Discussion
The IGF signaling system plays an important role in tumorigenesis [36]. Human IGF1 and IGF2 share 62% sequence identity and have overlapping functions: both IGF1 and IGF2 can activate the IGF1R which can drive tumor cell proliferation. A remarkable feature of IGF2 (but not IGF1) is that it can also bind to the IR and enhance cell proliferation. To further explore novel cancer therapeutics based on human mAbs targeting components of the IGF system, we identified three human antibodies specific for IGF1 utilizing phage display technologies. One of them, m708, exhibited also significant binding to human IGF2 and was further affinity matured by light-chain shuffling, mutagenesis and yeast display. This antibody, m708.5, potently inhibited both IGF1- and IGF2-induced phosphorylation of IGF1R as well as IGF2-induced phosphorylation of the IR, and less potently growth of MCF-7 cancer cell expressing IGF1R.
The inhibitory activity of the antibodies was cell type dependent (data not shown) with a likely major determinant being the surface concentration of the IGF1R and the IR. To date, many IGF1R-specific antibodies have been under preclinical studies, and several are being evaluated in clinical trials [1]. We previously reported a human mAb (m610) with high affinity to IGF2 that potently blocked the growth/migration of human cancer lines in vitro [11], and significantly suppressed the growth of prostate cancer cells in a human bone environment [37]. A murine mAb cross-reactive to human IGF1 and IGF2 inhibited the development of new bone tumors, and the progression of established tumors [20]. Recently, a new mAb against IGF1 and IGF2 was reported which was effective in a mouse model of caner with cells expressing high levels of IGF1R [22]. Based on the data described in our article and those previously reported, we believe that m708.5 would also exhibit inhibitory activity in vivo. The finding that IgG1 m708.5 also binds IGF1,2 is important because it would allow to use the IgG antibody format, which is most stable and has long half-life in vivo.
m708.5 is fully human mAb cross-reactive to both human IGF1 and IGF2 with picomolar affinity and potently inhibits signal transduction mediated by IGF1,2 interacting with IGF1R and IGF2 interacting with IR. It also inhibits potently tumor cell growth in vitro and is likely to exert similar activity in vivo. Importantly, m708.5 cross-reacts to mouse IGF1 and IGF2 (supplementary Figure 3). Therefore, the activity of m708.5 can be tested with human xenografts in mouse models of cancer where it could inhibit both autocrine (human) and paracrine (mouse) IGF. Therefore, m708.5 offers a new and promising therapeutic strategy for treating tumor cells and a possible therapeutic addition to IGF1R targeted agents. It could be used in combination with other IGF1,2 or IGF1R targeting mAbs, and also with other cancer therapeutics. We do not know where exactly its epitope is located, although it inhibits binding of IGF1,2 to both IGF1R and IR and therefore its epitope may overlap with their binding sites although steric hindrance is also possible. It competes with our previously identified anti-IGF2 mAb m610 for binding to IGF2 and therefore their epitopes are likely to be overlapped although steric hindrance could not be excluded. It would be interesting to find out whether m708.5 competes with the recently described mAbs against IGF1,2. Further research is needed to exactly localize the epitope of m708.5 and evaluate its inhibitory properties in vivo as a candidate therapeutic.
Supplementary Material
Acknowledgements
We thank Dane Wittrup for providing reagents and members of our group for helpful discussions. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 2.Yee D. Targeting insulin-like growth factor pathways. Br J Cancer. 2007;96 Suppl:R7–R10. [PubMed] [Google Scholar]
- 3.Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, et al. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol. 2005;205:145–153. doi: 10.1002/path.1712. [DOI] [PubMed] [Google Scholar]
- 4.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Sun Y. Insulin-like growth factor receptor-1 as an anti-cancer target: blocking transformation and inducing apoptosis. Curr Cancer Drug Targets. 2002;2:191–207. doi: 10.2174/1568009023333863. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann F, Garcia-Echeverria C. Blocking the insulin-like growth factor-I receptor as a strategy for targeting cancer. Drug Discov Today. 2005;10:1041–1047. doi: 10.1016/S1359-6446(05)03512-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith TJ. Insulin-like growth factor-I regulation of immune function: a potential therapeutic target in autoimmune diseases? Pharmacol Rev. 2010;62:199–236. doi: 10.1124/pr.109.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez-Sanchez C, Blakesley V, Kalebic T, Helman L, LeRoith D. The role of the tyrosine kinase domain of the insulin-like growth factor-I receptor in intracellular signaling, cellular proliferation, and tumorigenesis. J Biol Chem. 1995;270:29176–29181. doi: 10.1074/jbc.270.49.29176. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Dimitrov DS. Monoclonal antibodies against components of the IGF system for cancer treatment. Curr Opin Drug Discov Devel. 2008;11:178–185. [PubMed] [Google Scholar]
- 10.Zha J, O'Brien C, Savage H, Huw LY, Zhong F, Berry L, et al. Molecular predictors of response to a humanized anti-insulin-like growth factor-I receptor monoclonal antibody in breast and colorectal cancer. Mol Cancer Ther. 2009;8:2110–2121. doi: 10.1158/1535-7163.MCT-09-0381. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Zhu Z, Xiao X, Choudhry V, Barrett JC, Dimitrov DS. Novel human monoclonal antibodies to insulin-like growth factor (IGF)-II that potently inhibit the IGF receptor type I signal transduction function. Mol Cancer Ther. 2006;5:114–120. doi: 10.1158/1535-7163.MCT-05-0252. [DOI] [PubMed] [Google Scholar]
- 12.Resnicoff M, Coppola D, Sell C, Rubin R, Ferrone S, Baserga R. Growth inhibition of human melanoma cells in nude mice by antisense strategies to the type 1 insulin-like growth factor receptor. Cancer Res. 1994;54:4848–4850. [PubMed] [Google Scholar]
- 13.Wang Y, Lipari P, Wang X, Hailey J, Liang L, Ramos R, et al. A fully human insulin-like growth factor-I receptor antibody SCH 717454 (Robatumumab) has antitumor activity as a single agent and in combination with cytotoxics in pediatric tumor xenografts. Mol Cancer Ther. 2010;9:410–418. doi: 10.1158/1535-7163.MCT-09-0555. [DOI] [PubMed] [Google Scholar]
- 14.Zhang MY, Feng Y, Wang Y, Dimitrov DS. Characterization of a chimeric monoclonal antibody against the insulin-like growth factor-I receptor. MAbs. 2009;1:475–480. doi: 10.4161/mabs.1.5.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su JL, Stimpson S, Edwards C, Van Arnold J, Burgess S, Lin P. Neutralizing IGF-1 monoclonal antibody with cross-species reactivity. Hybridoma. 1997;16:513–518. doi: 10.1089/hyb.1997.16.513. [DOI] [PubMed] [Google Scholar]
- 16.Dransfield DT, Cohen EH, Chang Q, Sparrow LG, Bentley JD, Dolezal O, et al. A human monoclonal antibody against insulin-like growth factor-II blocks the growth of human hepatocellular carcinoma cell lines in vitro and in vivo. Mol Cancer Ther. 2010;9:1809–1819. doi: 10.1158/1535-7163.MCT-09-1134. [DOI] [PubMed] [Google Scholar]
- 17.Rodon J, DeSantos V, Ferry RJ, Jr, Kurzrock R. Early drug development of inhibitors of the insulin-like growth factor-I receptor pathway: lessons from the first clinical trials. Mol Cancer Ther. 2008;7:2575–2588. doi: 10.1158/1535-7163.MCT-08-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolcher AW, Sarantopoulos J, Patnaik A, Papadopoulos K, Lin CC, Rodon J, et al. Phase I, pharmacokinetic, and pharmacodynamic study of AMG 479, a fully human monoclonal antibody to insulin-like growth factor receptor 1. J Clin Oncol. 2009;27:5800–5807. doi: 10.1200/JCO.2009.23.6745. [DOI] [PubMed] [Google Scholar]
- 19.Kurmasheva RT, Dudkin L, Billups C, Debelenko LV, Morton CL, Houghton PJ. The insulin-like growth factor-1 receptor-targeting antibody, CP-751,871, suppresses tumor-derived VEGF and synergizes with rapamycin in models of childhood sarcoma. Cancer Res. 2009;69:7662–7671. doi: 10.1158/0008-5472.CAN-09-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goya M, Miyamoto S, Nagai K, Ohki Y, Nakamura K, Shitara K, et al. Growth inhibition of human prostate cancer cells in human adult bone implanted into nonobese diabetic/severe combined immunodeficient mice by a ligand-specific antibody to human insulin-like growth factors. Cancer Res. 2004;64:6252–6258. doi: 10.1158/0008-5472.CAN-04-0919. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto S, Nakamura M, Shitara K, Nakamura K, Ohki Y, Ishii G, et al. Blockade of paracrine supply of insulin-like growth factors using neutralizing antibodies suppresses the liver metastasis of human colorectal cancers. Clin Cancer Res. 2005;11:3494–3502. doi: 10.1158/1078-0432.CCR-04-1701. [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Chesebrough JW, Cartlidge SA, Ricketts SA, Incognito L, Veldman-Jones M, et al. Dual IGF-I/II-neutralizing antibody MEDI-573 potently inhibits IGF signaling and tumor growth. Cancer Res. 2011;71:1029–1040. doi: 10.1158/0008-5472.CAN-10-2274. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z, Dimitrov DS. Construction of a large naive human phage-displayed Fab library through one-step cloning. Methods Mol Biol. 2009;525:129–42. doi: 10.1007/978-1-59745-554-1_6. xv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 25.Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 26.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci U S A. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boder ET, Wittrup KD. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, et al. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 29.Feng Y, Xiao X, Zhu Z, Streaker E, Ho M, Pastan I, et al. A novel human monoclonal antibody that binds with high affinity to mesothelin-expressing cells and kills them by antibody-dependent cell-mediated cytotoxicity. Mol Cancer Ther. 2009;8:1113–1118. doi: 10.1158/1535-7163.MCT-08-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, Chan YW, Lee SS, Cheung WT. One-step expression and purification of single-chain variable antibody fragment using an improved hexahistidine tag phagemid vector. Protein Expr Purif. 2009;68:190–195. doi: 10.1016/j.pep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, Bossart KN, Bishop KA, Crameri G, Dimitrov AS, McEachern JA, et al. Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J Infect Dis. 2008;197:846–853. doi: 10.1086/528801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaise L, Wehnert A, Steukers MP, van den Beucken T, Hoogenboom HR, Hufton SE. Construction and diversification of yeast cell surface displayed libraries by yeast mating: application to the affinity maturation of Fab antibody fragments. Gene. 2004;342:211–218. doi: 10.1016/j.gene.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Yeung YA, Wittrup KD. Quantitative screening of yeast surface-displayed polypeptide libraries by magnetic bead capture. Biotechnol Prog. 2002;18:212–220. doi: 10.1021/bp010186l. [DOI] [PubMed] [Google Scholar]
- 36.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 37.Kimura T, Kuwata T, Ashimine S, Yamazaki M, Yamauchi C, Nagai K, et al. Targeting of bone-derived insulin-like growth factor-II by a human neutralizing antibody suppresses the growth of prostate cancer cells in a human bone environment. Clin Cancer Res. 2010;16:121–129. doi: 10.1158/1078-0432.CCR-09-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.