Abstract
In mammals, circadian oscillators exist not only in the suprachiasmatic nucleus, which harbors the central pacemaker, but also in most peripheral tissues. It is believed that the SCN clock entrains the phase of peripheral clocks via chemical cues, such as rhythmically secreted hormones. Here we show that temporal feeding restriction under light–dark or dark–dark conditions can change the phase of circadian gene expression in peripheral cell types by up to 12 h while leaving the phase of cyclic gene expression in the SCN unaffected. Hence, changes in metabolism can lead to an uncoupling of peripheral oscillators from the central pacemaker. Sudden large changes in feeding time, similar to abrupt changes in the photoperiod, reset the phase of rhythmic gene expression gradually and are thus likely to act through a clock-dependent mechanism. Food-induced phase resetting proceeds faster in liver than in kidney, heart, or pancreas, but after 1 wk of daytime feeding, the phases of circadian gene expression are similar in all examined peripheral tissues.
Keywords: Circadian clock, restricted feeding, SCN, liver, clock genes, mouse
In mammals, most physiology and behavior are subject to well-controlled daily oscillations. Thus, sleep–wake cycles, heartbeat frequency, blood pressure, body temperature, renal activity, liver metabolism, and the secretion of many hormones are controlled by an endogenous time measuring system called the circadian clock (Portaluppi et al. 1996; Rabinowitz 1996; Hastings 1997; Schibler and Lavery 1999). On the basis of surgical ablation and transplantation experiments, it is believed that the suprachiasmatic nucleus (SCN) in the hypothalamus coordinates most if not all daily rhythms in behavior and physiology (Rusak and Zucker 1979; Ralph et al. 1990). The SCN consists of two small, bilateral groups of neurons and, as indicated by its name, is located directly above the optical chiasma. SCN neurons receive information about the light intensity in the environment via direct synaptic connections with the retina, which adapts the phase of SCN oscillator to the photoperiod (Takahashi 1995; Hastings 1997). The SCN clock then synchronizes overt rhythms in physiology and behavior, probably through both synaptic connections and humoral signals (Shibata and Tominaga 1991; Silver et al. 1996).
The molecular makeup of circadian clocks is subject to intense genetic and biochemical investigation in various organisms, including cyanobacteria, Neurospora, higher plants, Drosophila, and mammals (for review, see Reppert 1998; Dunlap 1999). In most examined systems, autoregulatory feedback loops of gene expression are believed to provide the rhythm-generating mechanisms. As several clock genes are homologous in flies and mammals, it is likely that these mechanisms have been largely conserved during animal evolution (Young 2000). In mammals, Clock and Bmal1 are part of the positive limb of the feedback circuitry, whereas two cryptochrome isoforms, Cry1 and Cry2, and perhaps the period isoforms, Per1, Per2, and Per3, are part of the negative limb (Brown and Schibler 1999; Jin et al. 1999; Shearman et al. 2000). At present, the negative feedback loop in the expression of timekeeper genes is the most widely used molecular model in explaining how circadian clockworks operate. Although it cannot be formally excluded that circadian gene expression is a manifestation, rather than the motor of, rhythm generation (Lakin-Thomas 2000), the cyclic mRNA and protein accumulation profiles can be used as convenient reporters for the phase of circadian oscillators.
Molecular oscillators do not exist solely in pacemaker cells such as lateral head neurons in Drosophila, pinealocytes in cold-blooded vertebrates and birds, and SCN neurons in mammals (for review, see Schibler and Lavery 1999). Rather, oscillators capable of generating several consecutive cycles of circadian gene expression are also found in peripheral, nonneuronal tissues of several animals, including Drosophila (Emery et al. 1997; Krishnan et al. 1999; Giebultowicz et al. 2000), zebrafish (Cahill 1996; Whitmore et al. 2000), and mammals (Balsalobre et al. 1998; Yamazaki et al. 2000). In Drosophila and zebrafish embryos, which are semitransparent, these peripheral clocks can be entrained directly by light via nonocular mechanisms (Plautz et al. 1997; Whitmore et al. 2000). However, in mammals, which are opaque, such mechanisms are unlikely to be operative. Rather, light resets the time of the central pacemaker in the SCN via ocular mechanisms (see above), and the SCN clock then synchronizes peripheral oscillators via neuronal connections and/or chemical signals (Shibata and Tominaga 1991; Sakamoto et al. 1998; Yamazaki et al. 2000). Experiments with tissue-culture cells are in support of a role of blood-borne substances as time-resetting cues. Thus, a brief treatment of immortalized fibroblasts with high concentrations of serum induces circadian gene expression persisting for several days (Balsalobre et al. 1998). Similar results are obtained with cells incubated for a short time period with chemicals activating a variety of known signal transduction pathways. Thus, TPA, a tumor promoter activating protein kinase C and MAP kinases; FGF, a chemokine activating MAP kinases; forskolin and butyryl cAMP, substances that activate protein kinase A; and dexamethasone, a glucocorticoid hormone analog, all provoke circadian gene expression in tissue-culture cells (Akashi and Nishida 2000; Balsalobre et al. 2000b; Yagita and Okamura 2000). Given the responsiveness of these oscillators to the multiple signals, it appears likely that the central pacemaker may exploit several chemical entrainment pathways to synchronize peripheral clocks.
A major question in mammalian chronobiology concerns the physiological purpose of circadian gene expression in peripheral cells. In liver, most known genes with rhythmic expression encode enzymes or regulatory proteins involved in food processing and energy homeostasis. These include cholesterol 7α hydroxylase (Mitropoulos et al. 1972; Noshiro et al. 1990; Lavery and Schibler 1993), the rate-limiting enzyme in the synthesis of bile acids, a number of cytochrome P450 enzymes involved in detoxification and elimination of food components (e.g., coumarin hydroxylase, Cyp2a5; Lavery et al. 1999), enzymes involved in carbohydrate metabolism (e.g., PEPCK, glycogen synthase, glycogen phosphorylase; Ishikawa and Shimazu 1976; Roesler and Khandelwal 1985; Frederiks et al. 1987), and transcription factors governing fatty acid metabolism (e.g., PPAR and spot 14; Kinlaw et al. 1987; Lemberger et al. 1996). At least in liver, the coordination of physiological needs during the absorptive and postabsorptive phase may be the major function of circadian oscillators. Previous reports on restricted feeding are in keeping with this conjecture. For example, in the liver of (nocturnal) rats, the phase of the daily Dbp mRNA accumulation profile is severely altered when food is offered exclusively during the day (Ogawa et al. 1997).
As nocturnal animals, mice consume most of their food during the night. Here we show that feeding of mice exclusively during the day completely inverses the phase of circadian oscillators in peripheral cells, but has little if any effect on the central oscillator in the suprachiasmatic nucleus. Interestingly, feeding during the subjective day under constant dark (DD) conditions uncoupled peripheral from central oscillators to a similar extent as under light–dark (LD) conditions. Hence, the feeding-time-induced uncoupling of circadian phases in peripheral cells and SCN neurons cannot be explained by a dominant Zeitgeber effect of light in the SCN. Rather, feeding elicits entrainment cues that act specifically on oscillators in peripheral tissues and that are ineffective in resetting time in the SCN.
Results
Restricted feeding during the day uncouples circadian liver gene expression from circadian gene expression in the suprachiasmatic nucleus
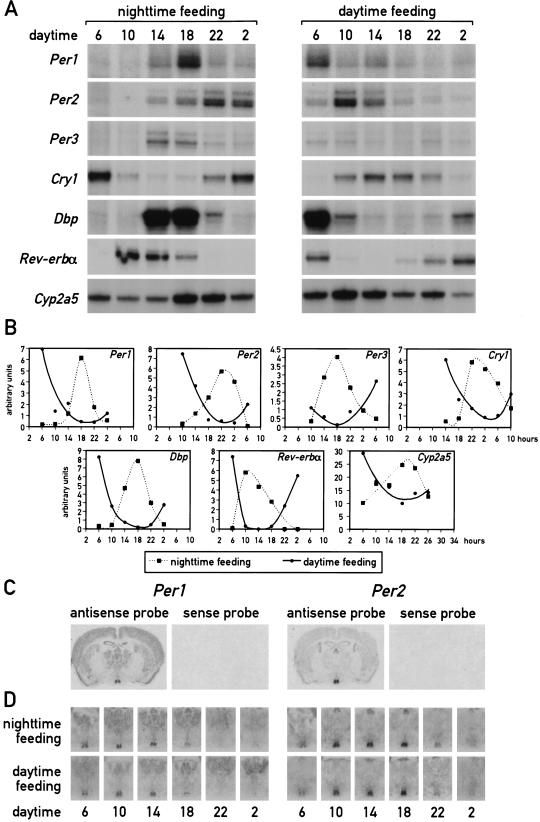
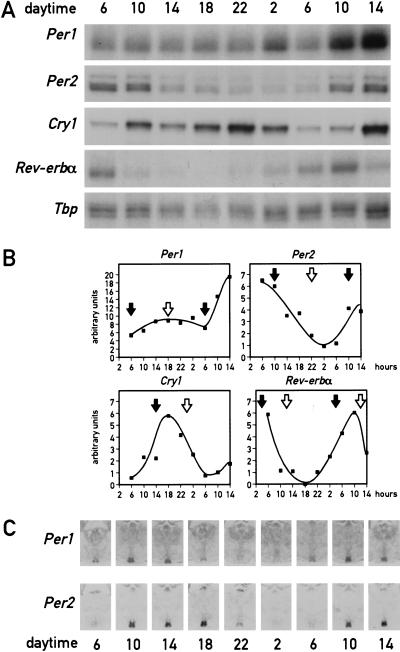
As mentioned above, a major task of circadian oscillators in liver cells (and perhaps other peripheral cell types) may be to anticipate and adapt the physiological conditions required for food processing. As nocturnal animals, mice consume ∼80% of their food during the active dark phase if kept under a 12-h light/12-h dark cycle (LD) and if food is offered ad libitum. To examine whether feeding time can affect the phase of circadian liver gene expression, mice were fed for 9 d exclusively during the night or during the day. At the conclusion of the entrainment period, the phase of mRNA expression for the four clock components, PER1, PER2, PER3, and CRY1, the two circadian transcription factors DBP and Rev-erbα, and the cytochrome P450 enzyme coumarin 7-hydroxylase (Cyp2a5) was determined. As shown in Figure 1A and B, the phases of all examined mRNA accumulation profiles differ by 8–12 h between mice fed during the day or during the night. As expected, livers of mice fed exclusively during the night (Fig. 1A,B) or ad libitum displayed a similar phase angle of cyclic liver gene expression (for mice fed ad libitum, see Lopez-Molina et al. 1997; Lavery et al. 1999; Balsalobre et al. 2000a). However, feeding during the day almost entirely inversed the phase of the liver oscillator. Hence, feeding time appears to be a potent Zeitgeber for circadian liver gene expression.
Figure 1.
Daytime feeding changes the phase of circadian gene expression in the liver but not in the suprachiasmatic nucleus (SCN). Mice, kept under a light-dark regimen (lights on 6 a.m., lights off 6 p.m.), were fed for eight consecutive days exclusively during the light phase (6 a.m. to 6 p.m.) or during the dark phase (6 p.m. to 6 a.m.). During the ninth day, animals were killed at 4-h intervals to prepare whole-cell RNA from liver and tissue sections from the brain. (A) Circadian accumulation of clock and clock-controlled genes in the liver. The levels of transcripts from the genes indicated to the left of the panels were determined by ribonuclease protection assays (Cyp2a5 stands for coumarin hydroxylase). Tbp mRNA or β-actin mRNA were included as standards for transcripts whose expression is constant throughout the day (data not shown). (B) The mRNA signals obtained in the ribonuclease protection assays shown in panel A were measured by scanning the autoradiography and normalized to the signals generated by β-actin mRNA (for Cyp2a5) or Tbp mRNA (for all other transcripts). (C) Serial coronal brain sections taken above the optical chiasma were hybridized in situ to 35S-labeled Per1 and Per2 antisense and sense RNA strands. Note that no detectable hybridization signals were obtained with the sense probe. (D) Circadian accumulation of Per1 and Per2 mRNA in the SCN of mice after 8 d of restricted feeding. Coronal sections from brains harvested at 4-h intervals around the clock were hybridized in situ to Per1 and Per2 antisense RNA probes as described in panel C. Note that the phases of rhythmic Per1 and Per2 expression are not affected by restricted feeding.
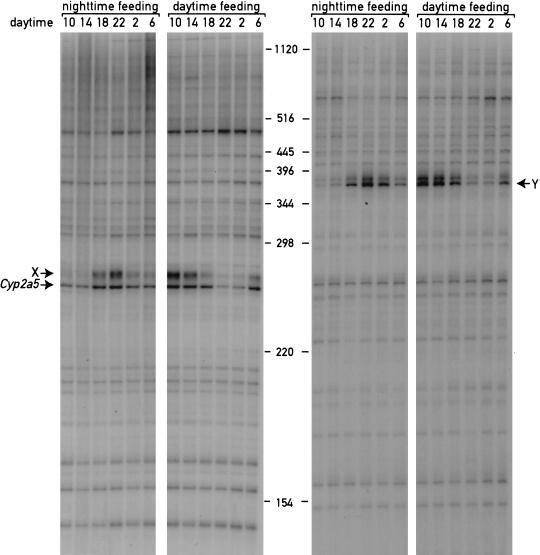
From the data presented thus far, the effects of feeding regimen on circadian phase could be either caused by a specific change in circadian gene expression or by a more general perturbation of liver transcription. To differentiate between these possibilities, we examined the expression profiles of ∼110 random genes using a novel cDNA display method, dubbed ADDER (amplification of double-stranded cDNA end restriction fragments; B. Kornmann, N. Preitner, D. Rifat, F. Fleury-Olela, and U. Schibler, in prep.). This method involves the PCR amplification of cDNA fragments subpopulations using pairs of primers differing in their ultimate and penultimate nucleotides. The left panel of Figure 2 shows the display of the cDNA fragments populations containing the 3′-terminal cDNA fragment of coumarin 7-hydroxylase (Cyp2a5; see Fig. 1A,B), using the RNAs depicted in Figure 1A as templates for cDNA synthesis. A visual inspection of the ∼60 cDNA fragments contained in this subpopulation suggests that the large majority of mRNAs are expressed at constant levels, irrespective of the feeding regimen. In contrast, the phase of circadian Cyp2a5 mRNA expression is completely inversed, as expected from the ribonuclease protection study presented in Figure 1 (panels A and B). Another circadian cDNA fragment yet unidentified (labeled as X) is also present and shows an inverted pattern in daytime-fed mice. A second example of a cDNA population amplified with a different set of primers is shown in the right panel and reveals only one circadian- and feeding-dependent cDNA fragment (Y) among ∼50 others. Thus, restricted feeding appears to affect specifically the accumulation of circadian mRNAs and does not result in a gross alteration of liver gene expression.
Figure 2.
ADDER (amplification of double-stranded cDNA end restriction fragments) differential display of liver cDNAs from mice fed exclusively during the day or during the night. Double-stranded cDNA fragments encompassing mRNA sequences located between the poly A addition sites and the most proximal Mbo1 restriction sites were synthesized using total liver RNA as a template. The RNA was collected at 4-h intervals from mice fed either during the day or during the night. The left panel shows a subpopulation of cDNAs containing the 3′ sequences of coumarin 7 hydroxylase (Cyp2a5) transcripts and ∼60 other 3′-terminal mRNA sequences amplified and displayed on a 5% urea-polyacrylamide gel (see Materials and Methods). X is a yet unidentified circadian fragment. The right panel shows a subpopulation of cDNA amplified using a second set of primers (see Materials and Methods). Y is a yet unidentified circadian fragment. The positions of molecular size markers are indicated in the middle of the figure.
In principle, the resetting of circadian gene expression in the liver could be accomplished either by phase shifting the SCN clock, which then imposes the new phase upon the liver, or by uncoupling the liver oscillator from the SCN pacemaker. To discriminate between these two possibilities, circadian Per1 and Per2 mRNA accumulation was examined by in situ hybridization to coronal brain sections containing the SCNs from the same animals used in Figure 1A. The results displayed in Figure 1D clearly demonstrate that feeding time has little if any influence on circadian Per1 and Per2 expression in the SCN.
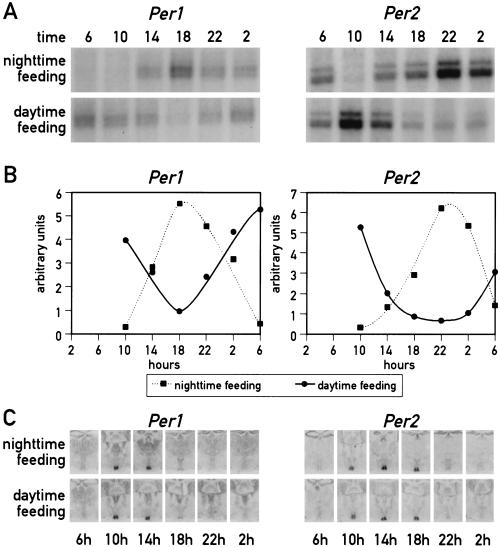
The experiments described above, in which the animals were kept under a LD regimen, could be interpreted in two ways. Either food entrainment cannot serve as an effective Zeitgeber for the SCN pacemaker or light entrainment is dominant over food entrainment in the SCN. To address this issue, Per1 and Per2 mRNA accumulation was monitored in the SCN and in the liver of mice kept in constant darkness (DD) during the time period of restricted feeding. As shown in Figure 3, the results obtained in this experiment are similar to the ones observed with animals kept under 12-h LD cycles. It thus appears that feeding time does not act as a potent Zeitgeber in SCN neurons.
Figure 3.
Circadian gene expression in the liver and the suprachiasmatic nucleus (SCN) of food-entrained mice kept in constant darkness. Mice, kept in constant darkness (DD) were fed for six consecutive days exclusively during the subjective day (6 a.m. to 6 p.m.) or during the subjective night (6 p.m. to 6 a.m.). During the seventh day, animals were killed at 4-h intervals to prepare whole-cell RNA from liver and tissue sections from the brain. (A) Circadian Per1 and Per2 mRNA accumulation in the liver. Transcript levels were determined as described in Figure 1. (B) The signals obtained in the ribonuclease protection assays shown in panel A were quantified and normalized to Tbp mRNA levels (not shown in panel A). (C) The accumulation of Per1 and Per2 mRNA in the SCN of food entrained animals was revealed by in situ hybridization as described in Figure 1.
Food entrainment of peripheral clocks requires several days
The adaptation of the mammalian circadian system to sudden and large changes in the photoperiod requires several days (Deacon and Arendt 1996). In contrast, mice with a defective clock, such as the ones homozygous for Cry1 and Cry2 null alleles, immediately adapt their behavior to new LD cycles. Apparently, their behavior is driven solely by environmental Zeitgebers, such as differences in light intensity. Hence, at least for light-induced phase resetting of circadian locomotor activity, a gradual temporal adaptation to abrupt and large changes in entrainment can be taken as a criterion for a clock-dependent mechanism (van der Horst et al. 1999).
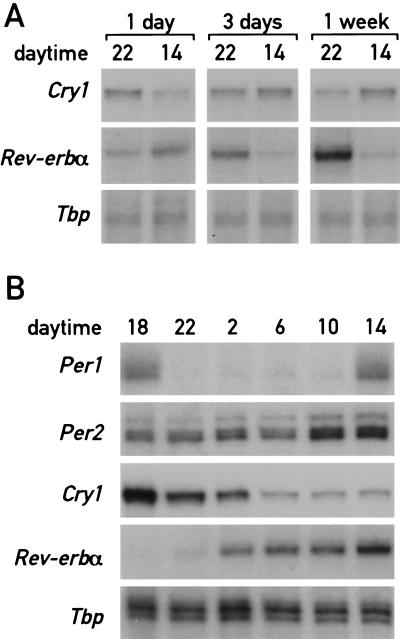
To examine whether shifting mice from unrestricted feeding to daytime feeding entrains circadian gene expression in liver gradually or abruptly, the accumulation of Cry1 mRNA and Rev-erbα mRNA was determined at 2 p.m. and 10 p.m. during day 1, day 3, and day 7 after initiation of the day-only feeding regimen. As seen in Figure 1 A and B, the accumulation cycles of these two transcripts exhibit roughly opposite phases during the daytime and nighttime feeding regimens after a long-term entrainment. The results displayed in Figure 4A clearly demonstrate that the phases of Cry1 and Rev-erbα expression do not change immediately after the onset of the restricted feeding regimen. This gradual phase shifting is reminiscent of light-mediated time resetting in the SCN (Van der Horst et al. 1999) and suggests that a clock-dependent mechanism is operative during the resetting of the liver oscillator. Further evidence for this conjecture was obtained by examining the expression of Dbp during the entrainment period (see below).
Figure 4.
Circadian gene expression changes gradually during restricted feeding. (A) Mice kept under a LD regimen (lights on at 6 a.m., lights off at 6 p.m.) were fed exclusively during the day for 1 d, 3 d, or 1 wk. The animals were then sacrificed at 2 and 10 p.m. to prepare whole-cell liver RNA. Transcript levels for Cry1 and Rev-erbα were determined as described in Figure 1. (B) Mice were fed exclusively during the day for seven consecutive days and, after a starvation of 24 h, switched to nighttime feeding by offering food during the night of the eighth day (from 6 p.m. to 6 a.m.). Animals were sacrificed at 4-h intervals (starting at 6 p.m. of the eighth day) for the analysis of mRNAs. Note that Per2 mRNA, Rev-erbα mRNA, and Cry1 mRNA are still at levels intermediate between those observed for mice fed during the night or during the day.
Since the food-induced phase reversal of circadian liver gene expression required several days, one would expect that the newly entrained oscillator would also need some time to readapt to the original phase on reversal of feeding time. As shown in Figure 4B, this appears to be the case. Switching from daytime to nighttime feeding with an interval of 24 h without food, resulted in only moderate phase changes for Cry1 and Per2 mRNA expression during the 24-h period following food readdition (cf. Fig. 1A with Fig. 4B). Moreover, Per2 and Rev-erbα mRNA levels still rise between 4 and 8 h earlier than in mice entrained to night feeding. Interestingly, food intake after a 24-h starvation period appears to down-regulate Per1 mRNA levels for several hours before they rise again. Conceivably, this altered Per1 expression may play a role in the readaptation of peripheral clocks to nighttime feeding.
We also wished to examine whether the phase of the food-entrained oscillator changed on food withdrawal. As shown in Figure 5 A and B, the phases of Rev-erbα, Per2, and Cry1 expression remain similar for 24–36 h when food is removed after 7 d of day-only feeding. Per1 expression behaves somewhat differently under this condition in that the amplitude of Per1 mRNA accumulation becomes severely dampened after prolonged food deprivation (Fig. 5A,B). A constitutively high expression of Per1 mRNA is generally observed in starved mice, irrespective of whether they have been fed during the day or ad libitum before food deprivation (F. Damiola, N. Preitner, and U. Schibler, unpubl.).
Figure 5.
Circadian gene expression in liver after food-entrainment. (A) Mice kept under a LD regimen (lights on at 6 a.m., lights off at 6 p.m.) were fed exclusively during the day for seven consecutive days. On the morning of the eighth day, food was not given back, and animals were sacrificed at 4-h intervals during the following 36 h. Thus, after daytime feeding the mice were starved between 12 and 48 h before they were killed for the analysis of the various mRNAs (depicted to the left of the panels). (B) mRNA levels obtained for starved mice (panel A) were quantified and normalized to Tbp mRNA. The times at which zenith mRNA levels were reached in daytime- and nighttime-fed mice are indicated by black and white arrows, respectively (see Fig. 1). (C) Per1 and Per2 mRNA accumulation in the suprachiasmatic nucleus (SCN) of starved animals (see panel A) has been determined by in situ hybridization to coronal brain sections as described in Figure 1. Note that daytime feeding followed by starvation has no effect on circadian Per1 and Per2 mRNA accumulation in the SCN.
Figure 5C shows the Per1 and Per2 mRNA accumulation profiles in the SCN of mice that were starved during the 48-h time period following day-only feeding. A comparison of these data with those presented in Figures 1D and 3C reveals that neither starvation nor restricted feeding affect the phase of the SCN pacemaker to any significant degree.
Food entrainment proceeds with different kinetics in different tissues
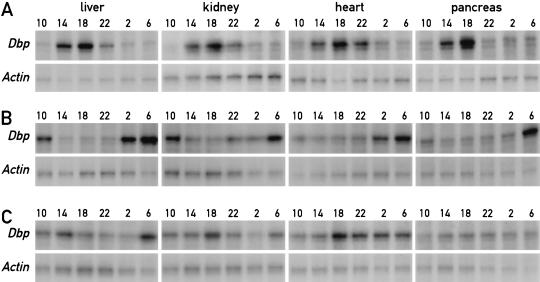
In mice fed ad libitum, the phases of all peripheral oscillators appear to be synchronous. We thus wanted to determine whether restricted feeding resets the time in peripheral tissues other than liver, which plays a particularly important role in food processing. To this end, we recorded Dbp mRNA accumulation in kidney, heart, and pancreas, in addition to liver, in mice fed exclusively during the night or during the day for 1 wk. Dbp transcription is directly controlled by the molecular oscillator (Ripperger et al. 2000) and displays a high amplitude of circadian expression in several tissues (Wuarin et al. 1992; Lopez-Molina et al. 1997). Figure 6A shows that the phases of Dbp mRNA accumulation cycles are similar in all examined tissues of animals fed during the night for 1 wk. As already pointed out earlier, this phase is nearly identical to that observed in animals fed ad libitum (Lopez-Molina et al. 1997; Ripperger et al. 2000). Importantly, the phase of Dbp expression is inversed in all four examined tissues of mice fed exclusively during the day (Fig. 6B). Hence, restricted feeding resets the phase of circadian gene expression in peripheral tissues other than the liver and, perhaps, in all tissues.
Figure 6.
Restricted feeding resets the phase in various peripheral tissues. Mice kept under a light-dark regimen (lights on 6 a.m., lights off 6 p.m.) were fed for three or six consecutive days exclusively during the light phase (6 a.m. to 6 p.m.) or for six days exclusively during the dark phase (6 p.m. to 6 a.m.). During the fourth or seventh day, respectively, animals were sacrificed at 4-h intervals to prepare whole-cell RNA from liver, kidney, heart, and pancreas. Dbp mRNA levels were determined by ribonuclease protection assays in the organs and at the daytimes indicated on top of the panels. β-actin mRNA was included as a transcript whose accumulation does not oscillate during the day. (A) Temporal Dbp expression in mice fed during 6 d exclusively during the night. (B) Temporal Dbp expression in mice fed during 6 d exclusively during the day. (C) Temporal Dbp expression in mice fed during 3 d exclusively during the day.
We wished to examine whether daytime feeding shifts the phase of circadian gene expression with the same or different kinetics in different tissues. To this end, Dbp mRNA accumulation was recorded in liver, kidney, heart, and pancreas on the fourth day after the onset of restricted feeding, at which time the phase of the liver oscillator is not yet fully reversed (see above). As shown in Figure 6C, different temporal Dbp expression patterns were obtained for liver, kidney, heart, and pancreas. In liver, Dbp mRNA levels reach peak values around 2 p.m. on day 3 and rise around 6 a.m. on day 4. In contrast, the phase of circadian Dbp expression in kidney is still similar to that observed in animals fed ad libitum or during the night only (see Fig. 6A). In heart and pancreas, Dbp mRNA levels are relatively high throughout the examined time period. We do not know whether the low-amplitude accumulation profiles on day 3 in heart and pancreas reflect intermediate states of phase resetting in all cell types composing these tissues or whether they are the results of different superimposed accumulation profiles with different phases in the different cell types of these tissues.
Daytime feeding changes circadian body temperature fluctuations
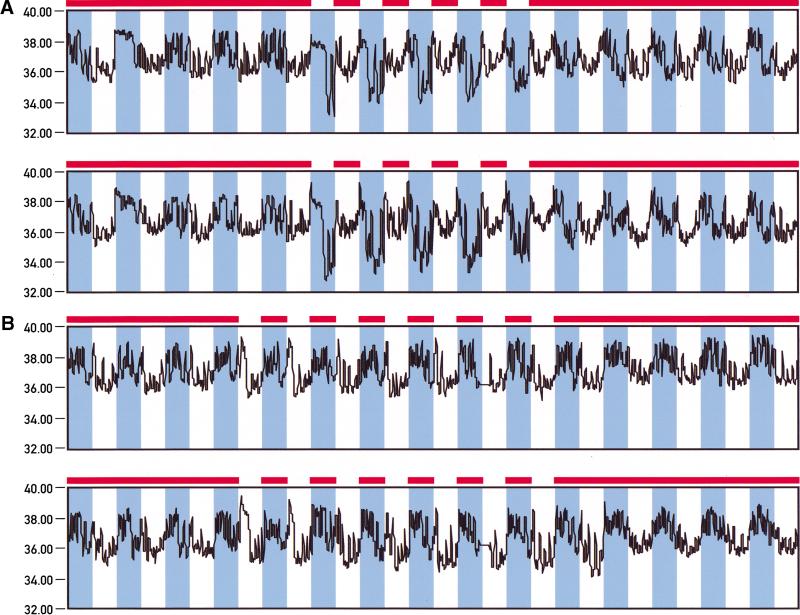
In mammals, body temperature oscillates in a daily fashion with a difference between highest and lowest values of 1°–3°C, depending on the species (Refinetti 1996). Temperature cycles may not only be an output of the pacemaker but may actually also feed back as input cues for circadian oscillators. We thus recorded body temperature fluctuations by telemetry to determine whether restricted feeding during the night or during the day affects diurnal body temperature fluctuations. Figure 7 shows temperature profiles recorded for animals fed ad libitum, during the night, and during the day. As shown in panel B of Figure 7, nighttime feeding has little influence on the amplitude, phase, or shape of daily temperature profiles. Irrespective of whether the mice are fed throughout the 24-h day or during the night only, zenith values are reached during the dark phase. These peak values are about 3°–4°C higher than the trough values observed during the light phase. In contrast, switching from unlimited food supply to day-only feeding alters the temperature cycles dramatically (Fig. 7A). Already, toward the end of the first night after food deprivation, a strong transient depression of up to 3°C below normal nadir values is observed. This transient temperature depression occurs somewhat earlier during the second day of restricted feeding and then repeats itself during the following days. Starting at the second day after food limitation, nadir temperatures are reached during the dark phase and zenith temperatures during the dark–light and light–dark transitions.
Figure 7.
Daytime feeding causes body temperature depressions during the night. Temperature probes were implanted in the abdominal cavities of mice and body temperature rhythms were recorded by telemetry in animals fed either ad libitum or exclusively during the night or during the day. The feeding periods are indicated by solid bars on top of the panels. The dark phases are depicted in blue. (A) Body temperature recordings from two individuals fed during 5 d exclusively during the day. Note the dramatic temperature depressions during the dark phases of daytime feeding. (B) Recordings from two individuals fed during 7 d exclusively during the night. In some animals (e.g., the one shown in the bottom panel), the minimal temperature values observed during the light phase became slightly lower after several days of nighttime feeding.
In conclusion, daytime feeding dramatically affects the phase, shape, and amplitude of body temperature profiles. During the first 24 h after food deprivation, a difference between zenith and nadir levels of up to 6°C could be observed. Even more dramatic daily drops in body temperature have previously been observed in mice that were starved for extensive time periods or subjected to chronic caloric restriction (Hudson and Scott 1979; Nelson and Halberg 1986).
Discussion
Daytime feeding uncouples peripheral circadian oscillators from the central SCN pacemaker
Recent evidence obtained in mouse and rat suggests that many peripheral cell types of mammalian organisms harbor circadian oscillators (Balsalobre et al. 1998; Zylka et al. 1998; Akashi and Nishida 2000; Yamazaki et al. 2000). According to the currently held model, these oscillators are synchronized via chemical cues by a master pacemaker, residing in the suprachiasmatic nucleus (SCN) of the hypothalamus, which itself is entrained by the photoperiod (Takahashi 1995; Hastings 1997; Yamazaki et al. 2000). The results presented in this study demonstrate that feeding mice exclusively during the day inverses the phase of circadian expression of putative clock genes (Per1, Per2, Per3, and Cry1) and clock-controlled genes (Dbp, Rev-erbα, and Cyp2a5) in peripheral tissues. A similar phase resetting of peripheral circadian oscillators has also been observed in daytime-fed rats (data not shown), suggesting that food is a dominant Zeitgeber for peripheral clocks in various mammalian species. As suggested by differential mRNA display, daytime feeding appears to influence specifically the accumulation of circadian liver mRNAs and does not result in gross changes of gene expression (Fig. 2; B. Kornmann, N. Preitner, and U. Schibler, unpubl.). Interestingly, restricted feeding does not affect the phase of circadian gene expression in the SCN, irrespective of whether the animals are kept under LD or DD conditions. Hence, it appears that the unresponsiveness of the SCN clock to feeding time reflects an inherent property of the central pacemaker, rather than a dominance of photic entrainment over food entrainment.
In contrast to restricted feeding during the day, nighttime feeding has little effect on the phase of circadian gene expression. This may not be surprising, given that mice (as nocturnal animals) consume ∼80% of the food during the night (N. Preitner and F. Damiola, unpubl.) when they are fed ad libitum. Nighttime feeding also does not significantly alter circadian body temperature rhythms. Thus, in mice fed exclusively during the night, there is no sign of a core temperature depression below the nadir values recorded for animals fed ad libitum.
We also examined whether restricted feeding influences voluntary locomotor activity, as measured by circadian wheel-running activity (data not shown). Mice kept in a 12-h light/dark regimen displayed similar actograms irrespective of whether food was offered throughout the day, during the dark phase, or during the light phase. However, when mice were kept under DD conditions, we did observe some notable differences between the actograms of animals fed ad libitum or exclusively during the subjective night or during the subjective day. In constant darkness, restricted feeding during the subjective night kept the animals on a precise 24-h schedule and prevented them from free running with the period dictated by their endogenous pacemaker. Hence, in accordance with previously reported data (White and Timberlake 1995; Holmes and Mistlberger 2000), feeding time can serve as a Zeitgeber in the daily resetting of the central circadian clock by a few minutes. The wheel-running data obtained for mice kept under DD conditions and fed exclusively during the day were more difficult to interpret. While most of the wheel running was still restricted to the subjective night, most examined animals were more active during the second half of the night after ∼3 d. As this altered behavior is not accompanied by phase changes in circadian Per1 and Per2 gene expression in the SCN, the shifting of wheel-running activity toward the second half of the night may be brought about by noncircadian mechanisms of behavior (e.g., homeostatic sleep components; see Borbely and Achermann 1999).
How does restricted feeding entrain peripheral oscillators?
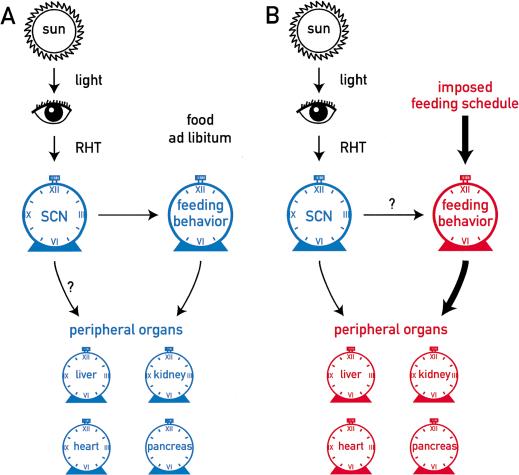
A hypothetical model summarizing the observations presented in this study is shown in Figure 8. We demonstrate that restricted feeding entrains peripheral oscillators without affecting the molecular clock in the SCN. Therefore, the food-induced phase resetting of peripheral clocks is probably not controlled solely by the SCN pacemaker. More likely, feeding operates through pathways that counteract the synchronization of peripheral clocks by the central SCN pacemaker. While the input signaling pathways for peripheral oscillators remain to be identified, it is tempting to speculate that body temperature cycles participate in the synchronization of peripheral oscillators. Indeed, in other systems there is ample evidence for the entrainment of circadian clocks by temperature cycles with very small differences between peak and nadir values (Liu et al. 1998; Sidote et al. 1998). As shown in Figure 7A, daytime feeding elicits a dramatic and transient depression of body temperature during the night, when maximal temperatures are reached in mice fed ad libitum. The lowest value of ∼33°C is reached during the first night after food removal. It is conceivable that this alteration in daily temperature profile is critically involved in the rephasing of peripheral clocks. If true, this would beg the question of why the SCN pacemaker is not reset by daytime feeding. Conceivably, the temperature fluctuations elicited by restricted feeding are less dramatic in the SCN region, or the central pacemaker is more resistant to temperature variations than peripheral oscillators.
Figure 8.
Hypothetical model on the entrainment of peripheral oscillators. The central circadian pacemaker in the suprachiasmatic nucleus (SCN) is entrained by solar cycles via the retino-hypothalamic tract (RHT). (A) When food is available throughout the day or during the normal activity phase (dark phase in nocturnal animals), the SCN synchronizes peripheral clocks via cycles in the secretion of blood-borne factors (e.g., hormones) or temperature rhythms. Both of these time cues may ultimately be controlled by the feeding behavior governed by the SCN. (B) When food is only available during the phase at which the animals are normally inactive (light phase in nocturnal animals), signals triggered by food processing and/or the lack of food processing act as dominant Zeitgebers on the oscillators of peripheral tissues.
Obviously, it is also possible that restricted feeding acts on peripheral oscillators via chemical cues that can only signal to peripheral cells. Glucocorticoid hormones would be excellent candidates for such signals, as they cause phase shifts exclusively in peripheral cells types (Balsalobre et al. 2000a). However, experiments with mice carrying liver-specific mutations in the glucocorticoid receptor gene indicate that glucocorticoid-signaling counteracts rather than facilitates the rephasing of peripheral clocks by daytime feeding (N. Le Minh, F. Damiola, F. Fleury-Olela, F. Tronche, G. Schütz, and U. Schibler, unpubl.).
The purpose of circadian gene expression in peripheral cell types
The work presented here suggests that circadian gene expression in peripheral cells is intimately connected to feeding. In keeping with this conjecture, liver is the organ reacting most rapidly to the temporal feeding regimen. Indeed, the liver plays a dominant role in the metabolism and processing of food components, such as carbohydrates, proteins, and lipids. For example, during the absorptive phase, liver cells polymerize glucose into glycogen stores, while during the postabsorptive phase they degrade glycogen into glucose. This helps in establishing a constant supply of glucose to cells that are incapable of gluconeogenesis. The temporal regulation of glucose homeostasis demands that the glycogen synthase activity is high during the absorptive phase and low during the postabsorptive phase and that the converse applies to the activity of glycogen phosphorylase. At least in part, this temporal regulation is under the control of the circadian time-keeping system (N. Preitner and U. Schibler, unpubl.; Ishikawa and Shimazu 1976). Another example is the conversion of cholesterol to bile acids, detergents required for the emulsification of food lipids. Thus, the expression of the gene encoding cholesterol 7α hydroxylase, the rate-limiting enzyme in bile acid synthesis, is under stringent circadian control (Lavery and Schibler 1993, and references therein). Rhythmic expression has also been observed for enzymes involved in protein and amino acid metabolism, such as serine dehydratase (Ogawa et al. 1994), and transcription factors involved in fat metabolism, such as PPARα and spot 14 (Kinlaw et al. 1987; Lemberger et al. 1996; Kersten et al. 2000). As food also contains numerous toxins, the modification and elimination of hazardous substances demand a high activity of detoxifying enzymes during the absorptive phase. Indeed, several genes specifying cytochrome P-450 enzymes involved in such processes or enzymes involved in the synthesis and regeneration of cytochrome P-450 enzymes are subject to circadian regulation. Examples include coumarin hydroxylase (Lavery et al. 1999; this article), NADPH-cytochrom-p450-reductase (Belanger 1996), and α-aminolevulinate synthase (Rodriguez et al. 1996).
A connection between circadian gene regulation and metabolism/energy homeostasis has been established for numerous organisms across many phyla, including cyanobacteria, marine algae, fungi, higher plants, and fruit flies (Portaluppi et al. 1996; Rabinowitz 1996; Hastings 1997; Roenneberg and Merrow 1999; Schibler and Lavery 1999). In fact, during the evolution of lower organisms, the adaptation of the metabolism to solar cycles may have been the major selective force in establishing circadian physiology. We propose that in mammals, food processing and its anticipation are the major purposes of circadian gene expression in liver and perhaps in other peripheral organs. This rhythmic gene expression is an output of peripheral oscillators that, under normal conditions, are probably synchronized via signals governed by the central pacemaker in the SCN. However, when food availability becomes restricted and remains in a long-lasting temporal conflict with the activity phase dictated by the SCN, peripheral oscillators become uncoupled from the central pacemaker. We speculate that this uncoupling may be facilitated by changes in body temperature rhythms that accompany the altered feeding behavior. As soon as the food availability returns to normal, the SCN clock, whose phase angle remained unaffected by temporally restricted feeding, resynchronizes the peripheral oscillators. It will be an enticing challenge to identify the signal transduction pathways participating in the entrainment of peripheral clocks by the SCN and by feeding.
Materials and methods
Animal care and handling
All experiments were done with mice between 10 and 16 wk of age. The animals were kept in a 12-h light/dark regimen (light on at 6 a.m.). Mice fed during the day received food from 6 a.m. to 6 p.m., whereas mice fed during the night received food from 6 p.m. to 6 a.m.
Telemetric body temperature recordings
To introduce the temperature probes, mice were anesthesized for 30 min by intraperitoneal injection of kétarom (7 μL/g of body weight). Kétarom is prepared by mixing 2.4 mL of 50 mg/mL kétasol (E. Graub AG), 0.8 mL of 2% rompun (Provet AG), and 6.8 mL of 0.9 % NaCl. The temperature probe was introduced into the abdominal cavity behind the gut via a 1.5-cm incision made in the skin and linea abla. The muscle wall and the skin were then closed by five stitches and two staples, respectively. All the components of the telemetric system, including the probes (PDT-4000 E-mitters), the receivers detecting the probe signals (ER-4000 Energizer-Receivers), and the Vitalview Software and Hardware Interface Card for data acquisition and analysis, were purchased from Minimitter. The mice were housed in cages in ventilated light-tight cabinets. Inside the cabinets, the timing of light/dark regimens was controlled by the computer software Chronobiology Kit (Standford Software System).
Ribonuclease protection experiments
Mouse tissues, except pancreas, were removed within 4 min after decapitation, frozen in N2 liquid, and stored at −70°C until use. The pancreas (which contains exceedingly high levels of ribonuclease) was immediately homogenized in the guanidium thiocyanate lysis buffer used for RNA extraction. The extraction of whole-cell RNA and its analysis by ribonuclease protection assays were performed as described (Schmidt and Schibler 1995). The antisense Rev-erbα RNA probe was transcribed from a pBluescript-KS+ vector containing an RT–PCR product of mouse Rev-erbα mRNA (+376 to +614; N. Preitner and U. Schibler, unpubl.). The Dbp and Tef antisense RNA probes are complementary to rat Dbp mRNA (+1126 to +1221) and rat Tef mRNA (+598 to +693; Fonjallaz et al. 1996). The Tbp probe is complementary to mouse Tbp mRNA (+36 to +135; Schmidt and Schibler 1995). The Per3 and Cry1 probes (kindly provided by A. Balsalobre) are complementary to mouse Per3 mRNA (+367 to +572) and mouse Cry1 mRNA (+83 to +312), respectively (Balsalobre et al. 2000a). The Per2 probe is complementary to mouse Per2 mRNA (+165 to +287; Balsalobre et al. 1998). The Per1 probe is complementary to mouse Per1 mRNA (+2397 to +2523; gift from J. Ripperger). The β-actin and coumarin hydroxylase (Cyp2a5) are described by Lavery et al. (1999).
In all cases, the plasmids were linearized with a suitable restriction enzyme and the antisense RNA probes were prepared by in vitro transcription of the linearized templates with T7 or T3 RNA polymerase using (α32P)-UTP. Autoradiography was performed with an intensifying screen (FUJI) at −70°C for 1–5 d.
The signals obtained in ribonuclease protection experiments were quantified by scanning the autoradiographies, using a HP ScanJet 6100C/T scanner, and normalized to the signals obtained for Tbp mRNA. Tbp mRNA accumulation is not subject to circadian regulation (Balsalobre et al. 1998).
Differential display of mRNA sequences by ADDER
The experimental details of the ADDER will be described elsewhere (B. Kornmann, N. Preitner, D. Rifat, F. Fleury-Olela, and U. Schibler, in prep.) and will be provided on request. Briefly, double-stranded cDNA fragments were synthesized using total liver RNAs as templates. These cDNA fragments encompassed mRNA sequences located between the poly A addition sites and the most proximal Mbo1 restriction sites and contained different PCR linker sequences at both ends. PCR amplification of these cDNA fragments with upstream and downstream primers that contain different ultimate and penultimate nucleotides yielded cDNA subpopulations complementary to ∼50–100 individual cDNA species each. These cDNA fragments were then visualized by electrophoretic size fractionation on 5% urea-polyacrylamide gels. The upstream primer 5′-AACGATCCC-3′ (Mbo1 site underlined) and the downstream primer 5′-AGCTTT TTTTTTTTTGC-3′ were used for the display shown in the left panel of Figure 2. The upstream primer 5′-AACGATCAG-3′ and the downstream primer 5′-AGCTTTTTTTTTTTTCC-3′ were used for the display shown in the right panel of the same figure.
In situ hybridization on coronal brain sections
Immediately after removal, brains were frozen in isopentane (4 min at −20°C) and stored at −70°C until use. Serial coronal brain cryosections of 12 microns above the optical chiasma were prepared using standard procedures. In situ hybridizations with sections though the central SCN were performed as described previously (Nef et al. 1996). The Per1 and Per2 riboprobes used in these experiments were prepared from the templates pKS-mPer1-Fl and pKS-mPer2-nqFl (gift from S. Brown), using RNA polymerases T3 and T7 for the antisense and sense strands, respectively. The pKS-mPer1-Fl and pKS-mPer2-nqFl were obtained by cloning the inserts of the plasmids pcDNA3.1-P1 and pcDNA3.1-P2, respectively, into the plasmid vector pBS-KS. To this end, the inserts of pcDNA3.1-P1 and pcDNA3.1-P2 were excised with Xba1 and Cla1/Not1, respectively. The plasmids pcDNA3.1-P1 and pcDNA3.1-P2 (Jin et al. 1999) were provided by Steven Reppert.
Acknowledgments
We thank Steven Reppert for his generous gift of the Per1 and Per2 cDNA plasmids pcDNA3.1-P1 and pcDNA3.1-P2, Steven Brown and Juergen Ripperger for their valuable discussions, and Nicolas Roggli for expert preparation of the illustrations. This work was supported by the Swiss National Science Foundation (grant no. 47314.96), the State of Geneva, the Bonizzi-Theler Stiftung, and the Louis-Jeantet Foundation for Medicine.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ueli.schibler@molbio.unige.ch; FAX 41-22-702-6868.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.183500.
References
- Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes & Dev. 2000;14:645–649. [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000a;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Curr Biol. 2000b;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Belanger PM. Circadian rhythms in hepatic biotransformation of drugs. Pathol Biol (Paris) 1996;44:564–570. [PubMed] [Google Scholar]
- Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- Brown SA, Schibler U. The ins and outs of circadian timekeeping. Curr Opin Genet Dev. 1999;9:588–594. doi: 10.1016/s0959-437x(99)00009-x. [DOI] [PubMed] [Google Scholar]
- Cahill GM. Circadian regulation of melatonin production in cultured zebrafish pineal and retina. Brain Res. 1996;708:177–181. doi: 10.1016/0006-8993(95)01365-2. [DOI] [PubMed] [Google Scholar]
- Deacon S, Arendt J. Adapting to phase shifts. I. An experimental model for jet lag and shift work. Physiol Behav. 1996;59:665–673. doi: 10.1016/0031-9384(95)02147-7. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Emery IF, Noveral JM, Jamison CF, Siwicki KK. Rhythms of Drosophila period gene expression in culture. Proc Natl Acad Sci. 1997;94:4092–4096. doi: 10.1073/pnas.94.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- Frederiks WM, Marx F, Bosch KS. Diurnal variation in glycogen phosphorylase activity in rat liver: A quantitative histochemical study. Eur J Cell Biol. 1987;43:339–341. [PubMed] [Google Scholar]
- Giebultowicz JM, Stanewsky R, Hall JC, Hege DM. Transplanted Drosophila excretory tubules maintain circadian clock cycling out of phase with the host. Curr Biol. 2000;10:107–110. doi: 10.1016/s0960-9822(00)00299-2. [DOI] [PubMed] [Google Scholar]
- Hastings MH. Circadian clocks. Curr Biol. 1997;7:R670–R672. doi: 10.1016/s0960-9822(06)00350-2. [DOI] [PubMed] [Google Scholar]
- Holmes MM, Mistlberger RE. Food anticipatory activity and photic entrainment in food-restricted BALB/c mice. Physiol Behav. 2000;68:655–666. doi: 10.1016/s0031-9384(99)00231-0. [DOI] [PubMed] [Google Scholar]
- Hudson JW, Scott IR. Daily torpor in the laboratory mouse Mus musculus. Physiol Zool. 1979;79:205–218. [Google Scholar]
- Ishikawa K, Shimazu T. Daily rhythms of glycogen synthetase and phosphorylase activities in rat liver: Influence of food and light. Life Sci. 1976;19:1873–1878. doi: 10.1016/0024-3205(76)90119-3. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- Kinlaw WB, Fish LH, Schwartz HL, Oppenheimer JH. Diurnal variation in hepatic expression of the rat S14 gene is synchronized by the photoperiod. Endocrinology. 1987;120:1563–1567. doi: 10.1210/endo-120-4-1563. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas PL. Circadian rhythms: New functions for old clock genes. Trends Genet. 2000;16:135–142. doi: 10.1016/s0168-9525(99)01945-9. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Schibler U. Circadian transcription of the cholesterol 7 α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes & Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C. Circadian expression of the steroid 15 α-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor α gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271:1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- Liu Y, Merrow M, Loros JJ, Dunlap JC. How temperature changes reset a circadian oscillator. Science. 1998;281:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitropoulos KA, Balasubramaniam S, Gibbons GF, Reeves BE. Diurnal variation in the activity of cholesterol 7-hydroxylase in the livers of fed and fasted rats. FEBS Lett. 1972;27:203–206. doi: 10.1016/0014-5793(72)80620-3. [DOI] [PubMed] [Google Scholar]
- Nef S, Allaman I, Fiumelli H, De Castro E, Nef P. Olfaction in birds: Differential embryonic expression of nine putative odorant receptor genes in the avian olfactory system. Mech Dev. 1996;55:65–77. doi: 10.1016/0925-4773(95)00491-2. [DOI] [PubMed] [Google Scholar]
- Nelson W, Halberg F. Meal-timing, circadian rhythms and life span of mice. J Nutr. 1986;116:2244–2253. doi: 10.1093/jn/116.11.2244. [DOI] [PubMed] [Google Scholar]
- Noshiro M, Nishimoto M, Okuda K. Rat liver cholesterol 7 α-hydroxylase: Pretranslational regulation for circadian rhythm. J Biol Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- Ogawa A, Yano M, Tsujinaka T, Morimoto T, Morita S, Taniguchi M, Shiozaki H, Okamoto K, Sato S, Monden M. Modulation of circadian expression of D-site binding protein by the schedule of parenteral nutrition in rat liver. Hepatology. 1997;26:1580–1586. doi: 10.1053/jhep.1997.v26.pm0009398001. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Pitot HC, Fujioka M. Diurnal variation of the serine dehydratase mRNA level in rat liver. Arch Biochem Biophys. 1994;308:285–291. doi: 10.1006/abbi.1994.1040. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Portaluppi F, Vergnani L, Manfredini R, Fersini C. Endocrine mechanisms of blood pressure rhythms. Ann NY Acad Sci. 1996;783:113–131. doi: 10.1111/j.1749-6632.1996.tb26711.x. [DOI] [PubMed] [Google Scholar]
- Rabinowitz L. Aldosterone and potassium homeostasis. Kidney Int. 1996;49:1738–1742. doi: 10.1038/ki.1996.258. [DOI] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Refinetti R. Comparison of the body temperature rhythms of diurnal and nocturnal rodents. J Exp Zool. 1996;275:67–70. doi: 10.1002/(SICI)1097-010X(19960501)275:1<67::AID-JEZ10>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Reppert SM. A clockwork explosion! Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes & Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C, Kotler M, Antolin I, Sainz RM, Menendez-Pelaez A. Regulation of the aminolevulinate synthase gene in the Syrian hamster Harderian gland: Changes during development and circadian rhythm and role of some hormones. Microsc Res Tech. 1996;34:65–70. doi: 10.1002/(SICI)1097-0029(19960501)34:1<65::AID-JEMT9>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. Circadian systems and metabolism. J Biol Rhythms. 1999;14:449–459. doi: 10.1177/074873099129001019. [DOI] [PubMed] [Google Scholar]
- Roesler WJ, Khandelwal RL. Diurnal variations in the activities of the glycogen metabolizing enzymes in mouse liver. Int J Biochem. 1985;17:81–85. doi: 10.1016/0020-711x(85)90089-8. [DOI] [PubMed] [Google Scholar]
- Rusak B, Zucker I. Neural regulation of circadian rhythms. Physiol Rev. 1979;59:449–526. doi: 10.1152/physrev.1979.59.3.449. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, et al. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- Schibler U, Lavery DJ. Circadian timing in animals. In: Russo E, et al., editors. Development-genetics, epigenetcs, and environmental regulation. Heidelberg: Springer; 1999. pp. 487–505. 1999. [Google Scholar]
- Schmidt EE, Schibler U. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development. 1995;121:2373–2383. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shibata S, Tominaga K. Brain neuronal mechanisms of circadian rhythms in mammalians. Yakugaku Zasshi. 1991;111:270–283. doi: 10.1248/yakushi1947.111.6_270. [DOI] [PubMed] [Google Scholar]
- Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Molecular neurobiology and genetics of circadian rhythms in mammals. Annu Rev Neurosci. 1995;18:531–553. doi: 10.1146/annurev.ne.18.030195.002531. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- White W, Timberlake W. Two meals promote entrainment of rat food-anticipatory and rest-activity rhythms. Physiol Behav. 1995;57:1067–1074. doi: 10.1016/0031-9384(95)00023-c. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Falvey E, Lavery D, Talbot D, Schmidt E, Ossipow V, Fonjallaz P, Schibler U. The role of the transcriptional activator protein DBP in circadian liver gene expression. J Cell Sci (Suppl.) 1992;16:123–127. doi: 10.1242/jcs.1992.supplement_16.15. [DOI] [PubMed] [Google Scholar]
- Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Young MW. Circadian rhythms: Marking time for a kingdom. Science. 2000;288:451–453. doi: 10.1126/science.288.5465.451. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]