Abstract
Background:
On the basis of the microRNA (miRNA) expression signature of maxillary sinus squamous cell carcinoma (MSSCC), we found that miR-874 was significantly reduced in cancer cells. We focused on the functional significance of miR-874 in cancer cells and identification of miR-874-regulated novel cancer networks in MSSCC.
Methods:
We used PCR-based methods to investigate the downregulated miRNAs in clinical specimens of MSSCC. Our signature analyses identified 23 miRNAs that were significantly reduced in cancer cells, such as miR-874, miR-133a, miR-375, miR-204, and miR-1. We focused on miR-874 as the most downregulated novel miRNA in our analysis.
Results:
We found potential tumour suppressive functions such as inhibition of cancer cell proliferation and invasion. A molecular target search of miR-874 revealed that PPP1CA was directly regulated by miR-874. Overexpression of PPP1CA was observed in MSSCC clinical specimens. Silencing of the PPP1CA gene significantly inhibited cancer cell proliferation and invasion.
Conclusion:
The downregulation of miR-874 was a frequent event in MSSCC, which suggests that miR-874 functions as a tumour suppressive miRNA, directly regulating PPP1CA that has a potential role of an oncogene. The identification of novel miR-874-regulated cancer pathways could provide new insights into potential molecular mechanisms of MSSCC oncogenesis.
Keywords: microRNA, miR-874 , tumour suppressor, PPP1CA , maxillary sinus
The maxillary sinuses are single pyramidal cavities in the body of the maxillae. Squamous cell carcinoma is the most common cancer of the maxillary sinus (60–70%), followed by adenoid cystic carcinoma (Million and Cassisi, 1994). Maxillary sinus squamous cell carcinoma (MSSCC) comprises 2–3% of all head and neck tumours and the annual incidence is 0.5–1.0 per 100 000 people (Tiwari et al, 2000; Dulguerov and Allal, 2006). Clinical symptoms of MSSCC present insidiously, and sometimes resemble those of chronic sinusitis. Although presentation of lymph node and distant metastasis were uncommon, primary tumours are often diagnosed as advanced disease. The 5-year survival rate of T4 tumours is ∼50%. Local recurrence is the most common cause of treatment failure and death (Konno et al, 1998; Tiwari et al, 2000; Dulguerov and Allal, 2006).
From an epidemiological standpoint, occupational exposures to leather, wood dust, nickel, arsenic, and formaldehyde have been implicated in the aetiology of MSSCC (Luce et al, 2002; Bornholdt et al, 2008). In contrast, tobacco, a major risk factor for head and neck squamous cell carcinoma (HNSCC) does not have an important role in MSSCC (Zheng et al, 1993; Holt, 1994). It has been suggested that molecular mechanisms of carcinogenesis might be different for these cancers (Lopez et al, 2011). Although analyses of major cancer-related genes, such as TP53 and K-ras, mutation were reported (Bornholdt et al, 2008; Holmila et al, 2010), relatively few genome-wide gene expression analyses of MSSCC have been conducted and no analyses of microRNAs (miRNAs) have been performed for this disease.
MicroRNAs are small non-coding RNAs of 20–22 nucleotides, and are involved in crucial biological processes, including development, differentiation, apoptosis, and proliferation through imperfect pairing with target mRNAs of protein-coding genes and the transcriptional or post-transcriptional regulation of their expression (Bartel, 2004). Bioinformatic predictions indicate that miRNAs regulate >30% of the protein coding genes (Filipowicz et al, 2008). Currently, 1424 human miRNAs are registered at miRBase release 17.0 (http://microrna.sanger.ac.uk/).
Recent studies suggest that miRNAs contribute to the initiation and development of various types of cancer (Calin and Croce, 2006). Some highly expressed miRNAs could function as oncogenes by repressing tumour suppressor genes, whereas low-expressed miRNAs could function as tumour suppressors by negatively regulating oncogenes (Esquela-Kerscher and Slack, 2006). We have conducted searches of tumour suppressive miRNAs based on analyses of expression signatures of various cancers (Ichimi et al, 2009; Kano et al, 2010; Kikkawa et al, 2010; Yoshino et al, 2011). These studies successfully identified several tumour suppressive miRNAs such as miR-1, miR-133a, and miR-145 (Chiyomaru et al, 2010b; Kano et al, 2010; Yoshino et al, 2011; Nohata et al, 2011b). The next major problem is the identification of the oncogenes that are controlled by these miRNAs. Our recent studies showed that several target genes did indeed function as oncogenes (Chiyomaru et al, 2010a, 2011; Fuse et al, 2011; Mutallip et al, 2011; Nohata et al, 2011a).
The aim of this study was to identify new tumour suppressive miRNAs revealed in our MSSCC expression analysis. We focused on miR-874, which was the most significantly downregulated miRNA in our signature. We found that it functioned as a tumour suppressor based on our findings of inhibited cancer cell proliferation and invasion. Genome-wide expression analysis identified several candidate target genes such as PPP1CA (protein phosphatase 1, catalytic subunit, a isozyme), PAAF1 (proteasomal ATPase-associated factor 1), and TGOLN2 (trans-Golgi network (TGN) protein 2). Insights into the association between tumour suppressive miR-874 and their target oncogene networks could enhance our understanding of the molecular mechanism of MSSCC carcinogenesis.
Materials and methods
Clinical MSSCC specimens
In all, 20 pairs of primary MSSCC and corresponding normal epithelial samples were obtained from patients with MSSCC in Chiba University Hospital (Chiba, Japan) from 2005 to 2010. The fresh specimens were immediately immersed in RNAlater (Qiagen, Valencia, CA, USA) and stored at −20 °C until RNA was extracted. The samples macroscopically considered normal were confirmed free of cancer cells by microscopic pathological examination. The patients' backgrounds and clinicopathological characteristics are summarised in Table 1. The patients were classified according to 2002 Union for International Cancer Control TNM staging criteria (Sobin and Wittekind, 2002) before treatment. Written consent of tissue donation for research purposes was obtained from each patient before tissue collection. The protocol was approved by the Institutional Review Board of Chiba University.
Table 1. Clinicopathological characteristics of 20 patients with maxillary sinus squamous cell carcinoma.
| No. | Age (years) | Gender | Differentiation | T | N | M | Stage |
|---|---|---|---|---|---|---|---|
| 1 | 65 | Female | Poor | 4b | 0 | 0 | IVB |
| 2 | 65 | Male | Moderate | 4a | 0 | 0 | IVA |
| 3 | 74 | Male | Well | 4a | 0 | 0 | IVA |
| 4 | 71 | Male | Moderate | 3 | 1 | 0 | III |
| 5 | 67 | Male | Moderate | 4a | 0 | 0 | IVA |
| 6 | 68 | Male | Well | 4b | 0 | 0 | IVB |
| 7 | 77 | Male | Poor | 3 | 0 | 0 | III |
| 8 | 76 | Male | Moderate | 3 | 0 | 0 | III |
| 9 | 61 | Male | Well | 3 | 0 | 0 | III |
| 10 | 54 | Male | Poor | 3 | 0 | 0 | III |
| 11 | 64 | Male | Poor | 4b | 0 | 0 | IVB |
| 12 | 64 | Male | Moderate | 4a | 0 | 0 | IVA |
| 13 | 80 | Male | Moderate | 4a | 0 | 0 | IVA |
| 14 | 66 | Female | Poor | 4a | 2c | 0 | IVA |
| 15 | 60 | Male | Poor | 4a | 0 | 0 | IVA |
| 16 | 66 | Female | Moderate | 4a | 0 | 0 | IVA |
| 17 | 85 | Male | Poor | 4a | 0 | 0 | IVA |
| 18 | 69 | Male | Well | 4a | 0 | 0 | IVA |
| 19 | 57 | Male | Poor | 4a | 0 | 0 | IVA |
| 20 | 69 | Male | Poor | 4a | 2b | 0 | IVA |
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RNA concentrations were determined spectrophotometrically, and molecular integrity was checked by gel electrophoresis. RNA quality was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
MicroRNA expression signatures and data normalisation
Tissue specimens for miRNA screening using a low density array (LDA) were from five MSSCC patients at Chiba University Hospital between 2005 and 2007 (Table 1; #1–#5). The miRNA expression patterns were evaluated using the TaqMan LDA Human microRNA Panel v2.0 (Applied Biosystems, Foster City, CA, USA). The assay was composed of two steps: generation of cDNA by reverse transcription (RT) and a TaqMan real-time PCR assay. Description of real-time PCR and the list of human miRNAs can be found on the company's website (http://www.appliedbiosystems.com). Analysis of relative miRNA expression data was performed using GeneSpring GX version 7.3.1 software (Agilent Technologies) according to the manufacturer's instructions. A cutoff P-value of <0.05 was used to narrow down the candidates after global normalisation of the raw data. After global normalisation, additional normalisation was carried out with RNU48.
Maxillary sinus squamous cell carcinoma cell culture
The human MSSCC cell line IMC-3 (Mizoguchi et al, 1991) was used. Cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37 °C.
Mature miRNA transfection and small-interfering RNA treatment
The following RNA species were used in this study: mature miRNAs, pre-miR miRNA precursors (hsa-miR-874; pre-miR ID: PM12355), negative control miRNA (P/N: AM17111) (Applied Biosystems), small-interfering RNA (Stealth Select RNAi siRNA; si-PPP1CA_1 Cat# HSS143413, si-PPP1CA_2 Cat# HSS143414) (Invitrogen), and negative control siRNA (Stealth RNAi Negative Control Medium GC Duplex; 12935–300) (Invitrogen). RNAs were incubated with Opti-MEM (Invitrogen) and Lipofectamine RNAiMax reagent (Invitrogen) as described previously (Ichimi et al, 2009). Transfection efficiency of pre-miR in cell lines was confirmed based on downregulation of TWF1 (PTK9) mRNA following transfection with miR-1 as previously reported (Ichimi et al, 2009).
Cell proliferation assays
Cells were transfected with 10 nM miRNA and siRNA by RT and plated in 96-well plates at 3 × 103 cells per well. After 72 h, cell proliferation was determined by the XTT assay, using the Cell Proliferation Kit II (Roche Molecular Biochemicals, Mannheim, Germany) as previously reported (Ichimi et al, 2009; Chiyomaru et al, 2010b). Triplicate wells were measured for cell viability in each treatment group. Furthermore, we also conducted counting number of cells with each treatment. Cells (1 × 104) were incubated in a 24-well plate for 24, 48, and 72 h. They were then treated with trypsin and stained with trypan blue. Viable cells, which excluded trypan blue dye, were counted in quadruplicate with a Countess (Invitrogen).
Cell invasion assays
A cell invasion assay was carried out using modified Boyden chambers containing transwell-precoated Matrigel membrane filter inserts with 8 μm pores in 24-well tissue culture plates at 1 × 105 cells per well (BD Biosciences, Bedford, MA, USA; Kano et al, 2010; Chiyomaru et al, 2010b). Triplicate wells were measured for cell invasion in each treatment group.
Target gene search for miR-874
Genome-wide screens using miR-874 transfectants were performed to identify target genes of miR-874 in IMC-3. Oligo-microarray human 44K (Agilent Technologies) was used for expression profiling of the transfectants in comparison with a miRNA-negative control transfectant. Hybridisation and wash steps were performed as previously described (Sugimoto et al, 2009). The arrays were scanned using a Packard GSI Lumonics Scan Array 4000 (Perkin Elmer, Boston, MA, USA). The data were analysed by means of DNASIS array software (Hitachi Software Engineering, Tokyo, Japan), which converted the signal intensity for each spot into text format. The log2 ratios of the median subtracted background intensities were analysed. Data from each microarray study were normalised by a global normalisation method. Predicted target genes and their target miRNA binding site seed regions were investigated using TargetScan (release 5.1, http://www.targetscan.org/). The sequences of the predicted mature miRNAs were confirmed using miRBase (release 17.0, http://microrna.sanger.ac.uk/).
Quantitative real-time RT–PCR
First-strand cDNA was synthesised from 1 μg of total RNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene-specific PCR products were assayed continuously using a 7900-HT Real-Time PCR System according to the manufacturer's protocol. The initial PCR step consisted of a 10 min hold at 95 °C, followed by 40 cycles consisting of a 15 s denaturation at 95 °C and a 1 min annealing/extension at 63 °C. TaqMan probes and primers for PPP1CA (P/N: Hs00267568_m1), PAAF1 (P/N: Hs00228523_m1), TGOLN2 (P/N: Hs00197728_m1) and GUSB (P/N: Hs99999908_m1) internal control were obtained from Applied Biosystems (Assay-On-Demand Gene Expression Products). The expression levels of miR-874 (assay ID: 002268) were analysed by TaqMan quantitative real-time PCR (TaqMan MicroRNA Assay; Applied Biosystems) and normalised to RNU48 (assay ID: 001006). The ΔΔCt method was adopted and applied to calculate relative quantity of subject genes. All reactions were performed in triplicate, and included negative control reactions that lacked cDNA.
Western blots
Cells were harvested at 72 h after transfection and lysates were prepared. A 50 μg of protein lysate was separated by NuPAGE on 4–12% bis-tris gels (Invitrogen) and transferred to PVDF membranes. Immunoblotting was performed with diluted (1:200) monoclonal protein phosphatase 1 α (PP1α) antibody (sc-7482; Santa Cruz Biotechnology, Santa Cruz, CA, USA), with β-actin antibody (sc-1615; Santa Cruz Biotechnology) used as an internal control. The membrane was washed and incubated with goat anti-mouse IgG (H+L)–HRP conjugate (Bio-Rad, Hercules, CA, USA). Specific complexes were visualised by echochemiluminescence (GE Healthcare Bio-Sciences, Princeton, NJ, USA), and the expression levels of these genes were evaluated by ImageJ software (version 1.44; http://rsbweb.nih.gov/ij/).
Plasmid construction and dual-luciferase reporter assay
The wild-type sequences of PPP1CA 3′-UTR and those with deleted miR-874 target sites (position 237–243) were inserted between the XhoI–PmeI restriction sites in the 3-UTR of the hRluc gene in psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). Sequences of oligonucleotides are described in the Supplementary Information. The synthesised DNA was cloned into the psiCHECK-2 vector. The IMC-3 cells were transfected with 15 ng of vector, 10 nM of miR-874 (Applied Biosystems), and 1 μl of Lipofectamine 2000 (Invitrogen) in 100 μl of Opti-MEM (Invitrogen). The activities of firefly and Renilla luciferases in cell lysates were determined with a dual-luciferase assay system (E1910; Promega). Normalised data were calculated as the quotient of Renilla/firefly luciferase activities.
Statistical analysis
The relationships between two groups and the numerical values obtained by real-time RT–PCR were analysed using the non-parametric Mann–Whitney U-test or the paired t-test. The relationship among three variables and numerical values was analysed using the Bonferroni adjusted Mann–Whitney U-test. Spearman's rank test was used to evaluate the relationships among the relative expression levels of miR-874, PPP1CA, PAAF1, and TGOLN2 mRNA. All analyses were performed using Expert StatView (version 4, SAS Institute Inc., Cary, NC, USA).
Results
Identification of downregulated miRNAs in MSSCC by miRNA expression signature: expression of miR-874 in MSSCC clinical specimens
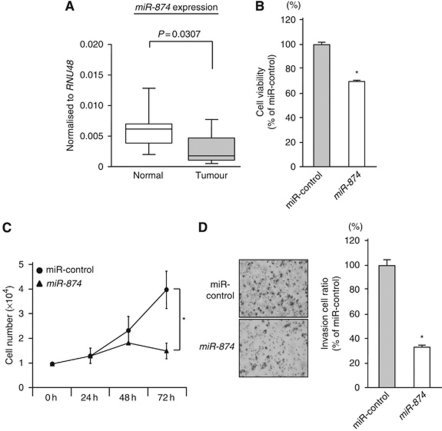
We evaluated mature miRNA expression levels of five pairs of normal epithelia and MSSCC by miRNA expression signature analysis. In all, 23 significantly downregulated miRNAs were selected after normalisation to RNU48 (Table 2). The miR-874, the most downregulated miRNA in the list, was selected for further study. Quantitative stem–loop RT–PCR demonstrated that the expression levels of miR-874 were significantly lower in 20 MSSCC specimens in comparison with normal tissues (P=0.0307, Figure 1A).
Table 2. Downregulated microRNAs in maxillary sinus squamous cell carcinoma (normalised to RNU48).
|
Normalised ratio
|
|||||
|---|---|---|---|---|---|
| MicroRNA | Accession no. | Fold change | Normal | Tumour | P-value |
| miR-874 | MIMAT0004911 | 0.011 | 3.05E-04 | 3.36E-06 | 0.0463 |
| miR-133a | MIMAT0000427 | 0.017 | 1.89E-02 | 3.14E-04 | 0.0033 |
| miR-375 | MIMAT0000728 | 0.035 | 3.95E-02 | 1.36E-03 | 0.0161 |
| miR-204 | MIMAT0000265 | 0.045 | 3.26E-02 | 1.47E-03 | 0.0055 |
| miR-1 | MIMAT0000416 | 0.054 | 1.88E-03 | 1.02E-04 | 0.024 |
| miR-139-5p | MIMAT0000250 | 0.132 | 3.61E-02 | 4.75E-03 | 0.0044 |
| miR-145 | MIMAT0000437 | 0.167 | 6.57E-01 | 1.10E-01 | 0.0367 |
| miR-143 | MIMAT0000435 | 0.177 | 2.58E-01 | 4.57E-02 | 0.019 |
| miR-486-3p | MIMAT0004762 | 0.183 | 2.26E-03 | 4.13E-04 | 0.0407 |
| miR-146a | MIMAT0000449 | 0.253 | 1.09E+00 | 2.76E-01 | 0.0041 |
| miR-410 | MIMAT0002171 | 0.254 | 4.92E-04 | 1.25E-04 | 0.002 |
| miR-126 | MIMAT0000445 | 0.262 | 3.16E+00 | 8.28E-01 | 0.0037 |
| miR-539 | MIMAT0003163 | 0.278 | 4.00E-03 | 1.11E-03 | 0.005 |
| miR-134 | MIMAT0000447 | 0.308 | 1.69E-03 | 5.19E-04 | 0.0158 |
| miR-218 | MIMAT0000275 | 0.316 | 1.62E-02 | 5.13E-03 | 0.0329 |
| miR-146b-5p | MIMAT0002809 | 0.337 | 5.18E-01 | 1.75E-01 | 0.0012 |
| miR-140-3p | MIMAT0004597 | 0.341 | 2.04E-02 | 6.94E-03 | 0.0111 |
| miR-30a-3p | MIMAT0000088 | 0.431 | 6.37E-02 | 2.75E-02 | 0.0089 |
| miR-191 | MIMAT0000440 | 0.489 | 6.22E-01 | 3.04E-01 | 0.0251 |
| miR-186 | MIMAT0000456 | 0.51 | 1.75E-01 | 8.90E-02 | 0.0309 |
| miR-148a | MIMAT0000243 | 0.523 | 3.30E-02 | 1.73E-02 | 0.0334 |
| miR-30e-3p | MIMAT0000693 | 0.525 | 4.95E-02 | 2.60E-02 | 0.0048 |
| miR-29c | MIMAT0000681 | 0.552 | 7.94E-02 | 4.39E-02 | 0.0305 |
Figure 1.
Expression of miR-874 in MSSCC clinical specimens and gain-of-function study using miR-874 in the IMC-3 cell line. (A) The miR-874 expression levels in clinical specimens. Real-time RT–PCR showed that miRNA expression in tumour tissues was lower than that of normal tissues. RNU48 was used as an internal control. (B) Cell proliferation determined by the XTT assay in the IMC-3 cell line transfected with 10 nM of miR-874 or miR-control. (C) Cell number was counted after transfection with 10 nM of miR-874 or miR-control at 24, 48, and 72 h. (D) Cell invasion activity determined by the Matrigel invasion assay in IMC-3 cell lines transfected with 10 nM of miR-874 or miR-control. *P<0.05.
Effect of miR-874 transfection on the proliferation and invasion of IMC-3
To investigate the functional roles of miR-874, we performed gain-of-function studies using miRNA transfection of IMC-3.
The XTT assay showed significant inhibition of cell proliferation in miR-874 transfectants in comparison with the miR-control transfectants (% of cell proliferation, 69.6±0.8 and 100.0±3.3, respectively; P<0.05; Figure 1B). This result was also confirmed by performing cell counting assay (cell number, 1.5 × 104±2.9 × 103 and 3.7 × 104±6.9 × 103, respectively; P<0.05; Figure 1C).
The Matrigel invasion assay demonstrated that invading cell numbers were significantly decreased in miR-874 transfected IMC-3 cells in comparison with the controls (% of cell invasion, 31.3±6.9 and 100.0±11.2, respectively, P<0.05; Figure 1C).
Gene expression profile identifies downregulated genes in miR-874 transfectants
To gain further insight into which genes were affected by miR-874 transfection, we performed gene expression analysis with miR-874 transfectants and the controls in IMC-3 cells. Signal values of raw data in miR-control transfectants <5000 were cutoff. A total of 18 genes were downregulated less than −1.0 (log2 ratio) in miR-874 transfectants compared with the controls. The TargetScan programme showed that seven of the genes had putative target sites of miR-874 in their 3′-UTR (Table 3). Entries from the microarray data were approved by the Gene Expression Omnibus (GEO) and were assigned GEO accession number GSE19714.
Table 3. The 18 downregurated genes in miR-874 transfectants.
| Entrez gene ID | Gene name | Gene symbol | Log2 ratio | miR-874 target |
|---|---|---|---|---|
| 1373 | Carbamoyl-phosphate synthetase 1, mitochondrial | CPS1 | −1.85 | 2 |
| 3939 | Lactate dehydrogenase A | LDHA | −1.75 | — |
| 1915 | Eukaryotic translation elongation factor 1 α 1 | EEF1A1 | −1.45 | — |
| 5499 | Protein phosphatase 1, catalytic subunit, α isozyme | PPP1CA | −1.25 | 1 |
| 5660 | Prosaposin | PSAP | −1.23 | — |
| 80227 | Proteasomal ATPase-associated factor 1 | PAAF1 | −1.22 | 1 |
| 567 | β-2-microglobulin | B2M | −1.21 | — |
| 303 | Annexin A2 pseudogene 1 | ANXA2P1 | −1.18 | — |
| 5223 | Phosphoglycerate mutase 1 (brain) | PGAM1 | −1.15 | — |
| 1303 | Collagen, type XII, α 1 | COL12A1 | −1.09 | 1 |
| 3486 | Insulin-like growth factor binding protein 3 | IGFBP3 | −1.08 | — |
| 2778 | GNAS complex locus | GNAS | −1.08 | — |
| 55536 | Cell division cycle associated 7-like | CDCA7L | −1.06 | — |
| 8667 | Eukaryotic translation initiation factor 3, subunit H | EIF3H | −1.05 | 1 |
| 10916 | Melanoma antigen family D, 2 | MAGED2 | −1.05 | — |
| 10618 | Trans-Golgi network protein 2 | TGOLN2 | −1.03 | 2 |
| 4077 | Neighbor of BRCA1 gene 1 | NBR1 | −1.02 | 1 |
| 343477 | Heat shock protein 90 kD β (Grp94), member 3 (pseudogene) | HSP90B3P | −1.00 | — |
Expression levels of candidate target genes of miR-874 in MSSCC clinical specimens
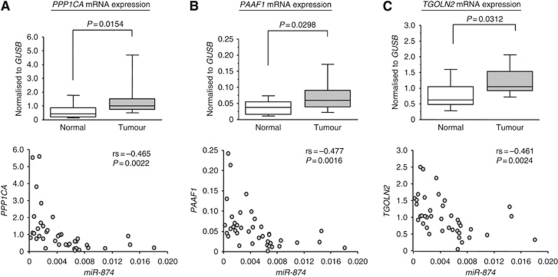
We measured the mRNA expression levels of seven candidate genes in MSSCC clinical specimens by quantitative real-time RT–PCR. Three genes, PPP1CA, PAAF1, and TGOLN2 were significantly upregulated in cancer tissues (P=0.0154, P=0.0298, and P=0.0312 respectively; Figure 2A, B and C, upper panel). The other four genes (CPS1, COL12A1, EIF3H, and NBR1) were not upregulated in the tumour region of MSSCC (Supplementary Figure). There were significant inverse correlations between each of the genes and the level of miR-874 expression (Figure 2A, B and C, lower panel).
Figure 2.
Expression levels of three candidate genes of miR-874 target were measured by real-time RT–PCR. (A, upper) PPP1CA mRNA expression levels in MSSCC clinical specimens. (B, upper) PAAF1 mRNA expression levels in MSSCC clinical specimens. (C, upper) TGOLN2 mRNA expression levels in MSSCC clinical specimens. Real-time RT–PCR showed that each of the three genes in tumour tissues was expressed at higher levels than that in the normal tissues. GUSB was used as an internal control. (A, B, and C, lower) Significant inverse correlations between each of the genes and the level of miR-874 expression were shown.
PPP1CA is directly regulated by miR-874
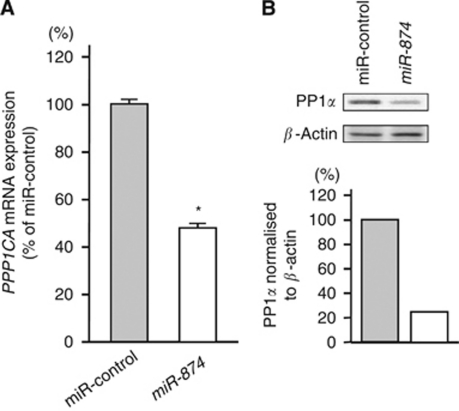
PPP1CA mRNA and PP1α protein expression levels were markedly downregulated in the miR-874 transfectants in comparison with the controls (Figure 3A and B). We performed a luciferase reporter assay to determine whether PPP1CA mRNA had a target site for miR-874. We used a vector encoding either the total sequence of the 3′-UTR of PPP1CA mRNA, including the predicted miR-874 target site (positions 237–243), or a vector lacking the miR-874 target site. We found that the luminescence intensity was significantly reduced by transfection of the entire 3′-UTR of PPP1CA, whereas deletion of positions 237–243 blocked the decrease in luminescence (Figure 4).
Figure 3.
PPP1CA mRNA and PP1α protein expression in the IMC-3 cell line. (A) PPP1CA mRNA expression 48 h after transfection with 10 nM with miR-874. PPP1CA mRNA expression was significantly repressed in miR-874 transfectants. GUSB was used as an internal control. (B) PP1α protein expression 72 h after transfection with miR-874. β-Actin was used as a loading control. The protein expression level of PP1α was also repressed in miR-874 transfectants.
Figure 4.
Schematic of conserved binding sites for miR-874 (upper). Putative conserved target site in the PPP1CA 3′-UTR was identified with the TargetScan database: the one miR-874 target site is indicated. IMC-3 cells were transfected with 15 ng of PPP1CA 3′-UTR in a vector construct and 10 nM of miR-874 or miR-control. Vectors were used with encoding the entire sequence of 3′-UTR of PPP1CA mRNA or that with a deletion of the miR-874 target (position 237–243). Renilla luciferase activity was measured after a 24 h transfection. The results were normalised against firefly luciferase values (lower). *P<0.05.
Effect of PPP1CA silencing on cell proliferation and invasion in IMC-3
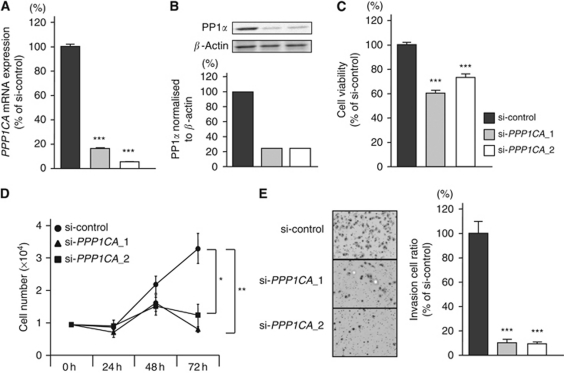
To examine the functional role of PPP1CA, we performed loss-of-function studies using two different si-PPP1CAs transfected into the IMC-3 cell line. The PPP1CA mRNA and PP1α protein expression levels were markedly reduced by the two different si-PPP1CA transfectants (Figure 5A and B).
Figure 5.
Effect of si-PPP1CA in the IMC-3 cell line. (A) PPP1CA mRNA expression at 48 h after transfection with 10 nM of si-PPP1CA_1, si-PPP1CA_2, or si-control. PPP1CA mRNA expression was repressed in si-PPP1CA_1 and si-PPP1CA_2 transfectants. GUSB was used as an internal control. (B) PP1α protein expression at 72 h after transfection of the siRNAs. β-Actin was used a loading control. The protein expression level of PP1α was also repressed in si-PPP1CA_1 and si-PPP1CA_2 transfectants. (C) Cell proliferation determined with the XTT assay in the IMC-3 cell line transfected with 10 nM of si-PPP1CA_1, si-PPP1CA_2, or si-control. (D) Cell number was counted after transfection with 10 nM of si-PPP1CA_1, si-PPP1CA_2, or si-control at 24, 48, and 72 h. (E) Cell invasion activity determined with the Matrigel invasion assay in the IMC-3 cell line transfected with 10 nM of si-PPP1CA_1, si-PPP1CA_2, or si-control. *P<0.0018, **P<0.0005, and ***P<0.0001.
The XTT assay revealed significant inhibition of cell proliferation in the two different si-PPP1CA transfectants in comparison with growth of the si-control transfectants (% of cell proliferation: 60.4±1.2, 73.2±1.0, and 100.0±5.0, respectively; P<0.0001; Figure 5C). This result was also confirmed by performing cell counting assay (cell number, 8.0 × 103±1.2 × 103, 1.4 × 104±4.6 × 103, and 3.2 × 104±6.0 × 103, respectively; P=0.0005 and P=0.0018, respectively; Figure 5D).
The Matrigel invasion assay demonstrated that the number of invading cells was significantly decreased in the two different si-PPP1CA transfectants compared with their counterparts (% of cell invasion, 11.5±2.8, 8.1±1.4, and 100.0±11.7, respectively; P<0.0001; Figure 5E).
Discussion
This is the first article to investigate aberrant miRNA expression in MSSCC clinical specimens. The miR-133a and miR-1 were among the top five downregulated miRNAs in our expression analysis. Interestingly, miR-1-1/miR-133a-2, and miR-1-2/miR-133a-1 are clustered on different chromosomal regions in the human genome, 20q13.33 and 18q11.2, respectively. Recently, our analyses of oesophageal cancer and bladder cancer expression signatures confirmed downregulation of both miRNAs (Kano et al, 2010; Chiyomaru et al, 2010b), and we demonstrated that miR-1 and miR-133a function as tumour suppressors in many types of cancers regulating several oncogenes (Chiyomaru et al, 2010a, 2010b; Kano et al, 2010; Mutallip et al, 2011; Nohata et al, 2011a, 2011b; Uchida et al, 2011; Yoshino et al, 2011). When we consider other miRNAs in this signature, miR-145 downregulation has frequently been reported in cancers, including prostate, bladder, colon, ovarian, and oesophageal cancers as well as B-cell malignancies (Akao et al, 2007; Iorio et al, 2007; Arndt et al, 2009; Kano et al, 2010; Chiyomaru et al, 2010b; Zaman et al, 2010). The miR-145 is located on chromosome 5q32–33 within a 4.09 kb region (http://microrna.sanger.ac.uk/). Of interest, 5q31.1 is a well-known fragile site in the human genome (http://www.genenames.org/) and is often deleted in cancers. Increasing evidence and our data indicate that miR-145 functions as a tumour suppressive miRNA and inhibits cell growth, invasion, and migration in cancer cells (Kano et al, 2010; Chiyomaru et al, 2010b; Fuse et al, 2011). Our present analysis generated a list of sequences that could be involved in the pathology of MSSCC. Analysis of miRNAs included in this signature could enhance our understanding of MSSCC carcinogenesis.
In this study, we focused on the functional significance of miR-874, because it was the most downregulated miRNA in our signature and functional analysis of miR-874 had not yet been reported. The miR-874 was recently identified based on small RNA library sequencing and is conserved across most, but not all mammals (Landgraf et al, 2007; Lui et al, 2007). Our results showed that miR-874 was downregulated in MSSCC cells and ectopic expression of miR-874 significantly inhibited cell proliferation and invasion in IMC-3 cells. These results indicated that miR-874 might function as a tumour suppressor in IMC-3 cells. Further studies are required to elucidate the precise mechanisms of miR-874 regulation for initiation and development of MSSCC oncogenesis.
We performed a genome-wide analysis using miR-874 transfected IMC-3 cells to elucidate the target genes regulated by miR-874. From the microarray analysis, we identified seven candidate genes (CPS1, PPP1CA, PAAF1, COL12A1, EIF3H, TGOLN2, and NBR1) containing miR-874 target sites. To validate the mRNA expression levels of seven candidate genes in MSSCC clinical specimens by quantitative real-time RT–PCR, we narrowed down to three genes (PPP1CA, PAAF1, and TGOLN2) whose expression levels were significantly upregulated in MSSCC clinical specimens compared with normal tissues.
PAAF1 inhibits proteasome 26S assembly and proteolytic activity by impairing the association of the 19S regulatory complex with the 20S core. The 26S proteasome consists of a 20S proteolytic core particle and 19S regulatory complexes. The 26S proteasome has an important role in ubiquitin-dependent proteolysis, which regulates many biological processes, such as cell cycle progression and signal transduction (Park et al, 2005). TGOLN2, TGN protein 2, is a cargo protein of retrograde transport, in which proteins and lipids are moved between endosomes and the TGN. Although the functions of several cargo proteins have been elucidated in retrograde transport, the role of TGOLN2 remains unknown (Johannes and Popoff, 2008; Pfeffer, 2009). These two genes currently have little association with cancer development. Therefore, we focused on PPP1CA as a subject of further experiment.
PPP1CA encodes the catalytic subunit of PP1α. The PP1α catalytic subunit can form complexes with many regulatory subunits, which regulate various cellular activities such as the cell cycle, apoptosis, and signal transduction (Cohen, 2002; Ceulemans and Bollen, 2004). Previous analysis of the protein showed that PP1α dephosphorylates the BRCA1 protein, coded by the tumour suppressor BRCA1, in breast and ovarian cancer. Those findings indicate that PP1α may have an oncogenic role (Liu et al, 2002). In addition, it has been reported that overexpression of PP1α is observed in pre-malignant hepatic cells and oral squamous cell carcinoma (Saadat et al, 1995; Imai et al, 1999; Hsu et al, 2006). In contrast, PP1α may function as a tumour suppressor by activating tumour suppressor protein pRB (Alberts et al, 1993). Our present data suggest that the PPP1CA gene functions as an oncogene in MSSCC. A molecular network search for downstream targets of PPP1CA in MSSCC will be necessary. Unfortunately, there was no significant relationship between miR-874 or PPP1CA expression and clinicopathological parameters in this study. Our cohort was too small to evaluate this relationship. In addition, our samples are mostly at the late stage. Hence, a large-scale clinical test including the early stage samples will be necessary.
It is also of interest that PPP1CA is located at chromosomal region 11q13. Amplification of the chromosomal region on 11q13 is frequently observed in human cancers including HNSCC and breast cancer, and it is well known that CCND1, encoding cyclin D1, is a putative oncogene in the 11q13 amplicon (Schuuring, 1995; Schwab, 1998; Gollin, 2001). We recently demonstrated that when chromosomal region 11q13 was gained, the expression levels of several genes were elevated (Sugimoto et al, 2009). The oncogenic function of PPP1CA and amplification of this region may be related closely, so it will be important to examine the structural changes in this region in MSSCC.
In conclusion, the reduction of miR-874 and increase of PPP1CA were frequent events in MSSCC cancer cells. The miR-874 may function as a tumour suppressor and may directly regulate PPP1CA. The miR-874 regulates novel cancer pathways and could provide new insights into molecular mechanisms in MSSCC and might contribute to the development of new therapeutic strategies for the disease.
Acknowledgments
This study was supported by the JSPS KAKENHI (C), 21592187.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T (2007) Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci 98: 1914–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts AS, Thorburn AM, Shenolikar S, Mumby MC, Feramisco JR (1993) Regulation of cell cycle progression and nuclear affinity of the retinoblastoma protein by protein phosphatases. Proc Natl Acad Sci U S A 90: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, Bittner A, Raponi M (2009) Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 9: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bornholdt J, Hansen J, Steiniche T, Dictor M, Antonsen A, Wolff H, Schlunssen V, Holmila R, Luce D, Vogel U, Husgafvel-Pursiainen K, Wallin H (2008) K-ras mutations in sinonasal cancers in relation to wood dust exposure. BMC Cancer 8: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84: 1–39 [DOI] [PubMed] [Google Scholar]
- Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K, Nishiyama K, Seki N, Nakagawa M (2010a) Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol; e-pub ahead of print 14 September 2010; PMID: 20843712; doi:10.1016/j.urolonc.2010.05.008 [DOI] [PubMed]
- Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M (2010b) miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer 102: 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T, Tatarano S, Kawakami K, Enokida H, Yoshino H, Nohata N, Fuse M, Seki N, Nakagawa M (2011) SWAP70, actin-binding protein, function as an oncogene targeting tumor-suppressive miR-145 in prostate cancer. Prostate; e-pub ahead of print 25 February 2011; doi: 10.1002/pros.21372; 10.1002/pros.21372 [DOI] [PubMed]
- Cohen PT (2002) Protein phosphatase 1--targeted in many directions. J Cell Sci 115: 241–256 [DOI] [PubMed] [Google Scholar]
- Dulguerov P, Allal AS (2006) Nasal and paranasal sinus carcinoma: how can we continue to make progress? Curr Opin Otolaryngol Head Neck Surg 14: 67–72 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Fuse M, Nohata N, Kojima S, Sakamoto S, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Naya Y, Ichikawa T, Seki N (2011) Restoration of miR-145 expression suppresses cell proliferation, migration and invasion in prostate cancer by targeting FSCN1. Int J Oncol 38: 1093–1101 [DOI] [PubMed] [Google Scholar]
- Gollin SM (2001) Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck 23: 238–253 [DOI] [PubMed] [Google Scholar]
- Holmila R, Bornholdt J, Suitiala T, Cyr D, Dictor M, Steiniche T, Wolff H, Wallin H, Luce D, Husgafvel-Pursiainen K (2010) Profile of TP53 gene mutations in sinonasal cancer. Mutat Res 686: 9–14 [DOI] [PubMed] [Google Scholar]
- Holt GR (1994) Sinonasal neoplasms and inhaled air toxics. Otolaryngol Head Neck Surg 111: 12–14 [DOI] [PubMed] [Google Scholar]
- Hsu LC, Huang X, Seasholtz S, Potter DM, Gollin SM (2006) Gene amplification and overexpression of protein phosphatase 1alpha in oral squamous cell carcinoma cell lines. Oncogene 25: 5517–5526 [DOI] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N (2009) Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 125: 345–352 [DOI] [PubMed] [Google Scholar]
- Imai Y, Kakinoki Y, Takizawa N, Nakamura K, Shima H, Kikuchi K (1999) Up-regulation of nuclear PP1alpha and PP1delta in hepatoma cells. Int J Oncol 14: 121–126 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM (2007) MicroRNA signatures in human ovarian cancer. Cancer Res 67: 8699–8707 [DOI] [PubMed] [Google Scholar]
- Johannes L, Popoff V (2008) Tracing the retrograde route in protein trafficking. Cell 135: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H (2010) miR-145, miR-133a and miR-133b: tumor suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer 127: 2804–2814 [DOI] [PubMed] [Google Scholar]
- Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N (2010) miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC). Br J Cancer 103: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno A, Ishikawa K, Terada N, Numata T, Nagata H, Okamoto Y (1998) Analysis of long-term results of our combination therapy for squamous cell cancer of the maxillary sinus. Acta Otolaryngol Suppl 537: 57–66 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Virshup DM, White RL, Hsu LC (2002) Regulation of BRCA1 phosphorylation by interaction with protein phosphatase 1alpha. Cancer Res 62: 6357–6361 [PubMed] [Google Scholar]
- Lopez F, Llorente JL, Garcia-Inclan C, Alonso-Guervos M, Cuesta-Albalad MP, Fresno MF, Alvarez-Marcos C, Suarez C, Hermsen MA (2011) Genomic profiling of sinonasal squamous cell carcinoma. Head Neck 33: 145–153 [DOI] [PubMed] [Google Scholar]
- Luce D, Leclerc A, Begin D, Demers PA, Gerin M, Orlowski E, Kogevinas M, Belli S, Bugel I, Bolm-Audorff U, Brinton LA, Comba P, Hardell L, Hayes RB, Magnani C, Merler E, Preston-Martin S, Vaughan TL, Zheng W, Boffetta P (2002) Sinonasal cancer and occupational exposures: a pooled analysis of 12 case-control studies. Cancer Causes Control 13: 147–157 [DOI] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A (2007) Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 67: 6031–6043 [DOI] [PubMed] [Google Scholar]
- Million RR, Cassisi NJ (1994) Management of Head and Neck Cancer: a Multidisciplinary Approach, 2nd edn. J.B.LIPPINCOTT COMPANY: Philadelphia [Google Scholar]
- Mizoguchi H, Komiyama S, Matsui K, Hamanaka R, Ono M, Kiue A, Kobayashi M, Shimizu N, Welgus HG, Kuwano M (1991) The response to epidermal growth factor of human maxillary tumor cells in terms of tumor growth, invasion and expression of proteinase inhibitors. Int J Cancer 49: 738–743 [DOI] [PubMed] [Google Scholar]
- Mutallip M, Nohata N, Hanazawa T, Kikkawa N, Horiguchi S, Fujimura L, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N (2011) Glutathione S-transferase P1 (GSTP1) suppresses cell apoptosis and its regulation by miR-133alpha in head and neck squamous cell carcinoma (HNSCC). Int J Mol Med 27: 345–352 [DOI] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Fujimura L, Yoshino H, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N (2011a) Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int J Oncol 38: 209–217 [PubMed] [Google Scholar]
- Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Shozu M, Okamoto T, Seki N (2011b) miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget 2: 29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Hwang YP, Lee JS, Seo SH, Yoon SK, Yoon JB (2005) Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol Cell Biol 25: 3842–3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR (2009) Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett 583: 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat M, Mizuno Y, Takizawa N, Kakinoki Y, Rikuchi R (1995) Neoplastic alterations in subcellular distribution of type 1 alpha protein phosphatase in rat ascites hepatoma cells. Cancer Lett 94: 165–170 [DOI] [PubMed] [Google Scholar]
- Schuuring E (1995) The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes--a review. Gene 159: 83–96 [DOI] [PubMed] [Google Scholar]
- Schwab M (1998) Amplification of oncogenes in human cancer cells. Bioessays 20: 473–479 [DOI] [PubMed] [Google Scholar]
- Sobin L, Wittekind C (2002) UICC TNM Classification of Malignant Tumors, 6th edn. Wiley-Liss: New York [Google Scholar]
- Sugimoto T, Seki N, Shimizu S, Kikkawa N, Tsukada J, Shimada H, Sasaki K, Hanazawa T, Okamoto Y, Hata A (2009) The galanin signaling cascade is a candidate pathway regulating oncogenesis in human squamous cell carcinoma. Genes Chromosomes Cancer 48: 132–142 [DOI] [PubMed] [Google Scholar]
- Tiwari R, Hardillo JA, Mehta D, Slotman B, Tobi H, Croonenburg E, van der Waal I, Snow GB (2000) Squamous cell carcinoma of maxillary sinus. Head Neck 22: 164–169 [DOI] [PubMed] [Google Scholar]
- Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M (2011) MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol; e-pub ahead of print 9 March 2011; doi: 10.1016/j.urolonc.2010.09.017 [DOI] [PubMed]
- Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M (2011) The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer 104: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MS, Chen Y, Deng G, Shahryari V, Suh SO, Saini S, Majid S, Liu J, Khatri G, Tanaka Y, Dahiya R (2010) The functional significance of microRNA-145 in prostate cancer. Br J Cancer 103: 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, McLaughlin JK, Chow WH, Chien HT, Blot WJ (1993) Risk factors for cancers of the nasal cavity and paranasal sinuses among white men in the United States. Am J Epidemiol 138: 965–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.