Abstract
Auxin plays a key role in lateral root formation, but the signaling pathway for this process is poorly understood. We show here that NAC1, a new member of the NAC family, is induced by auxin and mediates auxin signaling to promote lateral root development. NAC1 is a transcription activator consisting of an N-terminal conserved NAC-domain that binds to DNA and a C-terminal activation domain. This factor activates the expression of two downstream auxin-responsive genes, DBP and AIR3. Transgenic plants expressing sense or antisense NAC1 cDNA show an increase or reduction of lateral roots, respectively. Finally, TIR1-induced lateral root development is blocked by expression of antisense NAC1 cDNA, and NAC1 overexpression can restore lateral root formation in the auxin-response mutant tir1, indicating that NAC1 acts downstream of TIR1.
Keywords: Auxin, NAC, TIR1, AIR3, lateral root initiation
NACs are a family of genes specific to plants and are found to play a role in a diverse set of developmental processes. The encoded proteins are characterized by highly conserved domains in the N-terminal region, but their C-terminal sequences are divergent in both length and amino acid sequence. As a family, NACs show no definitive sequence homology to any other proteins, and computer-based searches detect no known functional motifs. The first NAC genes described were NAM of petunia (Souer et al. 1996) and CUC2 of Arabidopsis (Aida et al. 1997); both were recovered from screens for mutants defective in the shoot apical meristem (SAM) development. In addition to SAM defects and cotyledon fusions, both nam and cuc2 mutants also displayed fusions and duplications of floral organs. Another Arabidopsis NAC gene, NAP (Sablowski and Meyerowitz 1998), has been isolated as a target of Apetala3 and its role in controlling cell expansion in specific flower organs has been hypothesized. Other NACs have been found to be up-regulated during senescence in nonmeristematic tissues (John et al. 1997); thus, members of the NAC family seem to play different roles in plant development. Because some NAC proteins can bind and activate a 35S promoter in yeast (H. Hirt, unpubl.), a function as transcription factors has been hypothesized (Xie et al. 1999).
The process of lateral root development has been the subject of several studies, but the underlying genetic pathway is still largely undefined (Malamy and Benfey 1997a). Growth of a new lateral root is initiated from a subepidermal layer—the pericycle. Only the pericycle cells adjacent to the protoxylem poles have the capacity to generate lateral root. Environmental factors, such as the availability of water and nutrient, determine which of these properly positioned pericycle cells will form a new root. A considerable body of evidence has indicated that auxin plays a pivotal role in this process (for review, see Malamy and Benfey 1997b). Screens for mutants with lateral root formation resistant to the auxin signal have uncovered the involvement of several mediating genes. Only a few of these, however, have been cloned, and for most, the biological function of the protein products remains unknown. One exception comes from the molecular analysis of the auxin-resistant mutants tir1 (Gray et al. 1999) and axr1 (Leyser et al. 1993), whose wild-type gene products have been identified as members of a ubiquitin pathway. It is likely that activation of this pathway allows pericycle cells, otherwise arrested in G2-phase, to reenter the cell cycle. In addition to auxin-resistant mutants, only one other mutant, alf-4, has been found to have a specific defect in lateral root initiation (Celenza et al. 1995); the alf-4 gene remains uncloned. The genes activated downstream of lateral root initiation are even less characterized. One study identified few genes that are induced during lateral root formation, some of which encode products with characteristics of extracellular proteases (Neuteboom et al. 1999a,b).
In this study, we describe NAC1, a NAC family gene that plays an intermediary role in auxin-induced development of lateral roots. Whereas several other studies have shown that NACs participate in patterning of meristematic tissue in the SAM and floral meristem, this is the first report of a NAC influencing the initiation and development of lateral roots.
Results
NAC1 is expressed in root tips, cotyledons, and leaves
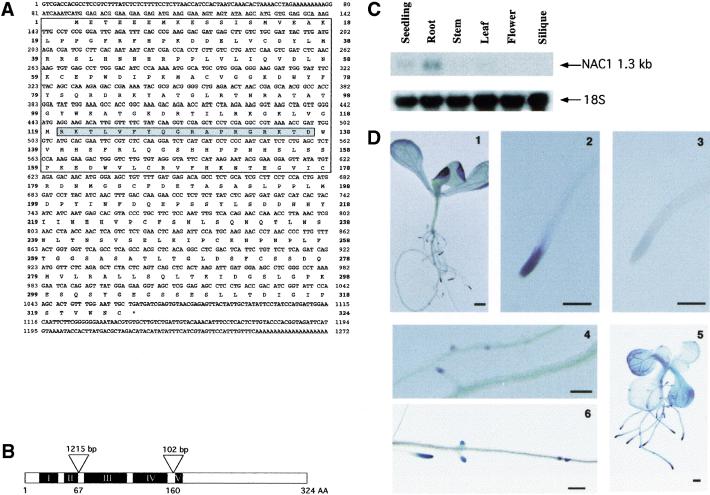
To determine genes of interest, several NACs (available as complete cDNAs from the ESTs stock center) were assayed throughout development by Northern blot and in situ hybridizations. Because these genes have a high degree of sequence conservation in the 5′ coding region, 3′ DNA fragments were used as specific probes. One family member, designated NAC1, displayed a particularly interesting expression pattern. NAC1 was expressed at a moderate level in 2-wk-old seedlings, however, in dissected tissues of mature plants it was expressed at high levels in roots, at low levels in stems and leaves, and not at all in flowers or siliques (Fig. 1C). Whole-mount in situ hybridization of 7–12-d-old seedlings detected three tissues actively expressing the NAC1 gene: the expanding cotyledons, the primordia of the developing leaves, and the root tip and lateral root initiation site (Fig. 1D). The pattern of expression in the root tips and lateral root initiation sites were the most striking. It was observed at a high level in root primordia, yet as the root elongated, NAC1 expression was confined to the meristem and the expansion region (Fig. 1D, panels 1,2,4).
Figure 1.
Nucleotide sequence and expression pattern of the NAC1 gene. (A) Nucleotide and amino acid sequences of NAC1 gene. The highly conserved region of NAC family members is boxed, and the putative bipartite nuclear localization signal is shaded (GenBank accession no. AF198054). (B) A schematic diagram showing the genomic structure of the NAC1 gene (GenBank accession no. AC002304). Open triangles represent introns. The first intron is 1215 nucleotides in length, with its 5′ splice junction at position bp 288 of the cDNA, and the second intron is 102 nucleotides, with its 5′ splice junction at position 568. The five conserved regions within NAC family members are indicated by I–V. AA, amino acid. (C) Organ-specific expression of the NAC1 gene. Seedlings were harvested from 2-wk-old plants. Cauline leaves (leaf), main and lateral stems (stem), flower clusters (flower), and siliques at different stages and maturity (silique) were harvested from 35–40-d-old plants grown in a growth chamber. Roots were taken from 2-wk-old seedlings grown vertically on MS medium. The filter was hybridized with a NAC1 cDNA 3′ fragment encoding the C-terminal region (amino acids 200–324) of NAC1. The 18S rRNA was used as a loading control. (D) Tissue-specific expression of NAC1. Whole-mount in situ hybridization was performed with a digoxigenin-labeled (DIG) NAC1 cDNA 3′ fragment encoding the C-terminal region (amino acids 200–324) of NAC1 (1,2,4) and with a DIG-labeled sense probe (3). NAC1–GUS expression in 10-d-old seedlings (5,6). Scale bar, 0.1 mm.
To confirm these results, a construct containing a 2-kb DNA fragment upstream of the NAC1 coding region was fused to a β-glucuronidase (GUS) reporter gene, and several independent transgenic lines were analyzed. GUS activity was observed in the root primordia and in lateral root initiation points (Fig. 1D, panel 6) and moderate activity was observed in the cotyledons (Fig. 1D, panel 5). In the leaf primordia, a signal, which diminished as the leaf matured, was also observed. This expression pattern correlated well with that observed by in situ hybridizations (Fig. 1D, cf. panels 1,2,4 with 5,6), indicating that NAC1 has a unique expression pattern compared with other NAC genes.
The structure of the NAC1 gene
The NAC1 cDNA is 1272 bp in length and encodes a protein of 324 amino acids (Fig. 1A). The N-terminal 172 residues contain the five conserved blocks of homology that characterize the NAC family. The divergent C terminus of 152 amino acids displays no homology to other known proteins. Computer analysis detected a putative bipartite nuclear localization signal sequence (NLS) between amino acids 120 and 137. Recently, the genomic region containing NAC1 has been sequenced, revealing that it lies on chromosome 1 and has two introns within its coding region (Fig. 1B). Southern blot analysis indicated that NAC1 is a single-copy gene (data not shown).
Nuclear localization of GFP–NAC1 fusion protein
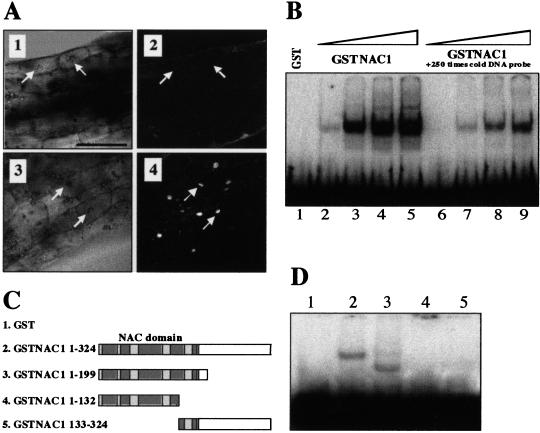
The detection of a putative NLS implies that NAC1 can be localized to the nucleus. To test this, transgenic lines carrying a GFP-NAC1 fusion gene expressed under the control of a dexamethasone-inducible GVG promoter (Aoyama and Chua 1997) were generated. Seeds of several independent transgenic lines were germinated in darkness in the absence or presence of dexamethasone (dex) and the hypocotyl of 7-d-old seedlings examined by confocal microscopy. In the absence of dex, no fluorescence was observed (Fig. 2A, panel 2), however, in the presence of dex, the GFP–NAC1 was localized exclusively to the nucleus (Fig. 2A, panel 4).
Figure 2.
Nuclear localization and DNA binding of NAC1 protein. (A) Nuclear localization of NAC1 protein. Transgenic plants carrying a GFP–NAC1 fusion gene under the control of a dex-inducible promoter (Aoyama and Chua 1997) were germinated on MS medium with 3% sucrose in the absence or presence of dexamethasone in the dark, as described in Materials and Methods. Hypocotyl regions of etiolated seedlings were analyzed by confocal microscopy. (1,2) No dexamethasone; (3,4) with 10 μM dexamethasone; (1,3) transmitted light reference image of hypocotyl cells; (2) no fluorescence was detected; (4) induced fluorescence expression of GFP–NAC1. Arrows indicate the position of nucleus in the cells. (B) Interaction of GST–NAC1 fusion protein with the −90 to +9 fragment of CaMV 35S promoter. Lane 1, 500 ng of GST; lanes 2,6, 10 ng of GST–NAC1; lanes 3,7, 100 ng of GST–NAC1; lanes 4,8, 250 ng of GST–NAC1; lanes 5,9, 500 ng of GST–NAC1. Lanes 6–9 contained 250-fold of unlabeled −90 to +9 fragment of 35S promoter. (C) A schematic diagram showing the different NAC1 protein derivatives used for the DNA gel shift experiment in D. Numbers indicate the amino acid residue at the N and C termini of the various NAC1 derivatives. (D) Interaction of the various NAC1 derivatives with the −90 to +9 fragment of the 35S promoter. (1) GST; (2) GST–NAC1 (1–324); (3) GST–NAC1 (1–199); (4) GST–NAC1 (1–132); (5) GST–NAC1 (133–324). 20 ng of each recombinant protein was used.
NAC1 is a DNA binding protein
Recently published studies suggest that NAC family members, such as NAM, CUC2, and GRAB1/2, may function as transcriptional factors (Aida et al. 1999; Xie et al. 1999). The wheat NACs, GRAB1 and GRAB2, bind to a 500-bp fragment of the 35S promoter (Q. Xie and C. Gutierrez, unpubl.). To test NAC1 for this activity, a recombinant GST–NAC1 fusion protein was expressed in Escherichia coli, purified and assayed for DNA binding. Figure 2B shows that NAC1 was indeed able to bind to the 35S −90 promoter fragment (Fig. 2B, cf. lanes 2–5 with 6–9). The −90 region of the 35S promoter is known to contain an AS1 element (TGACG), a binding site for bZIP transcription factors (Izawa et al. 1993). Therefore, the 20-bp segment (CTGACGTAAGGGATGACGCAC) from −83 to −63 that contains the AS1 element was used as probe and found to be recognized by GST–NAC1 (data not shown). However, when a fragment (CTGctGTA AGGGATGctGCAC) mutated in the AS1 element was tested, the binding was only partially affected (data not shown), whereas no binding activity was detected when the same mutant was tested with ASF-1, a member of bZIP transcription factors (Qin et al. 1994). These studies indicate that although the recognized sequence element is located within the −83 to −63 region, the sequence requirement is different from the AS1 element. Thus, the binding sequence determinants of NAC1 are distinct from those of bZIP transcription factors.
To establish the minimum length of NAC1 required for recognition of the 35S promoter, deletions of NAC1 (Fig. 2C) were tested. The conserved N-terminal 199 residues of the NAC domain did display the interaction (Fig. 2D, lane 3), whereas deletion of an additional 67 amino acids abolished binding activity (Fig. 2D, lane 4). However, these 67 residues alone were not sufficient to bind DNA (Fig. 2D, lane 5). Therefore, a significant portion of the NAC domain is required for DNA binding activity.
The C-terminal domain of NAC1 has transactivation activity
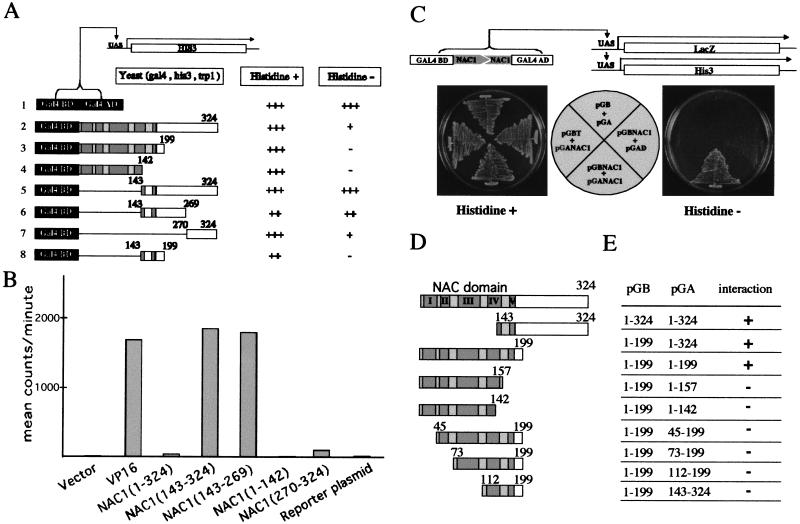
The evidence that NAC1 contains an NLS and a DNA binding domain suggests that NAC1 may be a transcription factor. We therefore tested for the presence of an activation domain in NAC1, first using yeast as an assay system (see Materials and Methods). Here, NAC1 fusions to the GAL4 DNA binding domain were assayed for the ability to activate transcription from the GAL4 upstream activation sequence (UAS) and promote yeast growth in the absence of histidine (Fig. 3A). Although the complete NAC1 protein showed only a modest level of activation capacity (Fig. 3A, row 2), when assayed in the absence of the N terminus, the C-terminal 181–amino-acid fusion protein was a potent activator (Fig. 3A, row 5). Deletion analysis of this C-terminal domain showed that amino acids 270–324 exhibited only a moderate level of activity (Fig. 3A, row 7), and residues 143–269 promoted somewhat more growth in the absence of histidine, yet not as robust growth as that observed with the intact C terminus (Fig. 3A, row 6). However, growth promoted by the 143–269 fragment cannot be directly compared to the others, as its expression had a deleterious effect on the yeast even in the absence of selective pressure (Fig. 3A, row 6).
Figure 3.
Localization of the transactivation and the dimerization domain of NAC1. (A) Transactivation analysis of NAC1 in yeast. NAC1 cDNA fragments encoding different portions of NAC1 were fused to DNA sequences encoding the Gal4 DNA binding domain in the yeast vector pGBT8 and transformed into yeast strain HF7c. The transformation mixture was plated on MM plates with or without histidine and the necessary amino acids. The plates were incubated for 3 d, and the results were scored. (B) Transactivation domain analysis of NAC1 in vivo. NAC1 cDNA fragments encoding different portions of NAC1 were cloned into the plasmid pGal and cobombarded into onion skin cells with the reporter gene plasmid pTALuc. Luciferase activities were measured by a highly sensitive camera CCS system-ST138S to determine luciferase bioluminscence (Millar et al. 1992). (C) Dimerization of NAC1 analyzed by yeast two-hybrid assay. Yeast HF7c cells were cotransformed with plasmids expressing the GAL4BD alone or GAL4BD–NAC1 fusion and the plasmids expressing the GAL4AD alone or fused to NAC1. Cells were streaked on plates with or without histidine plus 10 mM 3-AT (Sigma) according to the distribution shown in the center of picture. The ability to grow in the absence of histidine depends on the functional reconstitution of a GAL4 activity. The plates were incubated for 3 d, and the results were scored. (D) A schematic diagram showing the different NAC1 protein derivatives used for analysis of the NAC1 dimerization domain in yeast two-hybrid system. (E) Interaction between the different NAC1 derivatives. All conditions used were the same as indicated in C.
To further test the ability of the NAC1 fragments to activate transcription in plant cells, we performed similar studies using onion epidermal cells as an assay system and luciferase as a reporter enzyme (see Materials and Methods). The results confirmed and extended those described in the yeast-based experiments (Fig. 3B). Again, the full-length fusion NAC1 only displayed a moderate capacity to activate the reporter gene, yet the activity of the C-terminal half was comparable to that conferred by the VP16 activation domain. Interestingly, in onion cells we found that the 143–269 region had ninefold more activity than the 270–324 fragment, showing that the former fragment contained the most potent activation domain.
Our experiments identified a transcriptional activation domain in the NAC1 C terminus using two independent systems. These data, together with those showing DNA binding activity and the presence of an NLS that localizes the protein exclusively to the nucleus, provide strong evidence that NAC1 is a transcriptional activator.
The dimerization function of the NAC domain
We tested the ability of NAC1 to dimerize in a yeast two-hybrid assay. The presence of the NAC1-GAL4 activation domain fusion protein and the NAC1–GAL4 DNA binding domain fusion protein resulted in significantly more growth than the autoactivation seen with the latter alone (Fig. 3C), suggesting that NAC1 can indeed dimerize. To define the dimerization domain, NAC1 deletions were fused to the GAL4 activation domain and assayed for interaction with the 199-amino-acid N-terminal peptide of NAC1 fused to the GAL4 DNA binding domain (Fig. 3D). The NAC1 activation domain could not be tested for a possible dimerization function because it would autoactivate the system at a low level. Here, the conserved N-terminal NAC domain was able to dimerize; however, removal of ∼45 amino acids from both ends abolished the interaction (Fig. 3E). These data show that the NAC1 dimerization activity is located within the NAC domain and that more than one region of this domain participates in the association.
NAC1 overexpression affects plant size and lateral root development
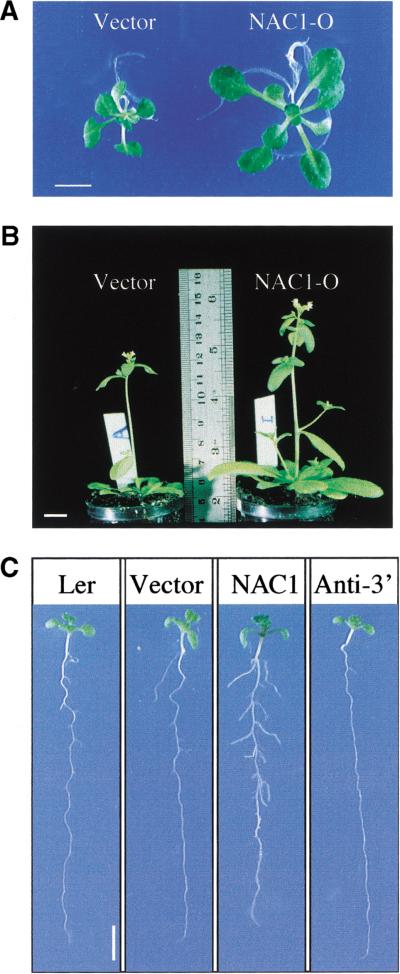
To investigate the biological function of NAC1, several independent transgenic lines overexpressing the NAC1 cDNA under the control of either the 35S or a dex-inducible promoter (Aoyama and Chua 1997) were analyzed. 35S::NAC1 overexpressing lines, either grown on plates or in soil, were bigger, with larger leaves, thicker stems, and more abundant roots compared with control plants (Fig. 4A,B, cf. transgenic control containing an empty vector with lines carrying the 35S::NAC1 transgene). Consequently, an increase in the total fresh weight, ranging from 1.6- to 2.6-fold, was observed in the overexpressing lines, though the dry weight increased only 1.2-fold with respect to control in 25-d-old seedlings (data not shown). Scanning electron microscopy analysis (SEM) showed that cells in older leaves of the 35S::NAC1 plants were significantly larger (data not shown). On vertically oriented plates it could be seen clearly that the greater root mass resulted from an increase in abundance and length of lateral roots (Fig. 4C). Similar results were obtained when Dex-inducible NAC1 transgenic plants were grown on induction medium with respect to uninduced plants (data not shown).
Figure 4.
Phenotypes of transgenic plants WT and transgenic plants were either germinated on MS medium with 3% sucrose or transferred to soil and grown in a growth chamber. (A) Constitutive overexpression of the NAC1 gene produces bigger plants. Transgenic plants carrying a 35S::NAC1 transgene (NAC1-O) and transgenic plants carrying an empty vector (Vector) were germinated and grown on MS agar medium with 3% sucrose for 25 d. Scale bar, 0.5 cm. (B) Soil-grown NAC1 transgenic plants also show an overall bigger size. Transgenic plants carrying a 35S::NAC1 transgene (NAC1-O) and vector control (Vector) plants grown in soil for 35 d. Scale bar, 1 cm. (C) Effects of over- and underexpression of NAC1 on lateral root development. Wild-type landsberg ecotype (Ler) and transgenic plants carrying an empty vector (Vector), a 35S::NAC1 transgene (NAC1), and a 35S-antisense 3′ NAC1 (Anti-3′) were grown on MS agar medium with 3% sucrose. Plants were photographed after 14 d. Scale bar, 0.5 cm.
Reduced lateral roots in NAC1 antisense plants
35S::NAC1 antisense lines were generated by expressing a 600-bp 3′ fragment of NAC1 cDNA in the antisense direction, encoding the NAC1 C terminus divergent region and the 3′ UTR. This antisense construct is specific for NAC1 gene, as a single band was detected in both Southern and Northern blot hybridizations. Also, this 600-bp DNA fragment showed no significant sequence homology with any other sequences in the Arabidopsis genome database.
Four independent transgenic lines, which showed a reduction in the endogenous level of NAC1 transcript levels, were analyzed (Fig. 6C, one representative line, see below). One-week-old seedlings displayed no clear phenotype either in the aerial part or in the root. However, at ∼10 d, a significant reduction in lateral root emergence was observed, although once the few lateral roots formed, there was neither a difference in the morphology nor length compared with wild-type lateral roots (Fig. 4, anti-3′ panel).
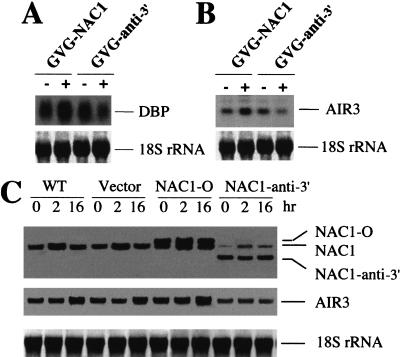
Figure 6.
Northern analysis of DBP, AIR3, and NAC1 gene expression in the roots of transgenic plants. (A) Expression of DBP in roots of GVG-transgenic plants. A 300-bp DNA fragment encoding the C terminus of DBP isolated from differential display was used as a probe. GVG–NAC1, transgenic GVG–NAC1 overexpression plants; GVG–anti-3′, transgenic GVG–NAC1 expressing dex-regulated 3′-specific NAC1 antisense. (B) Expression of AIR3 in roots of GVG-transgenic plants. A 280-bp DNA fragment encoding the C terminus of AIR3 isolated from differential display was used as probe. (C) Northern blot analysis of NAC1 and AIR3 gene expression in auxin-treated root sample of transgenic plants. Roots were separated from the appropriate transgenic seedlings and cultured as described in Materials and Methods. 1-NAA was added to the cultures at a final concentration of 2 μM, and samples were taken at different time for RNA preparation. The probe for NAC1 is the same as Figure 1C. The AIR3 probe is the same as used in panel B. WT, untransformed wild-type plant; Vector, transgenic plants carrying the empty vector; NAC1-O, transgenic plants with the 35S::NAC1 transgene; NAC1-anti-3′, transgenic plants carrying 35S::anti-3′ NAC1.
NAC1 is involved in the auxin-signaling pathway
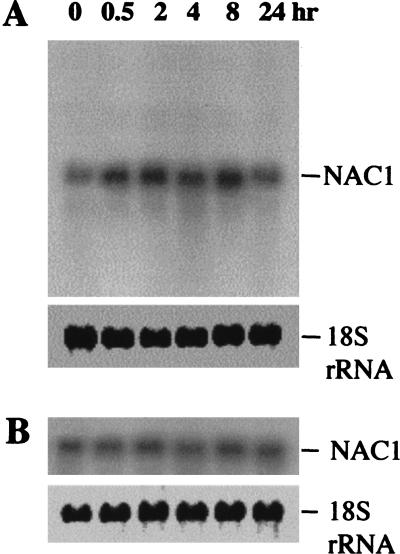
Several lines of evidence indicate that auxin plays a central role in lateral root initiation (Malamy and Benfey 1997b). To investigate whether NAC1 is involved in this pathway, we tested its expression in response to auxin treatment. Wild-type root cultures were treated with auxin at different times, and samples were analyzed by Northern blot hybridizations. An increase of NAC1 transcript level was indeed detected after only 30 min of auxin exposure and reached a maximum level after 2 h. The induction level was maintained for 8 h, whereas the steady-state expression was recovered after 24 h (Fig. 5, cf. A with untreated samples in panel B). This expression kinetics indicates that NAC1 may be an early auxin-responsive gene. Consistent with this notion, we found two TGTCTC elements, identified as early auxin-response elements (Ulmasov et al. 1997, 1999), at position −1397 to −1392 and −508 to −503 in the NAC1 upstream region.
Figure 5.
Induction of NAC1 gene expression by auxin. Roots were separated from wild-type seedlings and cultured as described in Materials and Methods. The active form of auxin, 1-NAA 2 μM was added to the culture, and samples were taken at different times for RNA preparation. The filter was hybridized with a NAC1 cDNA fragment encoding the NAC1 C-terminal region (amino acids 200–324). The 18S rRNA was used as a loading control. (A) 1-NAA dissolved in 50% ethanol was added to a final concentration of 2 μM. (B) As a control, the same volume of 50% ethanol was added.
We next compared root induction by auxin in wild-type and the different NAC1 transgenic lines. Plants were exposed to active auxin (1-NAA), inactive auxin (2-NAA), and antiauxin (PCIB), and the formation of lateral roots was scored (Table 1). We found that the 35S::NAC1 transgenic lines developed more lateral roots than wild-type control plants either in the presence or the absence of active auxin in the medium. Thus, NAC1 and auxin may have a synergistic effect on lateral root initiation process. Strikingly, the inductive effect of auxin was significantly inhibited in 35S::anti-3′ NAC1 transgenic lines.
Table 1.
Effects of auxin and antiauxin treatment on lateral root initiation in root cultures
| Plantsa
|
Treatmentb
|
Fold increase in the number of lateral roots
|
|||
|---|---|---|---|---|---|
| H2O
|
1-NAA
|
2-NAA
|
PICB
|
||
| WT | 1.3c | 16.8 | 1.4 | 1.2 | |
| Vector | 1.2 | 17.2 | 1.5 | 1.3 | |
| 35S-NAC1 | 4.7 | 24 | 5.2 | 4.4 | |
| 35S-anti-3′ NAC1 | 1.2 | 7.9 | 1.3 | 1.2 | |
WT, wild-type Landsberg; vector, transgenic line carrying the empty vector only; 35S-NAC1, transgenic line carrying 35S::NAC1 transgene; 35S-anti-3′ NAC1, 35S-antisense 3′ specific NAC1. The numbers of lateral roots per 30 root segments at the start of the treatments were: WT, 14; vector, 13; 35S-NAC1, 23; and 35S-anti-3′ NAC1, 11.
For details of hormone treatment, see Experimental Procedure.
Number refers to the fold increase in the number of lateral roots 48 h after the treatment compared with the number of lateral roots at 0 h. Data were obtained by counting all root tips in two independent treatments.
NAC1 regulates the expression of two auxin-responsive genes
Our data strongly indicate that NAC1 is a transcription activator. To identify genes downstream of NAC1, we used differential display (Fig. 6A) to compare the transcript expression profiles of transgenic lines in which NAC1 was overexpressed or silenced using the GVG dex-inducible system (Aoyama and Chua 1997). Among the several candidates isolated, two transcripts, AIR3 and DBP, showed an increase in the overexpression lines as well as a reduction in the antisense lines. Northern blot analysis confirmed the increased expression of both target genes in the NAC1-overexpressing lines and their reduction in NAC1 3′-specific antisense plants when incubated in the dex induction medium (Fig. 6A,B).
DBP encodes a ubiquitously expressed DNA binding protein. Its expression level increases fourfold in tissues undergoing rapid cell division and 10-fold in response to auxin (NAA) treatment (Alliotte et al. 1989). AIR3 encodes a protein with homology to subtilisin-like proteases (Neuteboom et al. 1999a,b). Interestingly, AIR3 was initially isolated in a screen devised to recover genes induced by auxin during lateral root development. AIR3 is a single-copy gene in Arabidopsis, although it belongs to a small gene family. AIR3 transcript is detected in all the plant tissues, but its expression in root increases on auxin (NAA) induction after 4–8 h of treatment (Neuteboom et al. 1999a). Our finding that two auxin-responsive genes are up-regulated by an increase in NAC1 expression, together with the results showing that the overexpression of NAC1 promotes lateral root formation, strongly suggest that NAC1 mediates the auxin signal in this developmental process.
NAC1 can induce AIR3 gene expression in vivo
Because AIR3 has been shown to be up-regulated during lateral root development (Neuteboom et al. 1999a) and in response to NAC1 overexpression, it is reasonable to hypothesize that NAC1 may regulate AIR3 gene expression in this developmental process. To test this hypothesis, we analyzed the kinetics of AIR3 and NAC1 expression in response to auxin treatment in wild type and 35S::NAC1 and 35S::anti-3′ NAC1 transgenic (Fig. 6C). As expected, auxin treatment increased both NAC1 and AIR3 expression levels in wild-type plants but with different kinetics. NAC1 was induced earlier than AIR3, whose transcript increased only after 16 h (Fig. 6C, WT). When 35S::NAC1 lines were analyzed, higher AIR3 transcript levels were observed even in the absence of auxin (Fig. 6C, NAC1-O). Nevertheless, AIR3 expression still increased on auxin exposure in the transgenic plants (Fig. 6C, NAC1-O), consistent with the synergistic effect of NAC1 overexpression and auxin treatment observed in lateral root induction. Finally, in transgenic 35S::anti-3′ NAC1 plants with reduced NAC1 expression, AIR3 transcript was poorly or not induced at all in response to auxin, further confirming the results observed in the root-development assay (Fig. 6C, NAC1-anti3′).
To check whether NAC1 can activate AIR3 transcription, different combinations of plasmids harboring AIR3–GUS fusion, 35S::NAC1, or 35S::GUS were used to bombard onion peels. Table 2 shows the results obtained from three independent experiments. Whereas AIR3–GUS plasmid alone did not produce any GUS-positive cells, co-bombardment with the 35S::NAC1 construct resulted in a large number of cells expressing GUS. It should be noted that a lighter blue staining was observed when compared to cells expressing 35S::GUS, which was used as a positive control. As a negative control, we also examined the effect of 35S::NAC1 on LAT52::GUS activity; the latter is known to confer pollen cells specific expression (Twell et al. 1990). No GUS-positive cells were detected (data not shown), thus demonstrating a specific activation of AIR3 promoter by NAC1. When the same bombardment experiments were repeated using tobacco BY2 cells as an in vivo expression system, similar results were obtained (data not shown). Taken together, our results indicate that NAC1 can function as an intermediate between auxin and AIR3-mediated lateral root development.
Table 2.
Effect of NAC1 expression on AIR3 promoter activity in vivo
| Plasmid
|
GFP-positive cells number
|
GUS-positive cells number
|
|---|---|---|
| pPS022+pAIR3=pGFP2a | 176 | 0 |
| 153 | 1 | |
| 190 | 0 | |
| pPSNAC1+pAIR3+pGFP2b | 159 | 101 |
| 124 | 91 | |
| 179 | 117 | |
| pPS022+pBI121+pGFP2c | 162 | 146 |
| 127 | 104 | |
| 218 | 212 |
Numbers of positive cells from three independent experiments are given.
pGFP2, 1 μg; pPS022, 3 μg; pAIR3, 2 μg. The molar ratio of pPS022 to pAIR3 was 5 : 1.
pGFP2, 1 μg; pPSNAC1, 4 μg; pAIR3, 2 μg. The molar ratio of pPSNAC1 to pAIR3 was 5 : 1.
pGFP2, 1 μg; pPS022, 3 μg; pBI121, 2 μg. The molar ratio of pPS022 to pBI121 was 5 : 1.
NAC1 overexpression restores lateral root formation in tir1 mutant
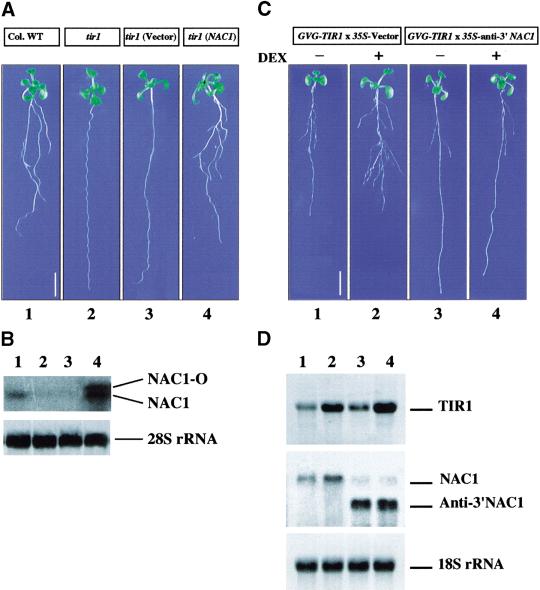
To study the site of action of NAC1 in the auxin signaling pathway leading to lateral root development, we introduced the 35S::NAC1 transgene (NAC1-O) into the Arabidopsis auxin-response mutant tir1. The tir1 mutant is deficient in a variety of auxin-regulated growth processes, including hypocotyl elongation and lateral root formation (Ruegger et al. 1998). TIR1 encodes the Transport Inhibitor Response 1 protein, which contains a series of leucine-rich repeats and a recently identified motif called F box. Sequence comparisons indicate that TIR1 is related to the yeast protein Grr1p and the human protein SKP2, which are involved in ubiquitin-mediated proteolysis processes. The overexpression of TIR1 promotes auxin response, suggesting that this protein is a limiting factor for appropriate hormonal response (Gray et al. 1999). Figure 7A shows that overexpression of NAC1 in tir1 can rescue the deficiency in lateral root formation, giving a root phenotype similar to that of the wild type (Table 3), whereas transgenic tir1 plants, carrying an empty vector, continued to display the mutant phenotype. Northern blot analysis showed that, in comparison to WT control, the NAC1 gene expression level is very much reduced in the tir1 mutant as well as in transgenic tir1 carrying an empty vector (Fig. 7B). Interestingly, overexpression of NAC1 in tir1 mutant plants restored the expression of the endogenous NAC1, suggesting that NAC1 could activate its own transcription.
Figure 7.
NAC1 acts downstream of TIR1 to promote lateral root development. (A) Wild-type Columbia ecotype (Col) plant (lane 1), tir1 mutant (lane 2), transgenic tir1 plants carrying an empty vector (lane 3), and transgenic tir1 plants carrying a 35S::NAC1 transgene (lane 4) were germinated and grown on MS agar medium with 3% sucrose. Plants were photographed after 12 d. Scale bar, 0.5 cm. (B) Northern blot analysis of NAC1 gene expression. RNA samples were collected from 2-wk-old seedlings. The filter was hybridized with a NAC1 cDNA fragment encoding the C-terminal region (amino acids 200–324) of NAC1. The 28S rRNA was used as a loading control. Lanes 1–4 were samples extracted from plants shown in lanes 1,2,3,4, respectively, in panel A. (C) Promotion of lateral root development by TIR1 is blocked by a NAC1 antisense transgene. A homozygous line carrying a GVG–TIR1 transgene (Gray et al. 1999) was crossed with a homozygous line carrying an empty vector control or a 35S-anti-3′ NAC1 transgene. F1 progeny from the cross was used in the experiments. Seven-day-old seedlings of GVG–TIR1 × 35S-vector and GV–TIR1 × 35S-anti-3′ NAC1 were incubated on MS or MS +30 μM DEX for 2 d and then transferred back to MS medium for an additional day. Plants were photographed (10 d old). Scale bar, 0.5 cm. The average number of lateral roots per seedling for GVG–TIR1 × 35S-vector (−Dex, lane 1) was 5.8; for GVG–TIR1 × 35S-vector (+Dex, lane 2) it was 9.8; for GVG–TIR1 × 35S-anti-3′ NAC1 (−DEX, lane 3) it was 2.2; and for GVG–TIR1 × 35S-anti-3′ NAC1 (+DEX, lane 4) it was 2.4. For each line, lateral roots from 30 seedlings were counted. (D) Northern blot analysis of TIR1 and NAC1 gene expression. RNA samples extracted from plants shown in panel C were used. A 1785-bp fragment covering the coding sequence of TIR1 was amplified by PCR and used as a probe. The probe for NAC1 was the same as that used in panel B. Lanes 1–4 were samples extracted from plants shown in lanes 1, 2, 3, and 4, respectively, in panel C.
Table 3.
Number of lateral roots in 12-d-old tir1 mutant and tir1-NACl transgenic seedlings
Col, Columbia wild type; tir1-vector, tir1 carrying empty vector; tir1-NAC1, tir1-NAC1 overexpressing plants.
Average number of lateral roots per seedling; for each line, lateral roots from 30 seedlings were counted.
To further investigate the role of NAC1 in the auxin signaling controlled by TIR1, transgenic plants that overexpress TIR1 under the control of a dexamethasone (dex)-inducible promoter (Aoyama and Chua 1997; Gray el al. 1999) were crossed with 35S::anti-3′ NAC1 transgenic plants, and F1 progeny was analyzed. Figure 7C–D shows that dex-inducible TIR1 overexpression resulted in an increase in NAC1 transcript level as well as in the number of lateral roots. However, these promoting effects of TIR1 were blocked by the 35S::anti-3′ NAC1 transgene.
The finding that NAC1 expression can rescue the tir1 mutant and that the promotion of lateral root formation by TIR1 can be blocked by antisense-3′ NAC1 places NAC1 downstream of TIR1 in transmitting the auxin signal to the AIR3 gene to promote lateral root development.
Discussion
NAC1 characterization identifies a novel family of transcription factors unique to plants
We report the isolation and characterization of NAC1, a new member of the NAC-domain gene family and the first NAC gene shown to be involved in root development. We demonstrate that a functional nuclear localization signal (NLS) and the DNA binding and dimerization domain of NAC1 are localized at the highly conserved N-terminal region that defines the NAC-domain family members. These findings confirm and extend the previous suggestion (Xie et al. 1999) that NAC-domain genes encode a novel family of transcription factors unique to plants. Moreover, the NAC1 C-terminal domain can function as a potent activator both in yeast and in plant, suggesting it has a role in the activation of downstream target genes in tissues in which NAC1 is present and/or induced.
NAC1 plays an important role in plant root development
In contrast to the other members of the family, whose expression is restricted to shoot meristem and flower (NAM, CUC2, NAP) or to vascular tissues (CmNACP), NAC1 is the first NAC family gene expressed in roots. Although some expression can be detected also in leaf primordia or in expanding cotyledons, the highest NAC1 expression is restricted to the root tip (meristem and elongation zone) and the lateral root initiation regions, where actively dividing cells are present. The phenotype of the transgenic lines, in which NAC1 is over- or under-expressed, is consistent with a main function of NAC1 in the root, as lateral root development is affected. Lateral roots originate from a small number of pericycle cells in the differentiated region of the primary root, which resume the ability to divide. The placement of lateral roots is not predetermined but it is strongly influenced by endogenous auxin levels and exogenous environmental conditions.
Overexpression of NAC1 results in a higher number of lateral roots than the control, whereas NAC1 antisense expression reduces lateral root initiation in the absence of any environmental change. However, once the lateral roots initiate in transgenic plants, no alterations in the further root development occurs. Thus, NAC1 is specifically involved in the control of lateral root initiation. Nonetheless, we cannot exclude that NAC1 may play additional roles in other aspects of development or in aerial parts of the plant. Attempts to isolate knockout mutations in NAC1 gene were unsuccessful, and NAC1 antisense lines show reduced but not null expression of the gene. This could account for a lack of clear additional phenotypes in antisense plants so that further studies are needed to fully evaluate the function of NAC1 in plant development.
NAC1 mediates auxin signaling to promote lateral root development
Very little is known about how lateral root placement is regulated and about the signals and mechanisms that direct lateral root formation. There is strong evidence for an involvement of an auxin-signaling pathway in this organogenesis process. In Arabidopsis, the application of a high concentration of exogenous auxin triggers nearly all of the pericycle cells adjacent to protoxylem poles to become part of a lateral root primordium (Boerjan et al. 1995; Laskowski et al. 1995). Mutants that accumulate high endogenous levels of auxin show an increase in lateral root formation (Boerjan et al. 1995; King et al. 1995), whereas mutants resistant to exogenous auxin application display drastically reduced numbers of lateral roots (Hobbie and Estelle 1995; Timpte et al. 1995). Moreover, inhibitors of auxin polar transport or mutations that affect auxin transport also block lateral root formation (Ruegger et al. 1997).
A reduction of NAC1 levels affects auxin response in roots, as auxin treatment is poorly effective in triggering lateral root formation in NAC1 antisense plants. In addition, in the NAC1 overexpressing plants, lateral root formation was not blocked by the antiauxin (PICB), suggesting that NAC1 functions downstream of the auxin signal cascade. The position of NAC1 in the auxin-response pathway is also reinforced by the finding that transgenic lines that overexpress NAC1 develop more lateral roots, even in the absence of any exogenous auxin, and the NAC1 gene itself is induced by auxin as early as 30 min after exposure to the hormone. The identification of two auxin-responsive genes, DBP and AIR3, whose transcripts are up-regulated in response to the NAC1 induction in transgenic plants, provides a hint on the possible mechanism by which NAC1 may transduce auxin signal to promote lateral root formation. DBP encodes a lysine-rich DNA binding protein, and its expression is enhanced in actively dividing tissues and in response to auxin, whereas AIR3 encodes a subtilisin-like protease that is thought to weaken cell-to-cell connections to facilitate lateral root emergence. We have demonstrated that NAC1 can activate AIR3 promoter in vivo, although we do not know whether this effect is direct or indirect via an intermediate protein, such as DBP. In vitro binding experiments of NAC1 to the AIR3 promoter region will be needed to address this question.
Several auxin-resistant mutants, axr1, axr4, and aux1, as well as tir (transport inhibitor resistant), have drastically reduced numbers of lateral roots (Hobbie and Estelle 1995; Timpte et al. 1995; Ruegger et al. 1997). One of the mutants, tir1, has been shown to be affected in auxin response rather than auxin transport (Ruegger et al. 1998); consequently, the defect in lateral root initiation displayed by this mutant depends on a reduced sensitivity to auxin and not on an altered distribution.
TIR1 is an F-box protein presumably involved in ubiquitin-mediated proteolysis of regulatory proteins required for auxin response. Here, we show that NAC1 overexpression can rescue lateral root initiation and growth in tir1 mutant, thus indicating that NAC1 acts downstream of TIR1 and plays a key role in the lateral root initiation process. As NAC1 transcript level is greatly reduced in tir1 with respect to wild type, TIR1 likely regulates NAC1 at the transcriptional level, perhaps through the auxin-dependent degradation of a negative regulator of NAC1. Moreover, the strong decrease in lateral root formation displayed by anti-3′ NAC1 transgenic plants could not be rescued by the controlled overexpression of TIR1. These results, taken together, establish a definite hierarchy in the auxin pathway in which TIR1 promotes the auxin response in roots through the regulation of NAC1, which in turn, controls the expression of auxin-responsive genes involved in lateral root initiation.
With respect to lateral root development, we note that NAC1 overexpression is not as efficient as auxin treatment and that there is, in fact, a synergistic effect between the two (Table 1). This is perhaps not surprising if an auxin-dependent posttranslational modification of NAC1 is needed for activity; in this case, overexpression of unmodified NAC1 cannot fully supplant the activity of the modified form.
Our data identify NAC1 as a new regulatory factor in the auxin-signaling pathway that activates genes encoding molecules involved in the specification of lateral root placement and initiation. Further studies will be needed to identify other components of the auxin pathway leading to lateral root organogenesis and to further investigate the relationship between the TIR1/SCF complex and NAC1. Also, isolation of NAC1 knockout mutants would be useful to investigate whether NAC1 can mediate the auxin-signaling pathway in other aspects of plant development.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana Landsberg ecotype was used for this study. Seeds were surface sterilized with 20% bleach and 0.01% Triton X-100 and washed three times with sterile water. Sterile seeds were suspended in 0.15% agarose and plated on MS or MS plus 3% sucrose, with varying concentrations of dexamethasone (dex). Plates were vernalized in darkness for 2 d at 4°C and then transferred to a tissue culture room at 22°C under a 16-h-light/8-h-dark photoperiod. After 2–3 wk, seedlings were potted in soil and placed in a growth chamber at 22°C and 75% humidity under a 16-h-light/8-h-dark photoperiod.
Dex (Sigma) was brought to 100 mM in DMSO and stored at −20°C. For growth on plates, dex was added to medium at 0.1, 1, and 10 μM and weekly reapplied to the surface of the medium in 1/25 volume of sterile water. For growth in soil, a sterile solution of 0.01% w/v Triton X-100, 30 μM dex was sprayed on the surface of all aerial portions of the plant every other day. As a control, plants were sprayed with 0.01% Triton X-100.
Transformation vectors and construction of transgenic plants
Transgenic plants carrying both dex-inducible and constitutively expressing transgenes were generated. For constitutive expression, we used the vector pVIP96 (van der Krol and Chua 1993), in which transgene expression is under the control of the CaMV 35S promoter. The plasmid pTA7002, containing the glucocorticoid-inducible system (Aoyoma and Chua 1997), was used to generate dex-inducible transgenic lines. For lines overexpressing NAC1, a 1287-bp SalI–NotI fragment containing the NAC1 cDNA was cloned into the appropriate vector. Antisense-expressing constructs were made by using a 605-bp BamHI–NotI fragment encoding the NAC1 C terminus. All ligations were blunt, and overhanging ends were filled in with the Klenow fragment of DNA polymerase I (Boeringher). Transgenic plants were generated via root transformation (Valvekens et al. 1988). T2 seeds were germinated on MS plates containing 20 μg/mL hygromycin B for pTA7002 constructs and 50 μg/mL kanamycin A for pVIP96 constructs, and resistant plants were transferred to soil to obtain homozygous T3 seeds. Two independent lines of homozygous T4 plants containing a single insertion of each construct were used for detailed analysis.
Gene expression analysis
For Northern blot hybridization, RNA was isolated using the QIAGEN RNeasy Kit, and 10 μg of total RNA was loaded per lane. To specifically detect the NAC1 transcript, blots were probed with a PCR fragment encoding the NAC1 C-terminal domain labeled with [α-32P]dCTP using a Ready-primed labeling kit (Amersham International).
Whole plants or flowers were fixed for whole-mount in situ hybridizations as described by Engler et al. (1994). RNA complementary to that encoding the NAC1 C-terminal domain was synthesized by T7 RNA polymerase (Promega) and labeled by incorporating digoxigenin UTP (Boehringer Roche). Hybridization of the probe to the NAC1 transcript was detected by an alkaline phosphatase–conjugated antidigoxigenin Fab fragment. As a control, the coding strand of the same fragment was used as a probe.
For histochemical localization of GUS activity, seedlings were incubated in a solution containing 50 mM Na-phosphate buffer at pH 7.0, 5 mM K3Fe(CN)6, 5mM K4Fe(CN)6, 0.1% Triton X-100, and 1 mM X-Gluc and incubated at 37°C for several hours. Bombarded cells were vacuum treated for 5 min before the staining.
For differential display, the same amount of total RNA, isolated from dex-treated and untreated root samples, was used. Roots were separated from 10-d-old seedlings and transferred to a flask containing liquid B5 medium. Root cultures were maintained by transferring the root materials every week. Dex dissolved in DMSO was added to root culture at a final concentration of 10 μM, and DMSO alone served as a control. Root samples were taken 48 h after treatment. RNAimageTM Kit 1 (GenHunter Corporation) was used according to the manufacture's instructions.
Yeast two-hybrid analysis
We used the yeast strain HF7c (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3::GAL4 17mers(×3)-CyC1TATA-LacZ; Feilotter et al. 1994), which contains the LacZ and HIS3 reporter genes. Two-hybrid analysis was performed as described previously (Bartel et al. 1993a). To corroborate the interaction between the two fusion proteins, β-galactosidase activity was assayed on replica filters (Bartel et al. 1993b).
Using the vector pGBT9 (Clontech), sequences encoding the Gal4 DNA binding domain were fused to either the full-length NAC1 open reading frame or to a 5′ fragment of NAC1 encoding the first 199 amino acids to give pGBNAC1.1-324 and pGBNAC1.1-199, respectively. Full-length NAC1 cDNA and several truncations were fused to sequences encoding the Gal4 activation domain vector pGAD424: pGANAC1.1-324, which contains the complete NAC1 open reading frame; pGANAC1.1-199 and pGANAC1.1-142, which express the first 199 and 142 amino acids of NAC1, respectively; and pGANAC1.143-324, which expresses a NAC1 truncation from amino acid 143 to the C terminus of the polypeptide.
Transient expression in onion epidermal cells
Inner onion peels (2 × 2 cm2) were placed on a filter soaked with liquid MS medium and bombarded with 2 μg of each plasmid under a vacuum of 27 in of Hg and a helium pressure of 1100 psi. After bombardment, plates containing the bombarded tissues were incubated in a growth chamber for 16 h and then sprayed with 50 mM luciferase (Promega). Luciferase activity was monitored after 10 min incubation in the dark (Aoyama and Chua 1997).
The AIR3 promoter–GUS reporter plasmid pAIR3 was constructed by cloning a PCR-amplified 1.5-kb fragment containing AIR3 5′ sequences (−1535 to −1), into the GUS-reporter plasmid vector pBI101.1 (Clontech). For transexpression of NAC1, the 1.3-kb fragment of NAC1 cDNA was cloned into the vector pPS022 (P. Spielhofer and N.H. Chua, unpubl.), a pUC19-based vector that contains a polylinker between the CaMV 35S promoter and the NOS terminator sequence, to produce the plasmid pPSNAC1. The plasmid pBI121.1 (Clontech), which expresses 35S::GUS, was used as a positive control. As an internal control, we used the plasmid pGFP-2 (Kost et al. 1998), which contains a 35S::GFP gene. After bombardment, onion peels were incubated with liquid MS medium for 16 h, GFP was checked, and GUS staining was preformed.
Production of GST-fusion proteins and DNA binding
pGEX-4–2T (Pharmacia) was used to construct plasmids expressing GST-tagged fusions of NAC1, and the resulting constructs were transformed into E. coli. Single colonies were selected, and cells were grown to an optical density at A600 of 0.6–0.9. After induction with 0.4 mM IPTG at 30°C for 4 h, cells were collected by centrifugation, washed once with PBS, and resuspended in a GST binding buffer (50 mM Tris-HCl at pH 8.0; 200 mM NaCl; 1 mM EDTA; 1% Triton X-100; 10 mM β-mercaptoethanol; 5 μg/mL each of leupeptin, pepstatin, and aprotinin; and 1 mM PMSF). Cells were lysed on ice by sonication (5 × 30 sec), and lysates were clarified by centrifugation (15 min at 12,000 rpm). The GST fusion protein was recovered by affinity chromatography on glutathione-Sepharose beads (Pharmacia).
DNA binding reactions contained 2 × 105 cpm DNA, 5 μL of 4× reaction buffer (100 mM NaCl, 40 mM HEPES at pH 7.5, 2 mM EDTA, 20% glycerol, 140 mM β-mercaptoethanol), 0.5 μg poly dIdC (Pharmacia), and 1 μL 1% Nonidet P40 plus specified concentrations of protein in a final volume of 20 μL. 10 μL of each reaction was resolved on a 4%–6% agarose 0.5× TBE native gel. A 100-bp BamHI fragment containing the 35S −90 region (−90 to +9) was end labeled by [α-32P]dCTP using a Klenow enzyme.
Auxin and inhibitor treatment
Roots from 14-d-old seedlings were cultured in liquid B5 medium under constant shaking in the dark. After 14 d, cultured roots were cut into 1-cm segments and then incubated with the auxin 1-naphthaleneacetic acid (1-NAA), the inactive auxin 2-naphthaleneacetic acid (2-NAA), and the antiauxin p-chlorophenoxyisobutyric acid (PCIB), all at a final concentration of 2 μM. After 48 h, root samples, as well as samples taken before the treatment (0 h), were submerged in 70% ethanol and stained by a fivefold diluted epoxy solution from Electronic Microscopy Science Company. Visible lateral root primordia and lateral roots from 30 root segments were determined under a binocular microscope.
Acknowledgments
We thank Mark Estelle for kindly providing seeds of the tir1 mutant and Col (GVG-TIR1); the ABRC for the ESTs encoding NAC family members; Wei-Ping Tang, EeLing Ng, and Qi-Wen Niu for general technical assistance; and Mathias Zeidler for helpful discussion and comments on the manuscript. This work was supported in part by NIH grant GM 44640 to N.-H.C. G.F. was supported by a fellowship from the Italian National Council of Research (CNR), Comitato di Biotecnologie e Biologia Molecolare.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chua@rockvax.rockefeller.edu; FAX (212) 327-8327.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.852200.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- Alliotte T, Tire C, Engler G, Peleman J, Caplan A, Van Montagu MM, Inze D. An auxin-regulated gene of Arabidopsis thaliana encodes a DNA-binding protein. Plant Physiol. 1989;89:743–752. doi: 10.1104/pp.89.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Bartel PL, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein–protein interactions. In: In: Hartley DA, editor. Cellular interactions in development: A practical approach. Oxford: Oxford University Press; 1993a. pp. 153–179. [Google Scholar]
- ————— Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993b;14:920–924. [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inze D. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes & Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Engler J, van Montagu M, Engler G. Whole-mount in situ hybridization in plants. In: In: Martinez-Zapataer J, Salinas J, editors. Methods in molecular biology. Vol. 82. Totowa: Humana Press; 1994. pp. 373–385. [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D. Construction of an improved host strain for two-hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes & Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 1995;7:211–220. doi: 10.1046/j.1365-313x.1995.7020211.x. [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. Plant bZIP protein DNA binding specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- John I, Hackett R, Cooper W, Drake R, Farrell A, Grierson D. Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol. 1997;33:641–651. doi: 10.1023/a:1005746831643. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. A mutant altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell. 1995;7:2023–2037. doi: 10.1105/tpc.7.12.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997a;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- ————— Down and out in Arabidopsis: The formation of lateral roots. Trends Plant Sci. 1997b;2:390–396. [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuteboom LW, Ng JM, Kuyper M, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ. Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol. 1999a;39:273–287. doi: 10.1023/a:1006104205959. [DOI] [PubMed] [Google Scholar]
- Neuteboom LW, Veth-Tello LM, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ. A novel subtilisin-like protease gene from Arabidopsis thaliana is expressed at sites of lateral root emergence. DNA Res. 1999b;26:13–19. doi: 10.1093/dnares/6.1.13. [DOI] [PubMed] [Google Scholar]
- Qin XF, Holuigue L, Horvath DM, Chua NH. Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell. 1994;6:863–874. doi: 10.1105/tpc.6.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M. Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell. 1997;9:745–757. doi: 10.1105/tpc.9.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray B, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to the human cell cycle protein SKP2. Genes & Dev. 1998;12:505–510. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the flower homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The No Apical Meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The axr1 and aux1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- Twell D, Yamaguch J, McCormick S. Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development. 1990;109:705–713. doi: 10.1242/dev.109.3.705. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcriptional factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- ————— Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Lusbettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol AR, Chua NH. Flower development in petunia. Plant Cell. 1993;5:1195–1203. doi: 10.1105/tpc.5.10.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Guo H, Garcia JA, Gutierrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol Biol. 1999;39:647–656. doi: 10.1023/a:1006138221874. [DOI] [PubMed] [Google Scholar]