Abstract
We have determined the crystal structure, at 3.2 Å, of a ternary complex containing an OCA-B peptide, the Oct-1 POU domain, and an octamer DNA site. The OCA-B peptide binds in the major groove near the center of the octamer site, and its polypeptide backbone forms a pair of hydrogen bonds with the adenine base at position 5 of the octamer DNA. Numerous protein–protein contacts between the OCA-B peptide and the POU domain are also involved in the ternary complex. In particular, the hydrophobic surface from a short α-helix of OCA-B helps to stabilize the complex by binding to a hydrophobic pocket on the POU-specific domain. The structure of this ternary complex is consistent with previous biochemical studies and shows how peptide–DNA and peptide–protein contacts from OCA-B provide structural and functional specificity in the regulation of immunoglobulin transcription.
Keywords: Oct-1, POU domain, OCA-B, octamer, crystal structure, transcription, immunoglobulin, protein–DNA complex, protein–DNA interface
The OCA-B (OBF-1, Bob1) coactivator protein functions in immunoglobulin transcription and B-cell differentiation by forming a complex with the Oct-1 or Oct-2 transcription factors and the octamer DNA sequence (ATGCAAAT) (Luo et al. 1992; Gstaiger et al. 1995; Luo and Roeder 1995; Strubin et al. 1995). OCA-B is expressed almost exclusively in B cells and is essential for high levels of immunoglobulin gene transcription (Luo et al. 1992; Luo and Roeder 1995; Strubin et al. 1995; Gstaiger et al. 1996; Kim et al. 1996; Schubart et al. 1996). Absence of OCA-B causes failures in the immune system, including the elimination of germinal centers, the reduction of some immunoglobulin isotypes in serum, and severe deficiency in B-cell-mediated responses to antigen stimulation (Kim et al. 1996; Schubart et al. 1996). The biological role and promoter specificity of transcription stimulated by OCA-B is correlated with binding data showing that it prefers an A in the fifth position of the octamer and suggesting that this peptide makes direct contact with the DNA (Cepek et al. 1996; Gstaiger et al. 1996). OCA-B will not bind to complexes of Oct-1 or Oct-2 when the octamer element contains a T at position 5. In addition, biochemical and mutagenesis studies of ternary complex formation suggest that OCA-B contacts both the POU-specific domain and the POU homeodomain (Babb et al. 1997; Gstaiger et al. 1996; Luo and Roeder 1995; Sauter and Matthias 1998). We have determined the structure of a ternary complex to help understand the structural basis for combinatorial control and sequence-specific recognition in the regulation of immunoglobulin gene expression.
Results
Previous studies have shown that the amino-terminal portion of OCA-B (residues 1–65 or 1–118 out of 256 residues in the intact protein) make all of the contacts required for stable complex formation with the POU domain and the octamer site (Luo et al. 1992; Gstaiger et al. 1995, 1996; Luo and Roeder 1995; Strubin et al. 1995; Cepek et al. 1996). Although there was no strong homology between the sequence of OCA-B and other sequences in the public databases, we noticed a putative internal repeat in OCA-B that aligns residues 60–130 with residues 131–200. One of the most striking features of this repeat is conservation of the sequence (YTT/VPGPS) at positions 60 and 130 (data not shown). Given this partial sequence repeat, which may indicate some type of domain structure, and the previous deletion analysis by others (Strubin et al. 1995; Cepek et al. 1996; Gstaiger et al. 1996), we first tested complex formation and crystallization trials with peptides containing residues 1–59 and 1–118 of OCA-B. Both peptides formed the ternary complex with high affinity and both preferred A over T at the fifth position of the octamer element (data not shown). Consistent with previous observations, these peptides could bind the octamer element with low affinity even in the absence of the Oct-1 POU domain, and this peptide binding required an A at the fifth position (data not shown; Cepek et al. 1996).
The first crystals of the ternary complex contained the Oct-1 POU domain, an octamer element, and the OCA-B 1–59 peptide, but were hard to grow reliably. We subsequently did binding studies with a series of peptides containing the first 34, 39, 44, 49, 54, or 59 residues of OCA-B to identify shorter peptides that could still form the ternary complex and improve the crystallization. The OCA-B 1–39 peptide was the smallest in this series to form the ternary complex with high affinity (data not shown). However, measurements of ternary complex formation (monitored by gel electrophoresis) suggested that the 1–44 peptide binds the preformed Oct-1 POU domain/octamer element complex with the highest affinity (K. Cepek, V. Wang, and P. Sharp, unpubl.). Binding studies using surface plasmon resonance techniques established that the OCA-B 1–44 peptide binds a preformed Oct-1 POU domain/octamer complex with an equilibrium dissociation constant of ∼20 nm. As observed with the full-length OCA-B protein, stable complex formation with this peptide required an A rather than a T at position five of the octamer element (K. Cepek, D. Chasman, and P. Sharp, unpubl.).
Crystals of a ternary complex with the OCA-B 1–44 peptide were grown and were solved by molecular replacement and multiple isomorphous replacement (MIR) methods. Although the diffraction of the crystals was limited to ∼3.2 Å, the structure solution and all of our key conclusions were confirmed with unbiased omit maps (see Materials and Methods). We find that ∼20 residues of the OCA-B peptide are well-ordered in the crystals and that the structure of the Oct-1 POU domain/octamer DNA portion of the ternary complex is very similar to that reported previously for the binary complex (Klemm et al. 1994). The ordered region of OCA-B begins at residue 16 with a section of extended polypeptide that is near the Oct-1 homeodomain and the sugar phosphate backbone (Figs. 1A, 2A,B, and 3A). A turn at Gly-21 redirects the polypeptide strand so that it traverses the major groove and passes right next to the critical AT base pair at position 5 (Fig. 1B). After crossing the major groove, there is a turn near Pro-26, and the remaining ordered region of OCA-B forms a two-turn α-helix that contacts the POU-specific domain. At the carboxy-terminal end of this helix, a few residues make contacts with symmetry-related complexes in the crystal, and the peptide becomes disordered after Gly-37 or His-38. (There are two complexes in the asymmetric unit, and they differ slightly in how well these terminal residues are ordered.) The three peptide segments nearly encircle the DNA (Figs. 3A and 4), with the POU-specific domain and POU homeodomain binding on opposite sides of the double helix, the OCA-B peptide stretching across the major groove near the center of the octamer site, and the amino-terminal arm of the POU homeodomain fitting into the minor groove near the center of the octamer site (Klemm et al. 1994).
Figure 1.
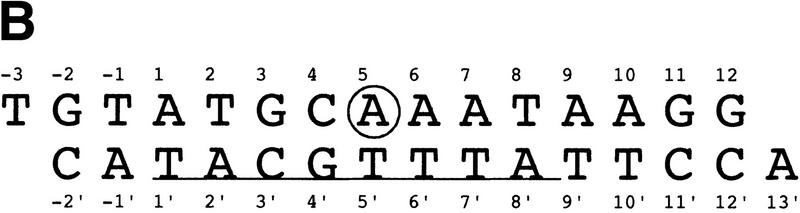
Octamer element and OCA-B 1-44 peptide used in crystallization of the ternary complex. (A) Sequence of the OCA-B 1-44 peptide used for crystallization. The boxed region indicates the portion of the peptide that is ordered in the structure. Lowercase letters below the amino acid sequence refer to elements of secondary structure in the crystal structure. (t) Turn; (H) helix. Boldface type highlights residues Val-22 and Val-24, which make base-specific contacts at the fifth and sixth positions of the octamer element. (B) Sequence of the octamer element and the oligonucleotide used for crystallization. The octamer site is underlined, and the A at position 5 of this site is circled.
Figure 2.
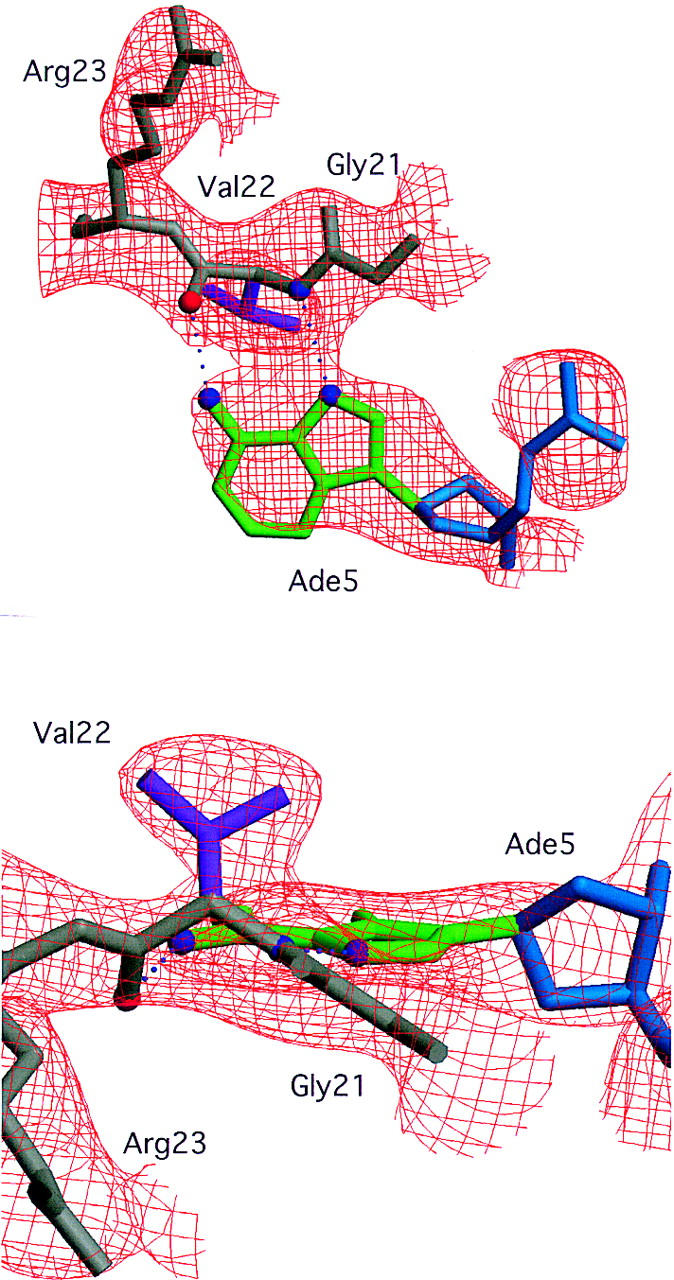
Electron density for the interaction between the adenine at the fifth position of the octamer element and Val-22. An SA-omit electron density map (which gives an unbiased way of checking a structure) was calculated with σA-weighted 2Fo-Fc coefficients (Read 1986) after omitting both Val-22 and the entire Ade-5 nucleotide from the final model and subjecting this partial model to simulated annealing in XPLOR (Brunger 1996). Gly-21 (gray), Val-22 (gray and purple), Arg-23 (gray), and Ade-5 (green and blue) from the final model are shown in the context of the SA-omit electron density contoured at 1.5σ. (A) The view perpendicular to the plane of the Ade-5 base (green, same view direction as in Fig. 3C) illustrates the electron density for the pair of hydrogen bonds (dotted lines) between the polypeptide backbone of Val-22 and the adenine ring. (B) The view looking into the major groove of the octamer element (same view direction as in Fig. 3A) illustrates the electron density for the Val-22 side chain (purple).
Figure 3.
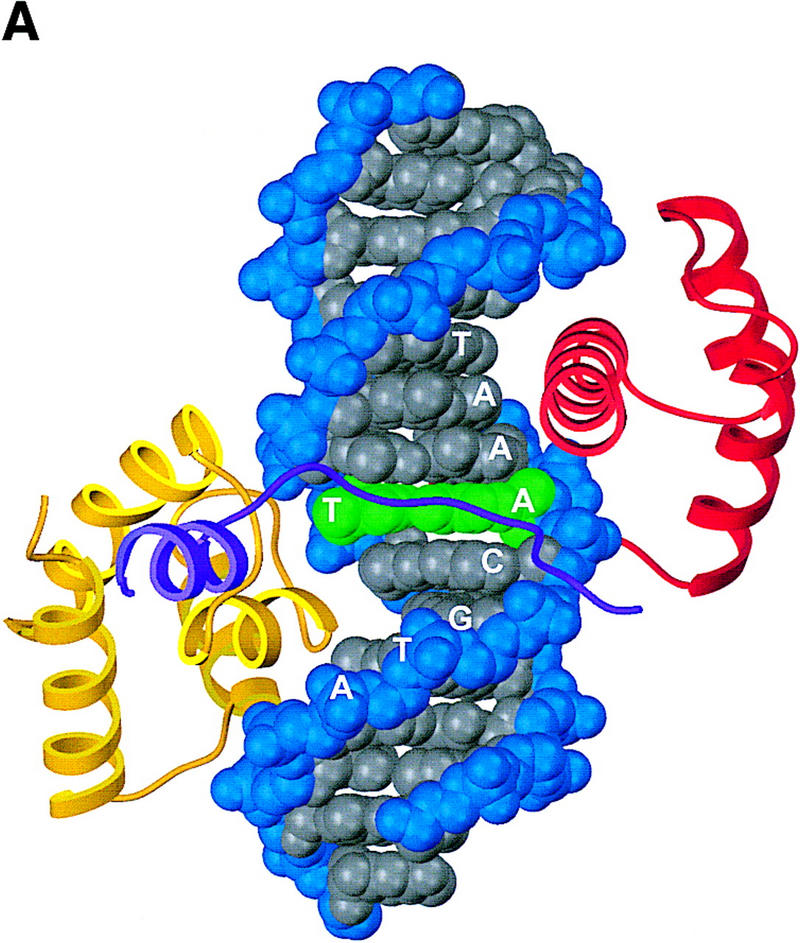
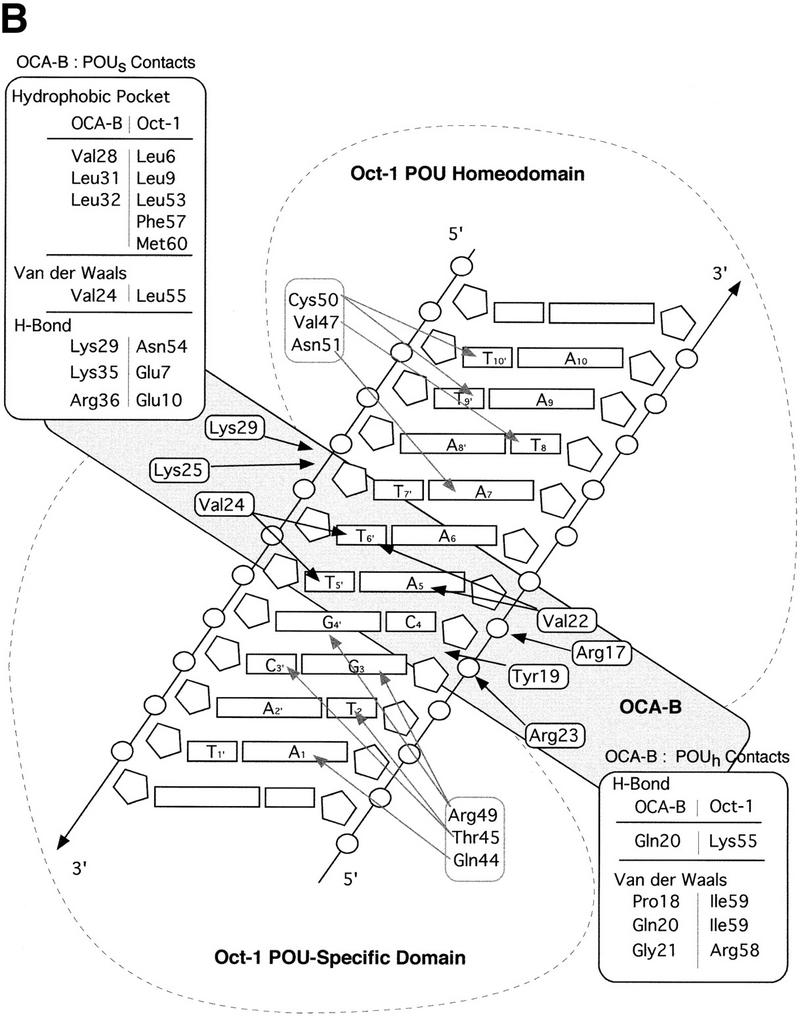
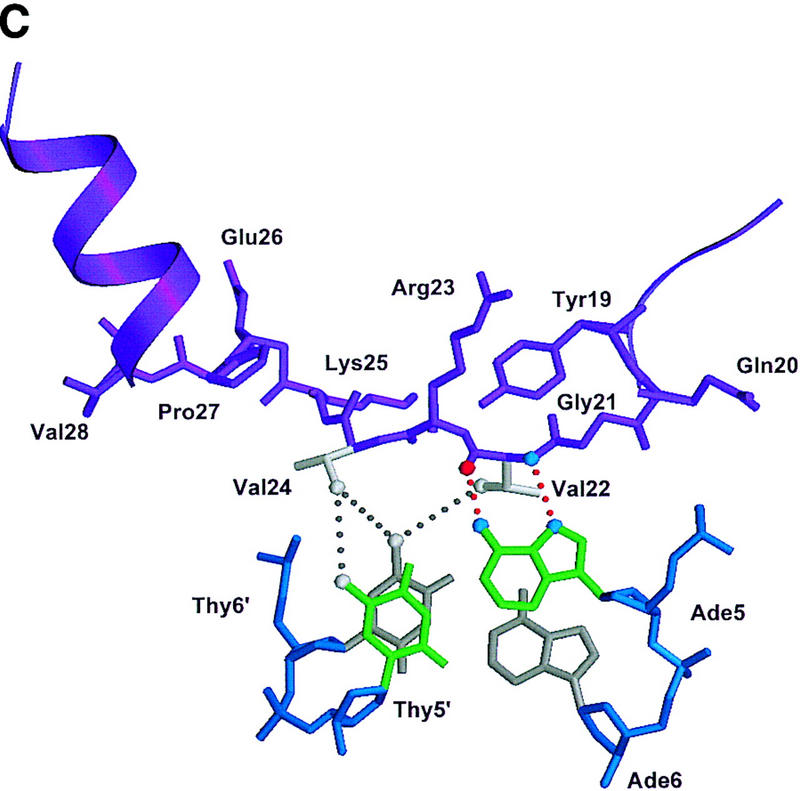
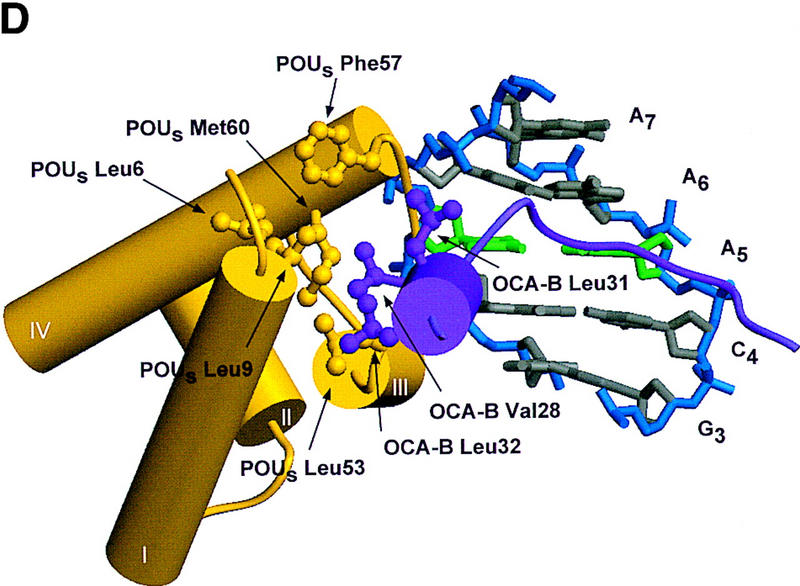
Structure of the ternary complex of the OCA-B 1–44 peptide, the Oct-1 POU domain, and an octamer element. (A) Overview of the ternary complex showing the OCA-B peptide (purple), the Oct-1 POU domain (the POU homeodomain is in red; the POU-specific domain is in yellow), and DNA containing the octamer sequence (position 5 is in green). About 20 amino acids of the OCA-B 1–44 peptide are well-ordered in the crystal. The OCA-B peptide traverses the major groove at the A:T base pair at position 5 of the octamer site. At the amino terminus of the ordered region, the OCA-B peptide makes contacts with the POU homeodomain; at the carboxyl terminus of the order region the OCA-B peptide forms a short α-helix and makes extensive contacts with the POU-specific domain. (B) Schematic summary of protein–DNA and protein–protein contacts in the ternary complex. The OCA-B 1–44 peptide is represented by the shaded rectangle across the center of the DNA; the positions of the POU-specific domain and the POU homeodomain are suggested by dashed outlines. Close contacts (<4.0 Å) between the OCA-B peptide and the POU-specific domain are summarized in the box in the upper left; close contacts with the POU homeodomain are summarized in the box in the lower right. All major DNA contacts are shown for the OCA-B peptide, but only key base contacts between the Oct-1 POU domain and the DNA are shown. The complete set of DNA contacts for the Oct-1 POU domain is very similar to those observed previously in its structure with an octamer element only (Klemm et al. 1994). (C) Contacts between the OCA-B 1–44 peptide (purple) and bases 5 (green) and 6 (gray) of the octamer element. The POU-specific domain, the POU homeodomain, and other bases in the octamer are not shown. Hydrogen bond contacts between the polypeptide backbone of the OCA-B peptide residue Val-22 and the exocyclic amine and N7 of adenine at position 5 are shown with red dots [the relevant atoms at these positions are blue (N), red (O), and gray (C)]. Hydrophobic interactions between OCA-B Val-22 and Thy-6′, and between OCA-B Val-24 and both Thy-5′ and Thy-6′ are indicated with gray dots. Residues at the amino terminus (Ala-16–Pro-18, extended chain) and at the carboxyl terminus (Lys-29-His-38, helix) of the ordered part of the OCA-B peptide are represented according to their secondary structure. (D) Interaction between the hydrophobic surface of the OCA-B peptide helix and the hydrophobic pocket of the Oct-1 POU-specific domain. POU-specific domain (yellow) residues at the amino terminus of helices one (Leu-6, Leu-9) and four (Phe-57, Met-60), and at the carboxyl terminus of helix three (Leu-53) form a shallow hydrophobic pocket. The surface of the helix from the OCA-B peptide (purple; Val-28, Leu-31, Leu-32) docks against the hydrophobic pocket of the Oct-1 POU-specific domain. The helices of the POU-specific domain are labeled with Roman numerals. Bases for one strand of the octamer element are also indicated.
Figure 4.
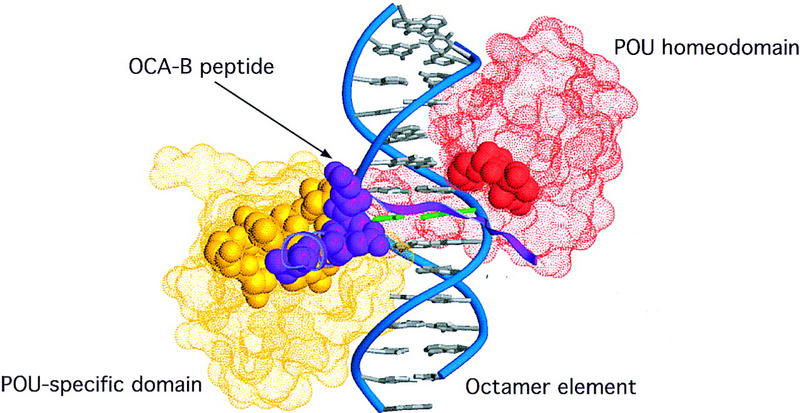
Mutations that attenuate the interaction of OCA-B with the complex of the Oct-1 POU domain and the octamer element are located at the peptide–protein interface. The positions of the POU-specific domain (yellow) and the POU homeodomain (red) are indicated by illustration of their Connolly solvent accessible surfaces (Connolly 1983). The OCA-B peptide (purple) is indicated by a ribbon. Residues deemed essential—via previous mutagenesis studies (Gstaiger et al. 1996; Babb et al. 1997; Sauter and Matthias 1998)—for ternary complex formation are shown in space-filling representation, and all of these are clustered at the peptide–protein interface seen in our crystal structure.
Surprisingly, the crystal structure shows that the specificity for an A at position 5 of the octamer site is primarily due to a pair of hydrogen bonds made by the polypeptide backbone from Val-22 of the OCA-B peptide (Fig. 3B,C). The electron density in this region of the map is very clear (Fig. 2), and both Ade-5 and Val-22 have some of the lowest B factors in the entire structure. We find that there is one hydrogen bond between the backbone amine group of residue 22 and the N7 of adenine 5, and there is another hydrogen bond between the backbone carbonyl group of residue 22 hydrogen and the exocyclic amine of adenine 5. Although these base contacts are made by the polypeptide backbone of OCA-B, the arrangement may be stabilized by contacts that the Val-22 and Val-24 side chains make with neighboring groups in the major groove. In particular, one of the methyl groups from Val-22 makes hydrophobic contact with the methyl group of thymine at position 6′ (4.2 Å). Val-24 (two residues later in the sequence) makes additional hydrophobic contacts with the methyl groups of Thy-5′ (4.5 Å) and Thy-6′ (4.5 Å). Val-24 also makes van der Waals contacts with Leu-55 of the Oct-1 POU-specific domain. [Some striking similarities between the helix-turn–helix units of the λ repressor and the Oct-1 POU-specific domain have been noted elsewhere (Assa-Munt et al. 1993; Dekker et al. 1993; Klemm et al. 1994). However, it is remarkable that Leu-55 of Oct-1, which has important contacts with the OCA-B peptide, corresponds to Asn-55 of the λ repressor, which in turn has important contacts with repressor's amino-terminal arm.]
Numerous other peptide–DNA and peptide–protein contacts help fix the position of the polypeptide strand of OCA-B in the major groove and help stabilize the base specific contacts made by Val-22 and Val-24 (Fig. 3B,C). At the amino-terminal end of the ordered region of OCA-B, Arg-17 makes salt bridges and hydrogen bonds with the phosphate of Cyt-4 of the octamer and the amide of Gln-20 of OCA-B. The side chain of Gln-20 may make a second hydrogen bond to Lys-55 of the POU homeodomain, and its peptide bond is positioned so that it could form a hydrogen bond to the phosphate of Cyt-4. Interesting van der Waals contacts are made by Tyr-19, which wedges between the deoxyribose groups of Gua-3 and Cyt-4 and has its ring parallel to the bases. Arg-23 (which is between the two valine residues that make base contacts) may adopt two conformations: In the better ordered complex of the asymmetric unit, it reaches away from the major groove to contact both the aromatic ring of Tyr-19 and the carboxyl terminus (in a symmetry related complex) of the Oct-1 POU homeodomain. In the other complex, the conformation of Arg-23 is less clear, but the density indicates that its guanidinium group is directed either toward Tyr-19 or toward the phosphate group of Ade-7 (4.9 Å). Lys-25 (which is just beyond the critical valine residues) forms simultaneous hydrogen bonds with the phosphate groups of nucleotides Thy-7′ and Ade-8′. This overall set of contacts helps to constrain the Val-22 and Val-24 residues of OCA-B to lie along the surface of the major groove.
The short α-helical segment of OCA-B (residues 28–36) makes extensive contacts with the Oct-1 POU-specific domain and thus firmly anchors the carboxyl-terminal end of the ordered peptide segment. Val-28, Leu-31, and Leu-32 of OCA-B form a hydrophobic surface on one face of the α-helix, and this surface binds to a complementary hydrophobic pocket in the Oct-1 POU-specific domain, which is lined by residues Leu-6, Leu-9, Leu-53, Phe-57, and Met-60 (Klemm et al. 1994) (Fig. 3D). Upon binding to the POU domain/DNA complex, the OCA-B peptide covers ∼500 Å2 of solvent accessible surface on the POU-specific domain, ∼350 Å2 of surface on the octamer element, and ∼170 Å2 of surface on the POU homeodomain. Solvent accessible residues on the hydrophilic face of this OCA-B helix also contribute to its interaction with Oct-1. For example, Lys-35 of OCA-B makes a hydrogen bond with Glu-7 of the Oct-1 POU-specific domain, while Arg-36 of OCA-B makes hydrogen bonds with both Glu-7 and Glu-10 of the Oct-1 POU-specific domain. Lys-29 of OCA-B also makes important contacts: the -NH3+ of this side chain reaches toward both the phosphate of Ade-8′ and the amide oxygen of Asn-54 from the Oct-1 POU-specific domain; the aliphatic portion of the Lys-29 side chain makes van der Waals contact with the deoxyribose of Ade-8′. Finally, we note that the short α-helical region is stabilized by several internal salt bridges involving interactions of OCA-B residues Arg-33 and Arg-34 with OCA-B residues Gln-26 and Gln-30.
Extensive mutational analysis previously had been used to characterize the OCA-B/Oct-1 interactions within the ternary complex (Gstaiger et al. 1996; Babb et al. 1997; Sauter and Matthias 1998), and the structure readily explains observations from these studies. Accordingly, mutations that substitute alanine for large hydrophobic side chains in the pocket region of the POU-specific domain compromise the interaction with OCA-B (Leu-6, Leu-9, Leu-53, and Met-60) (Babb et al. 1997). Similar effects are noted for POU homeodomain residue Ile-59, which is near the interface with OCA-B (Babb et al. 1997; Sauter and Matthias 1998), and OCA-B residue Leu-32, which makes contacts with residues in the POU-specific domain hydrophobic pocket (Gstaiger et al. 1996). The consequences of a Leu-55 → Ala mutation in the Oct-1 POU-specific domain can also be understood since this change would disrupt van der Waals contacts to Val-24 of OCA-B and thus reduce the affinity of OCA-B in the ternary complex (Gstaiger et al. 1996; Babb et al. 1997; Sauter and Matthias 1998). Mutations in Oct-1 that attenuate binding of OCA-B by eliminating hydrogen bonding and/or ionic interactions can also be explained by the structure. These include Glu-7 → Ala or Glu-10 → Ala changes in the POU-specific domain and Arg-58 → Ala, or Lys-55 → Ala changes in the POU homeodomain (Babb et al. 1997; Sauter and Matthias 1998). Mutation of certain DNA-contacting residues in the Oct-1 POU domain also seems to interfere with binding of OCA-B (Gstaiger et al. 1996; Babb et al. 1997). These effects can be rationalized since (1) Ser-56 makes a direct contact with both OCA-B and the DNA, (2) Asn-51 makes one of the most important homeodomain–DNA interactions, and mutation may affect the docking of the POU homeodomain (Klemm et al. 1994; Jacobson et al. 1997), and since (3) Arg-49 and Asn-59 of the POU-specific domain make critical contacts to the DNA. A double mutation of OCA-B (residues Glu-26 → Gly and Pro-27 → Leu) has been shown to interfere with ternary complex formation (Gstaiger et al. 1996). Our structure shows that these residues are at the interface with the POU-specific domain and the DNA, and the mutations may disrupt the turn immediately preceeding the OCA-B helix. Finally, it has been shown that an OCA-B protein missing residues 4–18 is unable to form a ternary complex (Luo et al. 1998), presumably due to the removal of the three amino acids (16–18) that form the first ordered region of OCA-B in the crystal structure.
Discussion
The OCA-B/POU domain/octamer ternary site complex shows how the OCA-B polypeptide chain can specifically recognize the POU domain/octamer DNA interface and generate a new level of structural and functional specificity. Stable binding of the OCA-B peptide involves both peptide–DNA contacts (with the octamer site) and peptide–protein contacts (with the POU domain). Oct-1 binding alone to an octamer site stimulates transcription, but the ternary complex with the coactivator OCA-B confers additional stimulatory effects on transcription to octamer sites with an A at position 5. The most important contact in OCA-B's reading of the octamer sequence has the most surprising structural arrangement: The polypeptide backbone makes a pair of hydrogen bonds with the critical adenine at position five of the octamer site. Thus, the specific binding of the small OCA-B peptide to the Oct-1 protein–DNA interface significantly alters the function of a large transcription complex. More generally, this structure and other recent structures illustrate the huge potential for highly specific regulation of transcription through combinatorial arrangements of transcription factors that are responsible for distinct regulatory schemes (Li et al. 1995; Batchelor et al. 1998; Chen et al. 1998a,b; Tan and Richmond 1998). Our structure shows a novel way that specificity in ternary complex formation may be achieved.
Examination of the structure also suggests that other proteins that interact with the Oct-1 POU domain and an octamer element may use contacts related to those used by OCA-B. In particular, some of the interactions between Oct-1 and the SNAP190 component of the SNAPc snRNA transcription complex may be analogous to the interactions with OCA-B. Here, the region of SNAP190 critical for binding to Oct-1 is homologous to part of the OCA-B peptide seen in the crystal structure, including a perfect match of the residues that form the hydrophobic surface on the short OCA-B α-helix (corresponding to Val-28, Leu-31, and Leu-32 in OCA-B) (Ford et al. 1998). Strengthening the analogy, it has been observed that a Glu-7 → Arg mutation of the POU-specific domain disrupts interaction with SNAP190 (Mittal et al. 1996); the mutation can be suppressed by changing Lys-900 → Glu in SNAP190. The proposed Lys900 ↔ Glu-7 interaction would align perfectly with the salt bridge observed between OCA-B Lys-35 and Glu-7 of the POU-specific domain.
It would not be surprising if related contacts are found in other systems as well. We note that key residues of the hydrophobic pocket of the POU-specific domain (Leu-6, Leu-9, Leu-53, Phe-57, and Met-60) are conserved in several members of the POU family. This set of proteins is characterized by a very high degree of sequence conservation throughout the POU-specific domain and includes not only the Oct-1 and Oct-2 homologs from a variety of vertebrate species, but also proteins with roles in early development in Drosophilia (pdm-1, pdm-2) and in the development of mammalian skin cells (skn-1). [SwissProt accession identifiers for these proteins: PDM1_DROME (pdm-1), PDM2_DROME (pdm-2), SK1I_ RAT and SK1A_RAT (skn-1)]. It will be interesting to see whether these closely related proteins exploit a capacity for peptide binding and use co-activators analogous to OCA-B for stimulating transcription and distinguishing closely related binding sites. Recognition of a protein–DNA interface, by a peptide making contacts with both the protein and the DNA, may provide a very effective basis for specificity in higher order complex formation.
Materials and methods
Expression and purification of proteins
A synthetic gene had been constructed previously for expression of residues 1–118 from the amino terminus of OCA-B, with all codons optimized for high levels of expression in Escherichia coli. This expression system, based on the pGEX-5X-3 vector (Cepek et al. 1996) (Pharmacia), was then modified with standard procedures to direct expression of peptides containing residues 1–44 or 1–59 of OCA-B. These peptides were expressed and purified as described for the OCA-B 1–118 peptide (Cepek et al. 1996) with an additional purification step by FPLC on Mono-S (Pharmacia). Purified peptides were dialyzed against 5 mm HEPES (pH 7.5), 25 mm NH4OAc, and 1 mm DTT and concentrated to 1.0 mm in a Centricon device (MWCO 1000, Amicon). The Oct-1 POU domain was expressed and purified as described (Klemm et al. 1994) except that the DNA–cellulose was replaced with chromatography by FPLC on Mono-S (Pharmacia) (Klemm et al. 1994). The Oct-1 POU domain was dialyzed and concentrated under the same conditions used for the OCA-B peptides. DNA for crystallization was prepared as described (Klemm et al. 1994).
Templates for coupled transcription and translation in vitro were constructed using the pCITE system (Novagen). The products of this system have the S-peptide of RNase fused to various amino-terminal peptides of OCA-B (ranging in 5 amino acid increments from 34 to 59 residues in length). Peptide products from these templates were quantified by reconstitution of RNase A activity in the presence of the RNase S-protein (as described by the manufacturer), and the ability of these peptides to form a ternary complex with the Oct-1 POU domain and an octamer element was assessed in a gel electrophoresis mobility shift assay (Cepek et al. 1996).
Crystallization and structure determination
The ternary complex was formed by mixing the OCA-B 1–44 peptide (1.0 mm), the Oct-1 POU domain (1.0 mm), and the octamer element oligonucleotide (∼10 mm, Fig. 1B) in a stoichiometry of 1.0:1.0:1.2. This sample was combined with a precipitant containing 33 mm Am2SO4, 50 mm HEPES (pH 7.5), 10 mm MgCl2, and 24%–26% PEG (av. MW, 3350). Crystals, which were grown by vapor diffusion, appeared in a few days and reached maximum size within 2 weeks. These crystals formed in the space group P41212 and had cell dimensions a = b = 92.722 Å and c = 152.651 Å. For cryoprotection, crystals were soaked to equilibrium (a few hours) in solutions of the precipitant buffer supplemented (successively) with 1%, 5%, and 15% glycerol before flash freezing in gaseous nitrogen at −170°C.
A highly redundant diffraction data set from frozen crystals was collected at the National Synchrotron Light Source (NSLS, beamline X4A). Diffraction data had a maximal resolution of 2.9 Å and a useful resolution of 3.2 Å (where 50% reflections have I/σI>2). All processing was done with the HKL, CCP4, or XPLOR suites of programs (Collaborative Computational Project 1994; Brunger 1996; Otwinowski and Minor 1997). An initial solution of the crystal structure, which contained two ternary complexes in the asymmetric unit, was obtained by molecular replacement using the Oct-1 POU domain bound to an octamer element as the search model and using the AMORE program (Navaza 1994). Improvement of the phases from this initial solution by iterative rounds of noncrystallographic symmetry (NCS) averaging, solvent flattening, and histogram matching with DM (Cowtan 1994) revealed electron density that did not correspond to either the Oct-1 POU domain or the octamer element . This new density could be traced across the major groove in the vicinity of the A:T base pair at position five and extended in both directions to contact the POU-specific domain and the POU homeodomain. To improve the maps, derivatives were prepared with iodo-uracil at specfic positions in the octamer site, and data from two derivatives (using two crystals for each) were collected to a resolution of 3.3–3.5 Å (Fig. 1B; Table 1). Iodine positions were located from different Patterson maps, and the positions of these heavy atoms confirmed the molecular replacement solution: All of the iodine positions were close to the 5-methyl groups of the corresponding thymidine bases in the preliminary model. An electron density map calculated from multiple isomorphous replacement using the anomalous signal (MIRAS) phases combined with solvent flipping density modification in SHARP (La Fortelle and Bricogne 1997) also confirmed the molecular replacement solution. In this experimental map, an α-carbon trace for ∼20 amino acids of the OCA-B peptide could be identified, and the trace revealed that the overall arrangement of the OCA-B peptide in the two complexes of the asymmetric unit were equivalent. Noncrystallographic averaging of the solvent flattened MIRAS map with RAVE (Jones 1992) allowed unambiguous assignment of the side chains in the OCA-B model. Side chain assignments were confirmed by simulated annealing omit maps in XPLOR. Several rounds of model building with O (v. 5.10.3) (Jones et al. 1991) into 2Fo-Fc maps, positional refinement with XPLOR, tightly restrained individual B-factor refinement, and a bulk solvent correction gave a final model (Rwork = 0.260, Rfree = 0.326) that included amino acids 16–37 and 16–38 of OCA-B for the two complexes in the asymmetric unit. Noncrystallographic symmetry restraints were used in the very early stages of refinement, but these were not helpful—and thus were not applied—in the later stages. At every stage of the refinement, the peptide backbone and side chain geometry was monitored with Ramachandran plots and PROCHECK (Laskowski et al. 1993). The two ternary complexes in the asymmetric unit differ by 0.70, 0.68, 0.58, and 0.51 Å rms difference (measured for Cα or P atoms) for the POU-specific domain, the POU homeodomain, the OCA-B peptide, and the octamer element, respectively. A B-factor refinement procedure that assigned only two B factors to each residue—one for the main chain atoms and one for the side chain atoms—gave essentially the same R factors (Rwork = 0.332, Rfree = 0.267). The same regions of the Oct-1 linker that were disordered in the binary complex (Klemm et al. 1994) remain disordered in the ternary complex. Figures were made with the program Setor (Evans 1993). Coordinates are being deposited with the Protein Data Bank (accession 1CQT).
Table 1.
Crystallographic data
| Preparation
|
Native
|
I-1a
|
I-1 + I-10‘a
|
||
|---|---|---|---|---|---|
| Data Collection and MIR Statistics | |||||
| Crystal and data set | A | A | B | A | B |
| Resolution (30.0 Å)b | 3.2 Å | 3.4 Å | 3.5 Å | 3.3 Å | 3.5 Å |
| Total (Unique) Reflections | 175486 (11803) | 124686 (8942) | 212021 (10180) | 151902 (10827) | 53730 (9067) |
| Completeness | 99.5% | 99.0% | 99.4% | 99.7% | 71.2% |
| Rmergec | 0.084 | 0.101 | 0.147 | 0.126 | 0.102 |
| Rcullisd | |||||
| centric | — | 0.727 | 0.642 | 0.604 | 0.585 |
| acentric | |||||
| Isomorphous | — | 0.682 | 0.572 | 0.598 | 0.540 |
| anomalous | — | 0.930 | 0.891 | 0.595 | 0.881 |
| Phasing Powere | |||||
| centric | — | 1.83 | 2.03 | 2.20 | 2.27 |
| acentric | |||||
| Isomorphous | — | 2.48 | 2.94 | 2.93 | 3.07 |
| Anomalous | — | 1.24 | 1.50 | 1.26 | 1.51 |
| Refinement | |||||
| Rworkf (30.0–3.2 Å) | 0.260 | ||||
| Rfreef (30.0–3.2 Å) | 0.326 | ||||
| rms deviation in bond lengths (protein/DNA): | 0.007 Å/0.006 Å | ||||
| rms deviation in bond angles (protein/DNA): | 1.395°/1.222° | ||||
| rms in the difference of B factors between bonded atoms: | 3.8 Åb | ||||
Native data collected at NSLS. All other data collected with a rotating anode source (Rigaku and MSC Corp.). All data collected on RAXIS-IV detectors (MSC) and reduced with the HKL suite of programs (Otwinowski and Minor 1997). Derivatives were prepared by substitution of thymine bases with 5-iodo-uracil in the octamer element either at position 1 or at position 1 and 10 (numbering as in Fig. 1B).
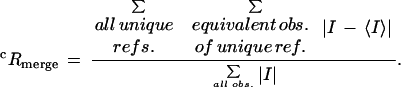 |
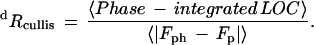 |
 |
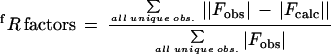 |
for reflections in the working (Rwork) or test (Rfree) data sets. Test set represented 10% of the total number of reflections selected at random. R factors were calculated with the bulk solvent model.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant GM31471 (C.O.P), by the Howard Hughes Medical Institute (C.O.P), and by U.S. Public Health Service grant PO1-CA42063 from the NIH (P.A.S.), by cooperative agreement CDR-8803014 from the National Science Foundation (P.A.S.), and by NCI Cancer Center Support (core) grant P30-CA14051. Some equipment was purchased with support from the PEW Charitable Trust (C.O.P). We thank James Berger (Whitehead Institute) and Craig Ogata (NSLS) for generous provision of x-ray facilities; Ernest Fraenkel and Tim Benson for assistance in data collection; and Mark Rould for suggestions during the refinement; Lena Nekludova for help with the Proteus database; Bob Grant for comments on the manuscript; Vicki Wang for help with some of the gel mobility shift analysis; and Robbyn Issner, Margaret Beddall, and Sandra Fay-Richard for superb technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pabo@mit.edu; FAX (617) 253-8728.
References
- Assa-Munt N, Mortishire-Smith RJ, Aurora R, Herr W, Wright PE. The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain. Cell. 1993;73:193–205. doi: 10.1016/0092-8674(93)90171-l. [DOI] [PubMed] [Google Scholar]
- Babb R, Cleary MA, Herr W. OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol Cell Biol. 1997;17:7295–7305. doi: 10.1128/mcb.17.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPalpha/beta: An ETS domain- ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Brunger A. XPLOR, v. 3.851. New Haven, CT: Yale University; 1996. [Google Scholar]
- Cepek KL, Chasman DI, Sharp PA. Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes & Dev. 1996;10:2079–2088. doi: 10.1101/gad.10.16.2079. [DOI] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998a;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998b;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Cowtan K. Joint CCP4 and ESF–EACBM Newsletter on Protein Crystallography 31: 34–38. Warrington, UK: Daresbury Laboratory; 1994. ‘dm’: An automated procedure for phase improvement by density modification. [Google Scholar]
- Dekker N, Cox M, Boelens R, Verrijzer CP, van der Vliet PC, Kaptein R. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature. 1993;362:852–855. doi: 10.1038/362852a0. [DOI] [PubMed] [Google Scholar]
- Evans S. Setor: Hardware lighted three-dimensional model representations of macromolecules. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Ford E, Strubin M, Hernandez N. The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes & Dev. 1998;12:3528–3540. doi: 10.1101/gad.12.22.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. The B-cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes & Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- Jones T. A set of averaging programs. In: Dodson E, Gover S, Wolf W, editors. Molecular replacement. Warrington, UK: SERC Daresbury Laboratory; 1992. pp. 91–105. [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjelgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kim U, Qin XF, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder RG. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD Methods. In: Carter C W Jr, Sweet R M, editors. Methods in Enzymology, Macromolecular Crystallography. New York, NY: Academic Press; 1997. pp. 472–494. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK—A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Li T, Stark MR, Johnson AD, Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- Luo Y, Roeder RG. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Fujii H, Gerster T, Roeder RG. A novel B-cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- Luo Y, Ge H, Stevens S, Xiao H, Roeder RG. Coactivation by OCA-B: definition of critical regions and synergism with general cofactors. Mol Cell Biol. 1998;18:3803–3810. doi: 10.1128/mcb.18.7.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V, Cleary MA, Herr W, Hernandez N. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol Cell Biol. 1996;16:1955–1965. doi: 10.1128/mcb.16.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology, Macromolecular crystallography. Vol. 276. New York, NY: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. 1986;A42:140–149. [Google Scholar]
- Sauter P, Matthias P. Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol Cell Biol. 1998;18:7397–7409. doi: 10.1128/mcb.18.12.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- Tan S, Richmond TJ. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]