Abstract
The BRCA1 gene encodes a tumor suppressor that is mutated in 50% of familial breast cancers. The BRCA1 protein has been implicated in the DNA damage response, as DNA damage induces the phosphorylation of BRCA1 and causes its recruitment into nuclear foci that contain DNA repair proteins. The ataxia-telangiectasia-mutated (ATM) gene product controls overall BRCA1 phosphorylation in response to γ-irradiation (IR). In this study, we show that BRCA1 phosphorylation is only partially ATM dependent in response to IR and ATM independent in response to treatment with UV light, or the DNA replication inhibitors hydroxyurea (HU) and aphidicolin (APH). We provide evidence that the kinase responsible for this phosphorylation is the ATM-related kinase, ATR. ATR phosphorylates BRCA1 on six Ser/Thr residues, including Ser 1423, in vitro. Increased expression of ATR enhanced the phosphorylation of BRCA1 on Ser 1423 following cellular exposure to HU or UV light, whereas doxycycline-induced expression of a kinase-inactive ATR mutant protein inhibited HU- or UV light-induced Ser 1423 phosphorylation in GM847 fibroblasts, and partially suppressed the phosphorylation of this site in response to IR. Thus, ATR, like ATM, controls BRCA1 phosphorylation in vivo. Although ATR isolated from DNA-damaged cells does not show enhanced kinase activity in vitro, we found that ATR responds to DNA damage and replication blocks by forming distinct nuclear foci at the sites of stalled replication forks. Furthermore, ATR nuclear foci overlap with the nuclear foci formed by BRCA1. The dramatic relocalization of ATR in response to DNA damage points to a possible mechanism for its ability to enhance the phosphorylation of substrates in response to DNA damage. Together, these results demonstrate that ATR and BRCA1 are components of the same genotoxic stress-responsive pathway, and that ATR directly phosphorylates BRCA1 in response to damaged DNA or stalled DNA replication.
Keywords: BRCA1, ATR, checkpoint, DNA damage, phosphorylation, replication
Damage to the genome, which can occur as a consequence of exposure to environmental agents or to the byproducts of oxidative metabolism, represents a persistent threat to genomic integrity and cellular viability. The deleterious effects of genotoxic stress are countered by surveillance mechanisms that monitor the genome for the presence of genetic lesions, including single-stranded DNA (ssDNA), DNA double-strand breaks (DSBs), and chemically modified nucleotide bases. The DNA damage response pathway that responds to these lesions consists of a cascade of protein kinases that control the phosphorylation of a number of effector proteins involved in DNA repair and cell cycle control. Central among these effectors is the BRCA1 protein. BRCA1 is a tumor suppressor gene mutated in 50% of familial breast and ovarian cancers (Easton et al. 1993). BRCA1 encodes an 1863-amino-acid nuclear phosphoprotein that is essential for viability in mice (Chen et al. 1996; Hakem et al. 1996; Bertwistle and Ashworth 1998; Zhang et al. 1998). Although BRCA1 has been reported to act as a transcription factor (Chapman and Verma 1996) and cell growth suppressor (Monteiro et al. 1996; Somasundaram et al. 1997; Aprelikova et al. 1999), the tumor suppressor functions of BRCA1 may be most closely related to its role in DNA repair and recombination (Zhang et al. 1998). BRCA1-deficient cells display spontaneous chromosomal abnormalities and defects in both homologous DNA recombination and transcription-coupled repair of oxidative base damage (Gowen et al. 1998; Moynahan et al. 1999; Xu et al. 1999). Cells that express a truncated version of BRCA1 (Tomlinson et al. 1998) are hypersensitive to DNA damaging agents and display slowed kinetics of DSB repair (Cortez et al. 1999; Scully et al. 1999; Zhong et al. 1999). BRCA1 physically associates with proteins implicated in homologous and nonhomologous DNA recombination, including Rad51 and the Rad50–Mre11–p95 DNA repair complex (Scully et al. 1997; Zhong et al. 1999; Wang et al. 2000). However, the precise contributions of BRCA1 to cell cycle checkpoint activation and DNA repair remain unclear.
BRCA1 is maximally expressed during S phase. Exposure of S-phase cells to γ-radiation (IR), UV light, or the DNA replication inhibitor hydroxyurea (HU) results in the rapid phosphorylation of BRCA1, indicating that BRCA1 is a target of the DNA damage response pathway (Scully et al. 1997a). Moreover, these agents induce dramatic alterations in the nuclear localization pattern of BRCA1. In the absence of damage, BRCA1 is localized to discrete nuclear foci during both S and G2 phases of the cell cycle (Scully et al. 1996, 1997a). Exposure of S-phase cells to IR or HU induces the relocalization of BRCA1 to new foci, some of which are sites of DNA synthesis (Scully et al. 1997a). Recent studies have demonstrated a role for ATM and the ATM-regulated kinase Chk2 in the phosphorylation of BRCA1 in IR-damaged cells (Cortez et al. 1999; Gatei et al. 2000; Lee et al. 2000). Whereas ATM controls the overall phosphorylation of BRCA1 in response to IR as judged by electrophoretic mobility-shift alterations, the regulated phosphorylation of individual sites in response to IR has not been examined. Furthermore, the protein kinase(s) responsible for BRCA1 phosphorylation in response to HU or UV light are unknown.
A candidate kinase in the DNA damage response pathway that may play a role in the ATM-independent regulation of BRCA1 is ATR. ATR is a member of a family of high molecular mass protein kinases whose catalytic domains bear sequence similarity to those of phosphoinositide 3-kinases (PI3-Ks) (Keith and Schreiber 1995; Bentley et al. 1996; Cimprich et al. 1996). Within the PIK family, ATR is most closely related to ATM, Rad3 and Tel1 (Schizosaccharomyces pombe), Mec1 and Tel1 (Sacharomyces cerevisiae), and Mei-41 (Drosophila melanogaster) (Zhou and Elledge 2000). Mutations in MEC1 and rad3 confer hypersensitivity to a variety of agents including IR, methylmethanesulfonate (MMS), UV light, and HU. These mutants are also defective in both the DNA damage-induced G2 checkpoint and the DNA replication checkpoint, which prevents mitotic entry in the presence of unreplicated DNA. Recent studies suggest that the HU sensitivity of mec1 mutant yeast strains is not solely the result of aberrant mitotic entry, but may also be a consequence of aborted chromosomal replication (Desany et al. 1998) and defects in suppression of late origin firing (Santocanale and Diffley 1998). Because Mec1 and Rad3 are the closest ATR homologs, ATR may perform similar S-phase surveillance functions in mammalian cells.
Accumulating evidence suggests that ATM and ATR carry out distinct as well as partially overlapping genome surveillance activities in mammalian cells (Keegan et al. 1996; Cliby et al. 1998; Tibbetts et al. 1999). Inactivating mutations in ATM are responsible for the chromosomal instability syndrome, ataxia-telangiectasia (A-T). Cells isolated from A-T patients (Kastan et al. 1992; Beamish and Lavin 1994; Canman et al. 1994; Beamish et al. 1996;), or from ATM-nullizygous mice (Barlow et al. 1996, 1997; Xu and Baltimore 1996; Xu et al. 1996; Westphal et al. 1997), display defective DNA damage checkpoints, hypersensitivity to IR and radiomimetic drugs, and elevated frequencies of chromosomal abnormalities. The IR-induced phosphorylation of replication protein A (Liu and Weaver 1993), c-Abl (Shafman et al. 1997), p53 (Siliciano et al. 1997), Mdm2 (Khosravi et al. 2000), hChk2/Cds1 (Matsuoka et al. 1998; Brown et al. 1999), and BRCA1 (Cortez et al. 1999; Gatei et al. 2000; Zhou and Elledge 2000) is compromised in A-T cell lines, indicating that ATM regulates the activities of multiple downstream effectors. In comparison to ATM, the cellular functions of ATR are less understood, due in part to the lack of ATR-deficient cell lines. Nonetheless, overexpression of a catalytically inactive ATR mutant protein (ATRKI) compromises G2 checkpoint function, and decreases cell survival in response to IR, alkylating agents, and HU (Cliby et al. 1998). The observation that overexpression of ATR complements the radioresistant DNA synthesis (RDS) defect of A-T cells provided the first genetic evidence that ATR and ATM can perform overlapping functions in mammalian cells (Cliby et al. 1998).
Despite considerable overlap in terms of substrate phosphorylation (Canman et al. 1998; Kim et al. 1999; Lakin et al. 1999; Tibbetts et al. 1999), the relative contributions of ATM and ATR vary with the type of genotoxic stress. Whereas cells that overexpress ATRKI are hypersensitive to HU, UV light, and IR (Cliby et al. 1998; Wright et al. 1998), ATM-deficient cells retain near normal resistance to UV and HU, but are extremely sensitive to IR. These findings and others support a model in which ATM and ATR respond to distinct, as well as partially overlapping types of genotoxic stress. Finally, functional distinctions between ATM and ATR are highlighted by the drastically different phenotypes associated with disruption of their respective genes. Whereas ATM-nullizygous mice survive to adulthood (Barlow et al. 1996; Xu et al. 1996), ATR deficiency results in early embryonic lethality (Brown and Baltimore 2000; de Klein et al. 2000). ATR−/− cells display accelerated chromosomal fragmentation, suggesting that functional ATR is essential for chromosome maintenance in the absence of environmentally imposed genotoxic stress (Brown and Baltimore 2000). Thus, ATR appears to carry out genome surveillance functions during both normal cell cycles and cellular exposure to DNA-damaging agents.
In this report, we provide evidence that BRCA1 is a direct target of the ATR kinase. Furthermore, we provide evidence supporting the hypothesis that ATM and ATR provide overlapping as well as distinct regulatory functions to control BRCA1 phosphorylation in response to genotoxic stress.
Results
DNA damage-induced phosphorylation of BRCA1 at Ser 1423
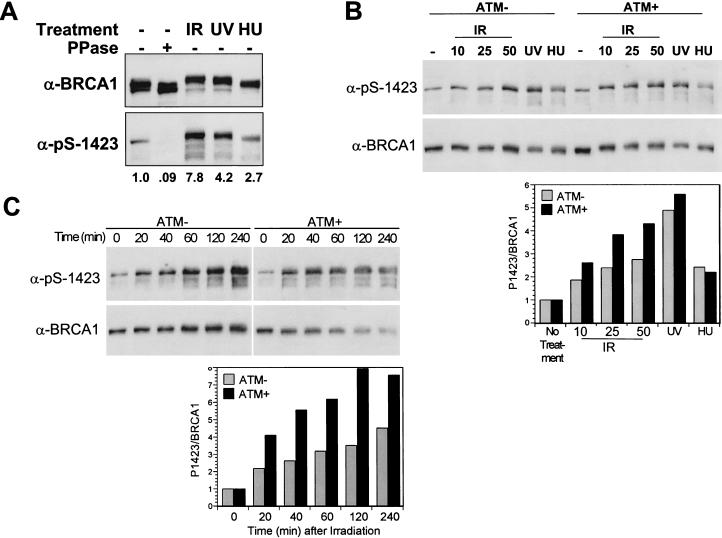
Recently, we demonstrated that ATM is required for the optimal phosphorylation of endogenous BRCA1 in γ-irradiated cells (Cortez et al. 1999). The results obtained with Ser→Ala substitution mutants strongly suggested that ATM phosphorylates BRCA1 on Ser 1423 in IR-damaged cells; however, ATM-dependent phosphorylation of this site was not determined directly in intact cells. In the present studies, we monitored the phosphorylation of BRCA1 at Ser 1423 in intact cells by generation of phosphoSer 1423-specific antibodies (α-pS-1423). In asynchronously cycling cells, the α-pS-1423 antibodies detected a 220-kD protein in human embyronic kidney (HEK) 293T cell extracts that comigrated with the BRCA1 protein and was absent in BRCA1-deficient HCC1937 cells (Fig. 1A; data not shown). Treatment of cell extracts with λ phosphatase (Fig. 1A), or mutation of Ser 1423 to Ala (data not shown) eliminated recognition by α-pS-1423, indicating that the α-pS-1423 antibody specifically recognizes BRCA1 phosphorylated on Ser 1423. In subsequent experiments, HEK 293T cells were treated with IR (25 Gy), UV light (50 J/m2), HU (2 mM), or the DNA polymerase inhibitor aphidicolin (APH), and cellular extracts were immunoblotted with α-pS-1423. All four genotoxic agents induced a rapid increase in the phosphorylation of BRCA1 at Ser 1423 (Fig. 1A; data not shown).
Figure 1.
DNA damage-induced phosphorylation of BRCA1 on Ser 1423. (A) Endogenous BRCA1 is phosphorylated at Ser 1423 in intact cells. HEK 293T cells were either left untreated or cultured for 1 h following cellular exposure to IR (25 Gy), UV light (50 J/m2) or HU (2 mM). Cellular lysates were prepared and analyzed by immunoblot analysis using α-BRCA1 and α-pS-1423 antibodies. Cell lysates were treated with λ phosphatase where indicated. The relative levels of Ser 1423 phosphorylation were determined by densitometric analysis of the α-pS-1423 immunoblot and are presented at bottom. Values were normalized to the total level of BRCA1 present in each lane. (B) Phosphorylation of BRCA1 on Ser 1423 in A-T cells. ATM-deficient AT22IJE-T cells that had been stably transfected with empty expression vector (ATM−) or wild-type ATM (ATM+) were left untreated or exposed to IR (10, 25, or 50 Gy), UV light (50 J/m2), or HU (2 mM) and harvested 1 h later. Cell lysates were analyzed by SDS-PAGE and immunoblot analysis with the α-pS-1423 antibody to detect Ser 1423-phosphorylated BRCA1. Total levels of BRCA1 were then determined by reblotting the stripped membrane with an α-BRCA1 mAb. Densitometry was performed on the α-pS-1423 and α-BRCA1 immunoblots and the α-pS-1423/α-BRCA1 density ratios were used to calculate normalized levels of Ser 1423 phosphorylation, which are plotted as histograms at bottom. (C) Comparison of the time courses of IR-induced Ser 1423 phosphorylation in ATM− and ATM+ AT22IJE-T cells. Cells were harvested at the indicated times following exposure to 25 Gy of IR and subjected to immunoblot analysis using α-pS-1423 and α-BRCA1 antibodies as described in B. Levels of normalized Ser 1423 phosphorylation are presented at bottom.
To determine whether functional ATM is required for DNA damage-induced phosphorylation of BRCA1 on Ser 1423, we compared the phosphorylation of this site in ATM-deficient and ATM-wild-type human fibroblasts. For this analysis, we utilized an A-T cell line (AT22IJE-T) that was stably transfected with plasmids encoding either wild-type ATM (ATM+) or an empty expression vector (ATM−) (Ziv et al. 1997). Exposure to IR (10–50 Gy), UV light (50 J/m2), or HU (2 mM) caused an increase in the levels of Ser 1423-phosphorylated BRCA1 in both the ATM− and ATM+ cell lines (Fig. 1B). Densitometric analyses of the α-pS-1423 immunoblots revealed that, although exposure to UV light or HU triggered Ser 1423 phosphorylation at comparable levels in ATM− and ATM+ cells, IR-induced phosphorylation at this site was consistently 30%–50% higher in ATM+ cells than in ATM− cells (Fig. 1B). A time-course analysis also revealed that ATM− fibroblasts were defective in IR-induced phosphorylation of Ser 1423 relative to their ATM-reconstituted counterparts (Fig. 1C).
Whereas AT fibroblasts including AT22IJE-T and GM05849B cells exposed to 25Gy of IR displayed a two-to threefold increase in Ser 1423 phosphorylation, cells with wild-type ATM alleles, including GM00637G and HEK 293T cells, consistently showed a seven- to eightfold increase in the phosphorylation of this site (Fig. 1; data not shown). The ATM-reconstituted AT22IJE-T cells showed a smaller, four- to sixfold, increase in Ser 1423 phosphorylation after IR treatment, suggesting that these cells may not be fully complemented for the A-T defect. Nevertheless, these results indicate that functional ATM is required for optimal Ser 1423 phosphorylation in response to IR; however, an additional protein kinase(s) contributes to the phosphorylation of this site in IR-damaged A-T cells. Furthermore, HU and UV light induce the phosphorylation of Ser 1423 through pathways that are largely independent of functional ATM.
ATR phosphorylates BRCA1 at Ser 1423 in vitro
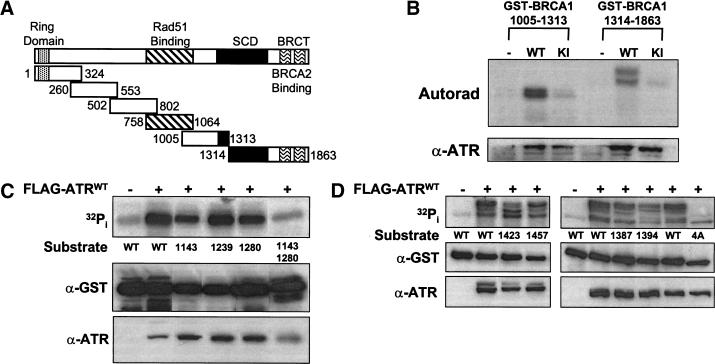
ATR has been shown previously to exhibit similar substrate specificity as ATM (Kim et al. 1999). Therefore, we examined whether BRCA1 might be a direct substrate for the ATR kinase. Six GST–BRCA1 fusion proteins, which collectively span the entire BRCA1 polypeptide (Scully et al. 1997b), were used as substrates in ATR immune complex kinase assays (Fig. 2A). These assays demonstrated that GST–BRCA1 (1005–1313) and GST–BRCA1 (1314–1863) were highly phosphorylated by wild-type ATR but not a catalytically inactive FLAG–ATRKI mutant (Fig. 2B). The four other GST–BRCA1 fragments were not phosphorylated (data not shown). These results suggest that phosphorylation of these fragments is catalyzed by ATR rather than by a coprecipitating protein kinase.
Figure 2.
ATR phosphorylates BRCA1 in vitro. (A) Schematic depiction of the six GST–BRCA1 fusion proteins tested as ATR substrates in immune complex kinase assays. SCD designates the S-Q cluster domain. (B) Phosphorylation of GST–BRCA1 fusion proteins requires a functional ATR kinase domain. HEK 293T cells were transiently transfected with either pcDNA3 (−), or plasmid constructs encoding FLAG-tagged wild-type ATR (WT), or FLAG-tagged catalytically-inactive ATR (KI). Cellular extracts were immunoprecipitated with α-FLAG mAb, and immune complex kinase assays were performed with GST–BRCA1 (1005–1313) or GST–BRCA1 (1314–1863) as the substrates. The levels of phosphorylated fusion protein and FLAG–ATR expression are presented at top and bottom, respectively. The other four GST–BRCA1 substrates depicted in A, were not phosphorylated by ATR above background levels (not shown). (C) Identification of ATR phosphorylation sites in GST–BRCA1 (1005–1313). ATR immune complex kinase assays were performed using cell lysates prepared from HEK 293T cells that had been transfected with either pcDNA3 (−) or FLAG–ATRWT (+). Wild-type (WT) or mutant GST–BRCA1 (1005–1313) fusion proteins containing either single or combination Ala substitutions at the indicated residues were used as substrates. (D) Identification of ATR phosphorylation sites in GST–BRCA1 (1314–1863). Wild-type (WT) or mutant GST–BRCA1 (1314–1863) fusion proteins containing single Ala substitutions at the indicated residues, or a GST–BRCA1 (1314–1863) mutant protein (4A) containing Ala substitutions at Ser 1387, Thr 1394, Ser 1423, and Ser 1457, were used as substrates in ATR kinase assays. Each panel represents a single experiment that is representative of the results obtained in independent trials.
Examination of the amino acid sequence of BRCA1 residues 1005–1863 revealed 14 S/T-Q sites that were candidates for phosphorylation by ATR (Canman et al. 1998; Lakin et al. 1999; Tibbetts et al. 1999). Ten of the fourteen identified S/T-Q sites were concentrated in a 300 amino acid region (amino acids 1250–1550), which has been referred to as an SQ cluster domain (SCD) (Cortez et al. 1999). To identify the sites phosphorylated by ATR in vitro, we mutated each of 14 candidate S/T-Q phosphoacceptor sites to Ala, and examined the phosphorylation of the resulting panel of fusion proteins in ATR immune complex kinase assays. Of the four potential phosphoacceptor sites in the BRCA1 (1005–1313) fragment (Ser 1143, Ser 1239, Ser 1280, Ser 1298), Ala substitutions at two sites, Ser 1143 and Ser 1280, reduced the in vitro phosphorylation of GST–BRCA1 (1005–1313) by ATR, whereas substitution of Ser 1239 or Ser 1298 with Ala had little or no effect (Fig. 2C; data not shown). A Ser 1143/Ser 1280 double mutant was a poor substrate for ATR, suggesting that these are the two major in vitro phosphorylation sites on this BRCA1 fragment.
In vitro phosphorylation by ATR of GST–BRCA1 (1314–1863) resulted in the appearance of at least four products with different electrophoretic mobilities after SDS-PAGE. Relative to the wild-type GST–BRCA1 (1314–1863) fragment, Ala substitutions at Ser 1387, Thr 1394, Ser 1423, or Ser 1457 altered the banding pattern of the phosphorylated GST–BRCA1 (1313–1863), and quantitatively reduced the level of 32Pi incorporation into the substrate (Fig. 2D, top). In contrast, Ala substitutions at Ser 1330, Ser 1466, Ser 1524, Thr 1720, or Ser 1755 had little or no effect on either the phosphorylation of the GST–BRCA1 (1314–1863) fragment, or the banding pattern of the phosphorylated fusion protein (data not shown). Although no single mutation eliminated the GST–BRCA1 (1314–1863) electrophoretic mobility shift, a quadruple mutant (GST–BRCA14A) containing Ala substitutions at Ser 1387, Thr 1394, Ser 1423, and Ser 1457 showed no alteration in electrophoretic mobility after phosphorylation by ATR-containing immune complexes (Fig. 2D). The total incorporation of 32Pi into the GST–BRCA14A substrate was reduced by 70% relative to that obtained with wild-type GST–BRCA1 (1314–1863), suggesting that these four residues account for most, but not all of the phosphorylation sites in this fragment.
ATR regulates BRCA1 phosphorylation in intact cells
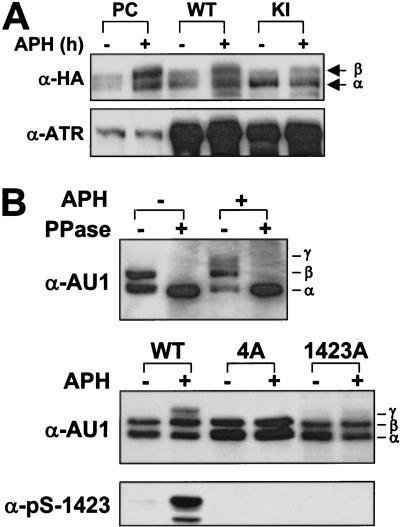
Because ATR phosphorylates BRCA1 in vitro within the SQ cluster domain, and BRCA1 phosphorylation in cells treated with DNA replication inhibitors or UV light is ATM independent, we examined whether ATR contributed to the phosphorylation of BRCA1 in intact cells. We examined BRCA1 phosphorylation in HEK 293T cells that were cotransfected with plasmids encoding full-length BRCA1–HA, and either wild-type ATR (ATRWT) or ATRKI. After transfection, cells were cultured in the absence or presence of APH, and α-HA immunoprecipitates were resolved by SDS-PAGE, followed by α-HA immunoblotting to detect changes in the electrophoretic mobility of BRCA1–HA. In cells cotransfected with vector alone or ATRWT, APH treatment for 24 h increased the level of a more slowly migrating species (β form) of BRCA1–HA (Fig. 3A). The electrophoretic mobility shift was inhibited when the immunoprecipitates were treated with λ phosphatase prior to SDS-PAGE, indicating that the β form represents a hyperphosphorylated species of BRCA1–HA (data not shown). In contrast, coexpression of ATRKI interfered with the appearance of the β form of BRCA1–HA in APH-blocked cells, suggesting that ATR kinase activity is required for maximal BRCA1 phosphorylation induced by a DNA replication block (Fig. 3A). We also noted that in the absence of APH treatment, overexpression of ATRKI caused an accumulation of faster-migrating, hypo-phosphorylated, BRCA1–HA relative to ATRWT-transfected cells (Fig. 3A). This finding suggests that ATR may regulate BRCA1 phosphorylation during the normal cell cycle, as well as in response to stalled DNA replication.
Figure 3.
Overexpression of kinase-inactive ATR inhibits BRCA1 phosphorylation. (A) HEK 293T cells were cotransfected with expression constructs encoding BRCA1-HA and either pcDNA3, wild-type ATR (WT) or kinase-inactive (KI). Cells were then cultured in the absence or presence of 5 μg/ml APH for 24 h. BRCA1-HA was immunoprecipitated from cell lysates and analyzed by SDS-PAGE and immunoblotting with α-HA. Levels of ATR overexpression were determined by α-ATR immunoblot. (B) BRCA1 is modified at one or more ATR phosphorylation sites in intact cells. (top) HEK 293T cells were transfected with an expression construct encoding an amino-terminally-truncated BRCA1 construct (AU1-BRCA1–ΔN) spanning BRCA1 amino acids 1317–1863 (see Materials and Methods). Cells were left untreated or cultured for 4 h in the presence of 5 μg/ml APH. Cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with α-AU1. Where indicated, cell lysates were treated with 400 units of λ phosphatase. The positions of α- β-, and γ-phosphorylated forms of AU1-BRCA1–ΔN are marked. (middle, bottom) HEK 293T cells were transfected with expression constructs encoding wild-type AU1-BRCA1–ΔN (WT), AU1-BRCA1–ΔN containing Ala substitutions at Ser 1387, Thr 1394, Ser 1423, and Ser 1457 (4A), or AU1-BRCA1–ΔN containing a single Ser 1423→Ala substitution (1423A). Transfected cells were then cultured for 4 h in the absence or presence of 5 μg/ml APH. Cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with α-AU1 (middle) or α-pS-1423 (bottom).
As a first step toward determining which BRCA1 residues were phosphorylated by ATR in vivo, we examined the impact of specific Ala mutations on BRCA1 phosphorylation in intact cells. To focus more specifically on the carboxy-terminal region of BRCA1, which contains the in vitro phosphorylation sites for ATR, we prepared an expression construct that encodes an amino-terminally truncated fragment of BRCA1 (BRCA1–ΔN, residues 1317–1863) linked to an AU1 epitope and two tandem copies of the SV40 NLS signal. HEK 293T cells were transiently transfected with the AU1-BRCA1–ΔN expression construct, and then were cultured for 4 h in the absence or presence of APH. Immunoblot analysis revealed that AU1-BRCA1–ΔN migrated as a 75-kD, α, β doublet after separation by SDS-PAGE. Cellular exposure to APH (Fig. 3B) or HU (not shown) induced the appearance of a third AU1-BRCA1–ΔN species (γ form), which displayed a further reduction in electrophoretic mobility. Treatment of extracts from either control or APH-treated cells with λ phosphatase eliminated both the β and γ forms of BRCA1, which confirmed that these electrophoretic variants represented phosphorylated forms of the BRCA1 fragment (Fig. 3B, top). Thus, like full-length BRCA1, AU1-BRCA1–ΔN undergoes multi-site phosphorylation in cells exposed to a DNA replication inhibitor. Next, we generated an AU1-BRCA1–ΔN4A mutant construct bearing Ala substitutions at Ser 1387, Thr 1394, Ser 1423, and Ser 1457. The wild-type or mutant AU1-BRCA1–ΔN constructs were transfected into HEK 293T cells, and the cells were cultured for 4 h in the absence or presence of APH prior to the preparation of cellular extracts. Immunoblot analysis with α-AU1 demonstrated that, in contrast to AU1-BRCA1–ΔNWT, AU1-BRCA1–ΔN4A completely failed to shift into the hyperphosphorylated γ form following APH treatment (Fig. 3B, middle). In addition, an AU1-BRCA1–ΔN Ser 1423→Ala single mutant was almost completely defective with respect to the APH-induced gel mobility shift. As expected, neither AU1-BRCA-AN4A nor AU1-BRCA1–ΔN Ser 1423→Ala was recognized by the α-pS-1423 antibody (Fig. 3B, bottom). Thus, phosphorylation of Ser 1423 appears essential for the generation of electrophoretic variant forms of BRCA1 during DNA replicational arrest.
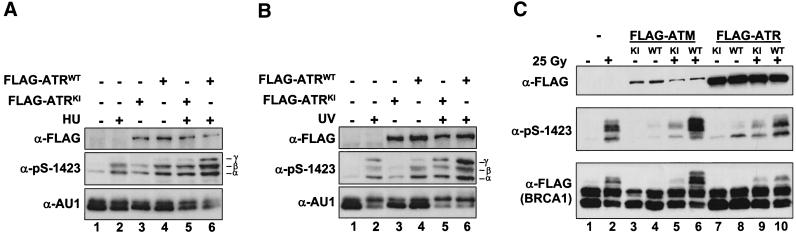
To assess the effects of ATR on the site-specific phosphorylation of BRCA1 on Ser 1423, we cotransfected HEK 293T cells with AU1-BRCA1–ΔN and either FLAG–ATRWT or FLAG–ATRKI expression constructs. After 24 h, the cells were left untreated or were cultured for 1 h in medium containing 2 mM HU. Immunoblot analysis with the α-pS-1423 antibody revealed that HU treatment caused an increase in the level of Ser 1423 phosphorylation in AU1-BRCA1–ΔN (Fig. 4A, lanes 1,2). A similar increase in phospho-Ser 1423 levels was observed following overexpression of FLAG–ATRWT in the absence of HU treatment, indicating that overexpression of ATR enhances the phosphorylation of Ser 1423 in normally cycling cells (Fig. 4A, lanes 1,4). Overexpression of FLAG–ATRKI also induced a variable, smaller increase in Ser 1423 phosphorylation in the absence of HU (Fig. 4A, lane 3; B, lane 3; C, lane 7). Although the actual mechanism is unknown, it is conceivable that expression of FLAG–ATRKI construct itself triggers a DNA damage response, which leads to the activation of a Ser 1423-directed kinase (e.g., ATM). Overexpression of FLAG–ATRWT, but not FLAG–ATRKI enhanced the phosphorylation of Ser 1423 in HU-treated cells (Fig. 4A, cf. lanes 2, 5, and 6). Overexpression of FLAG–ATRWT, but not FLAG–ATRKI, also enhanced Ser 1423 phosphorylation in UV light-treated cells (Fig. 4B). Taken together, the findings indicate that the level of Ser 1423-phosphorylated BRCA1 induced by HU or UV light exposure is related to the level of ATR kinase activity, and suggest that ATR directly phosphorylates BRCA1 at Ser 1423 when cells are exposed to these genotoxic agents.
Figure 4.
Overexpression of wild-type ATR potentiates the phosphorylation of BRCA1 at Ser 1423 in intact cells. (A) Overexpression of FLAG–ATRWT stimulates the HU-induced phosphorylation of Ser 1423 in HEK 293T cells. HEK 293T cells were transfected with an expression vector encoding AU1-BRCA1–ΔN together with empty expression vector (−) or plasmids encoding FLAG–ATRWT, or FLAG–ATRKI. After 24 h, the transfected cells were left untreated or exposed to 2 mM HU, and harvested 1 h later. Cellular extracts were separated by SDS-PAGE and subjected to immunoblot analysis with the α-pS-1423 antibody to detect Ser 1423-phosphorylated BRCA1. Immunoblotting with α-FLAG or α-AU1 antibodies was performed to monitor expression levels of the FLAG–ATR and AU1-BRCA1–ΔN constructs, respectively. α-, β-, and γ-phosphorylated forms of AU1-BRCA1–ΔN are marked. (B) Overexpression of FLAG–ATRWT stimulates UV light-induced phosphorylation of Ser 1423 in HEK 293T cells. The experiment was performed as described in A, except that the transfected HEK 293T cells were exposed to 50 J/m2 UV light, and then harvested 1 h later. Cell lysates were subjected to SDS-PAGE and immunoblot analysis with the indicated antibodies. (C) Relative contributions of ATM and ATR to the IR-induced phosphorylation of BRCA1 on Ser 1423. HEK 293T cells were cotransfected with expression plasmids encoding FLAG-tagged BRCA1 (1351–1552) together with an empty expression vector (−), FLAG–ATR, or FLAG–ATM plasmids. The FLAG–ATR and FLAG–ATM expression constructs encoded either wild-type (WT) or catalytically inactive (KI) proteins. After 24 h, the cells were either left untreated, or exposed to 25 Gy of IR and harvested 1 h later. Cell lysates were analyzed by SDS-PAGE and immunoblot analysis with the α-pS-1423 antibody to detect Ser 1423-phosphorylated BRCA1. Expression levels of FLAG–ATR, FLAG–ATM, and FLAG–BRCA1 (1351–1552) were monitored by immunoblotting with α-FLAG, and are presented at top and bottom, respectively.
Previous studies have suggested that ATR may participate in the signaling of IR-induced DNA damage, although ATM may be the predominant IR-activated kinase (Cliby et al 1998; Banin et al. 1998; Canman et al. 1998; Wright et al 1998; Tibbetts et al. 1999). To gain further insights into the relative roles of ATM and ATR in BRCA1 phosphorylation in IR-damaged cells, we measured the effects of overexpressing either ATM or ATR on the IR-induced BRCA1 mobility shift. For these experiments, we utilized a minimal FLAG–NLS-tagged BRCA1 fragment [FLAG–BRCA1 (1351–1552)] that undergoes a dramatic gel mobility shift following exposure of cells to IR (Cortez et al. 1999). When expressed in HEK 293T cells in the absence of ATM or ATR expression plasmids, FLAG–BRCA1 (1351–1552) was strongly phosphorylated on Ser 1423 at 1 h after cellular exposure to IR (25 Gy) (Fig. 4C, lane 2). Expression of ATMKI or ATRKI partially inhibited IR-induced Ser 1423 phosphorylation, whereas expression of ATMWT and ATRWT enhanced this response (Fig. 4C). Compared with ATRWT, ATMWT consistently showed a greater ability to increase Ser 1423 phosphorylation in IR-treated cells, even though ATM expression levels were 5- to 10-fold lower than the level of ATR (Fig. 4C, cf. lanes 6 and 10). This finding supports the conclusion that ATM is a major mediator of Ser 1423 phosphorylation in response to IR exposure. However, these results further suggest that ATR also plays a role in IR-induced phosphorylation of BRCA1 on Ser 1423.
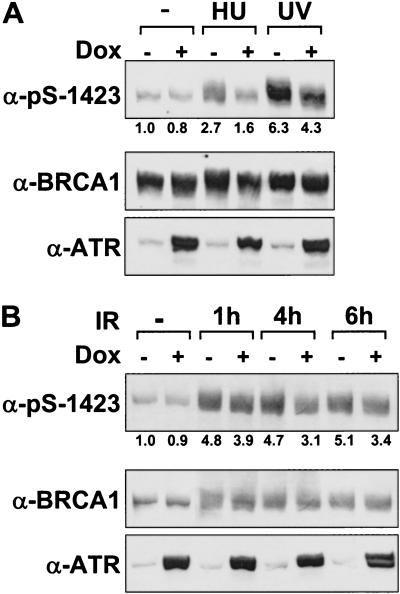
Overexpression of catalytically inactive ATR inhibits the damage-induced phosphorylation of endogenous BRCA1 on Ser 1423
We next sought to determine whether functional ATR is required for the phosphorylation of endogenous, full-length BRCA1 on Ser 1423 in DNA-damaged cells. For these studies, we examined BRCA1 phosphorylation in GM847 human fibroblasts engineered to overexpress FLAG–ATRKI when cultured in the presence of doxycycline (Dox) (Cliby et al. 1998). In the absence of Dox treatment, exposure of GM847 cells to either HU or UV light increased the levels of Ser 1423-phosphorylated BRCA1 by two- to fourfold (Fig. 5A). Dox-induced overexpression of ATRKI inhibited the phosphorylation of Ser 1423 in response to both treatments, but had little effect on basal Ser 1423 phosphorylation (Fig. 5A). Overexpression of ATRKI also partially inhibited Ser 1423 phosphorylation following exposure to IR (Fig. 5B). The strongest inhibitory effect of ATRKI overexpression was observed at 4 and 6 h post-irradiation, when Ser 1423 phosphorylation was reduced by ∼30% relative to the irradiated controls. Considered together, these results support a major role for ATR in the HU- and UV light-induced phosphorylation of BRCA1 on Ser 1423, and suggest that ATR also contributes to the phosphorylation of this site in IR-treated cells.
Figure 5.
Overexpression of catalytically inactive ATR inhibits the DNA damage-induced phosphorylation of BRCA1 on Ser 1423. (A) Inhibition of HU- and UV-induced Ser 1423 phosphorylation. GM847/ATRKI cells were cultured either in the absence (−) or presence (+) of Dox for 96 h to induce ATRKI. Cells were then either left untreated or exposed to 2 mM HU for 4 h, or harvested 1 h after exposure to 200 J/m2 UV light. Cell lysates were prepared and immunoblots were performed with the indicated antibodies. Autoradiograms were quantitated by densitometry and phospho-Ser 1423 levels were normalized to account for differences in total BRCA1 protein levels. (B) Time course of Ser 1423 phosphorylation in response to IR. GM847/ATRKI cells were cultured in the presence of Dox for 96 h and then left untreated or exposed to 20 Gy of IR and harvested at the indicated times. Analysis of Ser 1423-phosphorylated BRCA1 were determined as described in A.
ATR colocalizes with BRCA1 in DNA damage-induced nuclear foci
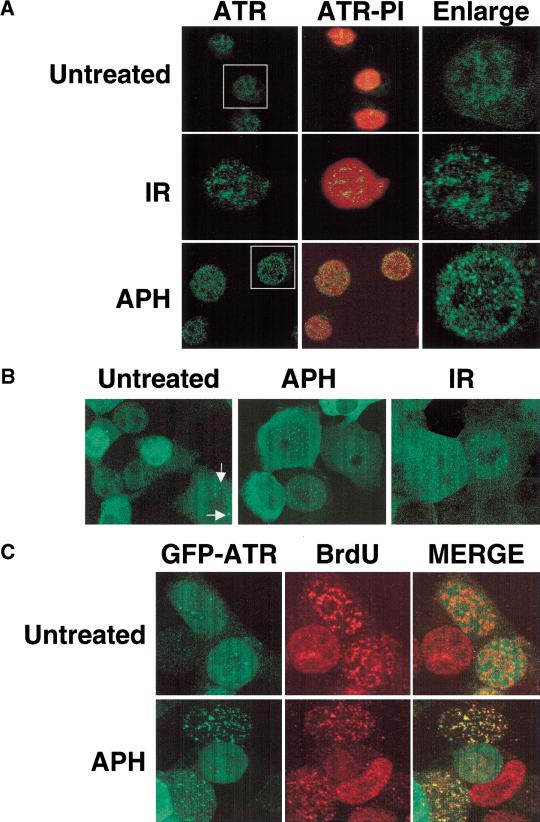
How ATR enhances BRCA1 phosphorylation in response to genotoxic stress is unknown. Whereas ATM kinase activity increases in response to DNA damage (Banin et al. 1998; Canman et al. 1998), the in vitro kinase activity of ATR is not detectably increased when cells are treated with a variety of DNA-damaging agents including IR, alkylating agents, and DNA replication inhibitors (data not shown). Therefore, we examined whether ATR might be altered in response to damage in a manner distinct from kinase activation. Immunostaining of K562 cells with α-ATR antibodies demonstrated that the subcellular localization pattern of ATR changed dramatically following cellular exposure to DNA-damaging agents or DNA replication inhibitors. Prior to treatment, most cells exhibited a uniform nuclear ATR localization pattern. Examination of K562 cells at 4 h after exposure to 10 Gy of IR revealed the presence of intense ATR nuclear foci, which numbered from 10 to more than 100 per cell (Fig. 6A). Exposure of K562 cells to APH caused an even more dramatic increase in the appearance of ATR nuclear foci (Fig. 6A). On average, ATR nuclear foci were observed in 68% of cells that had been cultured in the presence of APH for 24 h. Similar results were obtained with HU (data not shown). The dramatic relocalization of ATR provoked by the DNA replication inhibitors was not apoptosis related, as ATR nuclear foci were reversible, and cells maintained full viability over the course of the experiment. Furthermore, APH induced the formation of ATR nuclear foci in multiple cell lines, including Jurkat T cells, HEK 293T cells, and Raji B cells (data not shown), indicating that the appearance of ATR nuclear foci is a general cellular response to drug-induced S-phase arrest.
Figure 6.
DNA damage-induced alterations in ATR subcellular localization. (A) Immunolocalization of endogenous ATR. Exponentially growing K562 cells were left untreated, treated for 24 h with 5 μg/ml APH, or cultured for 4 h after exposure to 20 Gy IR. Cells were then immunostained with α-ATR antibodies and counterstained with propidium iodide (PI). (B) Localization of recombinant GFP–ATR. HEK 293T cells were transfected with a GFP–ATR expression vector and cultured for 24 h. Samples were then either left untreated, treated for 24 h with 5 μg/ml APH, or cultured for 4 h after exposure to 20 Gy of IR. Arrows highlight typical ATR nuclear foci found in a minority subpopulation of untreated HEK 293T cells. (C) ATR migrates to BrdU-positive sites of DNA replication in APH-blocked cells. The experiment was performed as in A, except that the cells were cultured in the presence of APH for 3 h, released from the APH block, pulse-labeled for 30 min with 100 μM BrdU, and then immunostained with α-BrdU. Foci demonstrating coincident GFP–ATR-and α-BrdU staining appear yellow in the merged image.
To confirm the localization results obtained by α-ATR antibody staining, we transfected HEK 293T cells with a plasmid vector encoding a green fluorescent protein (GFP)–ATR fusion protein, and compared the GFP–ATR localization patterns in untreated versus APH-treated cells. Although the GFP–ATR construct was not localized exclusively in the nucleus, the subnuclear pattern of GFP–ATR fluorescence strongly resembled that of endogenous ATR. In the absence of genotoxic stress, the majority of nuclei (∼90% in multiple experiments) demonstrated a diffuse GFP–ATR localization pattern (Fig. 6B, left), whereas ∼10% of cells demonstrated one or more GFP–ATR-containing dots (Fig. 6B, bottom, left, indicated by arrows). When GFP–ATR-expressing HEK 293T cells were exposed to APH for 24 h, we again observed a dramatic increase in the number of ATR-containing nuclear foci in ∼80% of the treated cells (Fig. 6B, middle). IR (right), and HU (data not shown) also induced the formation of GFP–ATR nuclear foci. Thus, both of the ATR localization assays indicate that cellular exposure to genotoxic stress induces the formation of ATR nuclear foci. In contrast, transfected HA-tagged ATM was not localized to nuclear foci under any of these treatment conditions (data not shown).
The staining pattern observed in APH-treated cells suggested that ATR nuclear foci might be localized near stalled replication forks, which contain regions of single-stranded and/or damaged DNA. To examine this possibility, we compared the pattern of GFP–ATR foci with that of BrdU-labeled DNA in cells that had been cultured in BrdU-containing medium for 30 min after release from a 3-h APH-induced DNA replication block. In untreated cells, the patterns of GFP–ATR and α-BrdU antibody staining were nonoverlapping, and the lack of colocalization was most striking in cells that displayed large, BrdU-positive foci, which are indicative of cells in mid-to-late S phase (Bravo and Macdonald 1987). The nuclear localization pattern of GFP–ATR in these cells was largely diffuse, with few or no foci apparent (Fig. 6C). In contrast, the GFP–ATR and BrdU staining patterns overlapped in APH-arrested cells, and the colocalization was most pronounced in cells arrested in mid-to-late S phase, as indicated by the large BrdU-positive foci (Fig. 6C, bottom). This finding supports the notion that ATR localizes to sites of stalled DNA synthesis in cells exposed to the DNA polymerase α inhibitor.
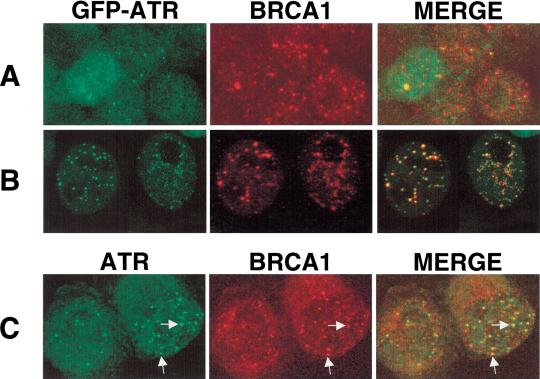
To determine whether ATR nuclear foci contain BRCA1, HEK 293T cells were transfected with GFP–ATR, treated with APH for 24 h, and then immunostained with an α-BRCAl antibody. In the absence of APH treatment, the GFP–ATR and BRCA1 localization patterns were largely distinct and, in the majority of cells that displayed BRCA1 nuclear foci, GFP–ATR was distributed diffusely throughout the nucleus (Fig. 7A). In APH-treated cells, the GFP–ATR and BRCA1 localization patterns overlapped extensively (Fig. 7B). The endogenous ATR and BRCA1 proteins were similarly colocalized in APH-arrested K562 cells, indicating that the previously observed colocalization was not a unique property of the ectopically expressed GFP–ATR (Fig. 7C). Thus, these results suggest that ATR localization is regulated in response to DNA damage or stalled DNA replication, and that ATR and BRCA1 may function together within nuclear foci induced by these agents. The increased phosphorylation of BRCA1 that is observed after treatment with genotoxic agents correlates with an increase in the colocalization of BRCA1 with ATR, and suggests that the regulated localization of these proteins may play an important role in coordinating their function.
Figure 7.
Colocalization of ATR and BRCA1 in APH-induced nuclear foci. HEK 293T cells were transfected with a GFP–ATR expression vector and then either left untreated (A), or cultured in the presence of APH for 24 h (B). Cells were then fixed, permeabilized, and immunostained with a BRCA1-specific monoclonal antibody. Foci that contain both ATR and BRCA1 appear yellow or orange in the merged image. (C) Colocalization of endogenous ATR and BRCA1 in K562 cells. K562 cells were treated with APH for 24 h, and then were fixed and immunostained with α-ATR and α-BRCA1 antibodies. Typical foci that demonstrate colocalization of ATR and BRCA1 are indicated by arrows.
Discussion
The results of this study support a role for ATR as an upstream regulator of BRCA1 phosphorylation during the cellular response to DNA damage and DNA replicational stress. These conclusions are based on the following data: (1) ATM controls only part of the IR-induced phosphorylation of BRCA1 and plays no detectable role in BRCA1 phosphorylation in response to UV, HU, or APH treatments; (2) ATR phosphorylates BRCA1 on six sites in vitro including Ser 1423; (3) overexpression of ATR enhances the basal level of BRCA1 phosphorylation on Ser 1423, and potentiates the induced phosphorylation of this site in response to HU and UV light; (4) interference with ATR function through expression of an inducible kinase-defective ATR mutant reduces endogenous BRCA1 phosphorylation on Ser 1423 in response to HU and UV light, and partially inhibits IR-induced Ser 1423 phosphorylation. These findings also confirm and extend the recent reports that ATM plays an important role in the induction of BRCA1 phosphorylation when cells are exposed to IR (Cortez et al. 1999, Gatei et al. 2000). Taken together, the results of these studies substantiate the hypothesis that ATM and ATR serve as proximate BRCA1 kinases in parallel cell cycle checkpoint pathways activated in response to different types of genotoxic stress. Moreover, the present findings lend further support to the idea that ATR and BRCA1 play continuous roles in genome surveillance during the normal process of DNA replication.
Functional ATR is necessary for BRCA1 phosphorylation in response to multiple forms of genotoxic stress
As noted, the evidence in support of an important role for ATR as an upstream effector of BRCA1 phosphorylation comes from two different experimental model systems. In cotransfection assays, we found that overexpression of wild-type ATR augmented the HU- and UV light-induced phosphorylation of Ser 1423 in HEK 293T cells. In the second approach, ATRKI overexpression was shown to substantially inhibit HU- or UV light-induced phosphorylation of BRCA1 at Ser 1423 in GM847 fibroblasts. The combined results strongly suggest that ATR phosphorylates BRCA1 in response to DNA replication blocks and UV light. In addition, because overexpression of ATRKI partially inhibited Ser 1423 phosphorylation in IR-treated cells, it is likely that ATR is at least partially responsible for the residual phosphorylation of BRCA1 observed in ATM-deficient cells (Fig. 1).
We have provided evidence that ATR phosphorylates BRCA1 in vitro on six Ser/Thr-Gln sites (Ser 1143, Ser 1280, Ser 1387, Thr 1394, Ser 1423, Ser 1457). It is presently unclear how many sites in BRCA1 are phosphorylated by ATR in vivo following exposure to genotoxic stress. However, the observation that a Ser 1423 →Ala substitution does not eliminate the APH-induced gel mobility shift of full-length BRCA1 (R. Tibbetts and D. Cortez, unpubl.) suggests that Ser 1423 is not the only site of ATR-dependent modification. A more direct assessment of the phosphorylation states of individual ATR phosphorylation sites in vivo awaits the development of suitable phospho-specific antibodies.
ATR and ATM may regulate BRCA1 phosphorylation in response to distinct and overlapping types of genotoxic stress
Given the fact that ATM and ATR phosphorylate BRCA1 on at least partially overlapping sites, it may be difficult to determine the relative contributions of these kinases to damage-induced BRCA1 phosphorylation in cell lines with intact ATM/ATR alleles. The results of the present study and two recent studies have indicated that functional ATM is required for optimal phosphorylation of BRCA1 in response to IR (Cortez et al. 1999; Gatei et al. 2000), which is consistent with accumulating evidence that ATM serves as the principal signal transducer during the response to DNA DSBs (Khanna and Lavin 1993; Siliciano et al. 1997; Matsuoka et al. 1998; Brown et al. 1999). In contrast, A-T cells did not display a defect in either the UV- or HU-induced phosphorylation of BRCA1 on Ser 1423 (Fig. 1), which is consistent with these earlier reports that failed to implicate ATM in damage responses initiated by these agents. The finding that HU- or UV light-induced phosphorylation of BRCA1 on Ser 1423 is enhanced by overexpression of wild-type ATR, and inhibited by overexpression of ATRKI, strongly suggests that ATR replaces ATM as the major Ser 1423 kinase when the genotoxic insult is supplied by DNA replication blocks or UV irradiation. Finally, the fact that both ATR and BRCA1 knockout mice display chromosomal abnormalities and die early during embryogenesis (Hakem et al. 1996; Xu et al. 1999; Brown and Baltimore 2000; de Klein et al. 2000) suggests that ATR may additionally regulate BRCA1 during the normal cell cycle. According to this model, cell death results from the accumulation of DNA damage, particularly as a result of uncorrected errors that occur during S phase.
It seems clear that the phosphorylation state of BRCA1 is under complex control by several protein kinases. In addition to cyclin-dependent kinases (Ruffner et al. 1999), ATM, and ATR, a recent report has shown that Chk2 phosphorylates BRCA1 at an independent site (Ser 988) in IR-damaged cells (Lee et al. 2000). The emerging picture is that BRCA1 lies at the intersection of multiple damage response pathways, and that specific phosphorylation events on BRCA1 may allow this protein to orchestrate specific responses to different types of genotoxic stress.
The carboxy-terminal residues in BRCA1 that are phosphorylated by ATR in vitro are proximal to several functional regions, including the transactivation domain, bipartite BRCT domains, and binding sites for p53 and BRCA2 (Chai et al. 1999; Chapman and Verma 1996; Bork et al. 1997; Chen et al. 1998). These phosphorylation events may also regulate interactions between BRCA1 and components of the DNA repair and recombination machinery, perhaps through the generation of binding sites for FHA domain-containing proteins (Sun et al. 1998; Durocher et al. 1999).
ATR nuclear foci: Implications for ATR function and regulation
The findings that cellular exposure to DNA-damaging agents or DNA replication inhibitors causes the formation of ATR nuclear foci offers the first evidence that ATR responds to genetic damage in somatic cells and may have implications for the regulation of ATR-dependent signaling pathways. In contrast to ATM, whose catalytic activity is up-regulated within minutes of cellular exposure to IR or radiomimetic drugs (Banin et al. 1998; Canman et al. 1998), we have been unable to detect an increase in the protein kinase activity of ATR in cells exposed to a variety of DNA-damaging agents (R. Tibbetts , unpubl.). Our failure to detect catalytic activation of ATR may reflect a requirement for cofactors that are lost during cellular lysis and immunoprecipitation. Alternatively, the phosphorylation of substrates by ATR may be regulated primarily through induced changes in proximity to these substrates as a consequence of stalled DNA replication forks or overt DNA damage. Because ATM does not localize to nuclear foci either in response to DNA damaging agents (Watters et al. 1997), or DNA replication inhibitors (R. Tibbetts, unpubl.), our findings raise the interesting possibility that the checkpoint activities of ATM and ATR are regulated by fundamentally different mechanisms.
The colocalization of ATR and sites of BrdU incorporation in APH-treated cells strongly suggests that ATR complexes assemble in the vicinity of stalled DNA replication forks. This conclusion is further supported by the finding that ATR colocalizes with replication fork-associated proteins, including replication protein A and Bloom's syndrome helicase, in HU-treated cells (R. Tibbetts and D. Cortez, unpubl.). An obvious, but as yet unproven possibility is that ATR is required to enforce the DNA replication checkpoint in S-phase-arrested cells. The localization of ATR to sites of arrested DNA synthesis may also reflect maintenance and/or stabilization functions for ATR at stalled DNA replication forks, analogous to those carried out by Mec1p in budding yeast (Desany et al. 1998; Santocanale and Diffley 1998). In addition, ATR nuclear foci may assemble at either potential or active sites of DNA recombination, which can occur as the result of DNA damage or paused DNA replication in somatic cells (Galli and Schiestl 1996; Essers et al. 1997). Consistent with a role in recombination, ATR and BRCA1 are localized to asynapsed regions of paired chromosomes during meiosis, which might reflect a role for the complex in monitoring or promoting homologous recombination (Keegan et al. 1996; Scully et al. 1997b). It is possible that the ATR complex also monitors recombinogenic lesions in somatic cells, which occur at low frequency during a typical S phase, and in much higher numbers when cells are exposed to genotoxic stress (Galli and Schiestl 1996; Takata et al. 1998). Loss of BRCA1 or ATR function may increase genetic instability in proliferating cells due to defective responses to replication fork stalling, base misincorporation errors, and other insults during the normal process of DNA replication. This hypothesis is compatible with the observation that ATR-containing nuclear foci are observed in both untreated, cycling cell populations and cells exposed to extrinsic DNA-damaging agents (see Fig. 7).
In summary, this study establishes a regulatory relationship between ATR and BRCA1 and raises the possibility that, like mutations in BRCA1 itself, alterations in ATR expression or activity may promote genetic instability, and increase susceptibility to the development of breast cancer and other tumors.
Materials and methods
Cell culture and antisera
K562 erythroleukemia and HEK 293T cells were maintained in RPMI-1640 and DMEM medium containing 10% FCS, respectively. ATM− and ATM+ AT22IJE-T fibroblasts were kindly provided by Dr. Yosef Shiloh (Tel Aviv University) and were maintained as described previously (Ziv et al. 1997). GM847 cells that express FLAG epitope-tagged catalytically inactive (ATRKI) ATR under control of the bacterial tet operator have been described previously (Cliby et al. 1998). Induction of ATRKI was accomplished by plating out 2 × 105 cells in 100-mm dishes in culture medium containing 1μg/ml Dox for 4 d. Where indicated, cells were γ irradiated with a 137Cs source or UV irradiated with a Stratalinker UV lamp (254 nm wavelength). For the generation of α-pS-1423 antisera, rabbits were immunized with a keyhole limpet hemocyanin-conjugated BRCA1 phosphopeptide (CVLEQHGpSQPSNS) derived from amino acids 1416–1428 of BRCA1. The resulting antiserum was passed over a column containing the nonphosphorylated peptide, and the unbound antibodies were subsequently affinity purified by binding to and elution from a column containing the phosphopeptide used for immunizations. Antibodies specific for ATR (Ab-1) and BRCA1 (Ab-1, Ab-2) were purchased from Oncogene Science. Other antibodies used in this study included α-BrdU-IU-4 (Caltag), α-AU1 (BAbCo), α-HA-12CA5 (BAbCo), and α-FLAGM2 (Sigma). Aphidicolin (APH, 5 mg/ml) was prepared as a stock solution in dimethylsulfoxide and stored at −80°C until use.
Plasmid constructs and transfections
ATR constructs containing an amino-terminal AU1 (DTYRYI) or FLAG (DYKDDDDK) epitope were generated by replacing a 1.0-kb ATR BamHI–SwaI fragment with a corresponding PCR-generated AU1- or FLAG-tagged ATR BamHI–SwaI fragment in the plasmid pcDNA3.1. GFP–ATR was constructed by subcloning a PCR-generated enhanced GFP cDNA (Clontech) into the unique BamHI site of AU1-ATR:pcDNA3.1. The catalytically inactive ATR mutant in this study contains a Lys-2327→Arg substitution in the ATP-binding pocket (Tibbetts et al. 1999). FLAG–NLS-tagged BRCA1 (1351–1552) and FLAG–ATM expression plasmids used in this study have been described (Cortez et al. 1999). The BRCA1 expression plasmids have been described (Scully et al. 1997b; Brown et al. 1999). AU1-BRCA1–ΔN was created by PCR amplification of full-length BRCA1 using the following primers: 5′-GGATCCATGGACACCTAC CGCTACATCTTGATTGGTTCTTCCAAAC-3′, 5′-GCGGC CGCGTAGTGGCTGTGGGGGATC-3′. The AU1-BRCA1–ΔN PCR fragment was subcloned into the BamHI–NotI sites of pcDNA3.1–2XNLS, which contains a bipartite SV40 nuclear localization sequence immediately downstream of the NotI site. Site-directed mutagenesis was carried out by the QuickChange method (Stratagene) according to the manufacturer's instructions. Transient transfection of HEK 293T cells was performed with the Fugene 6 lipid transfection reagent (Boehringer-Mannheim). All constructs were verified by sequencing.
Microscopy
For immunofluorescence microscopy of endogenous ATR, 1 × 104 K562 cells were collected, washed in PBS, and cytospun onto poly-L-lysine-coated slides. The cells were fixed in methanol at −20°C for 30 min, rehydrated in PBS, and incubated for 30 min with blocking solution (3% BSA, 2% goat serum in PBS). Samples were subsequently overlayed with protein G-purified α-ATR at a concentration of 1.0 μg/ml in blocking solution and incubated at 4°C for 1.5 h. Samples were then washed with PBS, 0.2% Tween-20, and overlayed for 1 h at room temperature with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibody (Caltag) that had been diluted 1:500 in blocking solution. Samples were then washed and incubated with 0.1 μg/ml propidium iodide, 100 μg/ml RNAse A in PBS for 30 min. After extensive washing, specimens were mounted with coverslips and an aqueous anti-fade mounting reagent (Vectashield, Vector Laboratories). For analysis of GFP–ATR localization, HEK 293T cells were plated onto 22-mm2 glass coverslips and transfected with 1 μg of GFP–ATR expression plasmid. Forty-eight hours after transfection, cells were fixed in 4% paraformaldehyde in PBS, and then either mounted onto slides or permeabilized in PBS containing 0.5% NP-40. The permeabilized cells were then immunostained with α-BRCA1 at a concentration of 0.5 μg/ml in blocking solution for 1.5 h at room temperature. Cells were washed and then overlayed for 1 h with a 1:500-diluted cyanin-3 (Cy3)-conjugated goat anti-mouse IgG antibody (Caltag). After staining, cells were washed and mounted as described above. For BrdU colocalization experiments, HEK 293T cells were incubated with 100 μM BrdU in fresh medium for 30 min prior to cell fixation. Cell staining was carried out by incubating cells with a 1:500 dilution of α-BrdU in blocking solution containing 20 mM MgCl2, 0.5% Triton X-100, and 20 units/ml DNAse I (Boehringer) for 2 h. Cells were then washed and incubated with Cy3-conjugated secondary antibody and mounted as described above. All fluorescence images were generated using a Carl Zeiss LSM410 scanning laser confocal microscope.
Protein analysis and kinase assays
Cell extracts were prepared by resuspending washed cell pellets in lysis buffer (25 mM HEPES, pH 7.4, 300 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 20 nM microcystin, 0.5% NP-40, 50 mM β-glycerophosphate) with protease inhibitors (10 μg/ml leupeptin, 5 μg/ml pepstatin A, 5 μg/ml aprotinin). Samples were incubated on ice for 10 min, and then clarified by centrifugation. For full-length BRCA1 electrophoretic mobility shift assays, 6 × 105 HEK 293T cells were plated onto 60-mm dishes and then transfected 16 h later with 4.5 μg of ATR, and 0.5 μg of BRCA1-HA plasmid DNAs. Twenty-four hours after transfection, APH was added to a concentration of 5 μg/ml, and the cells were cultured for an additional 24 h. Cells were resuspended in lysis buffer and immunoprecipitated with 2 μg of α-HA. α-HA immunoprecipitates were resolved by SDS-PAGE through 7.5% polyacrylamide gels, and BRCA1-HA was detected by immunoblotting with α-HA. Levels of ATR expression were determined by α-ATR immunoblotting from whole cell lysates. For AU1-BRCA1–ΔN mobility shift assays, HEK 293T cells were plated as described above and transfected with 2 μg of expression plasmid. Twenty-four hours after transfection, cells were cultured in the absence or presence of APH (5 μg/ml) for 4 h. Cell lysates were prepared and resolved on 10% SDS–polyacrylamide gels. Immunoblots were then carried out by use of α-AU1.
ATR kinase assays were carried out as described (Tibbetts et al. 1999). HEK 293T cells were transiently transfected with 5 μg of either pcDNA3.1, FLAG-tagged wild-type ATR, or catalytically inactive ATR expression constructs. Twenty-four hours following transfection, cell lysates were prepared and immunoprecipitated with 3 μg of α-FLAGM2. ATR immune complexes were washed and then incubated with 1 μg of GST–BRCA1 fusion protein test substrates. Kinase assays were then performed and quantitated as described previously (Tibbetts et al. 1999).
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA76193 and CA52995 to R.T.A. and GM44664 to S.J.E.), a Johnson and Johnson Focused Giving Award to R.T.A., and funding from the Baylor Breast Cancer SPORE to S.J.E.R.S. is supported by a Department of Defense IDEA award (DAMD17–97–1–7180) and a Howard Temin Award (K01 CA79576–01) from the National Cancer Institute. R.S.T is a Fellow of the Leukemia Society of America. D.C. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. K.M.B was supported by a postdoctoral fellowship from the National Cancer Institute (CA83319). S.J.E. is an investigator with the Howard Hughes Medical Institute. R.T.A. is a Glaxo-Wellcome Professor of Molecular-Cancer Biology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL abrah008@mc.duke.edu; FAX (919) 681-8463.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.851000.
References
- Aprelikova ON, Fang BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen RA, Liu ET. BRCA1-associated growth arrest is RB-dependent. Proc Natl Acad Sci. 1999;96:11866–11871. doi: 10.1073/pnas.96.21.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh SY, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: A paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Barlow C, Brown KD, Deng CX, Tagle DA, Wynshaw-Boris A. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat Genet. 1997;17:453–456. doi: 10.1038/ng1297-453. [DOI] [PubMed] [Google Scholar]
- Beamish H, Lavin MF. Radiosensitivity in ataxia-telangiectasia: Anomalies in radiation-induced cell cycle delay. Int J Radiat Biol. 1994;65:175–184. doi: 10.1080/09553009414550211. [DOI] [PubMed] [Google Scholar]
- Beamish H, Williams R, Chen P, Lavin MF. Defect in multiple cell cycle checkpoints in ataxia-telangiectasia postirradiation. J Biol Chem. 1996;271:20486–20493. doi: 10.1074/jbc.271.34.20486. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Bertwistle D, Ashworth A. Functions of the BRCA1 and BRCA2 genes. Curr Opin Genet Dev. 1998;8:14–20. doi: 10.1016/s0959-437x(98)80056-7. [DOI] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- Bravo R, Macdonald B. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: Association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Lee CH, Schwarz JK, Mitiku N, Piwnica-Worms H, Chung JH. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Wolff AC, Chen CY, Fornace AJ, Jr, Kastan MB. The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chai YL, Cui J, Shao N, Shyam E, Reddy P, Rao VN. The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene. 1999;18:263–268. doi: 10.1038/sj.onc.1202323. [DOI] [PubMed] [Google Scholar]
- Chapman MS, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–679. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- Chen Y, Farmer AA, Chen CF, Jones DC, Chen PL, Lee WH. BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res. 1996;56:3168–3172. [PubMed] [Google Scholar]
- Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Galli A, Schiestl RH. Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mut Res. 1996;354:69–75. doi: 10.1016/0027-5107(96)00037-1. [DOI] [PubMed] [Google Scholar]
- Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, Khanna KK. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 2000;60:3299–3304. [PubMed] [Google Scholar]
- Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- Hakem R, de la Pompa JL, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, Firpo E, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Keegan KS, Holtzman DA, Plug AW, Christenson ER, Brainerd EE, Flaggs G, Bentley NJ, Taylor EM, Meyn MS, Moss SB, et al. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes & Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL. ). PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Lavin MF. Ionizing radiation and UV induction of p53 protein by different pathways in ataxia-telangiectasia cells. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. ). Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci. 2000;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Lakin ND, Hann BC, Jackson SP. The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. ). hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- Liu VF, Weaver DT. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Monteiro AN, August A, Hanafusa H. Evidence for a transcriptional activation function of BRCA1 C-terminal region. Proc Natl Acad Sci. 1996;93:13595–13599. doi: 10.1073/pnas.93.24.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Ruffner H, Jiang W, Craig AG, Hunter T, Verma IM. BRCA1 is phosphorylated at Ser 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site. Mol Cell Biol. 1999;19:4843–4854. doi: 10.1128/mcb.19.7.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Brown M, De Caprio JA, Cannistra SA, Feunteun J, Schnitt S, Livingston DM. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–126. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997a;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997b;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Vlasakova V, Chen JJ, Sokolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- Shafman T, Khana KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes & Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram K, Zhang H, Zeng YX, Houvras Y, Peng Y, Wu GS, Licht JD, Weber BL, el-Deiry WS. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KB, Wiliams JW, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes & Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson GE, Chen TT, Stastny VA, Virmani AK, Spillman MA, Tonk V, Blum JL, Schneider NR, Wistuba II, Shay JW, et al. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Watters D, Khanna KK, Beamish H, Birrell G, Spring K, Kedar P, Gatei M, Stenzel D, Hobson K, Kozlov S, et al. Cellular localisation of the ataxia-telangiectasia (ATM) gene product and discrimination between mutated and normal forms. Oncogene. 1997;14:1911–1921. doi: 10.1038/sj.onc.1201037. [DOI] [PubMed] [Google Scholar]
- Westphal CH, Rowan S, Schmaltz C, Elson A, Fisher DE, Leder P. Atm and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat Genet. 1997;16:397–401. doi: 10.1038/ng0897-397. [DOI] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes & Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes & Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tombline G, Weber BL. BRCA1, BRCA2, and DNA damage response: Collision or collusion? Cell. 1998;92:433–436. doi: 10.1016/s0092-8674(00)80936-8. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- Zhou, Z. and Elledge, S.H. 2000. The DNA damage response: Putting checkpoints in perspective. Nature (in press). [DOI] [PubMed]
- Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]