PI-3K–mediated repression of FOXO1 and KLF2 promotes proinflammatory cytokine expression by lineage-committed human CCR6+ Th17/Th22 memory cells.
Abstract
Human memory T cells (TM cells) that produce IL-17 or IL-22 are currently defined as Th17 or Th22 cells, respectively. These T cell lineages are almost exclusively CCR6+ and are important mediators of chronic inflammation and autoimmunity. However, little is known about the mechanisms controlling IL-17/IL-22 expression in memory Th17/Th22 subsets. We show that common γ chain (γc)–using cytokines, namely IL-2, IL-7, and IL-15, potently induce Th17-signature cytokine expression (Il17a, Il17f, Il22, and Il26) in CCR6+, but not CCR6−, TM cells, even in CCR6+ cells lacking IL-17 expression ex vivo. Inhibition of phosphoinositide 3-kinase (PI-3K) or Akt signaling selectively prevents Th17 cytokine induction by γc-cytokines, as does ectopic expression of the transcription factors FOXO1 or KLF2, which are repressed by PI-3K signaling. These results indicate that Th17 cytokines are tuned by PI-3K signaling in CCR6+ TM cells, which may contribute to chronic or autoimmune inflammation. Furthermore, these findings suggest that ex vivo analysis of IL-17 expression may greatly underestimate the frequency and pathogenic potential of the human Th17 compartment.
In addition to Th1 and Th2 cells, it is now accepted that naive CD4+ T cells can differentiate into Th17 or Th22 cells that secrete IL-17 or IL-22, respectively (Annunziato and Romagnani, 2009; Zhou et al., 2009; Wolk et al., 2010). Th17 cells mediate proinflammatory immune responses against some species of extracellular bacteria and most fungal pathogens (Bettelli et al., 2006; Holland et al., 2007; Minegishi et al., 2007; de Beaucoudrey et al., 2008). Th17 cells are also broadly implicated in the pathogenesis of many common autoimmune disorders, including multiple sclerosis, rheumatoid arthritis (RA), psoriasis, and inflammatory bowel disease (Lock et al., 2002; Cua et al., 2003; Fujino et al., 2003; Langrish et al., 2005; Wilson et al., 2007). Th22 cells are involved in skin immunity and remodeling but are also implicated in cutaneous inflammatory conditions such as psoriasis (Eyerich et al., 2009; Wolk et al., 2010). Ex vivo analyses of human peripheral blood T cells indicate that nearly all IL-17–secreting cells, and the majority of IL-22–producing T cells, express the chemokine receptor CCR6 and the transcription factor RORC (Annunziato et al., 2007; Singh et al., 2008; Duhen et al., 2009; Romagnani et al., 2009; El Hed et al., 2010). Accordingly, ectopic expression of RORC in naive T cells or CCR6− memory cells is sufficient to induce IL-17 secretion (Manel et al., 2008). Currently, all effector T cell subsets, including Th17 and Th22 cells, are defined solely based on cytokine expression after ex vivo stimulation (Duhen et al., 2009). However, it is not clear whether this definition accurately reflects the full repertoire of memory T (TM) cells that have the capacity to produce IL-17/IL-22 at sites of inflammation. Moreover, the cytokines and downstream signaling pathways that control IL-17/IL-22 secretion in lineage-committed TM cells remain uncharacterized.
The IL-2 family of cytokines, which signal through multimeric receptors containing the shared common γ chain (γc) subunit, includes IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21. These cytokines, particularly IL-2, IL-7, and IL-15, play pivotal roles in promoting T cell development, homeostasis, and differentiation. In addition to activating Jak–Stat pathways, γc signaling induces the generation of lipid second messengers through activation of phosphoinositide 3-kinase (PI-3K; Rochman et al., 2009). One of these second messengers, PI(3,4,5)P3 (phosphatidylinositol-(3,4,5)-trisphosphate), binds to the PH (pleckstrin homology) domain of proteins and controls the activity and function of several signaling molecules, including the serine/threonine protein kinase Akt (Fruman, 2004). In turn, Akt directly phosphorylates the transcription factor FOXO1 (Forkhead box protein O1), thereby preventing its nuclear translocation and transcriptional activity (Brunet et al., 1999). Inhibition of FOXO1 by PI-3K has been shown to be essential for γc-cytokine signaling–mediated cell survival, proliferation, and glucose utilization in leukemia cells (Barata et al., 2004). In contrast, activation of FOXO1, by way of reduced PI-3K signaling, can lead to the expression of another transcription factor called KLF2 (kruppel-like factor 2; Sinclair et al., 2008; Kerdiles et al., 2009), which has been implicated in the modulation of IFN-γ and IL-4 production in human and mouse T cells (Weinreich et al., 2009, 2010; Bu et al., 2010). However, whether FOXO1 or KLF2 act downstream of PI-3K to regulate effector cytokine production in human TM cells is not yet known.
In searching for stimuli that regulate the production of IL-17 and IL-22 in human TM cells, we found that γc-cytokines, notably IL-2, IL-7, or IL-15, are sufficient to induce de novo expression of IL-17, IL-22, and other Th17-signature cytokines in CCR6+, but not CCR6−, TM cells. Treatment of cytokine-stimulated CCR6+ TM cells with small molecule inhibitors of PI-3K or Akt repressed γc-cytokine–driven IL-17/IL-22 expression, as did ectopic expression of FOXO1 or KLF2. These findings suggest that PI-3K signaling may amplify Th17- or Th22-associated tissue inflammation by promoting proinflammatory cytokine expression in lineage-committed human Th17/Th22 cells. Our results also demonstrate that the frequency of human TM cells harboring the capacity to secrete IL-17/IL-22 in vivo may be substantially higher than what is predicted based on ex vivo cytokine analysis.
RESULTS
De novo induction of IL-17 in CCR6+ IL-17− T cells is driven by γc-cytokines
Nearly all T cells defined as Th17, by virtue of IL-17 expression, also express CCR6, although only a fraction of CCR6+ T cells (∼5–10%) actively express IL-17 upon ex vivo stimulation. Given that both CCR6 and IL-17 are up-regulated by ectopic expression of RORC in human T cells (Manel et al., 2008) and that CCR6 is a direct target gene of the key Th17 transcription factor Stat3 (Durant et al., 2010), we hypothesized that expression of this chemokine receptor may broadly define human T cells that have the capacity to express IL-17, even if they lack IL-17 expression ex vivo. To address this possibility, we FACS sorted primary human CD4+CD45RO+CD25− TM cells stimulated either with beads conjugated to anti-CD3 and anti-CD28 (aCD3/aCD28) antibodies or with PMA and ionomycin into CCR6−IL-17−, CCR6+IL-17+, or CCR6+IL-17− subsets using a cytokine capture assay (Fig. 1 A and Fig. S1 A). Analyses of gene expression directly after sorting confirmed that Il-17a transcript levels were substantially higher in CCR6+IL-17+ cells compared with either CCR6− or CCR6+IL-17− cells, irrespective of the activating stimuli (i.e., aCD3/aCD28 or PMA and ionomycin). However, both CCR6+ T cell subsets displayed elevated RORC expression levels that ranged between 80- and 400-fold higher than those observed in CCR6− TM cells (Fig. 1 B and Fig. S1 B; Singh et al., 2008). Given that RORC was highly expressed in CCR6+ TM cells independent of ex vivo IL-17 production, we next cultured these T cell subsets in IL-2–supplemented medium for 6 d to ask whether CCR6+IL-17− cells could up-regulate IL-17 expression. As expected, a large majority of cells initially sorted as CCR6+IL-17+ maintained high-level IL-17 expression upon restimulation, whereas CCR6−IL-17− cells remained largely IL-17 negative (Fig. 1 C and Fig. S1 C). Remarkably, 20–40% of the CCR6+ cells initially sorted as IL-17− expressed IL-17 after culture with IL-2 (Fig. 1 C and Fig. S1 C). The appearance of IL-17–producing T cells within ex vivo–isolated CCR6+IL-17− cultures was not a result of selective outgrowth of residual IL-17+ cells, as FACS-sorted CCR6+IL-17− and CCR6+IL17+ cells proliferated equally well in response to IL-2 stimulation, as seen by fluorescent dye dilution and mixed co-culture experiments (Fig. S1, D–G).
Figure 1.
De novo expression of IL-17 in CCR6+IL-17− human TM cells is induced by γc-cytokines. (A) Total CD4+ TM cells (CD45RO+CD25−) were stimulated for 18–24 h with aCD3/aCD28 beads and then were FACS sorted into CCR6−IL-17−, CCR6+IL-17−, or CCR6+IL-17+ populations by cytokine capture assay. (B) Cells sorted as in A after aCD3/aCD28 stimulation were analyzed by quantitative (q) PCR for expression of Il-17a and Rorc gene expression. Data are presented as the mean fold change from duplicates from one representative donor and are derived from Ct values (qPCR) ± SD from duplicate samples. The data were obtained from a single donor that is representative of two to three independent experiments using T cells from different donors. (C) Live CCR6−IL-17−, CCR6+IL-17−, or CCR6+IL-17+ T cells were maintained in IL-2–containing medium for 6–7 d and were restimulated with PMA and ionomycin. Cytokine production was determined by intracellular staining. (D) Cytokine gene expression was determined using qNPA (see Materials and methods) in CCR6−IL17−, CCR6+IL-17−, or CCR6+IL-17+ populations expanded for 7 d in IL-2–supplemented medium and restimulated with PMA and ionomycin. Cytokine mRNA expression was normalized to a housekeeping gene (Rpl19) and normalized values are presented as a fold change on a scatter plot (where x axis = CCR6+IL-17−/CCR6−IL-17− and y axis = CCR6+IL-17+/CCR6−IL-17−). Data from A–D is representative of three independent donors. (E) FACS-sorted CCR6−IL-17− cells (white bars), or CCR6+IL-17− cells (black bars) sorted as in A, were cultured in different cytokine conditions as indicated for 6 d. IL-17 expression was determined by FACS analysis after restimulation with PMA and ionomycin. Data are presented as the mean percentage of cells ± SD from three different donors. (F) CCR6+IL-17− TM cells are specifically poised to express IL-17 in human RA patients. Total CD4+ TM cells isolated from the peripheral blood of RA patients were either stimulated directly ex vivo (day 0) with PMA and ionomycin or were stimulated and FACS sorted as in A into CCR6−IL-17− or CCR6+IL-17− cells. These cells were cultured in IL-2–supplemented medium for 7 d before restimulation with PMA and ionomycin (day 7). The frequencies of IL-17–producing cells were determined via intracellular staining and FACS analysis. Data represent the mean (horizontal bars) percentage of IL-17-producing cells from four individual RA patients.
A more comprehensive analysis of cytokine gene expression by human CCR6+IL-17− and CCR6+IL-17+ cells revealed that these two cell types were nearly indistinguishable after culture with IL-2. Specifically, several proinflammatory cytokines canonically associated with the Th17 lineage (IL-17a, IL-17f, IL-22, IL-26, Ccl20, and IL-21) were comparably expressed in both CCR6+IL-17− and CCR6+IL-17+ cells, although both CCR6+ cell types expressed substantially higher levels of these cytokines compared with CCR6− TM cells (Fig. 1 D). Importantly, differential expression of Th17-signature cytokines by both CCR6+ populations compared with CCR6− T cells was specific, as all three populations expressed roughly equivalent levels of other cytokines not related to the Th17 lineage, namely Tnf, Il4, and Ifng (Fig. 1 D).
Given that IL-2 is the prototype of the IL-2 family of cytokines, all of which signal through cytokine receptors comprised in part by the γc subunit, we next asked whether other γc-cytokines could also induce de novo IL-17 production by CCR6+IL-17− TM cells. Both IL-7 and IL-15 induced similar levels of IL-17 expression in CCR6+IL-17− T cells, whereas IL-23, which is known to enhance Th17 cell differentiation (Ivanov et al., 2007), did not influence IL-17 expression in the absence of IL-2 (Fig. 1 E). In contrast, and as observed for IL-2, neither IL-7 nor IL-15 induced IL-17 expression in CCR6− TM cells. In addition, CCR6+, but not CCR6−, IL-17− TM cells isolated from peripheral lymphoid organs of wild-type C57B/6 mice were capable of producing IL-17 after 6 d in culture with IL-2 (Fig. S2).
These findings suggest that ex vivo analyses of IL-17 expression underestimate the frequency of TM cells that can express IL-17 in inflammatory settings. Because several studies have investigated changes in Th17 frequencies within autoimmune patient cohorts, we asked whether CCR6+IL-17− TM cells isolated from the peripheral blood of patients with RA could be similarly induced to express IL-17 by IL-2 stimulation. Indeed, we observed that CCR6+, but not CCR6−, IL-17− TM cells from RA patients up-regulated IL-17 after culture with IL-2 to similar levels as those observed in healthy adult donors (Fig. 1 F). Collectively, these data demonstrate that CCR6+ TM cells are uniquely poised to express IL-17 in response to IL-2 stimulation irrespective of their IL-17 phenotype ex vivo. These results also indicate that this inflammatory feature of CCR6+ TM cells is conserved between humans and mice.
IL-17 induction in response to γc-cytokine stimulation is conserved in heterogeneous CCR6+ TM cell subsets
Human CCR6+ Th17 cells have been reported to be enriched within CXCR3− or CD161+ subsets as cells of these subphenotypes produce more IL-17 upon ex vivo stimulation compared with either CCR6+CXCR3+ or CCR6+CD161− cells (Acosta-Rodriguez et al., 2007; Cosmi et al., 2008). We confirmed that a significant proportion of human CCR6+ TM cells coexpressed either CD161 or CXCR3 and that CD161 expression was enriched within CCR6+ cells, whereas expression of CXCR3 was enriched within the CCR6− compartment (Fig. 2 A). As expected, CCR6+CD161+ TM cells were enriched for IL-17–producing cells upon ex vivo stimulation compared with CCR6+CD161− cells, whereas CCR6+CXCR3+ cells produced less IL-17 but more IFN-γ than CCR6+CXCR3− cells (Fig. 2 A). To address the possibility that IL-17 up-regulation by CCR6+IL-17− cells is restricted to predefined CCR6+ subpopulations, we fractionated CCR6+IL-17− cells based on either CD161 or CXCR3 expression and expanded these subsets in the presence of IL-2 (Fig. 2 A). After expansion and restimulation, we found that all CCR6+IL-17− subsets were capable of high-level IL-17 production in contrast to CCR6− TM cells (Fig. 2, B and C). Consistent with the ex vivo analyses of these subsets, however, the frequency of IL-17+ cells was somewhat higher (approximately twofold) in the CD161+ or CXCR3− subsets compared with CD161− or CXCR3+ counterparts, respectively (Fig. 2, B and C). Moreover, CCR6+CXCR3+IL-17− cells maintained their propensity to express IFN-γ as compared with CCR6+CXCR3− memory cells (Fig. 2 C). These data demonstrate that heterogeneous subpopulations of CCR6+ T cells share a capacity to express IL-17 in response to γc-cytokine stimulation.
Figure 2.
IL-17 induction in response to γc-cytokine stimulation is a conserved feature shared by heterogeneous CCR6+ TM cell subsets. (A) Ex vivo (day 0) isolated CD4+ TM cells were costained with CCR6, CD161, and CXCR3. The frequency of CCR6+ cells positive or negative for either CD161 (left) or CXCR3 (right) are shown. Ex vivo cytokine production was determined after PMA and ionomycin stimulation within each gated CCR6+ TM cell subset as indicated. FACS plots show the intracellular expression of IL-17, IL-22, and IFN-γ. (B) FACS-sorted CCR6−IL-17−, CCR6+CD161+/−IL-17− (left), or CCR6+CXCR3+/−IL-17− (right) cells were cultured in IL-2–supplemented medium for 7 d and restimulated with PMA and ionomycin. Cytokine production was determined by intracellular staining for IL-17, IL-22, and IFN-γ. Each set of data is representative of three donors.
PI-3K/Akt signaling mediates Th17 cytokine induction by γc-cytokines
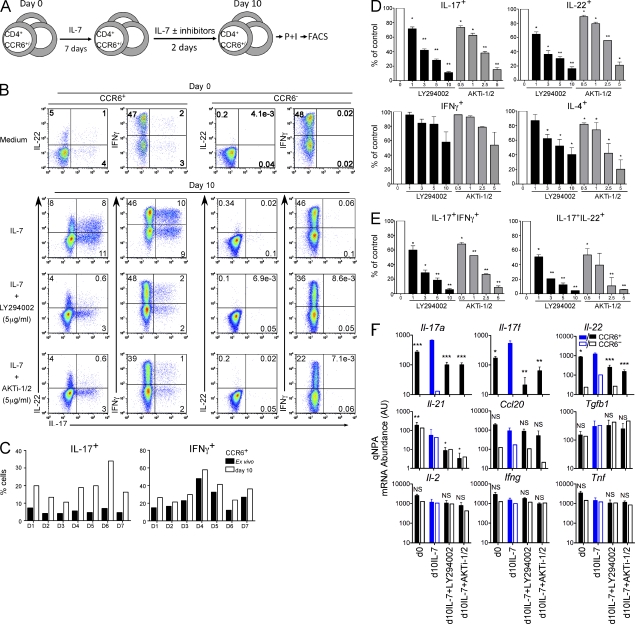
We have previously shown that γc-cytokines, such as IL-7, can promote T cell survival in the absence of TCR signaling and do so without inducing significant cell division (Unutmaz et al., 1994, 1999). Given that IL-7 and IL-15 could substitute for IL-2 in inducing Th17 cytokine expression in TCR-activated CCR6+ TM cells, we next asked whether γc-cytokine signaling was sufficient to promote IL-17/IL-22 expression in resting human CCR6+ TM cells not stimulated through the TCR (Fig. 3 A). Indeed, stimulation of resting CCR6+CD45RO+CD25− TM cells with IL-7 for 10 d consistently enhanced the expression of both IL-17 and IL-22 more than threefold compared with the same cells isolated ex vivo (day 0; Fig. 3, B and C). Importantly, IL-7–mediated induction of IL-17 and IL-22 expression was restricted to CCR6+ cells, as CCR6− TM cells stimulated with IL-7 failed to express IL-17 or IL-22 (Fig. 3 B). Moreover, culture of resting CCR6+ or CCR6− TM cells with IL-7 had almost no impact on IFN-γ production (Fig. 3, B and C). Stimulation of resting CCR6+ TM cells with IL-15 led to similar increases in IL-17/IL-22 expression (unpublished data), whereas another γc-cytokine, IL-4, did not promote Th17 cytokine production in resting CCR6+ TM cells or confer TM cell survival (Fig. S3 B and not depicted). Interestingly, IL-23 did not significantly synergize with IL-7 to further induce IL-17 or IL-22 expression (Fig. S3 B).
Figure 3.
PI-3K/Akt inhibitors suppress IL-17 and IL-22 production from IL-7–stimulated CCR6+ TM cells. (A) Resting CD4+CD45RO+CD25−CCR6+/− T cells were cultured in IL-7–containing medium for 7 d followed by culture with or without PI-3K signaling inhibitors for an extra 2 d. Cells were then stimulated with PMA and ionomycin and subjected to intracellular staining and FACS analysis as described in Materials and methods. (B) Blockade of PI-3K signaling suppresses IL-17 and IL-22 production induced by IL-7 from CCR6+ TM cells. Intracellular cytokine staining was performed on CCR6+ and CCR6− TM cells ex vivo (day 0) and after different culturing conditions (day 10) as described in A. CCR6+/− TM cells were cultured with IL-7 alone or plus 5 µg/ml LY294002 or 5 µg/ml AKTi-1/2. (C) Frequency of IL-17–producing T cells is increased by γc-cytokine stimulation across different donors. CCR6+ TM cells isolated from seven healthy donors (D) were stimulated either ex vivo or at day 10 after culture with IL-7. (D and E) PI-3K and Akt inhibitors inhibit IL-17, IL-22, and IL-4 expression in IL-7–activated CCR6+ TM cells in a dose-dependent manner. Cells cultured in IL-7 with or without LY294002 or AKTi-1/2 at different concentrations (µg/ml) were collected and analyzed. The frequency of single (D) or double (E) cytokine-producing cells without inhibitors treatment (control, 0) was normalized to 100%. Cytokine percentage of control was calculated accordingly. Data are presented as the mean percentage cytokine ± SD from at least three different donors. (F) Cytokine gene expression in CCR6+/CCR6− TM cells. Different cytokine gene expression was analyzed by qNPA from either ex vivo freshly isolated CCR6+ (filled rectangle) or CCR6− (empty rectangle) cells at day 0 (d0) and day 10 (d10) after culturing in IL-7–containing medium in the absence or presence of 5 µg/ml PI-3K/Akt inhibitors. Data from CCR6+ TM cells are presented as mean mRNA abundance (AU) ± SD from three different donors. The statistical analyses, either ex vivo or at day 10 with PI-3K inhibitor treatments, are relative to day 10 IL-7 only (filled blue rectangle). Data from CCR6− is representative out of three donors. *, P < 0.05; **, P < 0.005; ***, P < 0.001. NS, no significance.
Human CCR6+ T cells can express IL-17 alone or in combination with other proinflammatory cytokines such as IL-22 or IFN-γ. Cells coexpressing IFN-γ and IL-17 have been shown to share functional characteristics with those expressing IL-17 alone (Boniface et al., 2010), whereas the function of IL-17+IL-22+ cells is unclear. IL-7 stimulation had even more profound effects on the proportion of cells that coexpressed either IL-22 or IFN-γ with IL-17; these double cytokine-producing cells increased more than fivefold in response to IL-7 signaling (Fig. 3 B). Together, these results demonstrate that γc-cytokines, particularly IL-2, IL-7, and IL-15 potently enhance IL-17/IL-22 secretion by CCR6+ TM cell subsets.
Signaling through γc-containing cytokine receptors regulates T cell survival and homeostasis through activation of the PI-3K–Akt pathway (Rochman et al., 2009). To determine if this pathway also controls expression of IL-17 and IL-22, we treated CCR6+ TM cells that had been cultured for 1 wk in IL-7 with or without the PI-3K inhibitor LY294002 or the Akt inhibitor AKTi-1/2 for additional 48 h (Fig. 3 A). We confirmed that treatment of Jurkat cells with 1–10 µg/ml of either inhibitor down-regulated cytokine-dependent phosphorylation of Akt without adversely affecting cell viability (Fig. S3 A). Control- or compound-treated T cells were then restimulated to analyze their impact on cytokine production. Treatment with either LY294002 or AKTi-1/2 repressed γc-mediated IL-17/IL-22 induction in a dose-dependent manner, whereas expression of IL-4 or IFN-γ was either less reduced or not affected, respectively (Fig. 3, B, D, and E). Inhibitory effects of the PI-3K and Akt inhibitors were even more profound when considering the proportion of T cells coexpressing IL-22 or IFN-γ together with IL-17 (Fig. 3 E). Furthermore, treatment with PI-3K or Akt inhibitors also repressed IL-17 and IL-22 expression in TCR-activated CCR6+ TM cells cultured in IL-2 (Fig. S3, C–E). These effects were reversible because IL-17/IL-22 levels were restored upon removal of PI-3K or Akt inhibitors from culture medium (unpublished data). Moreover, IL-17/IL-22 repression by PI-3K or Akt inhibition was not associated with changes in Rorc or Foxp3 expression levels (Fig. S3 F).
We next evaluated the impact of PI-3K or Akt inhibition on the expression of a broader panel of both Th17-related and unrelated cytokines to determine how selectively this pathway regulates inflammatory cytokine expression. In addition to IL-17A, IL-17F, and IL-22, Th17 cells have been shown to preferentially express IL-21 (Nurieva et al., 2007), IL-26 (in human cells), CCL20 (Wilson et al., 2007), and, in some instances, IL-10 (Zhu and Paul, 2010). Although intracellular IL-10 production was not detectable in either CCR6+ or CCR6− TM cells upon stimulation ex vivo or after culture with IL-7, we found that, unlike IL-17 and IL-22, ex vivo production of IL-21 was equivalent in both human CCR6− and CCR6+ TM cell subsets (Fig. S4, A and B). Similarly, and in contrast to IL-17 and IL-22, expression of IL-21 was not increased in either CCR6+ or CCR6− TM cells upon stimulation with IL-7, and in fact was slightly reduced (Fig. 3 F; and Fig. S4, A and B). IL-21 expression was, however, sensitive to treatment with PI-3K or Akt inhibitors (Fig. S4, A and B). Ccl20 was preferentially expressed ex vivo by CCR6+ TM cells (Fig. 1 D) and was neither significantly induced by IL-7 stimulation nor repressed by PI-3K pathway inhibition (Fig. 3 F and Fig. S4 C). The expression of other non–Th17-related cytokines, such as Il2, Ifng, Tnf, and Tgfb1 were not influenced by IL-7 stimulation or by treatment with PI-3K or Akt inhibitors (Fig. 3 F and Fig. S4 C). Thus, PI-3K signaling downstream of γc-cytokine receptors promotes the expression of Th17-signature cytokines (Il-17a, Il-17f, and Il-22) within CCR6+ TM cells and does so with a high degree of specificity over other unrelated cytokines.
FOXO1 and KLF2 repress IL-17/IL-22 expression
PI-3K activation downstream of γc-cytokine stimulation negatively regulates FOXO1 nuclear localization and transcriptional activity (Birkenkamp and Coffer, 2003). Because we found cytokine-driven IL-17 and IL-22 expression by CCR6+ TM cells to be sensitive to PI-3K or Akt inhibition, we next asked whether ectopic expression of FOXO1 could also repress γc-mediated IL-17/IL-22 expression. Human CCR6+ TM cells were activated and transduced with control (empty vector [EV]) or FOXO1-expressing lentiviruses and then expanded in the presence of IL-2, FOXO1 overexpression was confirmed by intracellular staining (Fig. S5 A), and these cells were then restimulated to evaluate effects on cytokine production. Analogous to treatment of cells with PI-3K and Akt inhibitors, FOXO1 overexpression reduced both IL-17 and IL-22 secretion (Fig. 4, A and B).
Figure 4.
IL-17/IL-22 production from CCR6+ TM cells is suppressed by ectopic FOXO1 expression. (A) T cells activated through their TCR were transduced with FOXO1-GFP and cultured in IL-2 for 14 d then stimulated with PMA and ionomycin, stained for intracellular cytokines, and subjected to FACS analysis. (B) The frequency of single or double cytokine-producing cells from FOXO1 or EV-transduced cells is shown. The horizontal bars are presented as mean frequency of cytokine producing cells from four independent individuals. *, P < 0.05. NS, no significance.
Another transcription factor, KLF2, is also reported to be negatively regulated by PI-3K signaling (Sinclair et al., 2008). Unlike FOXO1, however, KLF2 is repressed by PI-3K signaling primarily at the level of gene expression (Weinreich et al., 2009). To investigate whether KLF2 may regulate IL-17/IL-22 secretion by CCR6+ TM cells, we first analyzed the kinetics of Klf2 mRNA expression in TM cells. Consistent with previous studies (Kuo et al., 1997a; Grayson et al., 2001; Wu and Lingrel, 2005), we found that Klf2 expression was rapidly down-regulated after TCR activation (>100-fold; Fig. 5 A). Klf2 mRNA expression rebounded somewhat after extended culture of TCR-activated T cells in IL-2, although even under these conditions Klf2 mRNA levels remained 10–20-fold lower than those seen in resting T cells (Fig. 5 A). Klf2 expression was also decreased in resting CCR6+ TM cells upon stimulation with IL-7 alone (Fig. 5 B). These data indicate that γc-dependent induction of IL-17/IL-22 parallels Klf2 down-regulation. Importantly, treatment of TM cells with PI-3K/Akt inhibitors reversed cytokine-mediated Klf2 down-regulation (Fig. 5 B and Fig. S5 B).
Figure 5.
Klf2 expression is regulated by γc-cytokine–induced PI-3K signaling pathway. (A) Klf2 message is abundant in resting CD4+ T cells and down-regulated upon activation through the TCR. Total cellular RNA was extracted from cell pellets of resting aCD3/aCD28 bead–activated CD4+ TM cells. The RNA was subjected to RT-qPCR for Klf2 and b-actin mRNA quantification. (B) Blockade of PI-3K signaling by LY294002 treatment up-regulates Klf2 transcripts in a dose-dependent manner in CCR6+ TM cells cultured in IL-7. CCR6+ TM cells were cultured in IL-7–containing medium as in Fig. 3 A. Total RNA from either ex vivo freshly isolated (day 0) or cultured in IL-7 (day 10) with or without LY294002 at the indicated concentrations was collected and subjected to RT-qPCR to detect Klf2 levels. (C) Ectopic transduction of KLF2 in TCR-activated IL-2–cultured TM cells maintains Klf2 message levels comparable to resting cells. Data shown is from one representative of three different donors.
To directly address the putative function of PI-3K–mediated Klf2 down-regulation on IL-17 and IL-22 expression, we restored KLF2 expression in cytokine-stimulated TM cells using lentiviral transduction (Fig. S5 C). Human TM cells transduced with KLF2-expressing lentiviruses displayed Klf2 mRNA levels comparable to either resting T cells (Fig. 5 C) or cytokine-stimulated T cells treated with LY294002 (Fig. 5 B). Upon restimulation, human CCR6+ TM cells ectopically expressing KLF2 produced less IL-17 or IL-22 compared with cells transduced with control lentiviruses (Fig. 6, A and B), whereas neither IFN-γ nor IL-4 was affected (Fig. 6 A). In contrast to KLF2 overexpression, shRNA-mediated silencing of endogenous KLF2 (Fig. S5 D) enhanced IL-17/IL-22 secretion by human CCR6+ TM cells (Fig. 6, A and B). Interestingly, T cells expressing KLF2 shRNA also displayed enhanced IL-4 expression, particularly within the CCR6+ subset (Fig. 6 A), suggesting that KLF2 may regulate expression of a broader set of cytokines in TM cell subsets.
Figure 6.
KLF2 suppresses IL-17 and IL-22 production from CCR6+ TM cells in the presence of ectopic RORC expression. (A and B) KLF2- or shKLF2-transduced CCR6+ TM cells cultured for 2 wk in IL-2 medium were treated with PMA and ionomycin and then subjected to intracellular IL-17, IL-22, IFN-γ, and IL-4 staining. shCont., control shRNA; shKLF2, KLF2 shRNA. The frequency of single or double cytokine-producing cells from either EV- or control shRNA–transduced cells was normalized to 100%. (C) RORC overexpression and silencing of KLF2 are synergistic on induction of IL-17. CCR6− TM cells co-transduced with shKLF2- and RORC-expressing lentiviruses or their corresponding controls were restimulated with PMA and ionomycin for 5 h. Cells were then stained with antibodies against IL-17, IL-22, IFN-γ, and IL-4 and shown as dot plots in C from a representative donor or, graphically, from multiple donors in D. Data are presented as mean percentage of cytokine ± SD from at least three individual donors and independent experiments. *, P < 0.05; **, P < 0.005. NS, no significance.
CCR6− TM cells also express KLF2, and shRNA-mediated knockdown of KLF2 in these cells also enhanced residual IL-17 or IL-22 expression (Fig. 6, C and D). Because high-level IL-17 expression by CCR6− TM cells can only be achieved by forced expression of RORC (Fig. 6, C and D; Manel et al., 2008), we next asked whether KLF2 regulates RORC-mediated IL-17 expression in CCR6− TM cells. Indeed, knockdown of KLF2 further enhanced RORC-induced IL-17 production in CCR6− TM cells (Fig. 6, C and D). These findings suggest that endogenous KLF2 restrains RORC-mediated induction of IL-17.
DISCUSSION
In this paper, we show that Th17-signature cytokines, particularly IL-17 and IL-22, are dynamically regulated within human CCR6+ TM cells by γc-cytokine signals. IL-17 and IL-22 expression by CCR6+ TM cells is controlled by PI-3K/Akt-dependent signaling that involves the repression of both FOXO1 and KLF2. We also report that CCR6 expression on nonregulatory (i.e., CD25−) TM cells broadly defines lineage-committed Th17 cells; phenotypically diverse subpopulations of CCR6+ (e.g., CD161+/−, CXCR3+/−) TM cells all share a capacity to express Th17 cytokines in response to γc-cytokine stimulation, irrespective of whether or not they express IL-17 ex vivo. Importantly, we show that the specific conversion of ex vivo–isolated CCR6+, but not CCR6−IL-17−, TM cells is conserved in mice (Fig. S2). Collectively, these results suggest that IL-17 and IL-22 production by CCR6+ TM cells may be influenced in a bystander fashion by γc-cytokines present at sites of inflammation or infection.
These results also suggest that current methods of calculating Th17 cell frequencies based solely on ex vivo IL-17 production may not represent the true size or inflammatory potential of the human Th17 memory cell compartment. This becomes particularly important for how Th17 cells are evaluated and quantified in clinical disease settings. Indeed, we have found that a substantial portion of CCR6+, but not CCR6−, IL-17− TM cells isolated ex vivo from the peripheral blood of RA patients convert into IL-17 producers upon culture with γc-cytokines (Fig. 1 F).
The ability of CCR6+ TM cells to express IL-17 may be a result of the fact that, in contrast to CCR6− TM cells, CCR6+ cells express high levels of RORC ex vivo, independent of IL-17. This is indeed likely given that ectopic expression of RORC can induce IL-17 expression in CCR6− TM cells (Fig. 6; Manel et al., 2008). These findings further suggest that RORC is necessary, but not sufficient, for IL-17 production by human TM cells. We propose that RORC synergizes with γc-mediated PI-3K and Akt signaling to trans-activate IL-17 expression. Two additional results support a model in which γc-cytokine signaling acts together with RORC to control IL-17 production by itself, as opposed to regulating Th17 lineage commitment. First, IL-7 stimulation, treatment with PI-3K or Akt inhibitors, and overexpression of FOXO1 or KLF2 did not affect RORC mRNA expression levels (Fig. S3 F and not depicted). Second, inhibition of IL-17 and IL-22 expression by treatment with LY294002 was completely reversible (unpublished data).
IL-23 is another cytokine thought to be important for the stability and proinflammatory functions of Th17 cells, particularly in the mouse (Zheng et al., 2007; Zhou et al., 2007). However, we found that IL-23 was not able to compensate for γc-cytokines in the induction of IL-17/IL-22 expression from CCR6+ human TM cells, even though these cells expressed Il23R mRNA (unpublished data). Conceivably, IL-23 could enhance IL-17–secreting cells but this effect may be attributed to preferential expansion of CCR6+IL-23R+ TM cells that are more likely to express IL-17 upon TCR triggering. It is also reasonable to speculate that a more dominant role of IL-23 in driving IL-17–associated pathology is through its reported effects on IL-17 expression within TCR-γ/δ lymphocytes or other innate immune cells (Moens et al., 2011).
PI-3K is activated in T cells in response to TCR engagement or γc-cytokine stimulation (Rochman et al., 2009; Smith-Garvin et al., 2009), which in turn results in the phosphorylation and activation of Akt (Coffer et al., 1998). On one hand, PI-3K/Akt signaling positively regulates cell metabolism through activation of mTORC1 (Gibbons et al., 2009). On the other hand, activated Akt also inhibits the transcriptional activity or expression of FOXO1 or KLF2, respectively (Fabre et al., 2008; Sinclair et al., 2008). We found that the induction of IL-17 and IL-22 expression by γc-cytokines was reversed by short-term (48 h) treatment with PI-3K or Akt inhibitors. Furthermore, overexpression of either FOXO1 or KLF2 recapitulated the effects of the PI-3K or Akt inhibitors on IL-17 and IL-22 induction. Conversely, shRNA-mediated silencing of endogenous KLF2 enhanced IL-17 and IL-22 expression by CCR6+ TM or RORC-expressing CCR6− TM cells. We therefore conclude that γc-dependent activation of PI-3K and Akt drives expression of IL-17 and IL-22, preferentially through a pathway that involves FOXO1 and KLF2 down-modulation (Fig. S6; Brunet et al., 1999; Barata et al., 2004; Fabre et al., 2005, 2008; Sinclair et al., 2008). However, as rapamycin treatment (Kopf et al., 2007) and mTOR deficiency (Delgoffe et al., 2009, 2011) have been reported to negatively regulate Th17 differentiation in naive mouse T cells, it will be of interest to determine whether mTOR signaling also controls Th17-signature cytokine production in human CCR6+ TM cells.
The mechanism by which FOXO1 and KLF2 negatively regulate IL-17 and IL-22 expression in human TM cells is unclear. The most obvious possibility is that both FOXO1 and KLF2 function nonredundantly as transcriptional repressors of IL-17 and IL-22 (Fig. S6). However, it is also plausible that FOXO1 regulates IL-17/IL-22 expression indirectly by inducing KLF2 expression. Indeed, previous studies have shown that genetic ablation of Foxo1 leads to down-regulation of Klf2 expression (Fabre et al., 2008; Sinclair et al., 2008; Kerdiles et al., 2009). Consistent with these studies, we found that lentiviral overexpression of FOXO1 in human TM cells led to increased Klf2 expression (unpublished data). It will be important in future studies to determine the mechanism by which these and other transcription factors acutely regulate IL-17/IL-22 expression in human CCR6+ TM cells.
The negative regulation of IL-17 and IL-22 expression represents a novel function of KLF2. KLFs are a large family of zinc-finger transcription factors (Kaczynski et al., 2003; van Vliet et al., 2006); they play important roles in cellular differentiation and homeostasis (Kuo et al., 1997b; Wani et al., 1999; Atkins and Jain, 2007; Jiang et al., 2008; Yusuf et al., 2008; Yamada et al., 2009). KLF2 specifically has been reported to maintain naive T cell quiescence and survival in the mouse (Anderson et al., 1995; Kuo et al., 1997a,b; Wani et al., 1998, 1999; Kuo and Leiden, 1999; Buckley et al., 2001; Di Santo, 2001; Jiang et al., 2008; Yamada et al., 2009). It also regulates T cell emigration from the thymus (Carlson et al., 2006; Sebzda et al., 2008) through regulation of S1P1 (sphingosine-1 phosphate-1 receptor), CD62L, and the chemokine receptor CCR7 (Carlson et al., 2006; Sebzda et al., 2008; Weinreich et al., 2009). T cells from Klf2-deficient mice display enhanced production of IL-4 (Weinreich et al., 2009, 2010). In support of these findings, we observed slightly elevated IL-4 production from CCR6+ TM cells after shRNA-mediated KLF2 silencing. Moreover, statins (HMG-CoA reductase inhibitors), which are clinically used to treat atherosclerosis, have also been shown to induce KLF2 expression (Parmar et al., 2005). More recently, statins have been found to be broadly antiinflammatory and capable of repressing IL-17 secretion (Zhang et al., 2008). In light of our findings, it will be important to determine whether the regulation of IL-17 expression by statins is achieved through a KLF2-dependent mechanism.
In conclusion, our data indicate that human CCR6+ TM cells represent a broad proinflammatory effector lineage that expresses IL-17 and IL-22 in a dynamic manner relying on synergy between γc-cytokine signaling and RORC. Local immune responses and subsequent accumulation of γc-cytokines may thus contribute to Th17/Th22-driven inflammation, which may be sensitive to PI-3K- or Akt-targeted therapies. Moreover, our findings strongly suggest that ex vivo analyses of IL-17 and IL-22 secretion may vastly underestimate the true frequency and proinflammatory potential of Th17/Th22 cells in vivo. These results shed new light on the regulation of proinflammatory cytokine production by human TM cells and also call for a broader phenotypic definition of Th17/Th22 subsets within both healthy human donors and autoimmune patient populations.
MATERIALS AND METHODS
Human T cell purification and activation.
PBMCs from healthy individuals (New York Blood Center, New York, NY) were prepared using Ficoll-plaque plus (GE Healthcare). Whole blood samples from RA patients were purchased from Bioreclamation and were processed similar to healthy donor blood. All healthy donor and patient samples were consented before purchase, were none-identifiable, and were approved by the institutional human subjects board. CD4+ T cells were isolated using Dynal CD4 Positive Isolation kit (Invitrogen) directly from purified PBMCs and were >99% pure. CD4+ T cells were sorted by a FACSAria cell sorter (BD) into CD45RO+CD25− TM cells. TM cells were activated either by anti-CD3– and anti-CD28 (aCD3/aCD28)–coated beads (1:4 bead/cell ratio; Invitrogen) for 18–24 h or by 20 ng/ml PMA and 500 ng/ml ionomycin for 4 h and further FACS sorted based on CCR6 and IL-17 expression into CCR6−IL-17−, CCR6+IL-17+, or CCR6+IL-17− subsets (as determined by secretion assay labeling in L-17 secretion assay). Sorted cells (>99% pure) were cultured in serum-free Xvivo-15 medium (Lonza) at 37°C and 5% CO2 for 6–7 d before restimulation with PMA and ionomycin. Culture conditions included: 10 U/ml recombinant human IL-2 (BD), 20 ng/ml IL-7, 20 ng/ml IL-15, or 20 ng/ml IL-23 (all from R&D Systems). To determine the fold expansion, CCR6+IL-17+ or CCR6+IL-17− subsets were labeled with CFSE (Invitrogen) or eFluor 670 (eBioscience), respectively, as previously described (Oswald-Richter et al., 2004). Alternatively, CCR6+ or CCR6− TM cells directly sorted from CD4+ T cells were either maintained in IL-7 or activated with aCD3/aCD28-coated beads and cultured in IL-2 containing RPMI 1640 medium with 10% FCS (Oswald-Richter et al., 2007). In the experiments using PI-3K signaling inhibitors, cells were treated with PI-3K–specific inhibitor, LY294002 (EMD) or the Akt-specific inhibitor AKTi-1/2 (EMD) for 2 d before PMA and ionomycin restimulation.
Mouse T cell isolation and culture.
Wild-type C57B6/J mice were purchased from The Jackson Laboratory, and all mice were used in accordance with a protocol approved by the animal care and use committee of Sirtris pharmaceuticals. Single cell suspensions were prepared from spleens and peripheral lymph nodes after red blood cell lysis (Sigma-Aldrich). CD4+ T cells were enriched by magnetic cell sorting using the mouse CD4+ T cell isolation kit II (Miltenyi Biotec) in accordance with manufacturer’s instructions, and these cells were stimulated for 18–24 h using plate-bound anti-CD3 (eBioscience) and anti-CD28 (BD) antibodies as previously described (Sundrud et al., 2009). Activated T cells were then stained for cell surface receptors and subjected to mouse IL-17 capture assay for FACS sorting into CD62LloCCR6−IL-17−, CD62LloCCR6+IL-17−, or CD62LloCCR6+IL-17+ subsets (see Staining and FACS analysis). Sorted mouse T cell populations were cultured in DME medium (Mediatech, Inc., USA) containing 10% FCS (Invitrogen). To expand sorted T cell populations, 10 U/ml recombinant human IL-2 (BD) and mouse CD3/CD28 T cell expander beads (1:2 bead/cell ratio; Invitrogen) were added and cells were cultured for 6 d before restimulation.
IL-17 secretion assay.
Manufacturer’s protocols (Miltenyi Biotec) for labeling human and mouse IL-17–secreting cells were followed. In brief, activated T cells were washed in cold buffer (PBS + 0.5% BSA + 2 mM EDTA) and labeled with IL-17 catch reagent for 5 min on ice. Prewarmed medium was then added and the cells were incubated at 37°C on a shaker for 45 min. For human IL-17 capture experiments, cells were washed once more with cold buffer and PE-conjugated IL-17 detection antibodies were added and incubated on ice for 10 min. For mouse IL-17 capture experiments, all steps were identical, except that the IL-17 detection antibody used was biotinylated, and a secondary staining step using APC-conjugated streptavidin was used. Cells were then washed and stained with additional cell surface antibodies at room temperature for 20 min. These cells were then washed once more with cold buffer, resuspended in serum-free RPMI 1640 medium, filtered, and FACS sorted. Antibodies used for human IL-17 secretion sorts were CCR6-PercpCy5.5 (cloneTG7/CCR6; BioLegend) and CD25–Alexa Fluor 700 (clone BC96; BioLegend). Antibodies for mouse IL-17 secretion sorts were CD4–Pacific orange (Invitrogen), CD62L–Pacific blue (eBioscience), CCR6-PE (BioLegend), IL-17A–biotin (Miltenyi Biotec), and streptavidin-APC (eBioscience).
Plasmids and gene cloning.
Full-length human KLF2 cDNA (NM_016270; pCMV6-XL4-KLF2; Origene) was cloned into a lentiviral vector at EcoRI sites. This vector consists of HIV-derived vector (HDV), an internal ribosome entry site (IRES), and a gene for RFP. The primers for KLF2 amplification were: 5′ forward, 5′-CCGGAATTCGCCATGGCGCTGAGTGAACCCATC-3′; and 3′ reverse, 5′-CCGGAATTCCTACATGTGCCGTTTCATGTGCAG-3′. Wild-type FOXO1 (FOXO1-GFP) was a gift from T.G. Unterman (University of Illinois, Chicago, IL). FOXO1-GFP fusion protein was engineered into HDV in frame with XbaI and XhoI sites. The primers for FOXO1 amplification were: 5′ forward, 5′-GCTCTAGAGCATTGCCATGGCCGAGGCGCCT-3′; and 3′ reverse, 5′-TTCTCGAGGCTTACTTGTACAGCTCGTC-3′. The lentivirus-encoding RORC gene, RORC-IRES-HSA, was a gift from D. Littman (Skirball Center, New York University School of Medicine, NY, NY). Short hairpin RNA against KLF2 (shKLF2; Invitrogen) was engineered into a lentiviral vector, as previously described (Antons et al., 2008), targeting position 922 in the cDNA: 5′-GGCAAGACCTACACCAAGAGT-3′.
Lentiviruses production and transductions.
The lentiviruses pseudotyped with VSV-G envelope were generated as previously described (Unutmaz et al., 1999). All lentiviruses expressed GFP, RFP, or heat stable antigen (HSA) as the marker in place of the nef gene, as previously described (Unutmaz et al., 1999; Oswald-Richter et al., 2004, 2007). Viral titers were measured as described (Unutmaz et al., 1999; Oswald-Richter et al., 2007) and ranged from 1–30 × 106 IFU/ml. Activated TM cells were transduced with different genes, such as FOXO1, KLF2, and RORC, or KLF2 shRNA and maintained in IL-2 containing 10% FCS RPMI 1640 medium for 14 d before reactivation with PMA and ionomycin.
Staining and FACS analysis.
Cells were stained as previously described (Oswald-Richter et al., 2007). For intracellular staining, T cell cultures were stimulated for 5 h at 37°C with PMA and ionomycin and GolgiStop (BD). Stimulated cells were washed with PBS and stained with Fixable Viability Dye eFluor450 (eBioscience) to gate on live cells. Cells were then fixed and permeabilized by commercially available Foxp3 intracellular staining kit (eBioscience) or Cytofix/Cytoperm kit (BD) as per the manufacturer’s protocol. Antibodies used for surface and intracellular staining included: CD45RO-FITC; CD25-PE; IFN-γ–FITC (clone 4S. B3), PE Cy7, or A700; IL-4-APC or PE; IL-17A (IL-17)–Pacific blue (clone BL168), FITC, or PercpCy5.5; IL-10–APC (BioLegend); IL-22–PE (R&D Systems), or APC (eBioscience); IL-21–PE (clone 22URT1; eBioscience); FOXO1 rabbit monoclonal antibody; phospho-AKT (Ser473)–Alexa Fluor 488; Rabbit IgG isotype control–Alexa Fluor 488 (Cell Signaling Technology); and CCR6-biotin (BD). Secondary antibodies used were: streptavidin-APC or PercpCy5.5 (BD); goat anti–mouse-APC (BD); and anti–rabbit–Alexa Fluor 647 (Invitrogen). All data were acquired on LSRII instrument (BD) using FACSDiva software. Data analyses were done using FlowJo software (Tree Star). For phospho-Akt staining, Jurkat cells were fixed with Cytofix buffer (BD) prewarmed to 37°C for 10 min. After spinning down, the cells were permeabilized with ice-cold Phosflow Perm buffer III (BD) on ice for 30 min and were stained with phospho-antibody.
RNA isolation and quantitative PCR.
105 purified T cells were snap frozen in liquid nitrogen for total RNA isolation. Total RNA was isolated using RNeasy isolation kit (QIAGEN) according to the manufacturer’s protocol. The extracted RNA was then reverse transcribed into cDNA and quantified using TaqMan Cell to CT kit (Applied Biosystems). The following TaqMan primers and probe sets were purchased from Applied Biosystems: human KLF2 (Hs00360439_g1), RORC (Hs01076120_g1), Il-17a (Hs00174383_m1), b-Actin (Hs03023880_g1), and Foxp3 (Hs00203958_m1). Data were acquired and analyzed on an ABI 7500 sequence detection system. Quantitative real-time PCR data were analyzed by normalizing Ct values of genes of interest to the Ct values of the housekeeping gene b-actin.
Quantitative nuclease protection assay (qNPA).
T cell cultures were collected after PMA and ionomycin activation. Cells were resuspended at 4 × 106/ml in sample lysis buffer (High Throughput Genomics, Inc.), heated to 95°C for 10 min and stored at −80°C. mRNA levels were quantified in sample lysates by qNPA (High Throughput Genomics, Inc.) using four probes per transcript (see below for sequences), and samples were normalized based on abundance of a control (housekeeper) gene (e.g., Rpl19). Assay quality was monitored by a negative control (Arabidopsis thaliana DNA binding/transcription factor (Ant). Human oligonucleotide probes were generated against: the housekeeper gene ribosomal protein L19 (Rpl19; NM_000981), IL-17A (Il17a; NM_002190), and the negative control Arabidopsis thaliana DNA binding/transcription factor (Ant). Raw expression levels were normalized to a housekeeping gene (Rpl19). Probe sequences are listed below: Rpl19 (1), 5′-AATGAAATCGCCAATGCCAACTCCCGTCAGCAGATCCGGAAGCTCATCAA-3′; Rpl19 (2), 5′-TGATCATCCGCAAGCCTGTGACGGTCCATTCCCGGGCTCGATGCCGGAAA-3′; Rpl19 (3), 5′-CAGAGAAGGTCACATGGATGAGGAGAATGAGGATTTTGCGCCGGCTGCTC-3′; Rpl19 (4), 5′-CAAGCGGATTCTCATGGAACACATCCACAAGCTGAAGGCAGACAAGGCCC-3′; Ant (1), 5′-CAGTCACTGAGCTTATCCATGAGCCCTGGGTCACAATCTAGCTGCATCAC-3′; Ant (2), 5′-CTCCAAAGGTGGAGGATTTCTTTGGGACCCATCACAACAACACAAGTCAC-3′; Ant (3), 5′-CGAAGGAACAACAACAGCATTGTCGTCAGGAATACTGAAGACCAAACCGC-3′; Ant (4), 5′-GGGAGGTTATGATATGGAGGAGAAAGCTGCTCGAGCATATGATCTTGCTG-3′; Il-17a (1), 5′-CCCTCAGGAACCCTCATCCTTCAAAGACAGCCTCATTTCGGACTAAACTC-3′; Il-17a (2), 5′-TAACACTTGGCCAAGATATGAGATCTGAATTACCTTTCCCTCTTTCCAAG-3′; Il-17a (3), 5′-TGATGGTCAACCTGAACATCCATAACCGGAATACCAATACCAATCCCAAA-3′; Il-17a (4), 5′-CCTGGTCCTGCGCAGGGAGCCTCCACACTGCCCCAACTCCTTCCGGCTGG-3′; Il-17f (1), 5′-GTCATCCACCATGTGCAGTAAGAGGTGCATATCCACTCAGCTGAAGAAGC-3′; Il-17f (2), 5′-ATGACAGTGAAGACCCTGCATGGCCCAGCCATGGTCAAGTACTTGCTGCT-3′; Il-17f (3), 5′-TCAAGGAAAGGAAGACATCTCCATGAATTCCGTTCCCATCCAGCAAGAGA-3′; Il-17f (4), 5′-GGACTCTTAATAAGACCTGCACGGATGGAAACAGAAAATATTCACAATGT-3′; Il-22 (1), 5′-CACTGCAGGCTTGACAAGTCCAACTTCCAGCAGCCCTATATCACCAACCG-3′; Il-22 (2), 5′-GCTAAGGAGGCTAGCTTGGCTGATAACAACACAGACGTTCGTCTCATTGG-3′; Il-22 (3), 5′-CTTCTCTTGGCCCTCTTGGTACAGGGAGGAGCAGCTGCGCCCATCAGCTC-3′; Il-22 (4), 5′-CTCGAGTTAGAATTGTCTGCAATGGCCGCCCTGCAGAAATCTGTGAGCTC-3′; Il-4 (1), 5′-GGCGGGCTTGAATTCCTGTCCTGTGAAGGAAGCCAACCAGAGTACGTTGG-3′; Il-4 (2), 5′-GCTGATAAGATTAATCTAAAGAGCAAATTATGGTGTAATTTCCTATGCTG-3′; Il-4 (3), 5′-TTTGTCAGCATTGCATCGTTAGCTTCTCCTGATAAACTAATTGCCTCACA-3′; Il-4 (1), 5′-TTTCCTATTGGTCTGATTTCACAGGAACATTTTACCTGTTTGTGAGGCAT-3′; Il-21 (1), 5′-CTGAAGAGGAAACCACCTTCCACAAATGCAGGGAGAAGACAGAAACACAG-3′; Il-21 (2), 5′-GAAGGCCCAACTAAAGTCAGCAAATACAGGAAACAATGAAAGGATAATCA-3′; Il-21 (3), 5′-GGATCTAACTTGCAGTTGGACACTATGTTACATACTCTAATATAGTAGTG-3′; Il-21 (4), 5′-CCAGTCCTGGCAACATGGAGAGGATTGTCATCTGTCTGATGGTCATCTTC-3′; Il-26 (1), 5′-CCCAAGCTGTTGACGCTCTCTATATCAAAGCAGCATGGCTCAAAGCAACG-3′; Il-26 (2), 5′-TAGTCACTCTGTCTCTTGCCATTGCCAAGCACAAGCAATCTTCCTTCACC-3′; Il-26 (3), 5′-CGGCATGTTAGGTGATTCAGAATAGACAAGAAGGATTTAGTAAATTAACG-3′; Il-26 (4), 5′-AAGTACATTGTGTCAACTTAATTTAAAGTATGTAACCTGAATTAACTCGT-3′; Ifng (1), 5′-CCAGTGCTTTAATGGCATGTCAGACAGAACTTGAATGTGTCAGGTGACCC-3′; Ifng (2), 5′-CTGGTGCTTCCAAATATTGTTGACAACTGTGACTGTACCCAAATGGAAAG-3′; Ifng (3), 5′-AAGAACTACTGATTTCAACTTCTTTGGCTTAATTCTCTCGGAAACGATGA-3′; Ifng (4), 5′-AAATGAATATCTATTAATATATGTATTATTTATAATTCCTATATCCTGTG-3′; Tnf (1), 5′-CAAGACCACCACTTCGAAACCTGGGATTCAGGAATGTGTGGCCTGCACAG-3′; Tnf (2), 5′-GGACCTCTCTCTAATCAGCCCTCTGGCCCAGGCAGTCAGATCATCTTCTC-3′; Tnf (3), 5′-GTGTCTGTAATCGCCCTACTATTCAGTGGCGAGAAATAAAGTTTGCTTAG-3′; Tnf (4), 5′-TGTAGCCCATGTTGTAGCAAACCCTCAAGCTGAGGGGCAGCTCCAGTGGC-3′; Ccl20 (1), 5′-CACTTGCACATCATGGAGGGTTTAGTGCTTATCTAATTTGTGCCTCACTG-3′; Ccl20 (2), 5′-TTTGTTTAAGCATCACATTAAAGTTAAACTGTATTTTATGTTATTTATAG-3′; Ccl20 (3), 5′-ATAAAATTATATTTGGGGGGGAATAAGATTATATGGACTTTCTTGCAAGC-3′; Ccl20 (4), 5′-GCGAATCAGAAGCAGCAAGCAACTTTGACTGCTGTCTTGGATACACAGAC-3′; Il-2 (1), 5′-CACAGAACTGAAACATCTTCAGTGTCTAGAAGAAGAACTCAAACCTCTGG-3′; Il-2 (2), 5′-AAACTTTCACTTAAGACCCAGGGACTTAATCAGCAATATCAACGTAATAG-3′; Il-2 (3), 5′-CTGCTGGATTTACAGATGATTTTGAATGGAATTAATAATTACAAGAATCC-3′; Il-2 (4), 5′-GCTACCTATTGTAACTATTATTCTTAATCTTAAAACTATAAATATGGATC-3′; Tgfb1 (1), 5′-CGAGCCCTGGACACCAACTATTGCTTCAGCTCCACGGAGAAGAACTGCTG-3′; Tgfb1 (2), 5′-AACACATCAGAGCTCCGAGAAGCGGTACCTGAACCCGTGTTGCTCTCCCG-3′; Tgfb1 (3), 5′-CCTACATTTGGAGCCTGGACACGCAGTACAGCAAGGTCCTGGCCCTGTAC-3′; and Tgfb1 (4), 5′-GTGACAGCAGGGATAACACACTGCAAGTGGACATCAACGGGTTCACTACC-3′.
Western blot analysis.
KLF2 transient transfected HEK293T cells were lysed in RIPA buffer (Sigma-Aldrich) and mixed with 2× Laemmli sample loading buffer (Bio-Rad Laboratories), boiled, and separated on 10.0% polyacrylamide gels containing SDS (Thermo Fisher Scientific). After electrophoresis, proteins were transferred to a PVDF membrane by electroblotting and incubated for 1 h at room temperature in blocking buffer (5% nonfat dry milk in PBS). The blocked blot was exposed to the primary antibody rabbit anti-KLF2 (Invitrogen) 1:500 or anti-tubulin (Sigma-Aldrich) in blocking buffer with constant mixing. After extensive washing, bound antibodies were detected by chemiluminescence using horseradish peroxidase–conjugated species-specific secondary antibodies as described by the manufacturer (GE Healthcare).
Statistical analysis.
Data were analyzed using Prism software (GraphPad Software) using a Student’s t test for statistical analysis. P < 0.05 is considered significant.
Online supplemental material.
Fig. S1 shows the induction of IL-17 from CCR6+IL-17− TM cells stimulated with PMA/ionomycin. Fig. S2 demonstrates that mouse CCR6+IL-17− TM cells cultured ex vivo with γc-cytokines are able to express IL-17, similar to human CCR6+IL-17− TM cells described in Fig. 1. Fig. S3 demonstrates that CCR6+ TM cells activated through TCR and cultured in IL-2 express reduced level of IL-17 and IL-22 upon blockage of PI-3K/Akt signals. Fig. S4 presents more cytokine profiles upon PI-3K/Akt inhibitors treatment. Fig. S5 confirms the overexpression and the knockdown of FOXO1 and KLF2. Fig. S6 proposes a potential model of how γc-cytokine signals modulate IL-17 and IL-22 production from CCR6+ TM cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20102516/DC1.
Acknowledgments
We thank Frances Mercer, Angie Zhou, Stephen Rawlings, and Dr. Alka Khaitan for critical reading and suggestions, Dr. Terry G. Unterman for the FOXO1 plasmid, and Dr. Dan Littman for the RORC plasmid.
This work was supported by National Institutes of Health grants R21AI087973 and R01AI065303.
Disclosure: R. Ramesh, I.M. Djuretic, T.J. Carlson, and M.S. Sundrud are affiliated with a commercial organization, Tempero pharmaceuticals, which also provided financial support to the laboratory of D. Unutmaz at New York University. The authors have no conflicting or competing financial interests.
Footnotes
Abbreviations used:
- EV
- empty vector
- PI-3K
- phosphoinositide 3-kinase
- RA
- rheumatoid arthritis
- qNPA
- quantitative nuclease protection assay
References
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- Anderson K.P., Kern C.B., Crable S.C., Lingrel J.B. 1995. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: identification of a new multigene family. Mol. Cell. Biol. 15:5957–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F., Romagnani S. 2009. Heterogeneity of human effector CD4+ T cells. Arthritis Res. Ther. 11:257 10.1186/ar2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F., Cosmi L., Santarlasci V., Maggi L., Liotta F., Mazzinghi B., Parente E., Filì L., Ferri S., Frosali F., et al. 2007. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204:1849–1861 10.1084/jem.20070663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antons A.K., Wang R., Oswald-Richter K., Tseng M., Arendt C.W., Kalams S.A., Unutmaz D. 2008. Naive precursors of human regulatory T cells require FoxP3 for suppression and are susceptible to HIV infection. J. Immunol. 180:764–773 [DOI] [PubMed] [Google Scholar]
- Atkins G.B., Jain M.K. 2007. Role of Krüppel-like transcription factors in endothelial biology. Circ. Res. 100:1686–1695 10.1161/01.RES.0000267856.00713.0a [DOI] [PubMed] [Google Scholar]
- Barata J.T., Silva A., Brandao J.G., Nadler L.M., Cardoso A.A., Boussiotis V.A. 2004. Activation of PI3K is indispensable for interleukin 7–mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J. Exp. Med. 200:659–669 10.1084/jem.20040789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Birkenkamp K.U., Coffer P.J. 2003. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem. Soc. Trans. 31:292–297 10.1042/BST0310292 [DOI] [PubMed] [Google Scholar]
- Boniface K., Blumenschein W.M., Brovont-Porth K., McGeachy M.J., Basham B., Desai B., Pierce R., McClanahan T.K., Sadekova S., de Waal Malefyt R. 2010. Human Th17 cells comprise heterogeneous subsets including IFN-gamma-producing cells with distinct properties from the Th1 lineage. J. Immunol. 185:679–687 10.4049/jimmunol.1000366 [DOI] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 96:857–868 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- Bu D.X., Tarrio M., Grabie N., Zhang Y., Yamazaki H., Stavrakis G., Maganto-Garcia E., Pepper-Cunningham Z., Jarolim P., Aikawa M., et al. 2010. Statin-induced Krüppel-like factor 2 expression in human and mouse T cells reduces inflammatory and pathogenic responses. J. Clin. Invest. 120:1961–1970 10.1172/JCI41384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley A.F., Kuo C.T., Leiden J.M. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc—dependent pathway. Nat. Immunol. 2:698–704 10.1038/90633 [DOI] [PubMed] [Google Scholar]
- Carlson C.M., Endrizzi B.T., Wu J., Ding X., Weinreich M.A., Walsh E.R., Wani M.A., Lingrel J.B., Hogquist K.A., Jameson S.C. 2006. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 442:299–302 10.1038/nature04882 [DOI] [PubMed] [Google Scholar]
- Coffer P.J., Jin J., Woodgett J.R. 1998. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J. 335:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmi L., De Palma R., Santarlasci V., Maggi L., Capone M., Frosali F., Rodolico G., Querci V., Abbate G., Angeli R., et al. 2008. Human interleukin 17–producing cells originate from a CD161+CD4+ T cell precursor. J. Exp. Med. 205:1903–1916 10.1084/jem.20080397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua D.J., Sherlock J., Chen Y., Murphy C.A., Joyce B., Seymour B., Lucian L., To W., Kwan S., Churakova T., et al. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 421:744–748 10.1038/nature01355 [DOI] [PubMed] [Google Scholar]
- de Beaucoudrey L., Puel A., Filipe-Santos O., Cobat A., Ghandil P., Chrabieh M., Feinberg J., von Bernuth H., Samarina A., Jannière L., et al. 2008. Mutations in STAT3 and IL12RB1 impair the development of human IL-17–producing T cells. J. Exp. Med. 205:1543–1550 10.1084/jem.20080321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., Powell J.D. 2009. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 30:832–844 10.1016/j.immuni.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Pollizzi K.N., Waickman A.T., Heikamp E., Meyers D.J., Horton M.R., Xiao B., Worley P.F., Powell J.D. 2011. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 12:295–303 10.1038/ni.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Santo J.P. 2001. Lung Krüpple-like factor: a quintessential player in T cell quiescence. Nat. Immunol. 2:667–668 10.1038/90598 [DOI] [PubMed] [Google Scholar]
- Duhen T., Geiger R., Jarrossay D., Lanzavecchia A., Sallusto F. 2009. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 10:857–863 10.1038/ni.1767 [DOI] [PubMed] [Google Scholar]
- Durant L., Watford W.T., Ramos H.L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H.W., Kanno Y., Powrie F., O’Shea J.J. 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 32:605–615 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hed A., Khaitan A., Kozhaya L., Manel N., Daskalakis D., Borkowsky W., Valentine F., Littman D.R., Unutmaz D. 2010. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J. Infect. Dis. 201:843–854 10.1086/651021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich S., Eyerich K., Pennino D., Carbone T., Nasorri F., Pallotta S., Cianfarani F., Odorisio T., Traidl-Hoffmann C., Behrendt H., et al. 2009. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Invest. 119:3573–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre S., Lang V., Harriague J., Jobart A., Unterman T.G., Trautmann A., Bismuth G. 2005. Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J. Immunol. 174:4161–4171 [DOI] [PubMed] [Google Scholar]
- Fabre S., Carrette F., Chen J., Lang V., Semichon M., Denoyelle C., Lazar V., Cagnard N., Dubart-Kupperschmitt A., Mangeney M., et al. 2008. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J. Immunol. 181:2980–2989 [DOI] [PubMed] [Google Scholar]
- Fruman D.A. 2004. Phosphoinositide 3-kinase and its targets in B-cell and T-cell signaling. Curr. Opin. Immunol. 16:314–320 10.1016/j.coi.2004.03.014 [DOI] [PubMed] [Google Scholar]
- Fujino S., Andoh A., Bamba S., Ogawa A., Hata K., Araki Y., Bamba T., Fujiyama Y. 2003. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 52:65–70 10.1136/gut.52.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons J.J., Abraham R.T., Yu K. 2009. Mammalian target of rapamycin: discovery of rapamycin reveals a signaling pathway important for normal and cancer cell growth. Semin. Oncol. 36:S3–S17 10.1053/j.seminoncol.2009.10.011 [DOI] [PubMed] [Google Scholar]
- Grayson J.M., Murali-Krishna K., Altman J.D., Ahmed R. 2001. Gene expression in antigen-specific CD8+ T cells during viral infection. J. Immunol. 166:795–799 [DOI] [PubMed] [Google Scholar]
- Holland S.M., DeLeo F.R., Elloumi H.Z., Hsu A.P., Uzel G., Brodsky N., Freeman A.F., Demidowich A., Davis J., Turner M.L., et al. 2007. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357:1608–1619 10.1056/NEJMoa073687 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., Zhou L., Littman D.R. 2007. Transcriptional regulation of Th17 cell differentiation. Semin. Immunol. 19:409–417 10.1016/j.smim.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Chan Y.S., Loh Y.H., Cai J., Tong G.Q., Lim C.A., Robson P., Zhong S., Ng H.H. 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10:353–360 10.1038/ncb1698 [DOI] [PubMed] [Google Scholar]
- Kaczynski J., Cook T., Urrutia R. 2003. Sp1- and Krüppel-like transcription factors. Genome Biol. 4:206 10.1186/gb-2003-4-2-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdiles Y.M., Beisner D.R., Tinoco R., Dejean A.S., Castrillon D.H., DePinho R.A., Hedrick S.M. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10:176–184 10.1038/ni.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf H., de la Rosa G.M., Howard O.M., Chen X. 2007. Rapamycin inhibits differentiation of Th17 cells and promotes generation of FoxP3+ T regulatory cells. Int. Immunopharmacol. 7:1819–1824 10.1016/j.intimp.2007.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.T., Leiden J.M. 1999. Transcriptional regulation of T lymphocyte development and function. Annu. Rev. Immunol. 17:149–187 10.1146/annurev.immunol.17.1.149 [DOI] [PubMed] [Google Scholar]
- Kuo C.T., Veselits M.L., Barton K.P., Lu M.M., Clendenin C., Leiden J.M. 1997a. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11:2996–3006 10.1101/gad.11.22.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.T., Veselits M.L., Leiden J.M. 1997b. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 277:1986–1990 10.1126/science.277.5334.1986 [DOI] [PubMed] [Google Scholar]
- Langrish C.L., Chen Y., Blumenschein W.M., Mattson J., Basham B., Sedgwick J.D., McClanahan T., Kastelein R.A., Cua D.J. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240 10.1084/jem.20041257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock C., Hermans G., Pedotti R., Brendolan A., Schadt E., Garren H., Langer-Gould A., Strober S., Cannella B., Allard J., et al. 2002. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 8:500–508 10.1038/nm0502-500 [DOI] [PubMed] [Google Scholar]
- Manel N., Unutmaz D., Littman D.R. 2008. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 9:641–649 10.1038/ni.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Saito M., Tsuchiya S., Tsuge I., Takada H., Hara T., Kawamura N., Ariga T., Pasic S., Stojkovic O., et al. 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 448:1058–1062 10.1038/nature06096 [DOI] [PubMed] [Google Scholar]
- Moens E., Brouwer M., Dimova T., Goldman M., Willems F., Vermijlen D. 2011. IL-23R and TCR signaling drives the generation of neonatal Vgamma9Vdelta2 T cells expressing high levels of cytotoxic mediators and producing IFN-gamma and IL-17. J. Leukoc. Biol. 89:743–752 10.1189/jlb.0910501 [DOI] [PubMed] [Google Scholar]
- Nurieva R., Yang X.O., Martinez G., Zhang Y., Panopoulos A.D., Ma L., Schluns K., Tian Q., Watowich S.S., Jetten A.M., Dong C. 2007. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 448:480–483 10.1038/nature05969 [DOI] [PubMed] [Google Scholar]
- Oswald-Richter K., Grill S.M., Shariat N., Leelawong M., Sundrud M.S., Haas D.W., Unutmaz D. 2004. HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol. 2:E198 10.1371/journal.pbio.0020198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald-Richter K., Grill S.M., Leelawong M., Tseng M., Kalams S.A., Hulgan T., Haas D.W., Unutmaz D. 2007. Identification of a CCR5-expressing T cell subset that is resistant to R5-tropic HIV infection. PLoS Pathog. 3:e58 10.1371/journal.ppat.0030058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K.M., Nambudiri V., Dai G., Larman H.B., Gimbrone M.A., Jr, García-Cardeña G. 2005. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J. Biol. Chem. 280:26714–26719 10.1074/jbc.C500144200 [DOI] [PubMed] [Google Scholar]
- Rochman Y., Spolski R., Leonard W.J. 2009. New insights into the regulation of T cells by gamma(c) family cytokines. Nat. Rev. Immunol. 9:480–490 10.1038/nri2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnani S., Maggi E., Liotta F., Cosmi L., Annunziato F. 2009. Properties and origin of human Th17 cells. Mol. Immunol. 47:3–7 10.1016/j.molimm.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Sebzda E., Zou Z., Lee J.S., Wang T., Kahn M.L. 2008. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat. Immunol. 9:292–300 10.1038/ni1565 [DOI] [PubMed] [Google Scholar]
- Sinclair L.V., Finlay D., Feijoo C., Cornish G.H., Gray A., Ager A., Okkenhaug K., Hagenbeek T.J., Spits H., Cantrell D.A. 2008. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 9:513–521 10.1038/ni.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.P., Zhang H.H., Foley J.F., Hedrick M.N., Farber J.M. 2008. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J. Immunol. 180:214–221 [DOI] [PubMed] [Google Scholar]
- Smith-Garvin J.E., Koretzky G.A., Jordan M.S. 2009. T cell activation. Annu. Rev. Immunol. 27:591–619 10.1146/annurev.immunol.021908.132706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundrud M.S., Koralov S.B., Feuerer M., Calado D.P., Kozhaya A.E., Rhule-Smith A., Lefebvre R.E., Unutmaz D., Mazitschek R., Waldner H., et al. 2009. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 324:1334–1338 10.1126/science.1172638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unutmaz D., Pileri P., Abrignani S. 1994. Antigen-independent activation of naive and memory resting T cells by a cytokine combination. J. Exp. Med. 180:1159–1164 10.1084/jem.180.3.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unutmaz D., KewalRamani V.N., Marmon S., Littman D.R. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735–1746 10.1084/jem.189.11.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet J., Crofts L.A., Quinlan K.G., Czolij R., Perkins A.C., Crossley M. 2006. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 87:474–482 10.1016/j.ygeno.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Wani M.A., Means R.T., Jr, Lingrel J.B. 1998. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 7:229–238 10.1023/A:1008809809843 [DOI] [PubMed] [Google Scholar]
- Wani M.A., Wert S.E., Lingrel J.B. 1999. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 274:21180–21185 10.1074/jbc.274.30.21180 [DOI] [PubMed] [Google Scholar]
- Weinreich M.A., Takada K., Skon C., Reiner S.L., Jameson S.C., Hogquist K.A. 2009. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 31:122–130 10.1016/j.immuni.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M.A., Odumade O.A., Jameson S.C., Hogquist K.A. 2010. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11:709–716 10.1038/ni.1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.J., Boniface K., Chan J.R., McKenzie B.S., Blumenschein W.M., Mattson J.D., Basham B., Smith K., Chen T., Morel F., et al. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950–957 10.1038/ni1497 [DOI] [PubMed] [Google Scholar]
- Wolk K., Witte E., Witte K., Warszawska K., Sabat R. 2010. Biology of interleukin-22. Semin. Immunopathol. 32:17–31 10.1007/s00281-009-0188-x [DOI] [PubMed] [Google Scholar]
- Wu J., Lingrel J.B. 2005. Krüppel-like factor 2, a novel immediate-early transcriptional factor, regulates IL-2 expression in T lymphocyte activation. J. Immunol. 175:3060–3066 [DOI] [PubMed] [Google Scholar]
- Yamada T., Park C.S., Mamonkin M., Lacorazza H.D. 2009. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat. Immunol. 10:618–626 10.1038/ni.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I., Kharas M.G., Chen J., Peralta R.Q., Maruniak A., Sareen P., Yang V.W., Kaestner K.H., Fruman D.A. 2008. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int. Immunol. 20:671–681 10.1093/intimm/dxn024 [DOI] [PubMed] [Google Scholar]
- Zhang X., Jin J., Peng X., Ramgolam V.S., Markovic-Plese S. 2008. Simvastatin inhibits IL-17 secretion by targeting multiple IL-17-regulatory cytokines and by inhibiting the expression of IL-17 transcription factor RORC in CD4+ lymphocytes. J. Immunol. 180:6988–6996 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Danilenko D.M., Valdez P., Kasman I., Eastham-Anderson J., Wu J., Ouyang W. 2007. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 445:648–651 10.1038/nature05505 [DOI] [PubMed] [Google Scholar]
- Zhou L., Ivanov I.I., Spolski R., Min R., Shenderov K., Egawa T., Levy D.E., Leonard W.J., Littman D.R. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8:967–974 10.1038/ni1488 [DOI] [PubMed] [Google Scholar]
- Zhou L., Chong M.M., Littman D.R. 2009. Plasticity of CD4+ T cell lineage differentiation. Immunity. 30:646–655 10.1016/j.immuni.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Zhu J., Paul W.E. 2010. Heterogeneity and plasticity of T helper cells. Cell Res. 20:4–12 10.1038/cr.2009.138 [DOI] [PMC free article] [PubMed] [Google Scholar]