The Paf oncogene is highly expressed in cycling hematopoietic stem cells (HSCs) and is required for the development of long-term HSCs.
Abstract
Hematopoietic stem cells (HSCs) self-renew to maintain the lifelong production of all blood populations. Here, we show that the proliferating cell nuclear antigen–associated factor (Paf) is highly expressed in cycling bone marrow HSCs and plays a critical role in hematopoiesis. Mice lacking Paf exhibited reduced bone marrow cellularity; reduced numbers of HSCs and committed progenitors; and leukopenia. These phenotypes are caused by a cell-intrinsic blockage in the development of long-term (LT)-HSCs into multipotent progenitors and preferential loss of lymphoid progenitors caused by markedly increased p53-mediated apoptosis. In addition, LT-HSCs from Paf−/− mice had increased levels of reactive oxygen species (ROS), failed to maintain quiescence, and were unable to support LT hematopoiesis. The loss of lymphoid progenitors was likely due the increased levels of ROS in LT-HSCs caused by treatment of Paf−/− mice with the anti-oxidant N-acetylcysteine restored lymphoid progenitor numbers to that of Paf+/+ mice. Collectively, our studies identify Paf as a novel and essential regulator of early hematopoiesis.
The 15-kD proliferating cell nuclear antigen (PCNA)-associated factor (Paf) was originally identified in a yeast two-hybrid screen to identify novel PCNA-interacting proteins (Yu et al., 2001). PAF interacts with PCNA via a conserved PCNA binding motif and localizes to the nucleus in cell lines (Yu et al., 2001; Simpson et al., 2006; Hosokawa et al., 2007). PAF interacts with the DNA polymerase δ p125 subunit and flap endonuclease-1, and Paf expression triggers the DNA repair machinery after UV-induced DNA damage (Hosokawa et al., 2007; Turchi et al., 2009). Thus, it has been proposed that the biological function of Paf may be linked to one or more of the functions of PCNA, which has essential roles in DNA replication and repair and cell cycle control (Maga and Hubscher, 2003). However, cell line–based studies have also implicated Paf in such diverse processes as cell survival and proliferation. (Mizutani et al., 2005; Simpson et al., 2006; Guo et al., 2006; Hosokawa et al., 2007; Turchi et al., 2009).
Much effort has gone into identifying genes that are differentially expressed in cancer relative to normal tissues. These analyses have shown that Paf mRNA levels are overexpressed in many cancers (Yu et al., 2001; Petroziello et al., 2004; Mizutani et al., 2005; Marie et al., 2008) and increased Paf mRNA levels are part of a common cancer signature comprised of 46 genes (Xu et al., 2007). Paf also transforms NIH3T3 cells into tumor cells in a xenograft model (Hosokawa et al., 2007). Thus, Paf is overexpressed in tumors and has oncogenic potential. Cancer cells and hematopoietic stem cells (HSCs) share the ability to self-renew; thus, the genes and pathways associated with malignancies often also regulate HSC function (Reya et al., 2001). Here, we show that genetic disruption of Paf by homologous recombination in mice resulted in altered HSC function, disrupted progenitor development, and leukopenia.
RESULTS AND DISCUSSION
Paf−/− mice are leukopenic
We first examined Paf gene expression in various organs and cells by real-time PCR. Paf mRNA was most highly expressed in the thymus, although message expression was also detected in all other organs examined, albeit to different levels. CD8 single-positive thymocytes exhibited the highest level of mRNA expression among the analyzed lymphocyte populations. Furthermore, all thymocyte subsets expressed higher Paf transcript levels than splenic lymphocytes and DCs (Fig. S1 A). However, high mRNA expression levels did not correlate with high PAF protein levels, supporting that posttranslational mechanisms regulate PAF expression (Fig. S2 C).
To elucidate Paf function in hematopoietic and lymphocyte development, Paf−/− mice were generated (Fig. S1 B). Paf inactivation was confirmed by Southern blot, RT-PCR, immunoblot and FACS analysis of thymocytes from Paf+/+ and Paf−/− mice (Fig. 1 A; Fig. S1, C and D; and not depicted). Although the Paf gene is embedded within the thyroid hormone receptor interacting protein 4 locus, mRNA expression was not altered in Paf−/− mice (Fig. S1 E).
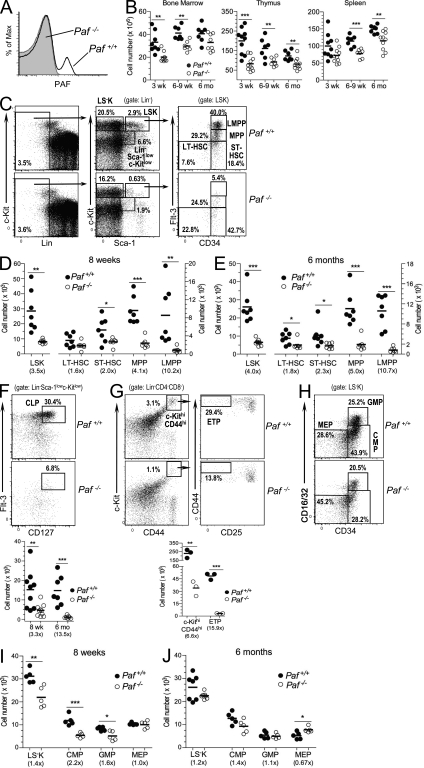
Figure 1.
Paf is necessary for proper HSC and progenitor development. (A) FACS of thymocytes from Paf+/+ and Paf−/− mice stained for intracellular PAF. (B) Total number of cells in BM, thymuses, and spleens of Paf+/+ and Paf−/− mice. (C) Representative staining profiles for BM HSC and progenitor populations. (D and E) Absolute cell numbers in the BM of 8-wk-old (D) or 6-mo-old (E) Paf+/+ and Paf−/− mice. (F) Representative staining profiles and absolute number of CLPs in the BM of 8-wk- and 6-mo-old mice. Plots are gated for Lin−Sca-1lowc-kitlow cells as defined in (C). (G) Representative staining profiles and absolute cell numbers of ETPs in the thymus of 8-wk-old mice. (H) Representative staining profiles for myeloerythroid progenitors. LS−K cells (defined in C) were subdivided into CMPs, GMPs, and MEPs as indicated. (I and J). Absolute cell numbers of LS−Ks, CMPs, GMPs, and MEPs in the BM of 8-wk-old (I) or 6-mo-old (J) mice. Each symbol represents a mouse combined from 3–6 experiments. Horizontal bars indicate the mean. Numbers in parentheses are the fold change of cell numbers for Paf−/− relative to Paf+/+ mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Paf−/− mice had reduced thymic, splenic and BM cellularity that did not self-correct with aging (Fig. 1 B). Total white blood cell counts were also reduced (Fig. S1 F).
Paf is necessary for proper HSC and progenitor development
The leukopenia in Paf−/− mice might be caused by insufficient hematopoietic BM progenitors, which mainly reside within the BM lineage− (Lin−) Sca-1+ c-KitHigh (LSK) population. Paf−/− mice (8 wk old) had lower percentages of LSKs among total BM cells, which resulted in a two to eightfold reduction in the absolute number of LSKs (Fig. 1, C and D). LSKs can be separated into HSCs and progenitors on the basis of CD34 and Flt-3 expression. CD34−Flt-3− cells are enriched for long-term (LT)-HSCs, CD34+Flt-3− cells comprise short-term (ST)-HSCs, cells with intermediate levels of Flt-3 are multipotent progenitors (MPPs), and the Flt-3bright cells are lymphoid-primed MPPs (LMPPs; Adolfsson et al., 2001). Paf−/− BM LSKs contained increased frequencies of LT- and ST-HSCs, but severely reduced frequencies of LMPPs. When the frequencies were converted to absolute cell numbers, there was a marked reduction of each population caused by the overall reduced BM cellularity. MPPs (4.1-fold) and LMPPs (10.2-fold) had the largest decrease in cell numbers (Fig. 1 D). The reduction in HSCs and progenitors in Paf−/− mice was maintained in 6-mo-old mice (Fig. 1 E). The BM of older Paf−/− mice showed a more severe and significant loss of LT-HSCs suggesting that Paf may be necessary for maintaining LT-HSCs.
Further differentiation of MPPs and LMPPs gives rise to common lymphoid progenitors (CLPs) in the BM (Kondo et al., 1997) and early thymic progenitors (ETPs) in the thymus (Chi et al., 2009). Consistent with the loss of MPPs and LMPPs in Paf−/− mice, a significant reduction in the frequency and absolute numbers of CLPs and ETPs was also observed (Fig. 1, F and G).
Myeloerythroid progenitors are derived from LSK cells and are contained in the Lin− Sca-1−c-Kit+ (LS−K) BM population. They can be further divided based upon CD34 and FcγRIII/II (CD16/32) expression into common myeloid progenitors (CMP; CD34+ FcγRII/IIlow), granulocyte-macrophage progenitors (GMP; CD34+ FcγRII/IIhigh), and megakaryocyte-erythroid progenitors (MEP; CD34− FcγRIII/IIlow; Akashi et al., 2000). BM from 8-wk-old Paf−/− mice had lower frequencies of LS−K cells (Fig. 1 C), which resulted in a small, but significant, decrease (1.4-fold) in the absolute numbers (Fig. 1 I). This decrease was caused by reduced numbers of CMPs (2.2-fold) and GMPs (1.6-fold), but not MEPs (Fig. 1, H and I). In older Paf−/− mice, the LS−K cells and GMPs and CMPs were present at near normal numbers (Fig. 1 J). However, MEPs were present at significantly higher numbers, although this difference was less than twofold (Fig. 1 J). Thus, myeloerythroid progenitors are also impacted by the loss of Paf, but to a lesser extent than lymphoid progenitors.
Paf restrains HSC, MPP, and LMPP proliferation and protects LMPPs and CLPs from p53-mediated apoptosis
An analysis of PAF protein expression in BM HSCs and progenitors showed that all LSK and LS−K subsets expressed PAF and that PAF protein expression was detected mainly in cycling cells (Fig. 2 A and Fig. S2, B and C). Consistent with the profound reduction of CLPs in Paf−/− mice, CLPs had the highest overall level of PAF (mean fluorescence intensity [MFI] = 12,392). LT-HSCs are, for the most part, quiescent cells. Thus, it was unexpected to find that the small number of cycling or cell cycle arrested LT-HSCs also expressed high PAF protein levels (MFI = 11,434).
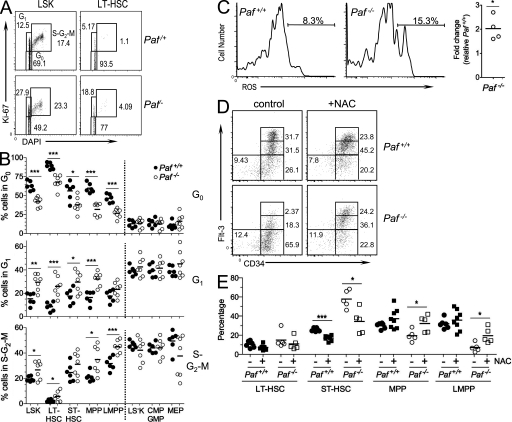
Figure 2.
Paf restricts HSC and progenitor cycling and protects LMPPs and CLPs from p53-mediated apoptosis. (A) PAF protein levels in BM HSC and progenitor populations relative to cell cycle (DAPI). Data are presented as an overlay of PAF staining obtained for each population from Paf+/+ and Paf−/− mice. Numbers in parentheses are the MFI obtained for PAF. Horizontal light blue line highlights the differences in PAF expression among the populations. LS−K cells were separated into MEPs (CD34−) and a population containing CMPs and GMPs (CD34+). (B) Percentage of BM Paf+/+ and Paf−/− HSC and progenitors that are in the S–G2–M phases of the cell cycle. Representative FACS profiles are shown in Fig. S2 D. (C) Representative staining profiles of Paf+/+ and Paf−/− BM cells from mice that were injected 24 h earlier with BrdU. The BrdU+ gate was established by using BM from PBS-injected mice (Fig. S2 E). (D) Percentage of each BM subset that incorporated BrdU. (E) Representative FACS analyses of HSC subsets showing Annexin V versus DAPI staining. Gating of apoptotic cells (AnnexinV+DAPI−) was determined as shown in Fig. S2 F. (F) Cumulative data showing the percentage of cells undergoing apoptosis. (G) Representative staining profiles for LT-HSC, ST-HSC, MPP, and LMPP from Paf+/+p53+/+, Paf+/+p53+/−, Paf−/−p53+/+, and Paf−/−p53+/− mice. (H) Cumulative data showing the LMPP frequency (left) and absolute cell numbers (right). Data were combined from 3–8 experiments on 1–2 mice/genotype. Each symbol represents a mouse, and horizontal bars mark the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Overexpression and knockdown studies in cell lines have shown that Paf promotes or impedes cellular proliferation, respectively (Petroziello et al., 2004; Mizutani et al., 2005; Hosokawa et al., 2007). Thus, progenitor populations might be diminished because of a requirement for Paf for progenitor cell expansion. Unexpectedly, the proportion of Paf−/− BM LSKs present in the S–G2–M phases of the cell cycle was significantly increased (Fig. 2 B and Fig. S2 D). All LSK HSC subsets and progenitor populations showed higher proportions of cycling cells. The largest difference was observed for Paf−/− LT-HSCs (1.8-fold). In contrast, Paf−/− and Paf+/+ LS−K subpopulations cycled at similar frequencies.
BM HSC populations were also examined after 24 h of in vivo BrdU labeling (Fig. 2, C and D and Fig. S2 E). Paf−/− LSK populations and CLPs incorporated significantly more BrdU than their Paf+/+ counterparts. As observed for the direct cell cycle analyses (Fig. 2 B), LT-HSCs showed the largest difference (twofold). Total Paf−/− LS−K cells, CMPs/GMPs and MEPs also incorporated significantly more BrdU than Paf+/+ LS−K populations, but the difference was not as large as that observed for LSK populations. Collectively, these data show that reduced HSCs and progenitors in Paf−/− BM was likely not caused by decreased cycling. Instead, these data support a role for Paf in restraining the cycling of BM HSCs and, in particular, in restricting LT-HSC proliferation.
These data suggested that the profound deficiency of LMPPs and CLPs in Paf−/− mice was more likely caused by increased apoptosis. The frequency of cells that were undergoing apoptosis (DAPI−AnnexinV+) were similar for Paf+/+ and Paf−/− total LSKs and LS−Ks (Fig. 2, E and F). However, analysis of HSCs and progenitors showed a significant increase in the frequency of apoptotic LMPPs (1.7-fold) and CLPs (2.5-fold). Thus, LMPPs and CLPs are likely diminished in the BM of Paf−/− mice because of increased apoptosis. The increased cell death was restricted to early lymphoid progenitors because enhanced cell death is not observed in other cell populations, including developing, mature and activated B and T cells (Fig. S2 G and not depicted).
To determine if the p53 pathway plays a role in Paf−/− progenitor cell death, Paf−/− mice were crossed with p53−/− mice to generate Paf−/−p53+/− mice. Paf−/− that lacked one copy of p53 had increased frequencies and absolute cell numbers of LMPPs when compared with Paf−/−p53+/+ mice (Fig. 2, G and H). Thus, Paf −/− LMPP death is caused, at least in part, by p53-mediated apoptosis.
Paf controls LT-HSC quiescence
Cellular quiescence is an essential mechanism by which the LT self-renewal potential of BM HSCs is maintained (Orford and Scadden, 2008). Because Paf−/− HSCs were more actively cycling, Paf might contribute to the maintenance of LT-HSC quiescence. The frequency of quiescent LT-HSCs (Ki-67−) was significantly reduced in Paf−/− mice (Fig. 3, A and B). Moreover, Paf deficiency resulted in reduced frequencies of Ki-67− (G0) cells for all other LSK progenitor subsets, but not for LS−Ks or LS−K subsets (Fig. 3, A and B; and Fig. S3 A). There were ∼3-fold more Paf−/− than Paf+/+ LT-HSCs in the resting G1 phase of the cell cycle, whereas there were ∼1.7-, ∼2-, and ∼1.2-fold more ST-HSCs, MPPs, and LMPPs, respectively (Fig. 3, A and B; and Fig. S3 A). Thus, Paf deficiency had a greater impact on the quiescence status of LT-HSCs than on other LSK subsets. Similar results were observed when cell cycle status was measured using Pyronin Y and Hoechst 33342 staining (Fig. S3, B and C). These data show that LT-HSCs from Paf−/− mice fail to maintain quiescence.
Figure 3.
Disruption of HSC quiescence in the absence of Paf. (A) Representative staining profiles for LSK and LT-HSC analyzed for Ki-67 and cell cycle (DAPI). (B) Cumulative data showing the percentage of cells in each BM subset that are quiescent (G0), resting (G1), or cycling (S–G2–M). (C) Representative histograms showing Lin− BM LT-HSCs loaded with the ROS detection reagent, 5-(and-6)-carboxy-2’,7’-difluorodihydrofluorescein diacetate. The gate shows the percentage of ROS+ cells. Cumulative data shows the fold difference in percentage of ROS positive LT-HSCs for Paf−/− relative to Paf+/+ LT-HSCs. (D) Representative FACS profiles for BM subsets from NAC-treated or control mice. (E) Cumulative data showing the frequency of each population as a percentage of the LSK gate from (D). Absolute cell numbers are shown in Fig. S3 D. Data were combined from 3–4 individual experiments of 1–3 mice/genotype. Each symbol represents a mouse and horizontal bars indicate the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The loss of LT-HSC quiescence frequently correlates with increased cellular reactive oxygen species (ROS), which is negatively associated with LT-HSC self-renewal (Pervaiz et al., 2009). Consistent with this, Paf−/− LT-HSCs displayed higher levels of ROS than Paf+/+ LT-HSCs (Fig. 3 C). In vivo treatment of Paf−/− mice with the antioxidant N-acetylcysteine (NAC) restored progenitor development (Fig. 3, D and E). When compared with progenitors from NAC-treated Paf+/+ mice, the frequency of Paf−/− ST-HSCs was significantly decreased, whereas MPPs and LMPPs were significantly increased. This resulted in a significant increase in the absolute number of LMPPs in the BM of NAC-treated Paf−/− mice (Fig. S3 D). Thus, the increased ROS levels observed for Paf−/− LT-HSC were functionally relevant.
Paf maintains the LT-HSC pool and controls HSC progenitor development
To determine if Paf deficiency altered the functional competence of HSCs, the competitive repopulating ability of Paf−/− BM cells was measured. Paf−/− and WT B6.SJL Lin− BM was mixed at a 1:1 or a 9:1 (Paf−/−/WT) ratio and transferred into lethally irradiated B6 x B6.SJL F1 recipients (Fig. S3E). The contribution of Paf−/− HSCs to hematopoiesis was monitored by FACS analysis of PBLs over 18 wk (Fig. 4 A). In 1:1 transplanted mice, Paf−/− HSCs did not contribute to hematopoiesis (<1%). However, in the 9:1 transplanted mice, a small fraction of granulocytes (5.1 ± 2.2), B cells (3.9 ± 1.7) and T cells (5.3 ± 1.7) were derived from Paf−/− HSCs at 6 wk. Reduced, but detectable levels of Paf−/− granulocytes (1.4 ± 0.8), B cells (2.0 ± 1.0) and T cells (1.7 ± 0.6) remained at 18 wk. Hematopoiesis at 16 wk post-transplant is derived from LT-HSCs (Dykstra et al., 2007). Therefore, Paf−/− HSCs were not only compromised in their ability to mediate ST hematopoiesis, but were also unable to mediate LT repopulation. These studies also show that Paf deficiency compromises HSC development and function in a cell-intrinsic manner.
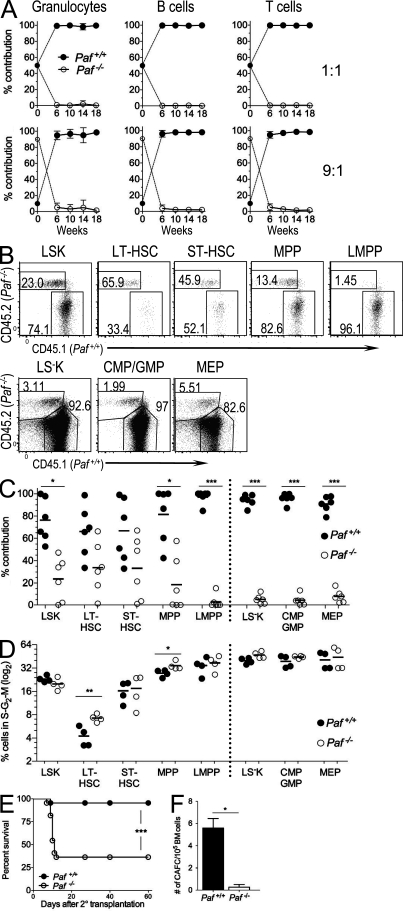
Figure 4.
Paf−/− HSCs are functionally compromised. (A) Peripheral blood from lethally irradiated mice transplanted with a 1:1 (n = 5) or 9:1 (n = 6) mix of Lin− BM from Paf −/− (CD45.2) and WT (CD45.1) donor mice were monitored for the contribution of Paf +/+ and Paf −/− HSCs to the granulocyte, T and B cell lineages. Data show mean ± SD and are representative of two identical experiments. (B) Representative FACS analysis of BM HSC populations at 20 wk after transplant for 9:1 BM-transplanted mice. (C) Cumulative data from B. Data were pooled from analyses of three mice/genotype for two independent chimera experiments. (D) Cumulative data showing the percentage of cycling cells in the BM of 9:1 transplanted mice (representative FACS plots are shown in Fig. S3 F). Data pooled for two mice from two independent experiments. (E) Paf−/− BM cells are defective in rescuing lethally irradiated recipients after a second transplant. Survival curves for recipients of secondary BM transplants. Data combined from two independent experiments of 10 mice/genotype each. (F) The number of cobblestone area forming cells in Lin− Paf+/+ and Paf−/− BM was determined by scoring colonies at 21 d. Data shown are means of two similar experiments ± SD of 1 mouse/genotype. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The inability of Paf−/− HSCs to contribute to hematopoiesis might be caused by the inability to home to hematopoietic niches of the BM. Although at 20 wk post-transplant LSKs were preferentially derived from WT cells (76.3.7 ± 19.4%) Paf−/− cells were also present (Fig. 4, B and C). Paf−/− cells made the largest contribution to the LT-HSC subset and the percentage of Paf−/− cells contributing to each developing subset decreased as the cells differentiated from LT-HSCs to LMPPs (Fig. 4, B and C). Reconstitution of myeloerythroid progenitors by Paf−/− cells was also diminished relative to LT-HSCs, but less impacted than LMPPs, which were undetectable in all but one mouse. Thus, when Paf−/− HSCs are placed in the same host environment as WT HSCs, Paf−/− deficient LT-HSCs engraft, but continue to display a preferentially impaired ability to generate MPPs and LMPPs. These data establish Paf−/− as a cell-intrinsic factor critical for HSC progenitor development, differentiation, and function.
Our data show that Paf controls LT-HSC quiescence. However, the need to replenish the leukopenic periphery of Paf−/− mice could potentially drive LT-HSCs out of quiescence. At 20 wk after transplant, LSKs and LS−Ks derived from Paf−/− and WT cells in the BM of the 9:1 recipient mice had similar cell cycle profiles (Fig. 4 D and Fig. S3 F). However, there was a significant increase in the percentage of Paf−/− LT-HSCs residing in the S–G2–M phases of the cell cycle (1.7-fold). Paf−/− MPPs also had a significantly increased percentage of cycling cells (1.2-fold). Thus, the cell cycle status of progenitor populations was, for the most part, normalized in the nonleukopenic environment of the chimeric hosts; however, LT-HSCs continued to disproportionally cycle. Collectively, these analyses support that Paf is essential for the maintenance of LT-HSC quiescence. However, progenitor proliferation in the BM of Paf−/− mice is likely caused by the need to replenish the leukopenic periphery and not directly caused by the loss of Paf.
The loss of LT-HSC quiescence often results in the loss of self-renewal potential. The self-renewal potential of Paf−/− HSCs was tested by serial BM transplantation under noncompetitive conditions. Lin− BM from Paf+/+ or Paf−/− mice rescued all lethally irradiated primary recipients. However, upon secondary transfer, Paf−/− BM protected only 30% of the secondary recipients from death, whereas nearly all the Paf+/+ recipients survived (Fig. 4 E). Thus, Paf−/− HSCs have reduced self-renewal potential. Finally, Paf−/− HSC function was tested with an in vitro cobblestone area-forming cell (CAFC) assay, an assay that identifies LT-HSCs (Breems et al., 1994). Lin− Paf−/− BM cells showed an ∼20-fold reduction in wk 3 CAFCs compared with Paf+/+ cells (Fig. 4 F). Collectively, our data shows that Paf has a pivotal role in maintaining the LT self-renewal potential of HSCs.
Despite being an active and clinically relevant area of research, the mechanisms that regulate HSC self-renewal and differentiation remain unclear. Excessive cycling of LT-HSCs eventually leads to premature exhaustion and loss of self-renewal. Thus, tight regulation of the cell cycle in LT-HSC is an important mechanism that contributes to ensure proper LT-HSC function. Studies have implicated cyclin-dependent kinases and their inhibitors, transcriptional regulators, and signaling proteins as being important for proper cell cycle control in LT-HSCs (Walkley et al., 2005; Jude et al., 2008). How these pathways interact at a molecular level to orchestrate the delicate balance between LT-HSC quiescence and proliferation remains an open question. Our studies demonstrate that Paf protects LT-HSCs from exhaustion through the maintenance of cellular quiescence, thus ensuring LT-HSC self-renewal and functional integrity. The identification of Paf as necessary for proper cell cycle regulation of LT-HSCs provides additional molecular insight into HSC biology and reinforces the tight linkage between LT-HSC function, progenitor development, and proper cell cycle control.
MATERIALS AND METHODS
FACS and antibodies.
BM single-cell suspensions were prepared from 1 tibia and femur for HSC subset analysis or from 2 tibias, femurs, pelvis, humerus, radius, and shoulder plates for cell cycle, Pyronin Y, BrdU, PAF, and Ki-67 analyses. Single-cell suspensions from hematopoietic organs were incubated with Fc receptor-blocking antibody (clone 2.4G2) before being stained on ice with specific antibodies conjugated to different fluorophores. Data were acquired on a LSR II cytometer (BD) and analyzed using FlowJo software (Tree Star). Cell doublets were excluded from all analyses and, when possible, dead cells were excluded by the use of DAPI. For BM and splenocyte preparations, RBCs were lysed before antibody staining. For intracellular Ki-67 or PAF staining, Lin− cells were enriched using a Lineage Cell Depletion kit (Miltenyi Biotec), stained for cell surface markers, fixed, and made permeable with an intracellular staining buffer set (eBioscience) or with ice-cold methanol, and then stained with specific antibody or isotype control. For cell cycle analysis, fixed and permeabilized cells were incubated with 10 µg/ml DAPI before analysis. ROS levels were measured by suspending Lin− BM in warm PBS containing and 5 µM 5-(and-6)-carboxy-2’,7’-difluorodihydrofluorescein diacetate. After incubation at 37°C for 15 min, cells were washed, surface stained to identify LT-HSCs, and analyzed by FACS.
The following antibodies (all from eBioscience or BD) conjugated to FITC, Alexa Fluor 488, PE, PE-Cy7, APC, Alexa Fluor 647, APC-Alexa Fluor 750, APC-Alexa Fluor-780, PercpCy5.5, Pacific Blue, Alexa Fluor 700, or PE-Cy5 were use for the flow cytometric analyses: α-CD3ε (145-2C11), -CD4 (RM4-5), -CD8α (53–6.7), -NK1.1 (PK136), -CD19 (1D3), -CD11c (N418), -CD11b (M1/70), -TCRβ (H57 597), -TCRγ (GL3), –Gr-1(RB6-8C5), -TER119 (TER-119), -CD25 (PC61), -CD44 (IM7), –c-Kit (2B8), –Flt-3 (A2F10.1), –Sca-1 (D7), -CD34 (RAM34), and –IL-7Rα (A7R34). A table listing all antibodies, lineage cocktails, and combinations of mAbs used to identify each progenitor population analyzed is presented in Tables S1 and S2.
Generation of PAF-specific mAbs
Armenian hamster mAbs specific for Paf were generated by the Monoclonal Antibody Core Facility of Memorial Sloan-Kettering Cancer Center (MSKCC) with standard approaches after immunization of animals with a GST-Paf fusion protein emulsified in Titermax. The specificity of two Paf-specific mAbs (called Jyld12 and Jyld15) was determined by flow cytometry, immunofluorescence, and Western blotting of CHO cells transfected with Paf or empty vector and also with lymphocytes derived from Paf+/+ and Paf−/− mice (Fig. S2 A). The α-Paf mAb, Jyld12, was produced, purified, and conjugated to Alexa Fluor 488 by the Monoclonal Antibody Core Facility of MSKCC.
Gene targeting and mice.
Paf−/− mice on the C57BL/6 background were generated using standard techniques as follows. A 436-bp NotI–BgII fragment ranging from 36 bp upstream of the Paf ATG in exon I to the HindIII site (destroyed), downstream of exon II, was generated by PCR and cloned into the pL451 vector using NotI and BamHI sites. The resultant plasmid was digested with RsaI and EcoRI to release an exon I–II fragment flanked by a Loxp site downstream of exon II. This fragment was subcloned into pL451 that had been digested with NotI and HindIII together with a 190-bp PCR fragment covering the rest of the 5′-UTR of exon I and extending to 15 bp upstream of the XhoI site and the Loxp-FRT-Neo-FRT cassette (from pL451) in a 4-piece ligation reaction. Finally, a 3.9-Kb XhoI–SalI PCR-generated fragment starting at the destroyed HindIII site downstream of exon II was cloned into the SalI site of the latter plasmid. The 2.4-Kb genomic DNA containing the inserted LoxP site and the LoxP-FRT-Neo-FRT cassette were released and successfully used for BAC recombineering to generate the BAC targeting vector. The RP23-177B-17 BAC clone containing the Paf locus was transformed into SW105, which was subsequently transformed with the 6.5-Kb genomic fragment mentioned above and activated for recombination (Lee et al., 2001; Liu et al., 2003). A DTA-AMP–negative selection cassette (from pKOSelectDT-840) was generated by PCR using hybrid primers containing genomic DNA sequences located upstream and downstream of the genomic region complementary to screening probe A (Fig. S1 B). The flanked DTA-AMP cassette was inserted into the RP23-177B-17 BAC containing the LoxP-FRT-Neo-FRT cassette by recombination to generate the BAC targeting vector. All fragments were sequenced to verify that mutations were not introduced. The BAC targeting vector DNA was purified using a QIAGEN BAC purification kit and used to target the Paf genomic locus of the CY2.4 ES parental cell line, derived from C57BL/6J-Tyr (c-2J) embryos, by the Gene Targeting Resource Center of The Rockefeller University. G418-resistant clones were screened by genomic Southern blot hybridization. Out of 96 clones, 32 were shown to have undergone homologous recombination at the Paf locus.
To derive Paf+/− ES cell clones, two independently targeted ES cell clones were transiently transfected with pCre-Pac a vector encoding the Cre-recombinase (Taniguchi et al., 1998). The resultant clones were screened for Paf and Neo cassette deletion by Southern hybridization of HindIII-digested DNA. Clones that underwent Paf deletion were checked for a normal karyotype and two of the clones exhibiting chromosomal integrity were used for injection into C57BL/6 blastocysts. The male offspring presenting a high level of chimerism were backcrossed to C57BL/6J-Tyr (c-2J) albino mice to derive Paf-null mice. The breeding of Paf +/− mice showed a Mendelian distribution of offspring, an absence of noticeable growth retardation, and comparable weight between Paf+/+ and Paf−/− mice. Both female and male Paf−/− mice were fertile. Analyses of mice derived from the two independent ES clones showed identical phenotypes. C57BL/6, C57BL/6.SJL, and p53+/− mice were obtained from The Jackson Laboratory. p53+/− mice were crossed with Paf−/− to generate Paf−/−p53+/− mice. All experimental mouse procedures were reviewed and approved by the IACUC of Memorial Sloan-Kettering Cancer Center.
Semiquantitative PCR.
RNA was extracted from whole organs or purified cell populations with TRIzol Reagent (Invitrogen). Splenic CD19+ B cells, CD4+ and CD8+ T cells were purified using MACS beads (Miltenyi Biotec). Splenic CD11c+ DC cells and thymocyte populations were by purified by cell sorting on a MoFlow Cytometer (Dako). cDNA was prepared with random primers and Superscript II reverse transcription (Invitrogen) and assayed by quantitative real-time PCR using SYBR Green Master Mix reagents (Roche) and a Light Cycler 1.0 (Roche). Gene expression was normalized to that of 18S RNA and expressed relative to the indicated reference sample. For semiquantitative Trip4 RT PCR, equivalent amount of thymus RNA from Paf+/+ and Paf−/− mice were converted to cDNA with Superscript II reverse transcription and titrated amounts of cDNA were assayed by PCR for Trip4 and 18S RNA expression. Primer sequences used were as follows: Paf, forward 5′-ACTACGTTCCAGGAGCCTAC-3′ and reverse 5′-CTgCTTCTTCAGGAGCCTG-3′; Trip4, forward 5′-ATGATTTCCTCGCTGACATCC-3′ and reverse 5′-TAGATTTGAAGGCTGTACCC-3′; 18S RNA, forward 5′-CCGCAGCTAGGAATAATGGAAT-3′ and reverse 5′-CGAACCTCCGACTTTCGTTCT-3′ (Akilesh et al., 2004).
Competitive repopulation assay.
Lin− BM cells (>95% Lin−) from Paf−/− mice (CD45.2) were mixed at a ratio of 1:1 or 9:1 with WT competitor Lin− BM cells from B6.SJL mice (CD45.1) or B6 x B6.SJL F1 mice (CD45.1/CD45.2), and 2 × 105 total cells were injected i.v. into lethally irradiated B6 x B6.SJL F1 or B6.SJL recipients, respectively.
Western blot analyses.
Whole-cell extracts were prepared by lysing cells at 100°C for 10 min in 50 mM Tris, pH 7.4, containing 1% SDS, 5 mM DTT, and 2 mM PMSF. Extracts were briefly sonicated, centrifuged for 10 min at 13,000 RPM, and the supernatants were transferred to a fresh tube. After separation of lysate by 15% SDS-PAGE and transfer, membranes were blocked in PBS containing 1% BSA and incubated with anti-Paf mAb Jyld12 or Jyld15. Primary antibodies were detected with HRP-conjugated goat anti–hamster IgG secondary antibodies (Jackson ImmunoResearch Laboratories), and blots were developed with SuperSignal West Pico chemiluminescent peroxidase substrate (Thermo Fisher Scientific).
In vivo BrdU incorporation.
Mice were injected i.p. with 1 mg BrdU in PBS/12 g of body weight. 24 h later, BM was isolated, Lin+ cells were depleted with a Lineage Cell Depletion kit (Miltenyi Biotech) and cells were stained for surface markers and α-BrdU with a BrdU Flow kit (BD) using the supplied protocol.
In vivo apoptosis assay.
BM cells enriched for Lin− cells were stained for surface markers and subsequently stained with Alexa Fluor 647–conjugated Annexin V (Invitrogen) and DAPI according to the supplied protocol.
In vitro apoptosis assay.
Paf+/+ and Paf−/− CD4 and CD8 single-positive thymocytes, CD4+ and CD8+ splenocytes and splenic B cells were purified by magnetic-based cell sorting (Miltenyi Biotech) and placed in culture in Bruff’s medium supplemented with 10% FBS. Cells were >80–95% pure as accessed by FACS. At 3, 24, and 48 h, cells were stained with Alexa Fluor 488–conjugated Annexin V (Invitrogen) and DAPI according to the supplied protocol and analyzed by FACS analysis. Apoptotic cells were defined as AnnexinV+, DAPI−.
Pyronin Y staining.
Enriched Lin− BM cells were surface stained, fixed, and permeabilized with Cytofix/Cytoperm solution (BD), and cells (2 × 106 cells/ml in PBS) were incubated at room temperature for 45 min with Hoechst 33342 (2 mM, Sigma) followed by a 45 min incubation with Pyronin Y (0.5 mm, SIGMA).
In vivo treatment of mice with NAC.
Cohorts of Paf+/+ and Paf−/− mice received 1 mg/ml NAC in the drinking water for 2 wk, and 500 mg/kg i.p. in saline daily for the last 7 d of the two weeks. Control mice were fed autoclaved water and received saline i.p. for the last 7 d.
In vitro HSC assay.
The number of CAFC was determined as previously described (Breems et al., 1994). In brief, Lin− BM or spleen cells from Paf+/+ (4 × 103 BM or 105 splenocytes) and Paf−/− (2.5 × 104 BM or 106 splenocytes) mice were plated in duplicate in T12.5 tissue-culture flasks (Becton Dickinson) containing confluent monolayers of MS5 cells. Cobblestone areas ≤10 phase-dark closely associated cells beneath the stroma were scored at day 21 as CAFC.
Serial BM transplantation.
Lin− BM cells from 11-mo-old Paf+/+ or Paf−/− (CD45.2) mice were transplanted by i.v injection (105 cells/recipient) into lethally irradiated B6.SJL mice (CD45.1; n = 10 each). For irradiation, 1,300 Rads of total body irradiation were administered as a split dose with a 3–4-h interval between doses. 16 wk after transplantation, BM cells were harvested from the Paf+/+ or Paf−/− primary recipient mice and transplanted into new cohorts (n = 10 each) of lethally irradiated B6.SJL recipient mice. Mice were monitored and scored for mortality during the next 60 d.
Statistics.
The unpaired, two-tailed Student’s t test was used for statistical confirmation of differences between experimental group means (P < 0.05) for most experiments. For the data shown in Fig. 2 F into Fig. 4 (C and D), paired t tests were used. Data were analyzed using GraphPad Prism 5.
Online supplemental material.
Fig. S1 shows the targeting strategy used to generate Paf−/− mice. Fig. S2 demonstrates the specificity of PAF-specific mAbs that were generated for these studies. Fig. S3 shows representative FACS analysis for HSC subsets stained with Ki-67 and DAPI. Tables S1 and S2 show the surface markers used to identify progenitor populations studies and the antibody-fluorophore combinations used for the each experiment. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20102170/DC1.
Acknowledgments
We thank G. Goldberg and A. Holland for providing experimental advice and mice; M. Curran and T. Merghoub for help with the gene targeting; and N. Draghi, W. Yi, D. Kovalovsky, G. Porter, H. Snoeck, and J. Chaudhuri for critical reading of the manuscript.
This work was supported by a Ruth L. Kirschstein National Research Service Award (F31CA130744 to E.S. Alonzo) and the US National Cancer Institute (P30-CA 08748).
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- ETP
- early thymic progenitor
- GMP
- granulocyte-macrophage progenitor
- HSC
- hematopoietic stem cell
- Lin−
- lineage−
- LSK
- Lin− Sca-1+, c-KitHigh
- LS−K
- Lin− Sca-1− c-KitHigh
- LT
- long-term
- LMPP
- lymphoid-primed MPP
- MEP
- megakaryocyte-erythroid progenitor
- MFI
- mean fluorescence intensity
- MPP
- multipotent progenitor
- NAC
- N-acetylcysteine
- Paf
- 15-kD PCNA-associated factor
- PCNA
- proliferating cell nuclear antigen
- ROS
- reactive oxygen species
- ST
- short-term
References
- Adolfsson J., Borge O.J., Bryder D., Theilgaard-Mönch K., Astrand-Grundström I., Sitnicka E., Sasaki Y., Jacobsen S.E. 2001. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 15:659–669 10.1016/S1074-7613(01)00220-5 [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Akilesh S., Petkova S., Sproule T.J., Shaffer D.J., Christianson G.J., Roopenian D. 2004. The MHC class I-like Fc receptor promotes humorally mediated autoimmune disease. J. Clin. Invest. 113:1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breems D.A., Blokland E.A., Neben S., Ploemacher R.E. 1994. Frequency analysis of human primitive haematopoietic stem cell subsets using a cobblestone area forming cell assay. Leukemia. 8:1095–1104 [PubMed] [Google Scholar]
- Chi A.W., Bell J.J., Zlotoff D.A., Bhandoola A. 2009. Untangling the T branch of the hematopoiesis tree. Curr. Opin. Immunol. 21:121–126 10.1016/j.coi.2009.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B., Kent D., Bowie M., McCaffrey L., Hamilton M., Lyons K., Lee S.J., Brinkman R., Eaves C. 2007. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 1:218–229 10.1016/j.stem.2007.05.015 [DOI] [PubMed] [Google Scholar]
- Guo M., Li J., Wan D., Gu J. 2006. KIAA0101 (OEACT-1), an expressionally down-regulated and growth-inhibitory gene in human hepatocellular carcinoma. BMC Cancer. 6:109 10.1186/1471-2407-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M., Takehara A., Matsuda K., Eguchi H., Ohigashi H., Ishikawa O., Shinomura Y., Imai K., Nakamura Y., Nakagawa H. 2007. Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res. 67:2568–2576 10.1158/0008-5472.CAN-06-4356 [DOI] [PubMed] [Google Scholar]
- Jude C.D., Gaudet J.J., Speck N.A., Ernst P. 2008. Leukemia and hematopoietic stem cells: balancing proliferation and quiescence. Cell Cycle. 7:586–591 10.4161/cc.7.5.5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 73:56–65 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476–484 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G., Hubscher U. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116:3051–3060 10.1242/jcs.00653 [DOI] [PubMed] [Google Scholar]
- Marie S.K., Okamoto O.K., Uno M., Hasegawa A.P., Oba-Shinjo S.M., Cohen T., Camargo A.A., Kosoy A., Carlotti C.G., Jr, Toledo S., et al. 2008. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int. J. Cancer. 122:807–815 10.1002/ijc.23189 [DOI] [PubMed] [Google Scholar]
- Mizutani K., Onda M., Asaka S., Akaishi J., Miyamoto S., Yoshida A., Nagahama M., Ito K., Emi M. 2005. Overexpressed in anaplastic thyroid carcinoma-1 (OEATC-1) as a novel gene responsible for anaplastic thyroid carcinoma. Cancer. 103:1785–1790 10.1002/cncr.20988 [DOI] [PubMed] [Google Scholar]
- Orford K.W., Scadden D.T. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9:115–128 10.1038/nrg2269 [DOI] [PubMed] [Google Scholar]
- Pervaiz S., Taneja R., Ghaffari S. 2009. Oxidative stress regulation of stem and progenitor cells. Antioxid. Redox Signal. 11:2777–2789 10.1089/ars.2009.2804 [DOI] [PubMed] [Google Scholar]
- Petroziello J., Yamane A., Westendorf L., Thompson M., McDonagh C., Cerveny C., Law C.L., Wahl A., Carter P. 2004. Suppression subtractive hybridization and expression profiling identifies a unique set of genes overexpressed in non-small-cell lung cancer. Oncogene. 23:7734–7745 10.1038/sj.onc.1207921 [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S.J., Clarke M.F., Weissman I.L. 2001. Stem cells, cancer, and cancer stem cells. Nature. 414:105–111 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Simpson F., Lammerts van Bueren K., Butterfield N., Bennetts J.S., Bowles J., Adolphe C., Simms L.A., Young J., Walsh M.D., Leggett B., et al. 2006. The PCNA-associated factor KIAA0101/p15(PAF) binds the potential tumor suppressor product p33ING1b. Exp. Cell Res. 312:73–85 10.1016/j.yexcr.2005.09.020 [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Sanbo M., Watanabe S., Naruse I., Mishina M., Yagi T. 1998. Efficient production of Cre-mediated site-directed recombinants through the utilization of the puromycin resistance gene, pac: a transient gene-integration marker for ES cells. Nucleic Acids Res. 26:679–680 10.1093/nar/26.2.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi L., Fareh M., Aberdam E., Kitajima S., Simpson F., Wicking C., Aberdam D., Virolle T. 2009. ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ. 16:728–737 10.1038/cdd.2009.2 [DOI] [PubMed] [Google Scholar]
- Walkley C.R., Fero M.L., Chien W.M., Purton L.E., McArthur G.A. 2005. Negative cell-cycle regulators cooperatively control self-renewal and differentiation of haematopoietic stem cells. Nat. Cell Biol. 7:172–178 10.1038/ncb1214 [DOI] [PubMed] [Google Scholar]
- Xu L., Geman D., Winslow R.L. 2007. Large-scale integration of cancer microarray data identifies a robust common cancer signature. BMC Bioinformatics. 8:275 10.1186/1471-2105-8-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Huang B., Shen M., Lau C., Chan E., Michel J., Xiong Y., Payan D.G., Luo Y. 2001. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene. 20:484–489 10.1038/sj.onc.1204113 [DOI] [PubMed] [Google Scholar]