M-phase DNA double-strand break repair differs from S-phase repair caused by the action of Cdk1, which prevents RPA-bound single-stranded DNA from activating classical DNA repair pathways.
Abstract
DNA double-strand break (DSB) resection, which results in RPA-bound single-stranded DNA (ssDNA), is activated in S phase by Cdk2. RPA-ssDNA activates the ATR-dependent checkpoint and homology-directed repair (HDR) via Rad51-dependent mechanisms. On the other hand, the fate of DSBs sustained during vertebrate M phase is largely unknown. We use cell-free Xenopus laevis egg extracts to examine the recruitment of proteins to chromatin after DSB formation. We find that S-phase extract recapitulates a two-step resection mechanism. M-phase chromosomes are also resected in cell-free extracts and cultured human cells. In contrast to the events in S phase, M-phase resection is solely dependent on MRN-CtIP. Despite generation of RPA-ssDNA, M-phase resection does not lead to ATR activation or Rad51 chromatin association. Remarkably, we find that Cdk1 permits resection by phosphorylation of CtIP but also prevents Rad51 binding to the resected ends. We have thus identified Cdk1 as a critical regulator of DSB repair in M phase. Cdk1 induces persistent ssDNA-RPA overhangs in M phase, thereby preventing both classical NHEJ and Rad51-dependent HDR.
Introduction
DNA double-strand breaks (DSBs) are potentially the most harmful form of DNA damage. DSBs are repaired by classical nonhomologous end joining (C-NHEJ), alternative nonhomologous end joining (Alt-NHEJ/microhomology-mediated end joining), or homology-directed repair (HDR). HDR and Alt-NHEJ pathways are initiated by degradation of the 5′ strand of the DSB to yield a 3′ single-stranded DNA (ssDNA) overhang, a process called DNA end resection (Symington, 2002). Resection allows RPA loading onto the ssDNA and subsequent repair by high-fidelity HDR pathways, which require Rad51 nucleoprotein filament formation and strand invasion into a homologous sequence. Resection in the absence of strand invasion may lead to mutagenic Alt-NHEJ, a source of chromosomal translocations (Zhang et al., 2010; Lee-Theilen et al., 2011; Zhang and Jasin, 2011).
At least two mechanistically distinct stages of DNA resection have been observed. Resection is initiated by the MRN (Mre11–Rad50–Nbs1) complex (Xrs2 is the budding yeast orthologue of Nbs1), which binds to DSB ends and facilitates activation of the ATM protein kinase. CtIP (Sae2 in budding yeast) is then recruited to the DSB-MRN complex (Lisby et al., 2004; Limbo et al., 2007), which promotes endonucleolytic cleavage of the 5′ strand, releasing short oligonucleotides (Jazayeri et al., 2008; Mimitou and Symington, 2008). In the second stage, the partially resected DSB recruits helicases and nucleases, including BLM (Sgs1 in budding yeast; both are RecQ homologues), DNA2, and Exo1, which catalyze extensive and processive resection (Gravel et al., 2008; Liao et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008; Budd and Campbell, 2009; Cejka et al., 2010; Niu et al., 2010). These pathways, however, are not independent: MRX (Mre11–Rad50–Xrs2) recruits Dna2 to budding yeast DSBs independent of its nuclease activity (Shim et al., 2010), and human MRN stimulates resection of linear DNA by Exo1 in vitro (Nimonkar et al., 2011).
Whether resection is initiated on a DSB is a critical determinant of repair pathway choice (Shrivastav et al., 2008). Resection enables HDR and Alt-NHEJ and prevents repair by C-NHEJ, which requires near-blunt double-stranded DNA ends. The mode of DSB repair depends on cell cycle status such that C-NHEJ is predominant in G0 and G1 when Cdk activity is low and no homologous template is available for repair, whereas DSBs are repaired primarily through HDR mechanisms in S and G2. This switch to HDR in S phase is controlled in part by Cdk-dependent phosphorylation/activation of Sae2/CtIP (Limbo et al., 2007; Huertas et al., 2008; Huertas and Jackson, 2009).
The ssDNA-RPA intermediates formed by resection also promote activation of the ATR-dependent damage checkpoint that acts through the Chk1 kinase (Costanzo et al., 2003; Zou and Elledge, 2003). Activated Chk1 inhibits Cdk1 activity by down-regulating Cdc25 phosphatases, which counteract inhibitory phosphorylation of Cdk1 by Wee1 kinase (Karlsson-Rosenthal and Millar, 2006). This G2/M checkpoint prevents entry into mitosis.
In contrast to interphase, little is known about signaling from and repair of DSBs in mitosis, which occurs in the context of condensed chromosomes and high Cdk activity. Chromosomes damaged at the onset of mitosis proceed through to anaphase without repair (Zirkle and Bloom, 1953). Therefore, canonical damage checkpoints that down-regulate Cdk1 are not fully operational after prophase (Morrison and Rieder, 2004). Indeed, Wee1 becomes inactive upon entry into mitosis, and damage-induced inactivation of Cdk1 does not occur (Okamoto and Sagata, 2007). However, more profound perturbation of chromatin structure or disruption of kinetochore–spindle attachments trigger the spindle assembly checkpoint (Rieder and Khodjakov, 1997), which significantly retards mitotic progression in an ATM-independent manner (Iwai et al., 1997; Mikhailov et al., 2002). Despite attenuated DNA damage checkpoints, phosphatidylinositol 3-kinase–like kinase family members are activated during mitosis, as indicated by ATM- and DNA-PK–dependent phosphorylation of histone H2AX (Giunta et al., 2010; Nakamura et al., 2010a). ATM-dependent phosphorylation also induces CEP63 dissociation from centrosomes and aberrant spindle assemblies (Smith et al., 2009).
In this paper, we (a) ask whether resection of DSBs occurs during vertebrate M phase, (b) assess the impact of Cdk1 activity on resection, and (c) characterize the consequences of resection in mitosis. We used cell-free extracts to monitor recruitment of signaling and repair proteins to restriction endonuclease–induced DSBs in a chromosomal context. We confirm that the two-step mechanism of S-phase resection in yeast is evolutionarily conserved in vertebrates, with one pathway dependent on MRN-CtIP and other, later pathways, independent of these factors. In addition, we report that DSB resection also occurs during mitosis, both in the condensed chromosomes of M-phase Xenopus laevis cell-free extracts and in cultured human cells. M-phase resection requires MRN-CtIP and phosphorylation of CtIP at a conserved Cdk consensus site. Remarkably, ssDNA-RPA generated by CtIP in M phase does not activate the ATR-Chk1 checkpoint or support Rad51 chromatin recruitment. Moreover, by showing that Cdk1 is responsible for the failure to recruit Rad51 to resected ssDNA-RPA in M-phase extracts, our experiments uncover a critical role for Cdk1 in regulating DSB repair during mitosis. Thus, by promoting CtIP-dependent resection of DSB ends while preventing Rad51 chromatin assembly, Cdk1 inhibits both the nonhomologous and homologous modes of DSB repair during mitosis. In this manner, Cdk1 can postpone DSB repair until chromosomes have been segregated and decondensed in the next cell cycle.
Results
Recruitment of proteins in response to chromosomal DSBs in S-phase Xenopus extract
We used cell-free extracts derived from the eggs of Xenopus to study the DNA damage response. These DNA-free extracts, which can be prepared as M-phase or interphase (S phase) extracts, contain all nuclear and cytoplasmic proteins and membrane lipids. When supplemented with demembranated sperm chromatin, S-phase extracts assemble nuclear envelopes and undergo DNA replication (Blow and Laskey, 1986; Hutchison et al., 1988). In contrast, chromatin added to M-phase extract remains highly condensed and supports formation of mitotic spindles (Verde et al., 1990). Significantly, upon exposure to endonuclease-generated chromosomal DSBs (Yoo et al., 2006), these extracts recapitulate many aspects of the cellular response to DSBs (Costanzo et al., 2000, 2001; Di Virgilio and Gautier, 2005; Dupré et al., 2006; You et al., 2007; Jazayeri et al., 2008).
We have induced DSBs with restriction endonuclease added to extracts supplemented with sperm chromatin. To monitor recruitment of proteins that respond to DSBs, aliquots of the treated samples were centrifuged through a dense sucrose cushion, which effectively separates soluble cytosolic and nuclear proteins from the insoluble chromatin fraction.
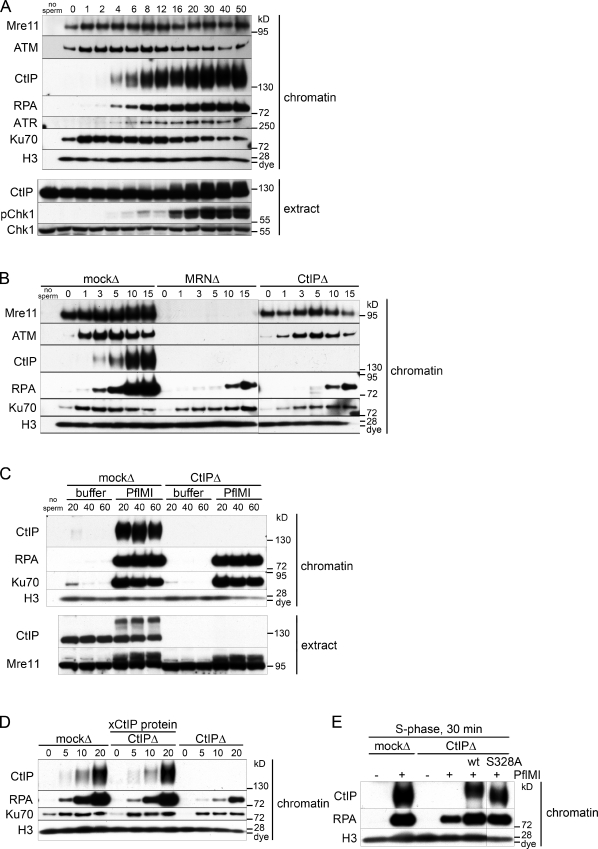
In S-phase extract, the response to chromosomal DSBs was extremely rapid (Fig. 1 A). Although basal levels of Mre11, ATM, and Ku70 bound to chromatin before treatment, these proteins were significantly enriched in the chromatin fraction by 1 min after addition of the restriction enzyme. CtIP and RPA bound to DSB-containing chromatin by 4 min followed by ATR (Fig. 1 A). Because RPA binds selectively to ssDNA, chromatin-associated RPA was used as a measure of the amount of ssDNA generated by DSB resection. Phosphorylation of the ATR substrate Chk1 was seen in the soluble extract 16 min after DSB induction, simultaneous with phosphorylation of soluble CtIP (Fig. 1 A, extract). In all chromatin-binding experiments, total histone H3 was used as a chromatin-loading control.
Figure 1.
DSB resection in S-phase Xenopus cell-free extract proceeds by CtIP-dependent and -independent pathways. (A) Kinetics of recruitment of proteins to DSB-containing chromatin in S-phase extract. S-phase extract was preincubated with sperm chromatin (5,000 sperm/µl). Aliquots of the sample were taken before (0 min) and at the indicated time (minutes) after addition of 0.05 U/µl PflMI restriction endonuclease. At each time point, 0.5 µl of the sample was removed and processed for Western blotting (extract), and 15 µl of the sample was removed and processed for chromatin isolation followed by Western blotting with the indicated antibodies (chromatin). A sample with no chromatin added (no sperm) serves as a chromatin purification control in A–C. (B) Early resection is dependent on MRN and CtIP in S-phase extract. Short kinetics of protein recruitment to chromatin in response to DSBs in mock-depleted, Mre11-depleted, and CtIP-depleted S-phase extracts as in A. (C) An MRN-CtIP–independent resection pathway operates in S-phase extract. Mock-depleted or CtIP-depleted S-phase extract was incubated with sperm chromatin and either buffer or PflMI restriction endonuclease for extended time points as in A. (D) Addition of recombinant xCtIP protein to CtIP-depleted S-phase extract restores resection activity to control levels. Experiment performed as in A but with an S-phase extract that was mock depleted, CtIP depleted supplemented with 110 nM recombinant xCtIP protein, or CtIP depleted as indicated. (E) The CtIP–BRCA1 interaction is not required for resection of endonuclease-induced chromosomal DSBs. Sperm chromatin was incubated in S-phase extract that was mock depleted, CtIP depleted, or CtIP depleted supplemented with 75 nM wt or S328A-xCtIP and was treated with buffer (−) or PflMI (+) for 30 min. Samples were processed for chromatin isolation as in A. The black line indicates that intervening lanes have been spliced out.
DSB resection occurs by CtIP-dependent and -independent pathways in S-phase extract
We next asked whether CtIP and MRN were required for resection in S phase. To this end, we immunodepleted CtIP or Mre11 (which removes the entire MRN complex) from S-phase extracts (Costanzo et al., 2001). Total mouse IgG was used in a mock depletion control. Depletion of MRN did not significantly codeplete CtIP or vice versa (Fig. S1 A). We determined that >98% of CtIP was depleted (Fig. S1 B) and that no residual protein was detected on damaged chromatin (Fig. S1 C). CtIP depletion was equally efficient in both S-phase and M-phase extracts (Fig. S1 B).
As expected, depletion of MRN abrogated ATM and CtIP recruitment to damaged chromatin (Fig. 1 B; Limbo et al., 2007; You et al., 2009). In contrast, ATM and Mre11 recruitment did not depend on CtIP. These data, along with the slower kinetics of CtIP recruitment compared with MRN and ATM (Fig. 1 A), suggests that MRN recognition of DSBs and ATM activation occur before and independently of CtIP recruitment. Importantly, depletion of either MRN or CtIP delayed but did not abolish DSB resection in S-phase extract (Fig. 1 B). This result reveals multiple distinct resection pathways in S phase, an MRN-CtIP–dependent pathway, and one or more pathways independent of the initial resection step. Initiation of resection was delayed by ∼7 min in the absence of CtIP-MRN. However, at later time points, similar amounts of ssDNA were formed in CtIP-depleted and control extract, suggesting that CtIP-independent mechanisms are able to initiate resection and continue to generate substantial amounts of ssDNA (Fig. 1 C).
Addition of excess purified Xenopus CtIP (xCtIP) protein to CtIP-depleted extracts (Fig. S1, D and E) restored CtIP chromatin binding to endogenous levels and reestablished resection kinetics to control rates (Fig. 1 D). This confirms that the resection defect in CtIP-depleted extract is caused by the specific removal of endogenous CtIP.
CtIP can interact with the BRCA1 tumor suppressor in a phosphodependent manner that requires Cdk-dependent phosphorylation of CtIP at Ser327 (in humans; S328 in Xenopus; Yu and Chen, 2004; Varma et al., 2005; Chen et al., 2008). Upon DNA damage, MRN is assembled into a larger protein complex that includes both CtIP and BRCA1 (Greenberg et al., 2006). To assess whether the CtIP–BRCA1 interaction influences DNA resection, we expressed and purified an xCtIP polypeptide (xCtIP-S328A) bearing a missense mutation known to abolish the CtIP–BRCA1 interaction in mammalian (Yu and Chen, 2004) and in chicken DT40 (Nakamura et al., 2010b) cells. As shown in Fig. 1 E, the xCtIP-S328A protein was as effective as the wild-type (wt) CtIP protein in stimulating resection when added to a CtIP-depleted S-phase extract. These results suggest that BRCA1 is not required for CtIP-dependent resection of nuclease-induced DSBs in the Xenopus extract.
Characteristics of CtIP-independent resection
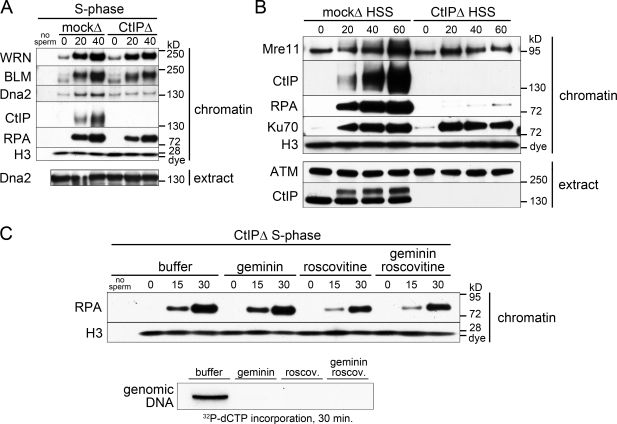
To characterize further the CtIP-independent pathway, we monitored the binding of BLM and WRN RecQ helicases and of DNA2 helicase/nuclease to endonuclease-treated chromatin. All three factors, which have been implicated in vertebrate resection (Yan et al., 2005; Toczylowski and Yan, 2006; Gravel et al., 2008; Liao et al., 2008; Mimitou and Symington, 2008; Nimonkar et al., 2008; Zhu et al., 2008; Budd and Campbell, 2009), were enriched on damaged chromatin in mock-depleted S-phase extracts (Fig. 2 A). Both RecQ helicases bound chromatin in the CtIP-depleted extract, strongly suggesting their participation in the CtIP-independent resection. In contrast to BLM and WRN, DNA2 recruitment was largely CtIP dependent. Note that CtIP depletion did not codeplete DNA2 from extracts (Fig. 2 A, extract).
Figure 2.
Characteristics of CtIP-independent resection. (A) Recruitment of late resection pathway components in the absence of CtIP. The mock- and CtIP-depleted S-phase extract was treated with PflMI restriction endonuclease, and chromatin was isolated at the indicated time points (minutes) followed by Western blotting with the indicated antibodies as in Fig. 1 A. (B) CtIP-independent resection does not occur in membrane-free HSS extract. Mock-depleted or CtIP-depleted membrane-free S-phase HSS extract was preincubated with sperm chromatin (5,000 sperm/µl). Aliquots were taken before (0 min) and at the indicated time after addition of PflMI restriction endonuclease (0.05 U/µl) and processed as in Fig. 1 A. (C) CtIP-independent resection does not require DNA replication. CtIP-depleted S-phase extract was supplemented with recombinant geminin protein, roscovitine (roscov.), both, or buffer and was preincubated with sperm chromatin. (top) 15-µl aliquots of the sample were taken before (0 min) and at the indicated times after addition of 0.05 U/µl PflMI restriction enzyme and processed for chromatin isolation followed by Western blotting. (bottom) In parallel, a 10-µl aliquot of each sample was taken before addition of the restriction enzyme supplemented with 0.1 µl [32P]deoxy-CTP (dCTP) and incubated for 30 min to monitor DNA replication.
Crude cell-free extracts contain lipids that assemble into nuclear envelopes around added sperm chromatin. This compartmentalization allows the import and concentration of nuclear factors essential for origin firing and DNA replication. Membrane-free extract obtained by high-speed centrifugation of crude S-phase extract (high-speed supernatant [HSS]) cannot form nuclear envelopes and fails to replicate chromatin. To determine the role of nuclear envelopes, we assayed resection in mock- or CtIP-depleted membrane-free HSS. CtIP, RPA, and Ku70 all associated with damaged chromatin in the HSS extract (Fig. 2 B). Resection, as seen by RPA binding, was undetectable in the CtIP-depleted extract (Fig. 2 B). This implies that late pathway resection requires chromatin compartmentalization. Failure to resect DNA in CtIP-depleted HSS was not caused by the absence of BLM, WRN, DNA2, or Exo1 proteins because all are present in HSS (Fig. S2 A).
Because a nuclear envelope is required for both chromosomal DNA replication and late pathway resection, we asked whether the latter depends on DNA replication. To test this idea, we inhibited prereplication complex formation with geminin (McGarry and Kirschner, 1998) or Cdk2 with roscovitine (Bresnahan et al., 1997) to prevent origin firing. Both inhibitors completely blocked incorporation of radiolabeled nucleotides into genomic DNA (Fig. 2 C, bottom). However, neither inhibitor abrogated late pathway resection, as determined by RPA recruitment in CtIP-depleted S-phase extract (Fig. 2 C). DNA replication, therefore, is not required for CtIP-independent resection.
DSBs generated in M phase undergo CtIP-dependent resection
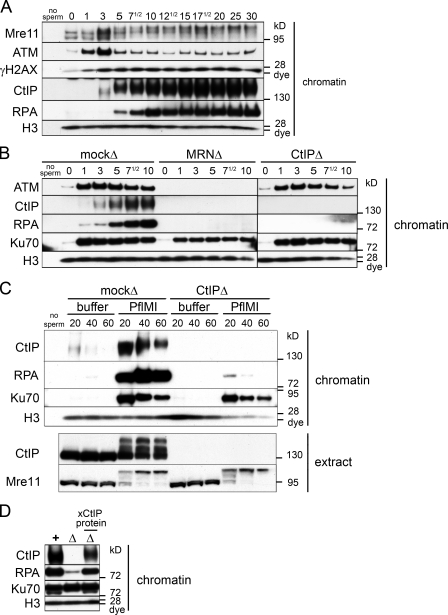
To determine whether DSB resection occurs in the context of highly condensed mitotic chromosomes, we induced DSBs in M-phase extract (cytostatic factor [CSF]–arrested extract in metaphase II of meiosis) and monitored recruitment of proteins to damaged chromatin. We confirmed that restriction endonuclease was able to generate DSBs in M-phase condensed chromosomes as indicated by (a) the rapid recruitment of Ku70 (a component of the Ku70/86 heterodimer that binds double-stranded DNA ends with high affinity; Fig. 3 B) and (b) TUNEL of endonuclease-treated chromatin isolated from M-phase extract (Fig. S2 B). ATM association with damaged chromatin peaked 1 min after endonuclease addition and then decreased (Fig. 3 A). A reduction in Mre11 gel mobility, indicating phosphorylation, and maximal histone H2AX phosphorylation was observed at 3 min. CtIP did not bind to undamaged M-phase chromatin but was recruited 3–5 min after damage. Chromatin-bound RPA was detected by 5 min, indicating that ssDNA is generated by DSB resection within M-phase chromatin (Fig. 3 A).
Figure 3.
DSBs generated in M phase undergo only CtIP-dependent resection. (A) Kinetics of recruitment of proteins to DSB-containing chromatin in M-phase extract. M-phase extract (meiotic and CSF arrested) was preincubated with sperm chromatin (5,000 sperm/µl). 15-µl aliquots of the sample were taken before (0 min) and at the indicated times (minutes) after addition of PflMI restriction endonuclease (0.05 U/µl) and processed for chromatin isolation followed by Western blotting with the indicated antibodies as in Fig. 1 A. (B) All resection is dependent on MRN and CtIP in M-phase extract. Short kinetics of protein recruitment to chromatin in response to DSBs in mock-depleted, Mre11-depleted, and CtIP-depleted M-phase extracts as in Fig. 1 B. (C) No resection occurs in the absence of CtIP in the M-phase extract. Long kinetics of protein recruitment to chromatin in response to DSBs. The mock-depleted or CtIP-depleted M-phase extract was incubated with sperm chromatin and either buffer or PflMI restriction endonuclease for extended time points as in Fig. 1 C. (D) Addition of recombinant xCtIP protein to CtIP-depleted M-phase extract restores resection activity to control levels. Experiment performed as in Fig. 1 D but with the M-phase extract that was mock depleted (+), CtIP depleted (Δ), or CtIP depleted supplemented with 50 nM recombinant xCtIP protein (Δ with xCtIP protein above) as indicated for a 30-min time point. The black line indicates that intervening lanes have been spliced out.
As in S phase, depletion of MRN in M phase abolished recruitment of ATM and, subsequently, CtIP to damaged chromatin (Fig. 3 B). Depletion of CtIP in M phase did not affect ATM or MRN recruitment and H2AX phosphorylation (Fig. 3 B and not depicted). Notably, M-phase extracts depleted of either MRN or CtIP were unable to resect DSBs. CtIP depletion did not affect ATM recruitment and, thus, MRN recognition of DSB; nevertheless, these breaks were not resected. Thus, MRN cannot support resection in the absence of CtIP.
In S phase, but not in M phase, we could detect resection by an MRN-CtIP–independent pathway by 10 min (compare Figs. 1 B and 3 B). However, even with prolonged incubation, we saw no resection in the absence of CtIP in M phase, despite the persistence of DSBs, as shown by Ku70 recruitment (Fig. 3 C). Furthermore, the amount of chromatin-bound RPA was proportional to the amount of endogenous CtIP in the extract, suggesting that CtIP is rate limiting for resection in M phase (Fig. S2 C). Rescue of CtIP-depleted M-phase extract with excess purified recombinant xCtIP protein (Figs. S1 D and S2 D) restored resection activity to mock-depleted levels (Fig. 3 D).
DSB resection also occurs in mitotic M-phase extract
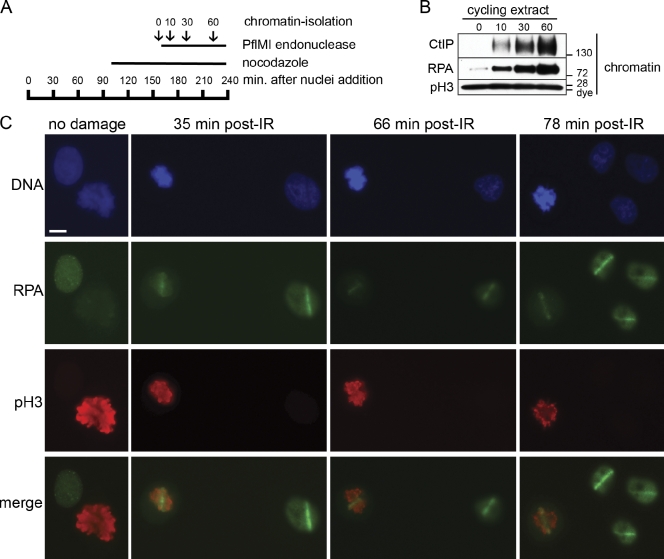
We considered the possibility that resection in M-phase extract might be specific for CSF-arrested meiotic M-phase extract. To address this concern, we prepared a cycling mitotic extract that was allowed to complete DNA replication. Nocodazole was then added to arrest the extract in mitosis, after which chromosomal DSBs were induced by restriction endonuclease (Fig. 4 A). Nuclear morphology was monitored by fluorescence microscopy to confirm cell cycle phase (unpublished data). Like CSF-arrested extract, mitotic extract supported CtIP binding to chromatin and DSB resection (Fig. 4 B). This confirms that resection can occur in both meiotic and mitotic M phases. We will continue to refer to CSF-arrested extract as M-phase extract for simplicity.
Figure 4.
DSB resection occurs in cycling mitotic Xenopus extract and mitosis of human cells. (A) Schematic timeline of the cycling mitotic extract experiment. Cycling extract was incubated with sperm chromatin (5,000 sperm/µl). Nocodazole was added at 96 min to trap the nuclei in the subsequent mitosis. Microscopy analysis 34 min later confirmed that the chromatin was in a highly condensed state indicative of mitosis. An aliquot was taken before addition of 0.05 U/µl PflMI restriction endonuclease at 163 min (time 0). Aliquots were also taken at 10, 30, and 60 min after addition of PflMI and processed for chromatin isolation and Western blotting. (B) Resection of DSBs occurs in nocodazole-arrested mitotic extract. After arrest with nocodazole, DSBs were induced in the mitotic chromatin, and aliquots were taken before and at the indicated time points (minutes) after addition of PflMI and processed for chromatin isolation and Western blotting with the indicated antibodies. The mitotic status of the extract was also confirmed by the presence of pSer10–histone H3. (C) Resection of DSBs occurs in mitosis of human cells. Asynchronous HeLa cells were grown on 8-well chamber slides and mock irradiated (no damage) or microirradiated using a high-energy UV laser microscope (PALM MicroBeam IV). After the indicated time (±4 min), slides were processed and stained with antibodies against human RPA34 and phospho–Ser10-histone H3. Bar, 10 µm.
As an independent means to monitor ssDNA formation directly, we adapted a BrdU detection assay in cycling extract. Chromosomal DNA was replicated in the cycling extract in the presence of BrdU and arrested in the subsequent mitosis. After generation of DSBs, chromatin was isolated, and ssDNA was detected using the anti-BrdU antibody on DNA isolated under nondenaturing conditions (Fig. S3 A). Detection of ssDNA in this way requires replication-dependent incorporation of BrdU (Fig. S3 A, first and second lanes). Notably, consistent with results obtained by monitoring RPA chromatin binding, generation of ssDNA as seen by BrdU staining in M phase is dependent on CtIP (Fig. S3 B, compare mock- or CtIP-depleted extracts).
Human cells undergo resection within mitotic chromosomes
Our finding that resection takes place in the M-phase extract was rather surprising because evidence for mitotic resection in mammalian cells has not been reported. To ascertain whether resection also occurs during mammalian mitosis, we used a focused high-power UV laser to generate discrete regions of DSBs within HeLa cell nuclei. Asynchronous cultures were irradiated and stained for both RPA and the mitotic marker pH3 (phosphorylated histone H3). A fraction of undamaged control cells showed highly condensed mitotic chromosomes that stained strongly for pH3 (Fig. 4 C, no damage). Distinct and persistent stripes of RPA were seen after irradiation in both interphase and mitotic cells (Fig. 4 C). 98% of cells irradiated in mitosis scored positive for γ-H2AX (98/100), and 80% scored positive for RPA (80/100). 98% of the RPA-positive cells were also positive for γ-H2AX (78/80; unpublished data). Because the G2/M damage checkpoint prevents irradiated G2 cells from entering mitosis, our data indicate that cells damaged during mitosis resect DSBs. Moreover, because undamaged HeLa cells proceed through mitosis in ∼45 min, cells that persist in metaphase at 66 and 78 min have presumably sustained kinetochore damage, causing arrest because of the spindle assembly checkpoint. We could not determine whether CtIP is required for resection in mammalian mitosis because CtIP is essential for cellular viability (Chen et al., 2005; Nakamura et al., 2010b) and because inactivation of CtIP induces rapid cell cycle arrest (unpublished data).
Phosphorylation of CtIP by Cdk1 is required for M-phase resection
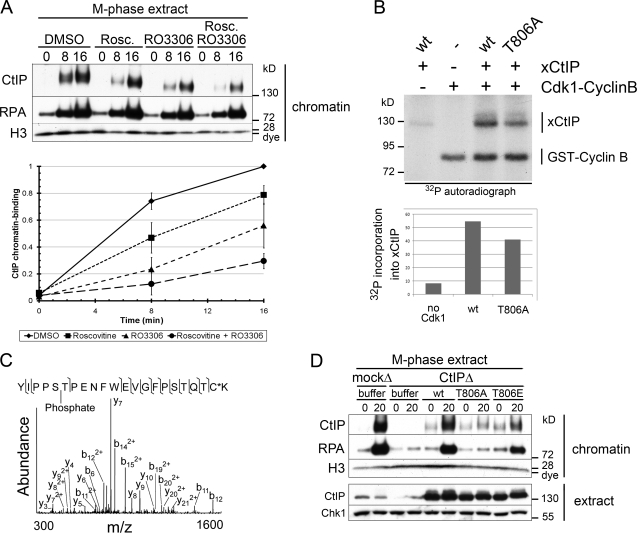
CtIP is phosphorylated by Cdc28 (in budding yeast) and Cdk2 (in mammals) at residues equivalent to T806 of xCtIP. This phosphorylation occurs at the onset of S phase and constitutes a critical switch from C-NHEJ to HDR (Limbo et al., 2007; Huertas et al., 2008; Huertas and Jackson, 2009). Cdk1–cyclin B is the prominent Cdk activity in M phase. To determine the role of Cdk in M-phase resection, we inhibited Cdk2 with roscovitine and Cdk1 with the highly specific small molecule RO-3306 (Vassilev et al., 2006). Treatment with both inhibitors simultaneously decreased CtIP binding to chromatin by ∼70% (Fig. 5 A). We next established that CtIP is an in vitro substrate for Cdk1–cyclin B (Fig. 5 B). We generated a nonphosphorylatable mutant of CtIP (CtIP-T806A) and purified the recombinant protein from baculovirus-infected insect cells (Fig. S4 A). Phosphorylation of xCtIP-T806A by Cdk1 was reduced relative to wt but not abolished because xCtIP contains several other Cdk sites, including the BRCA1 interaction site S328 (Fig. 5 B). To test whether CtIP was phosphorylated at T806 in M phase, we affinity purified endogenous CtIP from M-phase extract using the CtIP 11–1 mAb and subjected it to mass spectrometry. These results confirmed phosphorylation of CtIP at S805/T806 in M phase (Fig. 5 C).
Figure 5.
Phosphorylation of CtIP by Cdk1 is required for M-phase resection. (A) Cdk activity is required for CtIP function and M-phase resection. M-phase extract was treated with DMSO, 200 µM roscovitine (Rosc.), 200 µM RO-3306, or both, and chromatin was isolated at the indicated time points (minutes) after addition of PflMI restriction endonuclease. Quantification of relative CtIP chromatin binding from three independent experiments is shown on the bottom, with error bars representing one standard deviation. (B) Cdk1–cyclin B kinase phosphorylates recombinant xCtIP in vitro. Recombinant Cdk1–cyclin B was incubated with recombinant xCtIP (wt or T806A) as indicated in the presence of γ-[32P]ATP. Samples were analyzed by SDS-PAGE and autoradiography. Quantification of the autoradiograph signal for xCtIP in this experiment is graphed on the bottom. Note that xCtIP-T806A incorporation is reduced by ∼25% because of the presence of multiple Cdk phosphorylation sites. (C) Tandem mass spectrometry of endogenous M-phase CtIP reveals phosphorylation at S805/T806. Tandem mass spectrum of phosphopeptide spanning residues 801–822. The experimental molecular mass of the intact precursor ion (2664.1613 D) closely matches the theoretical mass of the tryptic peptide 801–822 plus a phosphate group (2664.1607 D; mass difference = 0.0006 D). The fragmentation pattern confirms the identity of this phosphopeptide and narrows the localization of the phosphate group to either S805 or T806. The asterisk indicates that the cysteine residue was alkylated using iodoacetamide. (D) Conserved residue T806 is required for CtIP activity and resection in M phase. M-phase extract was mock depleted or CtIP depleted. CtIP-depleted extract was supplemented with buffer (−), wt xCtIP (wt), xCtIP-T806A (T806A), or xCtIP-T806E (T806E). PflMI restriction endonuclease (+) or buffer (−) was added, and chromatin was isolated after 15 min. Time points above are in minutes. m/z, mass to charge ratio.
To address the consequence of T806 phosphorylation on M-phase resection, we rescued CtIP-depleted M-phase extract with purified recombinant wt CtIP, CtIP-T806A, or CtIP-T806E (Fig. S4 B). Importantly, CtIP-T806A failed to bind chromatin or to restore resection (Fig. 5 D). In contrast, the phosphomimic T806E mutant supported recruitment to damaged chromatin and resection (Fig. 5 D). This demonstrates that phosphorylation of CtIP by Cdk1 at T806 is required for resection of mitotic chromosomes. Phosphorylation of this same site is also required for CtIP-dependent resection in S-phase extract (Fig. S4, C and D).
M-phase resection does not lead to Rad51 chromatin recruitment or Chk1 activation
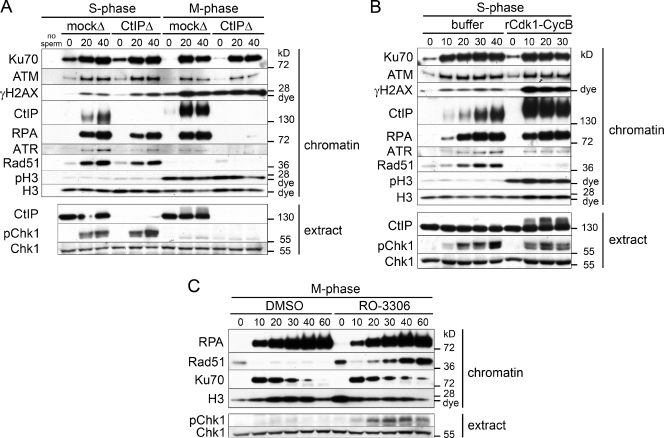
ssDNA generated in S-G2 sequentially activates ATR and Chk1, leading to inhibition of Cdk (Löbrich and Jeggo, 2007). Additionally, formation of ssDNA is the first step of the HDR pathway. Both of these outcomes pose problems to M-phase cells. Down-regulation of the mitotic kinase Cdk1, when chromosomes are highly condensed and under tension from spindles, could induce spontaneous exit from mitosis, with devastating consequences to genome integrity. In addition, condensed sister chromatids should not be available for homology search and HDR. To compare the consequences of DSB resection in S and M phase, we monitored Chk1 activation (phosphorylation on S344 in extract) and Rad51 chromatin binding in mock- or CtIP-depleted S- and M-phase extracts prepared from the same batch of eggs (Fig. 6 A). As also shown in Figs. 1 and 3, CtIP depletion delayed resection in S phase and abolished resection in M phase. As anticipated, ATR associated with chromatin, and Chk1 was activated in S phase after resection (Fig. 6 A). In contrast, despite generation of ssDNA-RPA, little chromatin-bound ATR was observed, and we failed to detect Chk1 activation in M phase. Consistent with this finding, damaged chromatin did not significantly reduce Cdk1 activity in M-phase extracts (Fig. S5 A). Chromatin-bound Rad51 was detected in control and CtIP-depleted S-phase extracts treated with a restriction enzyme, indicating that CtIP-independent resection can support Rad51 loading. In contrast, no chromatin-bound Rad51 was observed in M phase (Fig. 6 A).
Figure 6.
M-phase Cdk1 activity inhibits Rad51 loading on ssDNA-RPA. (A) Rad51 accumulates on resected chromatin, and Chk1 becomes activated in S- but not in M-phase extract. S- and M-phase extracts prepared from the same batch of eggs were either mock or CtIP depleted. Chromatin-binding time course in response to DSBs was performed as in Fig. 1 A. (B) Treatment of S-phase extract with Cdk1 inhibits Rad51 chromatin binding but not Chk1 activation. S-phase extract was supplemented with 100 nM recombinant Cdk1–cyclin B protein complex or buffer, and a chromatin-binding time course in response to DSBs was performed as in Fig. 1 A. (C) Inhibition of Cdk1 activity in M phase restores Rad51 accumulation in response to chromosomal DSBs. M-phase extract was supplemented with the specific Cdk1 inhibitor RO-3306 (200 µM) or DMSO, and a chromatin-binding time course in response to DSBs was performed as in Fig. 1 A. Time points above the blots are in minutes.
Cdk1 activity inhibits Rad51 chromatin assembly on RPA-ssDNA intermediates
We then asked whether Cdk1 inhibits Rad51 loading and/or Chk1 activation. In the S-phase extract, levels of chromatin-associated Rad51 increased with time after induction of DSBs (Fig. 6 B, buffer). Addition of recombinant Cdk1–cyclin B (Fig. S5 B) to endogenous levels (Fig. S5 C) abrogated Rad51 filament assembly on resected DNA (Fig. 6 B). S-phase extract incubated with Cdk1 resembled a mitotic extract, as indicated by robust H3 phosphorylation and damage-induced hyperphosphorylation of CtIP and H2AX (Fig. 6 B). Notably, Cdk1 activity abrogated Rad51 filament formation but did not prevent Chk1 activation.
Next, we examined the effect of Cdk1 inhibition in M phase by RO-3306 (Vassilev et al., 2006). As before, endonuclease treatment of M-phase extract induced robust resection and RPA binding but no Chk1 activation or Rad51 chromatin accumulation (Fig. 6 C, DMSO). Importantly, Cdk1 inhibition permitted Rad51 binding to damaged chromatin and partial activation of Chk1 (Fig. 6 C). Together, these data confirm that Rad51 chromatin loading is inhibited in a Cdk1-dependent manner downstream of the formation of ssDNA-RPA.
Discussion
Here, we show that vertebrate resection proceeds via distinct pathways in S phase (Fig. 1). We observe an early pathway that is dependent on both the MRN complex and CtIP. We also demonstrate robust but delayed resection in the absence of MRN-CtIP, which we denote as the late resection pathway. In Saccharomyces cerevisiae, resection also occurs by two distinct pathways (Mimitou and Symington, 2009). An initiating MRX- and Sae2-dependent endonucleolytic cleavage of 50–100-nt oligonucleotides from the 5′ strand is followed by a highly processive resection step involving helicases and nucleases, including Sgs1 (RecQ homologue), Dna2, and Exo1. In vertebrates, several factors have been implicated in resection of DSBs, including DNA2 (Liao et al., 2008), Exo1 (Bolderson et al., 2010), and the two RecQ homologues BLM (Gravel et al., 2008; Nimonkar et al., 2008) and WRN (Yan et al., 2005; Toczylowski and Yan, 2006). In our experiments, we show that both the BLM and WRN helicases are recruited independently of CtIP in S phase (Fig. 2 A) and are good candidates for late vertebrate resection factors. In contrast to WRN and BLM, recruitment of DNA2 to damaged chromatin was CtIP dependent. This is reminiscent of results obtained in S. cerevisiae, which show that Dna2 recruitment to DSBs is dependent on Mre11 (Shim et al., 2010). Our data indicate that the multistep process of DNA resection at DSBs is evolutionarily and functionally conserved from yeast and vertebrates.
Recent work by You et al. (2009) demonstrated that all resection in the Xenopus S-phase extract is exclusively dependent on CtIP. However, those experiments were performed in membrane-free HSS extracts. Our work shows that CtIP-independent resection does not occur in HSS extracts (Fig. 2 B) and that these investigators were, therefore, unable to detect late pathway resection, which is recapitulated only in complete extracts containing membranes.
Whether the BRCA1–CtIP interaction plays a significant role in DSB resection is presently unclear. A recent study using chicken lymphoblastoid DT40 cells reported that, although CtIP was dispensable for cell viability, the BRCA1–CtIP interaction was essential for DSB resection and subsequent HDR (Yun and Hiom, 2009). In contrast, Nakamura et al. (2010b) reported that CtIP was required for viability and that the BRCA1–CtIP interaction was dispensable for DSB resection and HDR in DT40 cells. We find that xCtIP-S328A, a point mutant that is defective for the BRCA1–CtIP interaction, is able to support resection of restriction endonuclease–induced DSBs when added to a CtIP-depleted extract (Fig. 1 E). Whereas our results suggest that BRCA1 is dispensable for resection of simple DSB ends, it is possible that BRCA1 is required to resect complex DNA ends. For example, cells harboring mutant CtIP defective in the BRCA1 interaction are sensitive to camptothecin, a topoisomerase inhibitor that creates DSBs with covalently attached protein adducts (Nakamura et al., 2010b). Additionally, Sae2/CtIP has been shown to process DNA ends containing adducts, including Spo11 and topoisomerase adducts (Keeney and Kleckner, 1995; Penkner et al., 2007; Hartsuiker et al., 2009; Nakamura et al., 2010b).
The fate of DSBs sustained during vertebrate mitosis is largely unknown. During this dynamic cell cycle phase, chromatin is highly condensed and may be relatively inaccessible to DNA metabolic enzymes. However, we show that DSBs are readily generated by restriction endonucleases in M phase and that resection of DSBs does indeed occur in Xenopus meiotic and mitotic M-phase extracts, as well as during mitosis in cultured human cells (Figs. 3 and 4). Previously, RPA was shown to relocalize in mitotic mammalian cells in response to ionizing radiation (Stephan et al., 2009).
Despite CtIP-dependent generation of ssDNA-RPA in M phase, we show that ATR does not accumulate to S-phase levels on damaged chromatin, and Chk1 is not activated, suggesting a defect in assembly of signaling complexes on ssDNA-RPA (Fig. 6 A). This is in agreement with a recent study showing that Chk1 was not phosphorylated in response to ionizing radiation in nocodazole-arrested human cells (Giunta et al., 2010). The mechanism of Chk1 repression in M phase is distinct from inhibition of Rad51. Thus, Cdk1 does not block Chk1 activation, whereas it completely prevents association of Rad51 with damaged DNA (Fig. 6 B). We propose that assembly of ATR-signaling complexes is inhibited during M phase because of a lack of an S-phase kinase activity, rather than Cdk1 activity itself.
Our experiments reveal a critical role for the mitotic kinase Cdk1 in regulating DSB repair in M phase. We establish that Cdk1–cyclin B phosphorylates CtIP at T806 and that the T806A phosphomutation completely abrogated resection in M phase (Fig. 5 D). T806 is also the target for Cdk2 phosphorylation at the onset of S phase, and phosphorylation of this site is required for S-phase resection in budding yeast and mammalian cells (Limbo et al., 2007; Huertas et al., 2008; Huertas and Jackson, 2009), as well as Xenopus extracts (Fig. S4, C and D). Moreover, we find that repression of Rad51 chromatin accumulation in M phase is caused by Cdk1 activity because addition of the recombinant Cdk1–cyclin B protein to S-phase extract abolished Rad51 binding (Fig. 6 B). In vitro and structural studies have shown that Cdk1-dependent phosphorylation of the C terminus of BRCA2 disrupts the interaction of that domain with Rad51, causing destabilization of Rad51 filaments and inhibition of HDR repair assays (Esashi et al., 2005, 2007). Moreover, human cells that have entered mitosis with DNA damage display γ-H2AX but not Rad51 foci (Ayoub et al., 2009). Our data support these findings by showing that Rad51 association with ssDNA generated from DSB resection was inhibited by Cdk1. It is also possible that Cdk1-dependent phosphorylation of RPA32 prevents exchange of RPA for Rad51 on ssDNA. In support of this hypothesis, serines 23 and 29 of RPA32 are phosphorylated specifically in mitosis (Oakley et al., 2003; Anantha et al., 2008). Together, our study reveals a role for Cdk1 in the DNA damage response to DSBs.
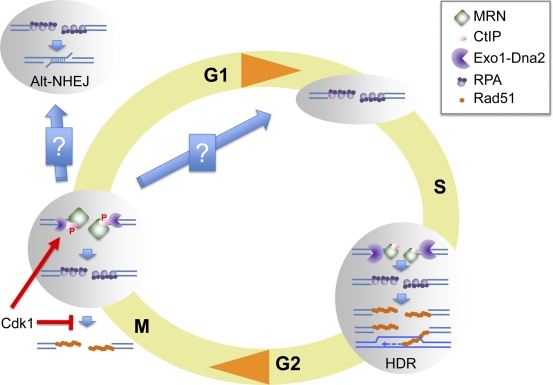
Our results demonstrate that Cdk1 promotes the initiation of DSB resection and accumulation of ssDNA-RPA while at the same time inhibiting Rad51, thus uncoupling resection from downstream high-fidelity HDR (Fig. 7). Chromosome condensation and dynamic chromosome movement may prevent strand invasion and homology search in M phase. Repression of Rad51 association with ssDNA, therefore, may prevent potentially abortive homologous recombination (HR) attempts. Additionally, by disrupting the double-stranded nature of DNA ends, resection inhibits Ku and thereby prevents C-NHEJ. However, short-range resection in the absence of Rad51 assembly may promote toxic, CtIP-dependent Alt-NHEJ, a repair pathway recently shown to be responsible for a majority of chromosomal translocations (Fig. 7; Lee-Theilen et al., 2011; Zhang and Jasin, 2011).
Figure 7.
Chromosomal DNA DSB resection in M phase. DNA DSBs generated in S phase are resected via the concerted action of MRN-CtIP and two additional pathways represented here as a single Exo1-DNA2 entity. S-phase resection generates ssDNA-RPA, which is competent for Rad51 filament assembly, a necessary step for HR. In contrast, DSBs generated in M phase are processed into ssDNA-RPA intermediates that do not support Rad51 chromatin assembly. Cdk1 promotes resection by phosphorylating CtIP while at the same time inhibiting Rad51 chromatin assembly. Resection initiation is dependent on MRN-CtIP in M phase as reflected by the relative size of the resection machinery components. M-phase resection generates ends that are not compatible for repair by NHEJ or by HR. These ends could be substrates for microhomology-mediated end joining in M or G1 phase. Alternatively, they could be transmitted to the next S phase and repaired by HR. P, phosphorylation.
Although we cannot directly assess the consequences of this CtIP- and M phase–dependent mode of resection, we posit that, if not immediately repaired by Alt-NHEJ, repair of DSBs incurred in M phase is delayed until the next cell cycle, at which point repair can proceed by high-fidelity mechanisms to preserve genome stability. In budding yeast, repair of a portion of DSBs sustained in G1 phase is delayed until HDR is possible in the following S phase (Lee and Petes, 2010). Moreover, budding yeast mitotic chromosomes harboring a DSB do not missegregate the chromosome fragment. Instead, the MRN complex facilitates maintenance of the broken DNA within a functional chromosome (Lobachev et al., 2004). Therefore, the same protein complex that is responsible for processing the DSB also ensures that the broken chromosome arm distal from the centromere is properly segregated.
In agreement with this notion, damaged DNA generated during mitosis in mammalian cells is marked by γ-H2AX, but these breaks persist and do not recruit repair factor 53BP1 until the cells exit mitosis (Giunta et al., 2010). Similarly, underreplicated DNA that escapes the G2/M damage checkpoint can form ultrafine bridges and chromosome breakage during mitosis (Chan et al., 2009). These breaks persist into the daughter cells, marked by 53BP1 after mitotic exit, until the subsequent S phase (Lukas et al., 2011). Together, these findings support a model in which damage is transmitted through mitosis to be repaired in the subsequent cell cycle after chromatin decondensation (Giunta and Jackson, 2011). A better understanding of how cells handle and process DSBs occurring in mitosis has important implications for evaluating the impact of ionizing radiation and radiomimetic drugs on genome integrity.
Materials and methods
Chemicals and reagents
Caffeine, roscovitine, and nocodazole were obtained from Sigma-Aldrich. Ku55933 was obtained from KuDOS Pharmaceuticals. RO-3306 was obtained from Enzo Life Sciences. PflMI and terminal transferase were obtained from New England Biolabs, Inc. Histone H1 (382150) was obtained from EMD. PKA inhibitor (PKI) peptide (TTYADFIASGRTGRRNAIH) was obtained from Sigma-Aldrich.
Extract and sperm chromatin preparation
For preparation of all types of extracts, adult female Xenopus frogs (Nasco) were injected subcutaneously with 50 U pregnant mare serum gonadotropin (EMD) 4–7 d before extract preparation. To induce ovulation, 800 U human chorionic gonadotropin (Sigma-Aldrich) was injected subcutaneously 16–20 h before extract preparation.
Mitochondria-free crude S-phase and M-phase egg extract (freezable extract) was originally described by Kubota and Takisawa (1993) and subsequently modified by Trenz et al. (2008). Accordingly, eggs were collected and rinsed in 0.25× MMR solution (20 mM Hepes-KOH, pH 7.5, 400 mM NaCl, 1 mM MgSO4, 2 mM CaCl2, and 0.1 mM EDTA). After dejellying with 5 mM DTT, 20 mM Tris, pH 8.5, and 110 mM NaCl, eggs were washed with 0.25× MMR. For S-phase extracts, eggs were activated with 1 µg/ml calcium ionophore A23187 (Sigma-Aldrich) and then washed several times with S buffer (50 mM Hepes-KOH, pH 7.5, 50 mM KCl, 2.5 mM MgCl2, and 250 mM sucrose) and once with S buffer plus 2 mM β-mercaptoethanol and 15 µg/ml leupeptin. For M-phase extracts, activation with A23187 was omitted, and S buffer was supplemented with 5 mM EGTA. Eggs were then spun at 160 g for 1 min in a swing-bucket Sorval rotor (HB-6; Thermo Fisher Scientific) to pack, and excess buffer was aspirated. Eggs were crushed at 16,500 g for 15 min at 4°C. The crude extract between the yolk top layer and pigmented granules was transferred to new tubes, supplemented with 20 µg/ml cytochalasin B (Sigma-Aldrich), and homogenized by rotation for 5 min at 4°C. The extract was then subjected to a high-speed spin in an ultracentrifuge (L8-80; Beckman Coulter) in a swing-bucket rotor (SW50.1; Beckman Coulter) for 15 min at 200,000 g at 4°C. The cytosolic and lipid membrane fractions were collected (excluding mitochondria directly below the membrane layer) and supplemented with 30 mM creatine phosphate, 150 µg/ml phosphocreatine kinase, and 20 µg/ml cycloheximide. Finally, extracts were mixed well with glycerol to 3% and were either immediately immunodepleted (see Immunodepletions) or flash frozen in 20-µl drops into liquid nitrogen to be later thawed and used for experiments and/or immunodepletion.
The membrane-free HSS extract was prepared as previously described (Smythe and Newport, 1991). In brief, eggs are washed, dejellied, packed, and crushed as in the previous paragraph. The crude extract was then spun at 46,000 rpm in a swing-bucket rotor (SW50.1) for 2.5 h. The clear, membrane-free HSS extract (top layer) was recovered, carefully excluding the cloudy membrane layer below. The HSS was either immediately immunodepleted or aliquoted to 100 µl into microcentrifuge tubes, flash frozen in liquid nitrogen, and stored at −80°C.
Cycling extract was prepared as previously described (Murray, 1991). Crude M-phase (CSF arrested) extract was activated with 0.4 mM CaCl2 for 15 min at 21°C in the absence of cycloheximide. Note that it is important that the CSF-arrested extract is not clarified, either by high-speed centrifugation (as with preparation of mitochondria-free crude low speed supernatant [LSS]) or by centrifugation in a table-top Eppendorf centrifuge, as the clarified CSF extract is unable to be “activated” by calcium and will remain CSF arrested.
Demembranated sperm nuclei (chromatin) were isolated as previously described (Murray, 1991). In brief, male frogs were injected with 25 U pregnant mare serum gonadotropin 3 d before and with 125 U human chorionic gonadotropin 16 h before sperm preparation. The testes were removed from anesthetized frogs, trimmed of fat and blood vessels, and placed in a dish with cold 0.25× MMR. Testes were washed three times in NPB (250 mM sucrose, 15 mM Hepes, pH 7.7, 1 mM EDTA, 0.5 mM spermidine, 0.2 mM spermine, and 1 mM DTT). Excess NPB was removed, and testes were macerated with a razor for 20 min. The material was diluted in 10 ml NPB, filtered through cheesecloth, and spun in a swing-bucket rotor (HB-6) at 6,000 g for 15 min. The supernatant was discarded, and the sperm pellet was washed and spun twice more. Sperm were demembranated by incubation in NPB with 0.2% Triton X-100 for 15 min at RT with gentle agitation. Demembranated sperm chromatin was then washed and recovered by spinning at 6,000 g in cold NPB + 3% BSA (with protease inhibitors) and then twice in cold NPB + 0.3% BSA and finally resuspended in NPB + 0.3% BSA + 30% glycerol. Small aliquots of 10 µl were flash frozen in liquid nitrogen and stored at −80°C.
Antibodies
Anti-CtIP mouse monoclonal antibody (11–1) was raised against a GST fusion protein containing the C-terminal 278 amino acids of human CtIP (residues 620–897) as described previously (Yu and Baer, 2000). The following rabbit antisera were generated in our laboratory as previously described: Xenopus Mre11 (Di Virgilio and Gautier, 2005), Xenopus ATM (Dupré et al., 2008), Xenopus ATR (Costanzo and Gautier, 2003), and Xenopus Cdk1 (Gautier et al., 1989).
The following antibodies were obtained from commercial sources: antiphospho–histone H2AX Ser139 (05-636; Millipore), anti–histone H3 (9715; Cell Signaling Technology), anti-Ku70 (SC-56129; Santa Cruz Biotechnology, Inc.), antiphospho-Chk1 Ser345 (2341; Cell Signaling Technology), anti–human RPA32 (MS-691-PO; NeoMarkers), antiphospho–histone H3 Ser10 (05-817; Millipore), and anti-Exo1 (NBP1-19709; Novus Biologicals). The following antibodies were received as gifts: RPA70 (P. Jackson, Genentech, South San Francisco, CA), Xenopus Rad51 (K. Maeshima, National Institute of Genetics, Shizuoka, Japan), total Xenopus Chk1 (J. Sible, Virginia Polytechnic Institute, Blacksburg, VA), Xenopus BLM (W. Dunphy, California Institute of Technology, Pasadena, CA), Xenopus DNA2, and Xenopus WRN (H. Yan, Fox Chase Cancer Center, Philadelphia, PA).
Immunodepletions
Immunodepletions were performed by prebinding washed protein–A Sepharose CL-4B beads (GE Healthcare) with serum or hybridoma supernatant overnight with constant rotation at 4°C in compact reaction columns (United States Biochemicals). The antibody beads were then washed extensively and resuspended in extract, rotated at 4°C for 40 min, and collected for a second round of depletion. A 3:1 ratio of extract/beads (bed volume) was used for all immunodepletions. The amount of antibody used for each depletion (extract/antibody volume per round) was as follows: mouse IgG (015-000-002; Jackson ImmunoResearch Laboratories, Inc.) for mock depletions (1:0.044), Xenopus Mre11 rabbit serum (1:1), CtIP 11–1 monoclonal hybridoma supernatant (1:1.5), and Xenopus Cdk1 rabbit serum (1:1).
Assay to monitor the response to chromosomal DSBs
Extracts were preincubated with demembranated sperm nuclei (5,000 sperm/µl) for 10 min at 21°C to allow the DNA to become chromatinized. Aliquots of the sample were taken before (0 min) and at time points (minutes) after addition of the PflMI restriction enzyme (0.05 U/µl; New England Biolabs, Inc.). As indicated, at each time point, 0.5 µl extract/chromatin was removed from the sample and diluted in 9.5 µl Laemmli buffer. Samples (extract) were fractionated on 7% SDS-PAGE minigels and processed for Western blotting according to standard procedures. In parallel, at each time point, chromatin was isolated. 15-µl aliquots were diluted with 800 µl ice-cold chromatin isolation buffer (50 mM Hepes-KOH, pH 7.8, 100 mM KCl, and 2.5 mM MgCl2) with 0.125% Triton X-100 and kept on ice for 10 min. 320 µl chromatin isolation buffer plus 30% sucrose (wt/vol) was added to low-retention microcentrifuge tubes (Thermo Fisher Scientific), and the diluted extract/chromatin was carefully layered on top of the sucrose cushion. Samples were spun at 8,500 g for 30 min at 4°C in a swing-bucket Sorval rotor (HB-6). The chromatin pellet was mixed with 15 µl Laemmli buffer, boiled, and fractionated on 3–8% Tris-acetate minigels (NuPAGE; Invitrogen) and processed for Western blotting according to standard procedures.
Cloning of xCtIP
xCtIP cDNA was acquired from Thermo Fisher Scientific (IMAGE [Integrated Molecular Analysis of Genomes and their Expression] ID 5514434; available from GenBank/EMBL/DDBJ under accession no. BC073395). The full-length cDNA was PCR amplified using cloning oligonucleotides encoding a 5′ 3×FLAG tag and a 3′ 6×His tag as well as unique restriction sites for subcloning: 5′ sense cloning oligonucleotide (3×FLAG; SalI), 5′-CGAGCTGTCGACCACCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGATTACAAGGATGACGATGACAAGGGAAGCATCACAGCATCCACTTGTGGCAGC-3′; and 3′ antisense cloning oligonucleotide (6×His; NotI), 5′-GCAGTGCGGCCGCCTAGTGGTGATGGTGATGATGTCCGGTCTTCTGCTCTTTAATCTTCG-3′.
The resulting PCR product was purified and digested with the engineered restriction sites and subcloned into pFastBac1. The xCtIP-pFastBac1 clone underwent transposition in host DH10Bac bacterial cells. The bacmid DNA was isolated and transfected into Sf9 insect cells to generate recombinant baculovirus following the manufacturer’s instructions (Bac-to-Bac; Invitrogen).
Site-directed mutagenesis of xCtIP
xCtIP mutants were generated by two-step PCR. In brief, wt xCtIP cDNA was used as a template for two separate PCR reactions, left and right. The left reaction contains the NotI-xCtIP forward oligonucleotide and the specific mutagenic sense oligonucleotide, which harbors the point mutation and also creates a novel restriction site for verification. The right reaction contains the xCtIP-EcoRI reverse oligonucleotide complementary to a naturally occurring EcoRI site in the middle of the xCtIP reading frame and the specific mutagenic antisense oligonucleotide, which harbors the point mutation and also creates a novel restriction site for verification. The PCR products are gel purified to remove parental template and mixed for use in a final PCR reaction containing only the NotI-xCtIP forward and xCtIP-EcoRI reverse oligonucleotide, to give a truncated mutant product. Digestion of the full-length wt clone with EcoRI and SalI and the truncated mutant PCR product with EcoRI and NotI, gel purification of the appropriate bands, and ligation yield the full-length mutant xCtIP coding sequence. The full-length coding sequence is then cloned into pBluescript (NotI and SalI sites) and sequenced for verification. The xCtIP sequence is then subcloned into pFastBac1, and the mutations were again verified by digestion with restriction enzymes corresponding to the novel sites introduced into the mutants.
The sequences for these primers were as follows: NotI-xCtIP forward, 5′-GCGGTGGCGGCCGCCTAGTGGTG-3′; xCtIP-EcoRI reverse, 5′-GTTGAATTCACTGAAGGTTCTATGG-3′; S328A sense (creates the AgeI site), 5′-GGAATAGAAGGGAAGCAGCACCGGTTTTTGGAGAACCTGTG-3′; S328A antisense (creates the AgeI site), 5′-CACAGGTTCTCCAAAAACCGGTGCTGCTTCCCTTCTATTCC-3′; T806A sense (creates the AfeI site) 5′-CGATACATTCCACCAAGCGCTCCTGAGAATTTTTGGGAG-3′; T806A antisense (creates the AfeI site), 5′-CTCCCAAAAATTCTCAGGAGCGCTTGGTGGAATGTATCG-3′; T806E sense (creates the BseYI site), 5′-GATTCCGATACATTCCACCCAGCGAGCCTGAGAATTTTTGGGAG-3′; and T806E antisense (creates the BseYI site) 5′-CTCCCAAAAATTCTCAGGCTCGCTGGGTGGAATGTATCGGAATC-3′.
Recombinant protein purification
3 d after infection with wt or mutant xCtIP baculovirus, Sf9 cells were harvested, and lysates were subjected to affinity purification. In brief, cells were washed in PBS and lysed in wash buffer (50 mM Tris-Cl, pH 7.5, 200 mM NaCl, 0.5% Triton X-100, and 10% glycerol) with protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitors (250 mM NaF, 50 mM sodium vanadate, 50 mM β-glycerophosphate, and 50 mM sodium pyrophosphate decahydrate). Lysate was dounced, cleared by centrifugation at 17,000 g, and incubated with equilibrated anti-FLAG M2 antibody-conjugated beads (Sigma-Aldrich) for 3 h at 4°C. Beads were then washed extensively in wash buffer and twice with elution buffer (25 mM Tris-Cl, pH 7.5, 50 mM NaCl, 0.01% Triton X-100, 20% glycerol, protease inhibitor cocktail, and 10 mM β-glycerophosphate). Protein was eluted with elution buffer plus 300 µg/ml 3×FLAG peptide (Sigma-Aldrich). Small aliquots were flash frozen in liquid nitrogen and stored at −80°C.
The Cdk1–cyclin B-GST complex was purified using glutathione beads (GE Healthcare) according to the manufacturer’s instructions. Kinase activity of the recombinant complex was verified and calibrated to endogenous Cdk1–cyclin B activity of M-phase extracts by performing the histone H1 kinase assay as described previously (Smythe and Newport, 1991).
DNA replication assay
10-µl aliquots of extract/chromatin were removed from the reaction, and 0.1 µl [32P]deoxy-CTP was added and incubated for 30 min at 21°C. DNA was then isolated by proteinase K digestion at 60°C for 1 h before phenol/chloroform extraction and ethanol precipitation. The DNA pellet was then resuspended in 20 µl water and run on a 0.8% agarose gel. The bottom third of the gel containing unincorporated radioactive nucleotide was cut off and discarded, and the remainder was fixed in 30% trichloroacetic acid, dried by pressing between Whatman paper and paper towels overnight, and exposed to x-ray film (Costanzo et al., 2000).
Immunofluorescence of UV laser-irradiated HeLa cells
HeLa cells were cultured on 8-well chamber slides (Thermo Fisher Scientific) and sensitized overnight with 10 µM BrdU (Sigma-Aldrich). Cells were microirradiated with a 355-nm solid-state UV laser using a microscope (PALM MicroBeam IV; Carl Zeiss). Slides were fixed in 4% paraformaldehyde for 5 min at RT, permeabilized with 0.1% Triton X-100 for 10 min, and blocked in a 50% FBS/5% milk solution. Slides were then stained with antibodies at a 1:1,000 dilution in 3% BSA overnight at 4°C, washed thoroughly with 0.2% PBS-Tween, and then incubated with the appropriately labeled secondary antibodies for 1 h at RT. Antibodies used were rabbit antiphospho-Ser10 histone H3 (05-817) with goat anti–rabbit Alexa Fluor 568 secondary fluorescent antibody and mouse anti-RPA34 (Ab-1 MS-691-PO; NeoMarkers) with goat anti–rabbit Alexa Fluor 488 secondary fluorescent antibody. After thorough washing with 0.2% PBS-Tween, cells were mounted with Vectashield medium with DAPI (Vector Laboratories). Slides were imaged on a microscope (Eclipse E400; Nikon) using a Plan Fluor 40× objective lens (0.75 numerical aperture, 0.72-mm working distance; Nikon) at RT. Images were captured with a camera (CoolSNAP EZ; Photometrics) and NIS-Elements software (F package, version 2.20; Nikon). Red (A568), green (A488), and blue (DAPI) images of the same cells were merged and assigned to R, G, or B channel in Photoshop CS (version 8.0; Adobe). Final images were produced in Photoshop CS by showing all three channels (merge) or turning down the output (levels) of two channels to show only one.
Histone H1 kinase assay (endogenous Cdk1 activity) and in vitro kinase assay of Cdk1–cyclin B and xCtIP
The assay to measure activity of endogenous (or added recombinant) Cdk1–cyclin B in extract was modified from Smythe and Newport (1991). 1 µl extract was diluted in 119 µl elution buffer (80 mM glycerol-2-phosphate, 10 mM MgCl2, and 5 mM EGTA) and flash frozen in liquid nitrogen until all samples were collected. Samples were thawed on ice, 10 µl diluted extract was mixed with 10 µl kinase mix (20 mM Hepes, pH 7.5, 5 mM EGTA, 10 mM MgCl2, 0.4 mg/ml histone H1, 20 µM PKI peptide, 350 µM ATP, and 0.25 µl γ-[32P]ATP) and incubated for 10 min at 21°C, and the reaction was stopped by mixing with 20 µl of 2× Laemmli buffer. Samples were fractionated by 12% SDS-PAGE, dried, and autoradiographed. For the in vitro kinase assay of recombinant xCtIP with Cdk1–cyclin B, 30 nM recombinant xCtIP was mixed with 6 nM recombinant Cdk1–cyclin B complex in a 20-µl reaction containing 10 µM PKI peptide, 0.25 mM ATP, 0.5 µl γ-[32P]ATP, 20 mM Hepes, pH 7.5, 5 mM EGTA, 10 mM MgCl2, and 40 mM glycerol-2-phosphate. This reaction was incubated for 90 min at 21°C and then fractionated on 7% SDS-PAGE, dried, and autoradiographed.
Isolation of endogenous M-phase xCtIP
Monoclonal anti–human CtIP 11–1 supernatant was purified using protein A–Sepharose beads in a high salt buffer (Harlow and Lane, 1988) and dialyzed into 0.1 M sodium phosphate buffer, pH 7.4. 60 µg antibody was conjugated to 6 mg paramagnetic M270 epoxy beads (Invitrogen) in 1 M ammonium sulfate in 1 ml total volume overnight at 30°C. The beads were extensively washed, blocked in 5% BSA, and used to isolate endogenous xCtIP from 3.5 ml freshly prepared M-phase extract for 3 h at 4°C. The beads were extensively washed and eluted with 0.5 N aqueous ammonium hydroxide and 0.5 mM EDTA, snap frozen in liquid nitrogen, and sublimated overnight in SpeedVac, and the dried pellet was resuspended in alkylating sample buffer (2% SDS, 10% glycerol, 100 mM DTT, 100 mM Tris, pH 8.8, 500 mM iodoacetamide [dissolved in 200 mM ammonium bicarbonate], and 0.05% bromophenol blue), heated to 70°C for 10 min, boiled briefly, run on 7% Tris-acetate gel (NuPAGE), and stained by colloidal Coomassie.
Tandem mass spectrometry analysis of endogenous M-phase xCtIP
The gel section corresponding to the expected size of CtIP was excised and subjected to in-gel digestion with proteomic-grade trypsin (Promega). The resulting digestion products were cleaned with a STAGE (stop and go extraction) tip (Rappsilber et al., 2003) and loaded onto a home-packed reverse-phase C18 column (75-µm internal diameter). The peptides were separated using a linear gradient (0–42% acetonitrile and 0.5% acetic acid for 120 min at 150 nl/min) and directly sprayed into a mass spectrometer (LTQ Orbitrap; Thermo Fisher Scientific) for analysis. The repetitive analytical cycle incorporated a high resolution mass scan in the Orbitrap (resolution = 60,000) followed by tandem mass spectrometry scans in the ion trap of the 10 most intense peaks observed in each Orbitrap mass spectrum. The raw mass spectral files were converted into the mzXML format, and CtIP peptides were identified with the X!tandem engine (The Global Proteome Machine Organization; Craig and Beavis, 2004). The spectra of the putative phosphopeptides were verified manually.
BrdU-based resection assay
The crude cycling extract (cycloheximide free) was activated with calcium and then briefly clarified by centrifugation at 4°C for 5 min at 14,000 rpm in a bench-top Eppendorf centrifuge. The extract was then incubated with 50 µM BrdU and 5,000 sperm nuclei per microliter for 60 min. Nocodazole was added to 12 µg/ml, and DNA was allowed to finish replication for an additional 2 h. Extract/nuclei were examined by fluorescence microscopy at intervals to monitor nuclear morphology and nuclear envelope breakdown. 30-µl samples were taken immediately before and at the indicated time points after addition of 0.05 U/µl PflMI restriction enzyme. DNA was isolated from the samples by proteinase K digestion for 30 min at 50°C, phenol/chloroform extraction, and ethanol precipitation. 50 µg DNA was spotted onto N+ charged nylon membranes (GE Healthcare), baked for 2 h at 60°C, and processed for Western blotting using the anti-BrdU antibody (B35128; Invitrogen). Immunodepletion requires clarified extract (microcentrifuge at 14,000 rpm for 5 min), which caused inefficient entry into mitosis and nuclear envelope breakdown (also see note under Extract and sperm chromatin preparation). To overcome this, recombinant Cdk1–cyclin B was added to 150 nM to force entry into mitosis at 3 h. 1 h later, samples were taken before and at the time points after PflMI addition.
Online supplemental material
Fig. S1 shows CtIP immunodepletion and recombinant protein purification. Fig. S2 shows that DSBs are generated in M-phase chromatin, and CtIP is rate limiting for their resection. Fig. S3 shows BrdU-based physical resection assay. Fig. S4 shows purification and assay of recombinant CtIP-T808A and -T806E. Fig. S5 shows endogenous and recombinant Cdk1–cyclin B kinase activity. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103103/DC1.
Acknowledgments
We gratefully acknowledge Dr. Theresa Swayne of the Herbert Irving Comprehensive Cancer Center Microscopy Shared Resource Center for assistance with UV laser microirradiation experiments. We thank Dr. Rebecca Burgess and Dr. Lorraine Symington for critical reading of the manuscript. We thank Drs. W. Dunphy, H. Yan, and K. Maeshima for antibodies.
This work was supported by grants RO1CA092245 and RO1GM077495 to J. Gautier and RR00862 and RR022220 to B.T. Chait.
Footnotes
Abbreviations used in this paper:
- Alt-NHEJ
- alternative nonhomologous end joining
- C-NHEJ
- classical nonhomologous end joining
- CSF
- cytostatic factor
- DSB
- double-strand break
- HDR
- homology-directed repair
- HR
- homologous recombination
- HSS
- high-speed supernatant
- LSS
- low speed supernatant
- NHEJ
- nonhomologous end joining
- PKI
- PKA inhibitor
- ssDNA
- single-stranded DNA
- wt
- wild type
- xCtIP
- Xenopus CtIP
References
- Anantha R.W., Sokolova E., Borowiec J.A. 2008. RPA phosphorylation facilitates mitotic exit in response to mitotic DNA damage. Proc. Natl. Acad. Sci. USA. 105:12903–12908 10.1073/pnas.0803001105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N., Rajendra E., Su X., Jeyasekharan A.D., Mahen R., Venkitaraman A.R. 2009. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr. Biol. 19:1075–1085 10.1016/j.cub.2009.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Laskey R.A. 1986. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 47:577–587 10.1016/0092-8674(86)90622-7 [DOI] [PubMed] [Google Scholar]
- Bolderson E., Tomimatsu N., Richard D.J., Boucher D., Kumar R., Pandita T.K., Burma S., Khanna K.K. 2010. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 38:1821–1831 10.1093/nar/gkp1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan W.A., Boldogh I., Chi P., Thompson E.A., Albrecht T. 1997. Inhibition of cellular Cdk2 activity blocks human cytomegalovirus replication. Virology. 231:239–247 10.1006/viro.1997.8489 [DOI] [PubMed] [Google Scholar]
- Budd M.E., Campbell J.L. 2009. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 4:e4267 10.1371/journal.pone.0004267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J.L., Kowalczykowski S.C. 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 467:112–116 10.1038/nature09355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11:753–760 10.1038/ncb1882 [DOI] [PubMed] [Google Scholar]
- Chen L., Nievera C.J., Lee A.Y., Wu X. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- Chen P.L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E.Y., Lee W.H. 2005. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25:3535–3542 10.1128/MCB.25.9.3535-3542.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V., Gautier J. 2003. Single-strand DNA gaps trigger an ATR- and Cdc7-dependent checkpoint. Cell Cycle. 2:17 10.4161/cc.2.1.290 [DOI] [PubMed] [Google Scholar]
- Costanzo V., Robertson K., Ying C.Y., Kim E., Avvedimento E., Gottesman M., Grieco D., Gautier J. 2000. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol. Cell. 6:649–659 10.1016/S1097-2765(00)00063-0 [DOI] [PubMed] [Google Scholar]
- Costanzo V., Robertson K., Bibikova M., Kim E., Grieco D., Gottesman M., Carroll D., Gautier J. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 8:137–147 10.1016/S1097-2765(01)00294-5 [DOI] [PubMed] [Google Scholar]
- Costanzo V., Shechter D., Lupardus P.J., Cimprich K.A., Gottesman M., Gautier J. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 11:203–213 10.1016/S1097-2765(02)00799-2 [DOI] [PubMed] [Google Scholar]
- Craig R., Beavis R.C. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 20:1466–1467 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- Di Virgilio M., Gautier J. 2005. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J. Cell Biol. 171:765–771 10.1083/jcb.200506029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré A., Boyer-Chatenet L., Gautier J. 2006. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 13:451–457 10.1038/nsmb1090 [DOI] [PubMed] [Google Scholar]
- Dupré A., Boyer-Chatenet L., Sattler R.M., Modi A.P., Lee J.H., Nicolette M.L., Kopelovich L., Jasin M., Baer R., Paull T.T., Gautier J. 2008. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4:119–125 10.1038/nchembio.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F., Christ N., Gannon J., Liu Y., Hunt T., Jasin M., West S.C. 2005. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 434:598–604 10.1038/nature03404 [DOI] [PubMed] [Google Scholar]
- Esashi F., Galkin V.E., Yu X., Egelman E.H., West S.C. 2007. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 14:468–474 10.1038/nsmb1245 [DOI] [PubMed] [Google Scholar]
- Gautier J., Matsukawa T., Nurse P., Maller J. 1989. Dephosphorylation and activation of Xenopus p34cdc2 protein kinase during the cell cycle. Nature. 339:626–629 10.1038/339626a0 [DOI] [PubMed] [Google Scholar]
- Giunta S., Jackson S.P. 2011. Give me a break, but not in mitosis: the mitotic DNA damage response marks DNA double-strand breaks with early signaling events. Cell Cycle. 10:1215–1221 10.4161/cc.10.8.15334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S., Belotserkovskaya R., Jackson S.P. 2010. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190:197–207 10.1083/jcb.200911156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767–2772 10.1101/gad.503108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M. 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20:34–46 10.1101/gad.1381306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: 726 pp [Google Scholar]
- Hartsuiker E., Neale M.J., Carr A.M. 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 33:117–123 10.1016/j.molcel.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Jackson S.P. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284:9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Cortés-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 455:689–692 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.J., Cox R., Ford C.C. 1988. The control of DNA replication in a cell-free extract that recapitulates a basic cell cycle in vitro. Development. 103:553–566 [DOI] [PubMed] [Google Scholar]
- Iwai M., Hara A., Andoh T., Ishida R. 1997. ICRF-193, a catalytic inhibitor of DNA topoisomerase II, delays the cell cycle progression from metaphase, but not from anaphase to the G1 phase in mammalian cells. FEBS Lett. 406:267–270 10.1016/S0014-5793(97)00282-2 [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Balestrini A., Garner E., Haber J.E., Costanzo V. 2008. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 27:1953–1962 10.1038/emboj.2008.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson-Rosenthal C., Millar J.B. 2006. Cdc25: mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 16:285–292 10.1016/j.tcb.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Keeney S., Kleckner N. 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA. 92:11274–11278 10.1073/pnas.92.24.11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Takisawa H. 1993. Determination of initiation of DNA replication before and after nuclear formation in Xenopus egg cell free extracts. J. Cell Biol. 123:1321–1331 10.1083/jcb.123.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.S., Petes T.D. 2010. From the Cover: mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc. Natl. Acad. Sci. USA. 107:7383–7388 10.1073/pnas.1001940107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Theilen M., Matthews A.J., Kelly D., Zheng S., Chaudhuri J. 2011. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 18:75–79 10.1038/nsmb.1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Toczylowski T., Yan H. 2008. Identification of the Xenopus DNA2 protein as a major nuclease for the 5′->3′ strand-specific processing of DNA ends. Nucleic Acids Res. 36:6091–6100 10.1093/nar/gkn616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 28:134–146 10.1016/j.molcel.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J.H., Burgess R.C., Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 118:699–713 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Lobachev K., Vitriol E., Stemple J., Resnick M.A., Bloom K. 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14:2107–2112 10.1016/j.cub.2004.11.051 [DOI] [PubMed] [Google Scholar]
- Löbrich M., Jeggo P.A. 2007. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 7:861–869 10.1038/nrc2248 [DOI] [PubMed] [Google Scholar]
- Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøfte M., Chan K.L., Hickson I.D., Bartek J., Lukas J. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 93:1043–1053 10.1016/S0092-8674(00)81209-X [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Cole R.W., Rieder C.L. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797–1806 10.1016/S0960-9822(02)01226-5 [DOI] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.). 8:983–995 10.1016/j.dnarep.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C., Rieder C.L. 2004. Chromosome damage and progression into and through mitosis in vertebrates. DNA Repair (Amst.). 3:1133–1139 10.1016/j.dnarep.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Nakamura A.J., Rao V.A., Pommier Y., Bonner W.M. 2010a. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle. 9:389–397 10.4161/cc.9.2.10475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. 2010b. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6:e1000828 10.1371/journal.pgen.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Ozsoy A.Z., Genschel J., Modrich P., Kowalczykowski S.C. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA. 105:16906–16911 10.1073/pnas.0809380105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–362 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Chung W.H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 467:108–111 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley G.G., Patrick S.M., Yao J., Carty M.P., Turchi J.J., Dixon K. 2003. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry. 42:3255–3264 10.1021/bi026377u [DOI] [PubMed] [Google Scholar]
- Okamoto K., Sagata N. 2007. Mechanism for inactivation of the mitotic inhibitory kinase Wee1 at M phase. Proc. Natl. Acad. Sci. USA. 104:3753–3758 10.1073/pnas.0607357104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Portik-Dobos Z., Tang L., Schnabel R., Novatchkova M., Jantsch V., Loidl J. 2007. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 26:5071–5082 10.1038/sj.emboj.7601916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Khodjakov A. 1997. Mitosis and checkpoints that control progression through mitosis in vertebrate somatic cells. Prog. Cell Cycle Res. 3:301–312 10.1007/978-1-4615-5371-7_24 [DOI] [PubMed] [Google Scholar]
- Shim E.Y., Chung W.H., Nicolette M.L., Zhang Y., Davis M., Zhu Z., Paull T.T., Ira G., Lee S.E. 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29:3370–3380 10.1038/emboj.2010.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L.P., Nickoloff J.A. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18:134–147 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- Smith E., Dejsuphong D., Balestrini A., Hampel M., Lenz C., Takeda S., Vindigni A., Costanzo V. 2009. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat. Cell Biol. 11:278–285 10.1038/ncb1835 [DOI] [PubMed] [Google Scholar]
- Smythe C., Newport J.W. 1991. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 35:449–468 10.1016/S0091-679X(08)60583-X [DOI] [PubMed] [Google Scholar]
- Stephan H., Concannon C., Kremmer E., Carty M.P., Nasheuer H.P. 2009. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 37:6028–6041 10.1093/nar/gkp605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630–670 10.1128/MMBR.66.4.630-670.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczylowski T., Yan H. 2006. Mechanistic analysis of a DNA end processing pathway mediated by the Xenopus Werner syndrome protein. J. Biol. Chem. 281:33198–33205 10.1074/jbc.M605044200 [DOI] [PubMed] [Google Scholar]
- Trenz K., Errico A., Costanzo V. 2008. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 27:876–885 10.1038/emboj.2008.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A.K., Brown R.S., Birrane G., Ladias J.A. 2005. Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex. Biochemistry. 44:10941–10946 10.1021/bi0509651 [DOI] [PubMed] [Google Scholar]
- Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 103:10660–10665 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F., Labbé J.C., Dorée M., Karsenti E. 1990. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 343:233–238 10.1038/343233a0 [DOI] [PubMed] [Google Scholar]
- Yan H., McCane J., Toczylowski T., Chen C. 2005. Analysis of the Xenopus Werner syndrome protein in DNA double-strand break repair. J. Cell Biol. 171:217–227 10.1083/jcb.200502077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.Y., Jeong S.Y., Dunphy W.G. 2006. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 20:772–783 10.1101/gad.1398806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Bailis J.M., Johnson S.A., Dilworth S.M., Hunter T. 2007. Rapid activation of ATM on DNA flanking double-strand breaks. Nat. Cell Biol. 9:1311–1318 10.1038/ncb1651 [DOI] [PubMed] [Google Scholar]
- You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 36:954–969 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Baer R. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275:18541–18549 10.1074/jbc.M909494199 [DOI] [PubMed] [Google Scholar]
- Yu X., Chen J. 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24:9478–9486 10.1128/MCB.24.21.9478-9486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M.H., Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jasin M. 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 18:80–84 10.1038/nsmb.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gostissa M., Hildebrand D.G., Becker M.S., Boboila C., Chiarle R., Lewis S., Alt F.W. 2010. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv. Immunol. 106:93–133 10.1016/S0065-2776(10)06004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 134:981–994 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkle R.E., Bloom W. 1953. Irradiation of parts of individual cells. Science. 117:487–493 10.1126/science.117.3045.487 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]