Nucleoli directly regulate p53 export and degradation rather than simply sequestering p53 regulatory factors.
Abstract
The correlation between stress-induced nucleolar disruption and abrogation of p53 degradation is evident after a wide variety of cellular stresses. This link may be caused by steps in p53 regulation occurring in nucleoli, as suggested by some biochemical evidence. Alternatively, nucleolar disruption also causes redistribution of nucleolar proteins, potentially altering their interactions with p53 and/or MDM2. This raises the fundamental question of whether the nucleolus controls p53 directly, i.e., as a site where p53 regulatory processes occur, or indirectly, i.e., by determining the cellular localization of p53/MDM2-interacting factors. In this work, transport experiments based on heterokaryons, photobleaching, and micronucleation demonstrate that p53 regulatory events are directly regulated by nucleoli and are dependent on intact nucleolar structure and function. Subcellular fractionation and nucleolar isolation revealed a distribution of ubiquitylated p53 that supports these findings. In addition, our results indicate that p53 is exported by two pathways: one stress sensitive and one stress insensitive, the latter being regulated by activities present in the nucleolus.
Introduction
The levels of the p53 tumor suppressor protein are regulated posttranscriptionally, with MDM2-mediated ubiquitylation and proteasomal degradation playing a major role (Ljungman, 2000; Marine and Lozano, 2010). p53 stabilization ensues by abrogation of this degradation, but the wide variety of cell stresses that can cause it has led to the proposal of a large number of activator proteins and pathways, all converging on the disruption of the p53–MDM2 interaction. In a radically different view of stress-induced p53 stabilization, we have proposed that functional nucleoli are required for MDM2 to promote p53 degradation (Rubbi and Milner, 2003). Because nucleolar function is extremely sensitive to cellular stresses, it can act as a unifying stress sensor signaling to p53: its impairment determines that p53 can no longer be degraded, and a p53 response ensues by default (Rubbi and Milner, 2003; Horn and Vousden, 2004; Olson, 2004; Mayer and Grummt, 2005). However, contemporary with our model, several transducers of nucleolar stress into p53 stabilization have been proposed, such as ribosomal L proteins, B23 (also called nucleophosmin), PML, etc., which are suggested to act mainly via the extensive relocalization of components caused by nucleolar disruption followed by their interaction with either p53 or MDM2 (Colombo et al., 2002; Lohrum et al., 2003; Zhang et al., 2003; Bernardi et al., 2004; Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004; Kurki et al., 2004). Hence, although the link between nucleolar/ribosomal stress and p53 stabilization is widely acknowledged, we now have two views for the underlying mechanism: one based on relocalization of diffusible components that can disrupt the p53–MDM2 interaction; the other based on a direct involvement of the nucleolus in p53 ubiquitylation and transport. This work aims to resolve these views by determining whether the nucleolus has a direct role in p53 regulation.
In addition, there is a more fundamental reason to study the nucleolar dependence of p53 regulation, which stems from the fact that nucleolar localization is conspicuous in both p53 and MDM2 biochemistry. Klibanov et al. (2001) have shown that p53 accumulates in a nucleolus-bound form after proteasomal inhibition. MDM2, on the other hand, has been proposed to transit through nucleoli and to be retained in nucleoli after actinomycin D treatment (Mekhail et al., 2005), as opposed to many nucleolar proteins whose mobility increases after ribosomal stress (Chen and Huang, 2001). Also, MDM2 appears to be exported to the cytoplasm via the nucleolus (Tao and Levine, 1999b). It is also possible that MDM2 may require its nucleolar localization signal to polyubiquitylate p53 (Lohrum et al., 2000). In addition, nucleolar sequestration of MDM2 by CDKN2A (p14ARF) is a well-documented p53 stabilization pathway (Sherr and Weber, 2000). Hence, nucleolar localization and trafficking are recurrent observations in p53 and MDM2 biology that can be expected to be of biological significance. Yet, surprisingly, to date, we do not have a model of p53 regulation that manages to account for these nucleolar localization and transport features. This work therefore addresses the fundamental question of whether nucleoli constitute a cellular compartment in which key steps in p53 regulation occur.
First, work was focused on determining whether the main p53 regulator was a stable nuclear structure (here hypothesized to be the nucleolus) or diffusible mediators. Heterokaryon (cell fusion) assays showed that the p53 level in each nucleus is a property intrinsic to the nucleus and that p53 stabilization is only local to a stressed nucleus. Furthermore, the presence of a nonstressed nucleus in a heterokaryon did not reduce the levels of p53 in a cocytoplasmic stressed nucleus, nor did a stressed nucleus raise p53 steady-state levels in a cocytoplasmic nonstressed nucleus. Next, micronucleation revealed that within a single cell, micronuclei without nucleoli have higher steady-state levels of p53, directly confirming the role of nucleoli. Furthermore, if the nucleolus directly regulates p53, p53 must transit through the nucleolus, such that all p53 molecules (or at least the vast majority) are nucleolar at some point. Such transit is demonstrated here by fluorescence loss in photobleaching (FLIP; Lippincott-Schwartz et al., 2003) using a cell line stably expressing p53-EGFP. Results show that clearance of the p53-EGFP fluorescence is comparably fast by nucleoplasmic or nucleolar photobleaching. In addition, subcellular fractionation experiments demonstrate that nucleolar p53 is ubiquitylated, with a distinctive pattern showing more polyubiquitylation than in the nucleoplasm and cytoplasm. Finally, our transport experiments reveal that there are two pathways of p53 nuclear export: one stress sensitive and one insensitive, the latter being regulated by the nucleolus.
Results
p53 steady-state level is a property local to a nucleus
We reasoned that the key to deciding which model best explained the nucleolar control of p53 levels was to address the fundamental question of whether signaling for p53 stabilization or degradation can be transmitted from one nucleus to another (i.e., whether or not it is diffusible). If two nuclei reside within a common cytoplasm (cocytoplasmic in a heterokaryon), would a stressed nucleus increase the p53 half-life in a nonstressed nucleus, or would the latter reduce the p53 half-life in the stressed one? This experiment has three possible outcomes: p53 stabilization in both nuclei, p53 degradation in both, or p53 stabilization only in the stressed nucleus. The first two (nonlocal control) would indicate that the mediators of p53 stability are free to move within and between nuclei, whereas the third (local control) would indicate that p53 stability is determined by a nontranslocating (nucleus bound) component.
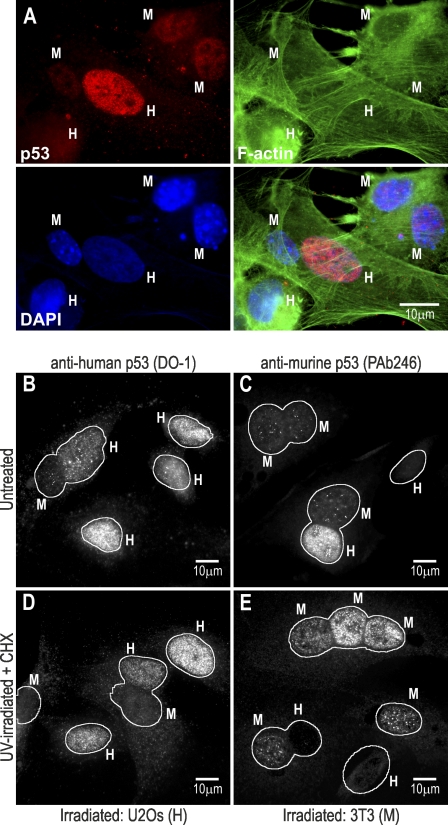
We generated heterokaryons (Fig. 1 A) between cells with different p53 half-lives (human U2Os and murine NIH3T3 [3T3] with t1/2 = 29.5 ± 4.6 and 13.7 ± 4.2 min, respectively; Fig. S1 A). Both human and murine p53, when examined in their homologous nuclei, showed no difference between nuclei inside a heterokaryon (fused) and nuclei outside a heterokaryon (nonfused; Fig. 1, B and C; and summarized in Table I). Thus, the presence of a human nucleus did not alter the murine p53 steady-state levels in a murine nucleus or vice versa. However, both human and murine p53 showed higher levels in human (U2Os) nuclei than in murine (3T3) nuclei (Fig. 1, B and C; Table I; and Fig. S1 B), correlating with the differences in p53 half-life in the individual cell lines. Cells had been fused for 6 h before fixation, so over that time, they shared the newly synthesized transcriptome and proteome (no protein synthesis inhibitor added). Thus, the p53 steady-state level in a nucleus is a property intrinsic to it (i.e., is locally controlled) and is not affected by the proteome contributed by a heterologous cocytoplasmic nucleus.
Figure 1.
p53 steady-state levels in heterokaryons. (A) Example of staining and identification of heterokaryons produced by fusion of U2Os (H, human) and 3T3 (M, murine) cells. Labeling: anti–murine p53 (PAb246). (B–E) Fusion of U2Os and 3T3 cells in normal growth and UV irradiation (20 J/m2). Cells were either left untreated (B and C) or UV irradiated (U2Os in D; 3T3 in E). Cells were fused 5 h after irradiation and cultured for a further 6 h (in D and E with 50 µg/ml CHX). Labeling antibodies are indicated. Cocytoplasmic nuclei are included within a perimeter.
Table I.
Analysis of mean nuclear p53 immunofluorescence intensities in human/mouse heterokaryons
| Irradiated cells | Treatment | p53 detected | Nuclei compared | p53 signal ratio | P-value |
| None | Untreated | Human | H inside/H outside | 1.20 | 0.4567 |
| H inside/M inside | 4.30 | 1.5 × 10−8 | |||
| None | Untreated | Murine | M inside/M outside | 2.83a | 0.1962 |
| H inside/M inside | 4.77 | 6.4 × 10−7 | |||
| Murine | Untreated | Human | H inside/M inside | 1.06 | 0.5735 |
| Human | CHX | Human | H outside/H inside | 2.16 | 0.0172 |
| Human | MG132 + CHX | Human | H outside/H inside | 1.34 | 0.0266 |
| H inside/M inside | 1.02 | 0.9985 | |||
| Human | LMB + CHX | Human | H outside/H inside | 1.17 | 0.1491 |
| H inside/M inside | 11.4 | 3.0 × 10−3 |
Human or murine p53 was detected with the species-specific mAbs DO-1 or PAb246, respectively. Inside and outside indicate being part or not of a heterokaryon, respectively, i.e., fused with a heterologous nucleus or not. p53 signal ratio indicates the average ratio of p53 mean intensities in each of the two types of nuclei compared. This value is only illustrative because the paired t test was used to establish the statistical difference between p53 mean intensities. P-value indicates the t test for the differences between the p53 nuclear fluorescence intensities under various conditions. Neighboring nuclei (both within and outside heterokaryons) from each field of view were compared. A paired t test was then applied to collate data from several fields. H, human; M, murine.
This quantitation is unreliable because it corresponds to basal levels of murine p53 in 3T3 fibroblasts. Nevertheless, the p-value indicates that the presence of a nonstressed human nucleus in a heterokaryon does not significantly alter the p53 levels in the murine nucleus.
p53 stabilization is a local property of a stressed nucleus
Because the experiment in Fig. 1 showed that p53 levels were a property intrinsic to each nucleus, we next examined whether this continued to be true in nuclei that had been subjected to genotoxic stress induced by UV irradiation. To analyze local versus nonlocal p53 stabilization under cell stress, heterokaryons were prepared with the seeded cell line UV irradiated and incubated for 5 h before fusion. After fusion with nonstressed cells, heterokaryons were incubated for a further 6 h with cycloheximide (CHX) to inhibit de novo p53 synthesis to reveal any p53 degradation that might occur. As shown in Fig. 1 (D and E), in the presence of CHX in heterokaryons, p53 is only detectable in irradiated nuclei irrespective of whether these are from 3T3 or U2Os cells, with nonirradiated nuclei showing levels similar to the extranuclear background. Fig. S1 (C and D) shows a gallery of images and quantitation for the experiments shown in Fig. 1 (D and E), confirming that fusion of murine or human irradiated nuclei with heterologous nonirradiated nuclei leads to a reduction in p53 levels (compare with nonfused irradiated nuclei). Thus, p53 stabilization is local to a stressed nucleus. This simple yet novel observation puts a constraint on any model of p53 regulation: it must be able to explain the asymmetric p53 stabilization between cocytoplasmic stressed and nonstressed nuclei (notice that in the presence of de novo synthesis p53 levels in stressed 3T3 nuclei are only slightly higher than in nonstressed U2Os nuclei, and thus, subsequent experiments used U2Os as the target for stress; Fig. S1 B).
The intranuclear and nucleocytoplasmic movement of components proposed to mediate stress signaling to p53 has been extensively documented both in normal and stressed conditions (see further confirmation regarding the translocation of factors previously suggested to be determinants of p53 stability in Fig. S1, E–H; Borer et al., 1989; Finch and Chan, 1996; Tao and Levine, 1999a,b; Chen and Huang, 2001; Leary et al., 2004; Tsai and McKay, 2005; Lam et al., 2007). Thus, local p53 stabilization is difficult to explain by models based on p53- or MDM2-interacting proteins being redistributed by nucleolar stress (the case of ribosomal protein L11 is analyzed in Fig. S1, K–N), and therefore, based upon our previous work demonstrating the central role of nucleolar stress in p53 stabilization, a model in which p53 regulation occurs in a functional nucleolus appears to be a better fit to the observations.
Distinct pathways determine p53 nucleocytoplasmic translocation
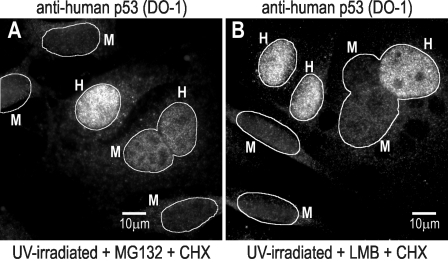
Irradiated nuclei show a reduction in p53 levels when they are fused with nonirradiated ones compared with nonfused irradiated nuclei (Fig. 1, D and E; Table I; and Fig. S1, C and D, images and plots). Thus, some p53 is stabilized by irradiation (i.e., persistent for ∼6 h in CHX), but some appears to be degradable if a nonstressed nucleus is present. This degradable fraction of p53 does in fact reach the nonstressed nucleus, as indicated by the even distribution of p53 in all nuclei within a heterokaryon when proteasome degradation is inhibited with MG132 (Fig. 2 A and Table I). Notice that when cells are treated with MG132, p53 levels (detected with an antibody specific for human p53) are slightly but significantly lower in U2Os nuclei in heterokaryons than in the nuclei of nonfused cells (Fig. 2 A and Table I), likely reflecting a dilution effect caused by transport to the fused nucleus. The translocation of p53 and its reaching a nonstressed nucleus (indicated by detection under conditions of proteasome inhibition) appear to be necessary for degradation because nuclear export inhibition by leptomycin B (LMB; Fornerod et al., 1997) preserves p53 stabilized levels in the irradiated nucleus (Fig. 2 B and Table I). It should be noted, however, that although p53 levels determined by immunofluorescence do not appear to show a statistical difference between LMB-treated U2Os fused and nonfused nuclei (Table I), it is possible that a small fraction of p53 (not resolved by immunofluorescence quantitation) may still be able to exit the U2Os nucleus under LMB treatment (see discussion of Fig. 3). In conclusion, a fraction of p53 that cannot be degraded by an irradiated nucleus can still be exported to the cytoplasm, and if it reaches a nonstressed nucleus, it can still be degraded.
Figure 2.
p53 transport in heterokaryons. (A) UV-irradiated U2Os fused to 3T3 cells as in Fig. 1 D with the addition of 10 µM MG132 and 50 µg/ml CHX after fusion. (B) UV-irradiated U2Os fused to 3T3 cells as in Fig. 1 D with the addition of 20 nM LMB and 50 µg/ml CHX after fusion. In both cases, labeling was for human p53 (DO-1). H, human U2O cells; M, murine 3T3 cells. Cocytoplasmic nuclei are included within a perimeter.
Figure 3.
FLIP analysis of p53 export. (A) Expression levels of p53-EGFP and endogenous p53 in U2Os–p53-EGFP cells compared with the parental cell line detected either with anti-p53 or anti-GFP antibodies (indicated). Cells were either untreated, UV irradiated (20 J/m2), or treated with 10 µM MG132 or 20 nM LMB for 6 h before lysis. Arrows indicate p53 bands. (B) FLIP by cytoplasmic photobleaching. U2Os–p53-EGFP cells were untreated, treated with UV (20 J/m2), 20 nM LMB, or 10 µM MG132 for 2–4 h, or fixed with PFA. Images show a selection of frames for one cell for each condition. Nuclei were surrounded by ROIs of similar dimensions (yellow lines; see legend and DIC images in Fig. S2 A), which were repeatedly photobleached at full power with a 488-nm Ar laser. (C) Fluorescence decay half-lives (t1/2) for each one of the nuclei analyzed in each treatment. Half-lives were obtained by fitting a single exponential decay to each photobleaching time course (the median correlation for all the fittings was r2 = 0.9972). P-values for the t tests for relevant comparisons between treatments are indicated. (D) Plots of time variation of nuclear fluorescence. Mean intensities were normalized to each initial intensity (t = 0). Plots show means (closed circles) ± SD of all nuclei analyzed for each treatment (numbers in C). Ctrl, control.
Because the level of p53 in a human irradiated nucleus in a heterokaryon does not increase in the presence of MG132 but the level in a cocytoplasmic (i.e., fused) murine nucleus does (Fig. 1 D and Fig. 2 A), it follows that a fraction of p53 is exported from a stressed nucleus and can be degraded in a location not exposed to the stress. Two modes of p53 nuclear export are therefore evident: one in which the export of a proportion of nuclear p53 is inhibited by cellular stress leading to nuclear accumulation, termed here stress-sensitive export (SSE), and one that appears to be constitutively active, termed here stress-independent export (SIE). This latter SIE leads to a proportion of p53 being exported even from a stressed nucleus, and this p53 can still be degraded if a nonstressed nucleus is present, as demonstrated by the heterokaryons of Fig. 1 (D and E) and Table I. In addition, the persistence of p53 after UV irradiation in the CHX-treated 6-h heterokaryons (Fig. 1, D and E) implies that commitment to SSE is extremely stable.
Nevertheless, the presence of a fraction of p53 that is still exportable after stress could be caused either by a persistent capacity to export p53 in irradiated nuclei (as proposed in the dual-export model) or by restoration of this capacity of the stressed nucleus by the presence of the fused normal nucleus. To discriminate between these possibilities, we used FLIP of (nonfused) U2Os cells stably expressing p53-EGFP (U2Os–p53-EGFP), which appears to behave similarly to endogenous p53 because it can be stabilized by UV irradiation and both MG132 and LMB treatment (Fig. 3 A). If a fluorescently labeled protein in any compartment in the imaged field spends some time in a compartment being bleached, its fluorescence will decay, and its transit across both compartments will thus be demonstrated (Lippincott-Schwartz et al., 2003). FLIP was performed by constantly bleaching a cytoplasmic region of interest (ROI) completely surrounding the nucleus (see details in Fig. S2 A) and collecting images at fixed intervals. The relatively large area photobleached in these experiments caused significant photobleaching inside nuclei. To account for this, PFA-fixed cells were imaged under the same conditions. As seen in Fig. 3 B, cytoplasmic bleaching rapidly depletes p53-EGFP from the nucleus of a nonstressed cell. Estimated nuclear p53 clearance half-life and decay profiles are shown in Fig. 3 (C and D), respectively (compare PFA-fixed cells). Both UV irradiation and LMB treatment reduce the rate of p53-EGFP fluorescence decay (significantly higher clearance half-life), yet p53 must still be exiting the nucleus because this decay is faster than that in fixed cells. Hence, loss of p53-EGFP fluorescence by cytoplasmic photobleaching confirms that some nuclear p53 is always exported, even after stress. Also, p53-EGFP is exported equally effectively after MG132 treatment, which agrees with data in Fig. 2 A. We also note that transiently expressed p53-EGFP in p53-null cells (Saos-2) is exported at a similar rate to that of U2Os–p53-EGFP cells, indicating that unconjugated p53 is not required for this export (Fig. S4, A–C).
The processes contributing to the net p53-EGFP fluorescence intensities measured are export and possibly nuclear degradation, the latter having only a minor contribution because the fluorescence decay half-lives are ∼10-fold shorter than our measured p53 decay rates. De novo synthesis and import do not contribute in these conditions because the bleaching ROIs completely surround the nuclei. Unfortunately, the FLIP method cannot be quantitated with sufficient accuracy to determine whether, in stressed conditions, all p53 is exported more slowly or only a fraction is, but results from Fig. 1 (D and E) and Fig. 2 A (6-h heterokaryons) support the latter conclusion because only part of the p53 is lost from the stressed nucleus. It is noteworthy that, as Fig. 3 shows, even in LMB-treated cells, some p53 is still exported, suggesting that the p53-stabilizing effect of LMB may not be fully explained by inhibition of its nuclear export (see Discussion).
Micronucleation shows that the nucleolus drives p53 degradation
Our data suggest that although p53 stability is determined by posttranslational modifications, a key step in this chain of events must be stably associated with a nuclear structure. On the basis of our previous experiments, we here suggest that this structure is the nucleolus, which we also propose to control SSE. Accordingly, a nucleus without nucleoli should only be able to perform SIE and would therefore show elevated p53 levels, providing that the relative contributions of SIE and SSE are comparable. Fig. 2 A suggests that this is the case. To test this prediction, we micronucleated cells stably expressing EGFP-tagged ribosomal protein L11 (U2Os–L11-EGFP; Fig. S3 A) by exposing them to colcemid for 48 h. Segregation of nucleolar-organizing regions in these cells produces nucleolated and nonnucleolated micronuclei (Fig. 4; Hernandez-Verdun et al., 1991) without detectable DNA damage (Fig. S3, B and C, shows no detectable γ-H2AX signal [Bonner et al., 2008] and restoration of cycling by removal of colcemid, respectively; Granetto et al., 1996). In agreement with the predicted effect of the presence of nucleoli, nonnucleolated micronuclei show significantly higher p53 levels than nucleolated ones (Fig. 4, A and E, plots), with the latter being similar to those of whole nuclei (not depicted). UV-irradiated cells, on the other hand, show a homogeneous distribution of p53 across all micronuclei, irrespective of the presence of nucleoli (Fig. 4, B and E, plots). This agrees with the abrogation of SSE by UV irradiation in nucleolated micronuclei, leaving only one active mode of export (SIE), which is independent of nucleoli and, thus, is common to both types of micronuclei.
Figure 4.
The presence of nucleoli determines p53 levels in micronuclei. (A–D) Examples of U2Os–L11-EGFP cells micronucleated by incubation with 0.1 µg/ml colcemid for 48 h either untreated (A, C, and D) or UV irradiated 5 h before fixation (B) followed by staining for p53 (A and B, red), MDM2 p53 (C, red), or p21 p53 (D, red) and DAPI (blue). (E) Relative expression intensities in a series of cells treated and imaged as in A–D. The normalization parameter for each micronucleated cell was the average of the mean intensity for the stained protein among all nucleolated (Yes) micronuclei. Untreated and UV-irradiated cells were analyzed separately. Plotted values are ratios, and therefore, no direct comparison can be made between the values for the untreated compared with the UV-irradiated cells. Means (closed circles) ± SD are indicated. P-values correspond to t tests for expression differences (null hypothesis: mean protein staining levels are equal between nucleolated and nonnucleolated micronuclei).
Interestingly, p53 stabilization by micronucleation has been observed before (for example, Granetto et al., 1996; Sablina et al., 1998), but although it was clear in these cases that DNA damage could not be the cause, to date, the mechanism of this stabilization has not been explained. Notably, the fact that not all micronuclei inside a cell show elevated p53 levels (Fig. 4 A; Granetto et al., 1996) makes p53 stabilization by micronucleation particularly difficult to explain by other stress signal transduction models, a phenomenon clearly explained by the nucleolar model of p53 regulation presented here. During a 48-h colcemid incubation, all micronuclei within a single cell will have shared a common proteome and transcriptome. This is observed when labeling for MDM2 and p21, transactivation targets of p53 (Ljungman, 2000), in which both proteins are equally distributed across both types of micronuclei (Fig. 4, C–E, plots). Importantly, although p53 levels in micronuclei strictly anticorrelate with the presence of nucleoli (Fig. 4, A and E), MDM2 levels are independent of it (Fig. 4, C and E), implying that p53 steady-state levels are not determined simply by the overall nuclear MDM2 levels as others have also observed (O’Hagan and Ljungman, 2004).
Nucleolar transit of p53
These results further support our proposal that the nucleolus is a critical regulator of p53 stability. Yet, the steady-state concentration of p53 in nucleoli is markedly lower than in the nucleoplasm. These two observations can only be reconciled if nucleoplasmic p53 rapidly transits through the nucleolus (see Olson and Dundr [2005] for a discussion on nucleolar protein abundance and residence times). This prediction was tested using FLIP on U2Os–p53-EGFP cells to determine whether p53-EGFP moves across nucleoli. Bleaching a nucleoplasmic ROI causes a progressive loss of p53-EGFP fluorescence across the nucleus, including nucleoli (Fig. 5 A and Fig. S2 B, position of the bleached ROIs). Placing the bleaching ROI inside a nucleolus eliminates all nucleoplasmic fluorescence at rates similar to nucleoplasmic bleaching (Fig. 5, A–C, comparison of decay half-life and decay profiles). When the bleaching ROI is placed within a nucleolus, some p53-EGFP photobleaching must occur in nucleoplasmic regions above and below the focused plane. However, in adherent U2Os cells, nucleoli span nearly all of the vertical cross section of the nucleus, leaving a minimal amount of nucleoplasm traversed by the photobleaching beam (Fig. S2 C). Thus, photobleaching occurring in this small, defocused nucleoplasmic region cannot account for a decay rate that is similar to nucleoplasmic photobleaching. Therefore, the majority of p53-EGFP photodestroyed by placing an ROI over a nucleolus must be nucleolar. Thus, these results demonstrate that nucleoplasmic p53 is in constant transit through nucleoli and that essentially all of the nucleoplasmic p53 passes through the nucleoli during the time period analyzed. Furthermore, because the steady-state level of nucleolar p53 is lower than the nucleoplasmic, the comparable photobleaching rates indicate that nucleolar transit of p53 is faster than nucleoplasmic transit.
Figure 5.
Nucleolar transit of p53. (A) FLIP by nucleolar/nucleoplasmic photobleaching. Galleries show a selection of images from FLIP analysis performed as in Fig. 3 with bleaching ROIs (yellow circles; see legend and DIC images in Fig. S2 B) located either in the nucleoplasm (Npl) or a nucleolus (Nol). UV irradiation and MG132 treatments were as in Fig. 3. (B) Plots of fluorescence decay half-life (fitting to single exponential plus constant; the median correlation for all the fittings was r2 = 0.9982) for each nucleus analyzed in each treatment. Means (closed circles) ± SD are indicated. (C) Plots of time variation of nuclear fluorescence. Mean intensities were normalized to each initial intensity (t = 0). Plots show means ± SD of all nuclei analyzed (numbers in B).
As with nuclear export (Fig. S4, A and B), we also tested p53-EGFP transiently transfected into p53-null Saos-2 cells. In this case, p53-EGFP is cleared very slowly by nucleolar photobleaching (in fact it is possible that it is not cleared at all, and we only see the effect of photobleaching the nucleoplasm above and below the targeted nucleoli; Fig. S4, D–F). We cotransfected wild-type p53 (at a plasmid ratio determined by Western blotting to give a p53/p53-EGFP protein ratio similar to that observed in stable U2OS–p53-EGFP; Fig. S4 G), and in these conditions, the clearance of nuclear p53 by nucleolar photobleaching was restored. We confirmed by Western blot analysis of MDM2 and p21 expression that p53-EGFP was transcriptionally active in Saos-2 cells, and because it induces massive cell death in transfected cells, we conclude that it is functional. However, as we show in Fig. S4 H, p53-EGFP has a longer half-life than wild-type p53, and this seems to be the main biochemical difference between the two species. This difference is entirely expected because EGFP-tagged proteins undergo reduced polyubiquitylation and proteasomal degradation (Baens et al., 2006). Therefore, these results show that p53-EGFP, which is transcriptionally competent but expected to be poorly polyubiquitylated, is both degraded more slowly and incapable of trafficking through nucleoli. Note that this reduced rate of clearance of p53-EGFP by nucleolar photobleaching confirms that nucleoplasmic photobleaching occurring above and below a targeted nucleolus cannot be responsible for the high FLIP decay rates observed in nucleolar photobleaching of untreated U2Os–p53-EGFP cells.
Nucleolar ubiquitylated p53 (Ub-p53)
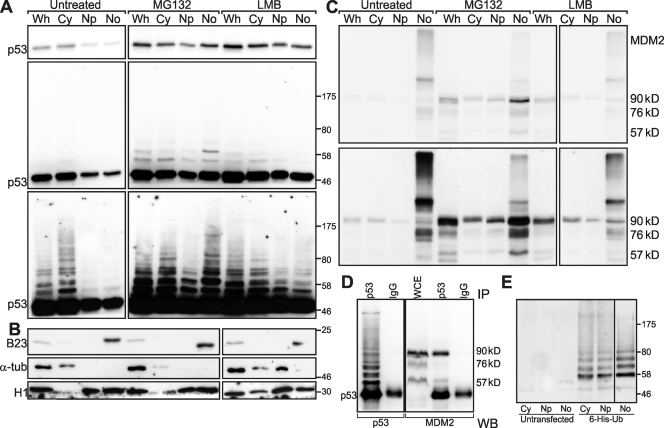
Our data show that p53 stability is directly regulated by the nucleolus. Ultimately, p53 degradation is a consequence of proteasome-mediated proteolysis of poly–Ub-p53, a process catalyzed by MDM2. Because MDM2 levels do not correlate with p53 levels in micronucleated nuclei, the role of nucleoli in regulating this process is likely to be either to promote p53–MDM2 interaction and/or to regulate the activity of p53 ubiquitylation by MDM2. However, to our knowledge, the presence of Ub-p53 in nucleoli has not previously been demonstrated. We therefore decided to fractionate cells into whole-cell, cytoplasmic, nucleoplasmic, and nucleolar fractions according to the protocol of Lam and Lamond (2006). Fig. 6 A (left column) shows the p53 content of all fractions in untreated U2Os cells (Fig. 6 B, fraction markers). Notably, p53 appears to be most abundant in the cytoplasm, which may reflect both the size of the cytoplasmic compartment and/or some leakage from nuclei during fractionation. Longer film exposure (Fig. 6 A, bottom) shows high molecular mass p53 bands consistent with ubiquitylation. To visualize Ub-p53, proteasomal degradation was inhibited with MG132 (Fig. 6 A, center column). As expected, p53 was stabilized, and more slowly migrating (likely ubiquitylated) forms were detectable in all fractions, including nucleoli. The nucleolar fraction in Fig. 6 A has a distinctive banding pattern, which ensures that the presence of Ub-p53 in nucleoli is genuine and not caused by contamination from other fractions (Fig. 6 A, center column). Fig. 6 E confirms the presence of Ub-p53 in all fractions by Ni-NTA pull-down of transfected His-tagged ubiquitin. Nucleoplasmic and cytoplasmic fractions show a largely similar Ub-p53 pattern, and this is the pattern represented in the whole fraction (note that the nucleolar fraction is ∼16 times concentrated by volume). Importantly, in the nucleolar fraction, the first Ub-p53 band has a lower abundance relative to higher molecular masses, in comparison with the nucleoplasmic and cytoplasmic fractions in which this pattern is inverted (Fig. 6 A, middle of MG132 column; and see profiles in Fig. S5). This indicates that p53 is relatively more abundant in the nucleoplasm and cytoplasm, whereas in nucleoli, more extensively ubiquitylated forms are relatively more abundant.
Figure 6.
Subcellular fractionation of U2Os cells. (A) Western blot of fractions of U2Os cells treated as indicated at the top developed with anti-p53. (top) Camera image. (middle) Scaled camera image. (bottom) Film long exposure. (B) Fraction markers B23, α-tubulin (α-tub), and histone H1 are as indicated. (C) Western blot as in A developed with anti-MDM2. (top) Camera image. (bottom) Film image. (A–C) Black boxes are used to indicate that the data are derived from two separate membranes. (D) Immunoprecipitation (IP) of MG132-treated U2Os cells. Cell lysates were immunoprecipitated with either anti-p53 or purified murine IgG and probed on a Western blot (WB) with either anti-p53 or anti-MDM2. Black boxes are used to indicate that pieces of a single membrane were probed in parallel with the indicated antibodies. (E) Ubiquitylated p53. U2Os cells were untreated or transfected with 6×His-tagged ubiquitin (Ub) as indicated. Subcellular fractions were absorbed with Ni-NTA resin, and eluates were analyzed by Western blotting and detected with anti-p53. Black boxes are used to indicate that the lane for the nucleolar fraction (rightmost) was taken from a shorter exposure. Cy, cytoplasmic; Np, nucleoplasmic; No, nucleolar; Wh, whole fraction; WCE, whole-cell extract.
To study the effect of nuclear export inhibition on nucleolar Ub-p53, U2Os cells were treated with LMB and fractionated in parallel with untreated and MG132-treated cells. LMB stabilizes p53 to similarly high levels as MG132 (Fig. 6 A, top). However, there is a noticeable reduction in p53 ubiquitylation, particularly in the nucleolar fraction in which there are hardly any high molecular mass forms (Fig. 6 A, middle and bottom). This surprising result suggests that an effect of LMB may be to prevent p53 from being ubiquitylated rather than preventing Ub-p53 from reaching proteasomes. We therefore analyzed the subcellular distribution of MDM2 under the same three conditions. As expected, MDM2 is abundant in nucleoli, in which it is detectable even without stabilization. In addition, and as expected, total MDM2 in intact cells is stabilized by both MG132 and LMB (Fig. 6 C). However, these inhibitors introduce two important changes: the 90-kD band is highly enriched in nucleoli after MG132 treatment, and although LMB increases total MDM2 levels, this increase is not paralleled in nucleoli in which it becomes less abundant. Hence, the increase in Ub-p53 induced by MG132 follows the observed increase in 90-kD MDM2 in nucleoli, whereas the p53 stabilization with relatively low ubiquitylation induced by LMB accompanies the relatively lower abundance of 90-kD MDM2 in nucleoli. The 90-kD isoform is the main p53-interacting isoform of MDM2 as shown by immunoprecipitation (Fig. 6 D), suggesting that nucleolar 90-kD MDM2 is critical for overall p53 ubiquitylation.
To confirm the presence of Ub-p53 in all the fractions, we applied pull-down of transiently transfected 6×His-tagged ubiquitin. Western blotting with anti-p53 (Fig. 6 E) reveals the expected high molecular mass bands. Next, we immunoprecipitated p53 from all fractions and probed the precipitates for polyubiquitylated forms (antibody FK1) or for both mono- and polyubiquitylated forms (antibody FK2). There are two striking features of this experiment. First, in Fig. 7 C, it is clear that polyubiquitylated forms of p53 are enriched in the nucleolar fraction. Second, as Fig. 7 (A and B) shows, proteasome-sensitive forms of p53 are greatly enriched in the nucleolus. Thus, our results provide evidence of nucleolar compartmentalization of p53 polyubiquitylation.
Figure 7.
Ubiquitylation of p53 immunoprecipitated from subcellular fractions. (A–D) Western blots of immunoprecipitated (IP) p53. (E) Western blot of whole-cell lysate. U2Os cells were grown untreated or treated with 10 µM MG132, fractionated as in Fig. 6, and immunoprecipitated with anti-p53 (DO-1). Antibody FK2 (A and B) detects mono- and polyubiquitylated proteins; FK1 (C) detects only polyubiquitylated proteins; ab2433 (D) is a rabbit antibody that detects p53, which avoids the signal from the murine antibody;and DO-1 (E) detects p53. Molecular masses (MM) are given in kilodaltons. Cyt, cytoplasm; Npl, nucleoplasm; Nol, nucleolus; IB, immunoblot; Ub, ubiquitin.
Discussion
Our previous study (Rubbi and Milner, 2003) demonstrated a critical role for the nucleolus in regulating p53 in response to stress. The aim of the present work was to determine whether regulation of p53 stability by nucleolar stress is likely exerted through diffusible (signaling) or stable (structural) components of nucleoli. To study the nature of the signals regulating p53 stability, heterokaryon assays were performed in which nuclei from cells with different p53 stability and steady-state levels coexisted within a common cytoplasm. The simplicity of the heterokaryon assay demonstrates unequivocally that the steady-state level of p53 in a nucleus, be it the normal or the DNA damage-induced level, is not affected by the status of p53 regulation in another nucleus (Fig. 1). For each nucleus studied, the proteome generated by another (cocytoplasmic) nucleus (with or without a different stress history) during a 6-h postfusion could not alter its p53 level, regardless of the origin (human or murine) of the p53 being monitored. To our knowledge, the question of whether p53 stability can be signaled from one nucleus to another had never been addressed before. This question is crucial because it imposes a constraint on models for p53 stabilization. In our system, we find that the p53 steady-state level is a property intrinsic and local to each nucleus. Mobile intermediates relaying nucleolar stress into inhibition of MDM2-mediated p53 ubiquitylation, which appear to be free to move between nucleus and cytoplasm (see Fig. S1), cannot satisfactorily explain the localized p53 stabilization observed in our heterokaryon assays.
From these findings, we went on to determine whether the nucleolus fulfils other requirements for p53 regulation, namely association with export and ubiquitylation. We have found that UV only partially abolishes export and degradation (Fig. 1), implying the existence of SSE and SIE pathways. If SSE depends on nucleolar function, nuclei without nucleoli would only perform SIE and show higher p53 levels. Micronucleation experiments confirmed this prediction (Fig. 4), demonstrating that nonnucleolated micronuclei express higher levels of p53, whereas levels across micronuclei become homogeneous after UV irradiation (SSE block). Interestingly, direct nucleolar control of p53 stabilization appears to be the only way to explain the paradoxical behavior of p53 in micronucleated cells, in which not only is it stabilized in the absence of any obvious DNA damage, but this stabilization occurs only in some, but not all, of the micronuclei within a cell (Granetto et al., 1996). The dual export and nucleolar regulation of p53 are summarized in the diagram presented in Fig. 8.
Figure 8.
p53 nucleolar control and dual-export model. Results suggest that MDM2 promotes monoubiquitylation of p53 in the nucleoplasm and that mono–Ub-p53 is exported via the SIE pathway (a possible consequence of this export may be p53 movement to mitochondria). When MDM2 localizes in nucleoli, p53 is polyubiquitylated, and this modification promotes export (SSE) and proteasomal degradation of p53. The proposed points of action of factors that lead to p53 stabilization are indicated. p53 may also be degraded by nuclear proteasomes.
Further evidence that the nucleolus is an integral part of the p53 regulation machinery comes from FLIP data revealing that essentially all nuclear p53 transits through nucleoli (Fig. 5) and from subcellular fractionation experiments that show that nucleolar p53 is enriched in ubiquitylated forms (Fig. 6 and Fig. 7), and particularly, in polyubiquitylated, proteasome-sensitive ones (Fig. 7). This association between p53 polyubiquitylation and nucleoli is further supported by first the poor (or null) nucleolar transit of p53-EGFP only (Fig. S4 D) and second the correlation between high p53 ubiquitylation and enrichment in the predominant p53-interacting form of MDM2 (90 kD) in the nucleolus observed under MG132 treatment (Fig. 6 C). Although the possibility of newly polyubiquinitylated p53 being rapidly recruited to nucleoli cannot be discarded, our data, especially the undetectable levels of poly–Ub-p53 in the nucleoplasmic fraction (Fig. 7), support the conclusion that p53 polyubiquitylation must occur in nucleoli (interestingly, Xirodimas et al. [2001] reported a need for an intact MDM2 nucleolar localization signal for p53 polyubiquitylation). Importantly, recent data suggesting that the nucleolus may be a site of ubiquitylation (Welcker et al., 2004; Mekhail et al., 2005; Stavreva et al., 2006; Latonen et al., 2011) strongly support this notion.
The existence of an export pathway that can operate under stress (SIE) can explain how cytoplasmic functions of p53 could be performed during a stress response in which nucleolus-dependent export (SSE) is impaired. The two-pathway model agrees with recent evidence suggesting that p53 mono- and polyubiquitylation constitute distinct events, possibly with different biological consequences. Monoubiquitylation appears to promote p53 nuclear export not leading to degradation, the latter requiring polyubiquitylation (Geyer et al., 2000; Li et al., 2003; Brooks and Gu, 2006; Carter et al., 2007), and more recently, p53 monoubiquitylation has been strongly linked to mitochondrial translocation and transactivation-independent apoptosis (Marchenko and Moll, 2007). Thus, it seems reasonable to hypothesize that poly–Ub-p53 follows a nucleolar route of export (SSE) destined for degradation, whereas mono–Ub-p53 is directly exported to the cytoplasm (SIE). Other p53 modifications, such as SUMOylation and NEDDylation, contribute to p53 export (Carter et al., 2007; Xirodimas, 2008), and clearly, the contributions of these to the present model will require investigation.
Recent findings in ribosome biogenesis are also consistent with the notion of nucleolar p53 regulation. Although nucleolar ubiquitylation of p53 may be a new concept, it has already been observed for c-Myc and the von Hippel-Landau tumor suppressor (Welcker et al., 2004; Mekhail et al., 2005). Importantly, ubiquitylated proteins accumulate in nucleoli (Stavreva et al., 2006), and it is likely that excess ribosomal proteins are cleared by ubiquitylation and nuclear proteasomal degradation (Lam et al., 2007). With respect to transport, Sherr and Weber (2000) have proposed that p53 and MDM2 may follow other proteins in exploiting the active export of ribosomal subunits by “riding the ribosome.” Interactions between p53 and ribosomal L proteins and ribosomal RNA (Fontoura et al., 1992, 1997; Marechal et al., 1994) provide a possible molecular basis for this transit. Therefore, the nucleolar role in transport of both p53 and MDM2 must be considered when interpreting changes in protein–protein interactions after ribosomal stress. The increased detection of proteins binding either p53 or MDM2 after nucleolar stress (Colombo et al., 2002; Lohrum et al., 2003; Zhang et al., 2003; Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004) is usually interpreted to reflect increased association rates enabled by relocalization of nucleolar proteins to the nucleoplasm (hence, signaling for cell stress). However, because recurring protein associations and dissociations accompany ribosome biosynthesis and export (Olson and Dundr, 2005; Hernandez-Verdun, 2006; Emmott and Hiscox, 2009), it should be noted that this increased binding may also reflect reduced dissociation rates caused by inhibition of transport by ribosomal stress.
The mechanism of p53 stabilization by inhibition of CRM1 may also need to be reconsidered. LMB treatment stabilizes p53 to the same extent that MG132 does but does so with a significantly lower level of ubiquitylation than the latter (Fig. 6), which suggests that under LMB treatment, MDM2 fails to reach p53 (and nucleoli) efficiently (Fig. 8). A possible precedent for an effect of CRM1 inhibition on intranuclear movement of MDM2 may be the accumulation of MDM2 in nuclear bodies induced by LMB (as well as p53 stabilization) reported by Laín et al. (1999). This effect of LMB on MDM2 is not unexpected in view of the recent work indicating that CRM1 has a role in intranuclear transport of nucleolar and preribosomal components (Sleeman et al., 2001; Boulon et al., 2004; Sleeman, 2007; Muro et al., 2008). Interestingly, it has been reported that human papilloma virus E6 expression reduces (Freedman and Levine, 1998) and even abolishes (Gray et al., 2007) p53 stabilization by LMB. This indicates that CRM1 inhibition by LMB prevents MDM2-dependent p53 degradation but is ineffective to prevent p53 degradation when p53 is polyubiquitylated by other ligases. Taking these data together, it seems reasonable to hypothesize that LMB stabilizes p53 through inhibition of nuclear transit of MDM2, rather than through nuclear retention of p53 (Fig. 8). In view of the interactions of MDM2 with ribosomal proteins (Colombo et al., 2002; Lohrum et al., 2003; Zhang et al., 2003; Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004) and ribosomal protein–ribosomal RNA complexes (Marechal et al., 1994) as well as nucleolar transit (Tao and Levine, 1999b), it is likely that intranuclear transport of MDM2 occurs in association with ribosomal biogenesis. Thus, the interpretation of data in which p53 levels are modulated by up/down-regulation of ribosomal components (hence, of ribosomal biogenesis) should take into account possible effects on MDM2 intranuclear transport and on its capacity to reach nucleoli.
The correlation between p53 export and degradation presented here and elsewhere appears to indicate that in normal growth conditions, p53 is degraded in the cytoplasm. However, it should be stressed that the evidence currently available for the subcellular compartment of proteasomal degradation of p53 is indirect and may well be confounded by the rapid nucleocytoplasmic exchange of p53. The present work highlights the problems with attempts to identify such compartments by either proteasome or nuclear export inhibition while looking only at p53 stabilization. Nuclear accumulation of p53 after MG132 treatment does not necessarily imply that p53 is degraded by nuclear proteasomes (Li et al., 2003); as shown here (Fig. 2 A), p53 continues to move between the nucleus and the cytoplasm after proteasome inhibition, and the observed nuclear accumulation of p53 appears simply to reflect nuclear import/export rates (Fig. 8). However, the correlation between nuclear export inhibition by LMB and p53 stabilization does not prove cytoplasmic degradation either because, as mentioned in the previous paragraph, CRM1 appears to participate in intranuclear transport, especially in association with ribosomal biogenesis. Thus, the question of cytoplasmic versus nuclear degradation of p53 in normal growth conditions is still open, and the answer may well be both.
Although the concept of p53 regulation occurring in association with nucleoli may be innovative to p53 biology, it perfectly matches many known features of MDM2/p53 biochemistry. As mentioned in this section, MDM2 transits through nucleoli (Mekhail et al., 2005) and appears to be exported to the cytoplasm via the nucleolus (Tao and Levine, 1999b), and it is also possible that MDM2 may require its nucleolar localization signal to polyubiquitylate p53 but not to monoubiquitylate p53 or autoubiquitylate (Lohrum et al., 2000). In addition, despite having its own nuclear export signal (NES), p53 nuclear export depends on the ubiquitylation capacity of MDM2 (intact RING finger domain) but not on the MDM2 NES (Boyd et al., 2000; Geyer et al., 2000), implying that the p53 NES is not fully effective for exporting p53 unless MDM2 ubiquitylates it and that MDM2 does not chaperone p53 out of the nucleus. Hence, although p53 ubiquitylation is intimately linked to its export, the transit of MDM2 through the nucleolus appears to be necessary for p53 ubiquitylation and export. Thus, this work introduces the novel concept that these biochemical events are associated with and depend on functional nucleoli. It also demonstrates that the nucleolus is a site of stress-dependent regulation of a protein not directly related to ribosomal biogenesis. Importantly, the direct nucleolar role in p53 regulation implies that the whole nucleolar function, including ribosomal biogenesis, rather than diffusible components, might define new targets for therapeutic p53 activation that need not depend upon inducing DNA damage.
Materials and methods
Cells and culture conditions
Human osteosarcoma cells (U2Os) and derived clones, Saos-2 and clone 9 cells (derived from p53-null H1299 cells stably overexpressing MDM2; Maguire et al., 2008), and murine 3T3 fibroblasts were cultured in DME + 10% FCS. For fixed-cell microscopy, cells were cultured in 13-mm-diam coverglasses and, for live imaging, in 42-mm coverglasses. U2Os–p53-EGFP and U2Os–L11-EGFP cells were generated by transfecting U2Os cells either with plasmid pp53-EGFP (Takara Bio Inc.) or L11-EGFP (Sundqvist et al., 2009; a gift from D. Xirodimas, Centre National de la Recherche Scientifique–Centre de Recherche de Biochimie Macromoléculaire, Montpellier, France), respectively, using transfection reagent (GeneJuice; EMD) at a 1:3 (wt/vol) DNA/reagent ratio. Stable clones were selected with 0.5 mg/ml G418 (EMD) and identified for cloning by fluorescence observation. Transient transfection in Saos-2 cells was performed similarly.
Heterokaryon and micronucleation assays
For cell fusion assays, seeded cells were grown for 24 h at a starting density of 4 × 104 cells per 13-mm coverglass (in 24-well plates). When applicable, UV irradiation was performed at 20 J/m2 5 h before fusion. Overlaid cells were seeded 3–5 h before fusion at 4 × 104 cells per 13-mm coverglass. Fusion was performed by washing cells in PBS, adding 50% polyethyleneglycol 3000 (Sigma-Aldrich) at 37°C in PBS for 2 min with gentle rocking, and washing twice with HBSS without phenol red (Invitrogen). Cells were then cultured in DME + FCS with the appropriate inhibitors added for a further 6 h before fixation. Immunofluorescence (see next section) included FITC-phalloidin (Sigma-Aldrich) to identify fused nuclei and DAPI to differentiate human and murine nuclei. Micronucleation was performed on U2Os–L11-EGFP cells because these offered the best identification of nucleolated and nonnucleolated micronuclei in both unstressed and UV irradiation conditions. Micronuclei were induced by adding 0.1 µg/ml colcemid (Sigma-Aldrich) to cultures and incubating for 48 h. UV irradiation was performed 6 h before fixation while still keeping colcemid present.
Immunofluorescence
Cells grown in coverglasses were washed twice in PBS and fixed for 10 min with 4% PFA (Sigma-Aldrich) followed by permeabilization for 2 min in 0.5% Triton X-100 in PBS and a wash in PBS + 0.01% Tween 20 (PBS-T). Cells were blocked in PBS-T + 10% normal donkey serum (PBS-TS) for 20 min. Primary and secondary antibodies were incubated in PBS-TS for 1 h with four washes in PBS-T. The primary antibodies used in this study were DO-1 (specific for human p53; EMD), PAb246 (specific for murine p53; EMD), anti-B23 mAb (EMD), goat polyclonal (Santa Cruz Biotechnology, Inc.), and antiphosphorylated histone H2AX (γ-H2AX; BioLegend). Secondary antibodies were raised in donkey, each one cross-absorbed for the species of the other primaries in the assay and labeled with either FITC or Cy3 (Jackson ImmunoResearch Laboratories, Inc.). FITC-phalloidin was added at 50 µg/ml together with the secondary antibody to visualize which nuclei were inside a common cytoplasm. Coverglasses were mounted in Mowiol 4–88 with 10 mg/ml 1,4-diazabicyclo[2.2.2]octane. Imaging was performed with a 100× 1.4 NA oil immersion objective in a microscope (TE300; Nikon) equipped with a camera (Orca AG; Hamamatsu Photonics). Images were acquired using Simple PCI software (Hamamatsu Photonics) and imported into ImageJ (National Institutes of Health) using the LOCI Bio-Formats Java library for ImageJ. Images were preprocessed by subtracting a dark current reference image and normalizing with an illumination reference obtained by averaging 10 images of a uniform layer of fluorescein in mounting medium. All processing and analysis was performed in ImageJ. Nuclear ROIs were either automatically or manually defined using masks from the corresponding DAPI images. Calculations used average p53 fluorescence intensity, hence reflecting p53 concentrations rather than total mass. However, integration over the total nuclear area did not alter the results. Statistical analyses were performed using SigmaPlot (Systat Software, Inc.). Because imaging exposure was optimized for each field, a paired t test was used, pairing the different categories of nuclei within each image. For flow cytometry, trypsinized cells were fixed with cold 70% ethanol, washed with PBS-T, resuspended in 50 µg/ml propidium iodide and 1 U/ml RNase A (Sigma-Aldrich), and after 1 h, analyzed in a flow cytometer (FACsort; BD).
FLIP
U2Os–p53-EGFP and transfected Saos-2 cells were cultured on 19-mm coverglasses bound to 35-mm plastic dishes and mounted in a heat (37°C)-, humidity (saturated)-, and CO2 (5%)-controlled stage (Carl Zeiss) with DME + 25 mM Hepes before observation. UV irradiation, MG132, or LMB was applied as before. Cells were imaged in a confocal scanning system (LSM 710; Carl Zeiss) using a 63× 1.4 NA oil immersion objective and Zen acquisition software (Carl Zeiss). Differential interference contrast (DIC) images were acquired using the 633-nm laser line to avoid GFP photobleaching. Bleaching was performed using the 488-nm line of a 25-mW Ar ion laser at 100% power on cycles of either 50 (nuclear photobleaching) or 70 repetitions (cytoplasmic photobleaching), each followed by imaging at 5% laser power. For cytoplasmic photobleaching, ROIs were manually defined at a distance of ∼1 µm from the cell nucleus, leaving nuclei unbleached but completely surrounding them. ROIs were inscribed in 26.4 × 26.4–µm squares (200 × 200 pixels; 694.7 µm2), thus ensuring identical bleaching times for all images (2.2 s between image collections for a total of 70 frames). Because of cell movement during acquisition, only time points at which the nucleus was completely included and not touching the bleaching regions were used for analysis. For nucleolar and nucleoplasmic photobleaching, ROIs were 0.75-µm2 circles (11-pixel diameter, a size that ensured full nucleolar inclusion and tolerance of cell movement) bleached for 1.7 s between image collections (for a total of 70 frames). Because of cell movement, DIC images were collected before and after FLIP acquisition to ensure that the bleaching ROIs remained within the target region (nucleolus or nucleoplasm) during imaging. Time series were imported into ImageJ using the LOCI Bio-Formats Java library. Bleaching correction (using reference, unbleached cells) and calculations (parameter fitting of exponential decay curves) used average fluorescence intensities over manually defined ROIs and were all performed in ImageJ.
FRAP
U2OS–L11-EGFP cells were placed in an incubation chamber in a confocal microscope (LSM 710) as for FLIP and photobleached with the 488-nm line of a 25-mW Ar ion laser at full intensity for 0.5 s over an ROI covering either a whole nucleolus, a 1.685-µm2 circular area within the nucleoplasm, or whole nuclei. Immediately after, images were collected at 3% laser power every 0.197 s (10 s for whole nuclear FRAP). Acquisition bleaching was compensated using whole nuclear fluorescence for nucleolar FRAP, a nucleoplasmic ROI for nucleoplasmic FRAP, or total fluorescence of a group of cells (whole nuclear FRAP). Nucleoli and nucleoplasm showed an extremely fast but partial recovery of fluorescence, with total recovery taking several minutes. For this reason, comparisons of recovery times and recovery fractions were made by fitting double exponential curves and analyzing decay rates (k) and fractions (F) from the fast recovery terms:
with immobile fractions calculated as 1 − Ffast. Recovery half-times were then calculated as
Subcellular fractionation
Cytoplasmic, nucleoplasmic, and nucleolar fractions of U2Os cells were prepared following Lam and Lamond (2006), using cultures from five 10 × 15–cm dishes for each treatment as required. In brief, cells were homogenized in hypotonic buffer (10 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, and 0.5 mM dithiothreitol, pH 7.9) using a Potter homogenizer, and the cytoplasmic fraction was recovered by centrifugation (218 g for 5 min). The nuclear pellet was resuspended in 0.25 M sucrose (plus 10 mM MgCl2) and cleared by centrifugation over a cushion of S2 solution (0.35 M sucrose and 0.5 mM MgCl2). Nuclei were resuspended in S2 solution and sonicated using a sonicator with a microtip (GEX600; Sonics & Materials, Ltd.) at 30% amplitude in 10-s intervals followed by phase-contrast monitoring for nuclear disruption. The sonicated sample was layered onto a cushion of 0.88 M sucrose and 0.5 mM MgCl2, and the nucleoplasmic upper phase and nucleolar pellet were recovered after centrifugation (3,000 g for 10 min). All steps were performed at 4°C. The different subcellular fractions were collected into a similar final volume, except for the nucleolar fraction (volume 16.5× smaller), so p53 band intensities are proportional to total p53 mass in each fraction but not to concentration. All buffers contained EDTA-free protease inhibitor (PI) cocktail (Set III; EMD) plus 10 mM N-ethylmaleimide (Sigma-Aldrich) as a deubiquitinase inhibitor (Brady et al., 2005). After fractionation, samples were added SDS to a final 1%, and the nucleolar pellet was dissolved in SDS-PAGE loading buffer with sonication if necessary. Samples from all three treatments corresponding to a same subcellular fraction were adjusted to have the same total protein concentration. Total protein was quantitated using protein reagent (DC Protein Assay; Bio-Rad Laboratories). To capture His-tagged ubiquitin, cells in 5 × 15–cm dishes were transfected (GeneJuice) with a plasmid expressing 6×His-ubiquitin for 36 h and subjected to subcellular fractionation, except that the cytoplasmic and nucleoplasmic fractions were precipitated with 10% trichloroacetic acid, centrifuged, and washed with acetone. Pellets were then processed to purify the 6×His-ubiquitin complexes as described by Xirodimas et al. (2001). In brief, pellets were dissolved in 6 M guanidinium-HCl, 0.1 M Na2HPO4, 0.01 M Tris/HCl, 5 mM imidazole, and 10 mM β-mercaptoethanol (BME), pH 8.0, and incubated with Ni-NTA beads (QIAGEN) for 4 h at RT. Beads were then sequentially washed for 5 min at RT with (a) 6 M guanidinium-HCl, 0.1 M Na2HPO4, 0.01 M Tris/HCl, and 10 mM BME, pH 8.0; (b) 8 M urea, 0.1 M Na2HPO4, 0.01 M Tris/HCl, and 10 mM BME, pH 8.0; (c) 8 M urea, 0.1 M Na2HPO4, 0.01 M Tris/HCl, and 10 mM BME, pH 6.3; (d) buffer c plus 0.2% Triton X-100; (e) buffer c plus 0.1% Triton X-100; and (f) 200 mM imidazole, 0.15 M Tris/HCl, pH 6.7, 30% glycerol, 0.72 M BME, and 5% SDS. Supernatants from step f were added SDS-PAGE loading buffer and analyzed by Western blotting.
Western blotting and immunoprecipitation
Cells were cultured in 60-mm Petri dishes, treated as required, trypsinized, and lysed in 1% SDS, 0.5% NP-40, 150 mM NaCl, 2 mM EDTA, and 50 mM Tris, pH 7.4. For p53 half-life determinations, cells were incubated in 6-well plates with 50 µg/ml CHX for the required times and then lysed with 200 µl of SDS-PAGE loading buffer and sonicated at 4°C. Protein content was determined using the DC protein reagent and used to normalize the loading amounts. After SDS-PAGE, Western blotting was performed by detection of p53 with DO-1 or rabbit polyclonal (ab2433; Abcam), MDM2 with IF-2 (EMD), B23 with goat polyclonal, histone H1 with mAb (Santa Cruz Biotechnology, Inc.), and α-tubulin with mAb (Abcam). For the chemiluminescence reaction, a reagent was used (SuperSignal West Dura; Thermo Fisher Scientific), and images were collected using either an imaging workstation (IS4000MM; Kodak) or autoradiographed. Immunoprecipitation was performed according to Brady et al. (2005). In brief, trypsinized cell pellets were lysed in standard lysis and immunoprecipitation (SLIP) buffer (50 mM Hepes, 150 mM NaCl, 10% glycerol, 0.1% Triton X-100, and 0.5 mg/ml BSA, pH 7.5) in the presence of PIs as for subcellular fractionation for 10 min on ice and centrifuged for 10 min at 4°C at 13,000 rpm. Volumes of lysate containing 4 mg total protein were supplemented to 0.5 ml with SLIP + PI. Lysates were precleared for 1 h with 50 µl of washed protein G–Sepharose beads (Sigma-Aldrich), and then, 2 µg antibody was added, and lysates were incubated for 1 h. Protein G–Sepharose beads were then added and incubated for 2 h with mixing. Finally, beads were washed three times with SLIP + PI and resuspended in SDS-PAGE loading buffer. For immunoprecipitation from subcellular fractions, the same procedure was used except that SLIP buffer was replaced by radioimmunoprecipitation assay buffer (150 mM NaC1, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) + PI (Lam and Lamond, 2006). The aqueous fractions (cytoplasmic and nucleoplasmic) were supplemented with 1:10 vol of 10× radioimmunoprecipitation assay buffer + PI.
Online supplemental material
Fig. S1 shows the half-life of p53 in cells used in heterokaryons, extends heterokaryon analyses for local p53 stabilization and leakage into nonstressed nucleus, addresses locality of putative p53 destabilizing factors in heterokaryons (MDM2, B23 translocation, and p53 Ser15 phosphorylation), and the transport of a ribosomal protein (FRAP with L11-EGFP). Fig. S2 shows controls for FLIP analyses. Fig. S3 shows controls for micronucleation experiments. Fig. S4 shows photobleaching behavior of transiently expressed p53-EGFP in a p53-null background (Saos-2). Fig. S5 shows intensity profiles for p53 Western blots from Fig. 6 A. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105143/DC1.
Acknowledgments
We are grateful to Prof. Jo Milner for making laboratory facilities available to C.P. Rubbi, to Dr. Joseph Slupsky for the use of a confocal microscope, and to Dr. Dimitris Xirodimas for the L11-EGFP plasmid.
This work was supported by Yorkshire Cancer Research grant Y248, University of Liverpool grant RDF 6349, and a North West Cancer Research Fund Fellowship to C.P. Rubbi and by funding from Cancer Research UK and Mersey Kidney Research to M.T. Boyd and N. Vlatković.
Footnotes
Abbreviations used in this paper:
- BME
- β-mercaptoethanol
- CHX
- cycloheximide
- DIC
- differential interference contrast
- FLIP
- fluorescence loss in photobleaching
- LMB
- leptomycin B
- NES
- nuclear export signal
- PI
- protease inhibitor
- ROI
- region of interest
- SIE
- stress-independent export
- SLIP
- standard lysis and immunoprecipitation
- SSE
- stress-sensitive export
References
- Baens M., Noels H., Broeckx V., Hagens S., Fevery S., Billiau A.D., Vankelecom H., Marynen P. 2006. The dark side of EGFP: defective polyubiquitination. PLoS ONE. 1:e54 10.1371/journal.pone.0000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R., Scaglioni P.P., Bergmann S., Horn H.F., Vousden K.H., Pandolfi P.P. 2004. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6:665–672 10.1038/ncb1147 [DOI] [PubMed] [Google Scholar]
- Bhat K.P., Itahana K., Jin A., Zhang Y. 2004. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 23:2402–2412 10.1038/sj.emboj.7600247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y. 2008. GammaH2AX and cancer. Nat. Rev. Cancer. 8:957–967 10.1038/nrc2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer R.A., Lehner C.F., Eppenberger H.M., Nigg E.A. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 56:379–390 10.1016/0092-8674(89)90241-9 [DOI] [PubMed] [Google Scholar]
- Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonné R., Bertrand E. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 16:777–787 10.1016/j.molcel.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Boyd S.D., Tsai K.Y., Jacks T. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563–568 10.1038/35023500 [DOI] [PubMed] [Google Scholar]
- Brady M., Vlatković N., Boyd M.T. 2005. Regulation of p53 and MDM2 activity by MTBP. Mol. Cell. Biol. 25:545–553 10.1128/MCB.25.2.545-553.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. 2006. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 21:307–315 10.1016/j.molcel.2006.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S., Bischof O., Dejean A., Vousden K.H. 2007. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9:428–435 10.1038/ncb1562 [DOI] [PubMed] [Google Scholar]
- Chen D., Huang S. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169–176 10.1083/jcb.153.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E., Marine J.C., Danovi D., Falini B., Pelicci P.G. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4:529–533 10.1038/ncb814 [DOI] [PubMed] [Google Scholar]
- Dai M.S., Lu H. 2004. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279:44475–44482 10.1074/jbc.M403722200 [DOI] [PubMed] [Google Scholar]
- Dai M.S., Zeng S.X., Jin Y., Sun X.X., David L., Lu H. 2004. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24:7654–7668 10.1128/MCB.24.17.7654-7668.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E., Hiscox J.A. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep. 10:231–238 10.1038/embor.2009.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch R.A., Chan P.K. 1996. ATP depletion affects NPM translocation and exportation of rRNA from nuclei. Biochem. Biophys. Res. Commun. 222:553–558 10.1006/bbrc.1996.0782 [DOI] [PubMed] [Google Scholar]
- Fontoura B.M., Sorokina E.A., David E., Carroll R.B. 1992. p53 is covalently linked to 5.8S rRNA. Mol. Cell. Biol. 12:5145–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura B.M., Atienza C.A., Sorokina E.A., Morimoto T., Carroll R.B. 1997. Cytoplasmic p53 polypeptide is associated with ribosomes. Mol. Cell. Biol. 17:3146–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 90:1051–1060 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Freedman D.A., Levine A.J. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R.K., Yu Z.K., Maki C.G. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569–573 10.1038/35023507 [DOI] [PubMed] [Google Scholar]
- Granetto C., Ottaggio L., Abbondandolo A., Bonatti S. 1996. p53 accumulates in micronuclei after treatment with a DNA breaking chemical, methylnitrosourea, and with the spindle poison, vinblastine. Mutat. Res. 352:61–64 [DOI] [PubMed] [Google Scholar]
- Gray L.J., Bjelogrlic P., Appleyard V.C.L., Thompson A.M., Jolly C.E., Lain S., Herrington C.S. 2007. Selective induction of apoptosis by leptomycin B in keratinocytes expressing HPV oncogenes. Int. J. Cancer. 120:2317–2324 10.1002/ijc.22591 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. 2006. Nucleolus: from structure to dynamics. Histochem. Cell Biol. 125:127–137 10.1007/s00418-005-0046-4 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D., Robert-Nicoud M., Geraud G., Masson C. 1991. Behaviour of nucleolar proteins in nuclei lacking ribosomal genes. A study by confocal laser scanning microscopy. J. Cell Sci. 98:99–105 [DOI] [PubMed] [Google Scholar]
- Horn H.F., Vousden K.H. 2004. Cancer: guarding the guardian? Nature. 427:110–111 10.1038/427110a [DOI] [PubMed] [Google Scholar]
- Jin A., Itahana K., O’Keefe K., Zhang Y. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24:7669–7680 10.1128/MCB.24.17.7669-7680.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klibanov S.A., O’Hagan H.M., Ljungman M. 2001. Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J. Cell Sci. 114:1867–1873 [DOI] [PubMed] [Google Scholar]
- Kurki S., Peltonen K., Latonen L., Kiviharju T.M., Ojala P.M., Meek D., Laiho M. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 5:465–475 10.1016/S1535-6108(04)00110-2 [DOI] [PubMed] [Google Scholar]
- Laín S., Midgley C., Sparks A., Lane E.B., Lane D.P. 1999. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp. Cell Res. 248:457–472 10.1006/excr.1999.4433 [DOI] [PubMed] [Google Scholar]
- Lam Y.W., Lamond A.I. 2006. Isolation of nucleoli. Cell Biology: A Laboratory Handbook. Third edition Vol. 2. J.E. Celis, editor Elsevier Academic Press, Amsterdam/Boston: 103–108 [Google Scholar]
- Lam Y.W., Lamond A.I., Mann M., Andersen J.S. 2007. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17:749–760 10.1016/j.cub.2007.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latonen L., Moore H.M., Bai B., Jäämaa S., Laiho M. 2011. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 30:790–805 10.1038/onc.2010.469 [DOI] [PubMed] [Google Scholar]
- Leary D.J., Terns M.P., Huang S. 2004. Components of U3 snoRNA-containing complexes shuttle between nuclei and the cytoplasm and differentially localize in nucleoli: implications for assembly and function. Mol. Biol. Cell. 15:281–293 10.1091/mbc.E03-06-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Brooks C.L., Wu-Baer F., Chen D., Baer R., Gu W. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 302:1972–1975 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Altan-Bonnet N., Patterson G.H. 2003. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Cell Biol. (Suppl.):S7–S14 [PubMed] [Google Scholar]
- Ljungman M. 2000. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia. 2:209–225 10.1038/sj.neo.7900073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum M.A., Ashcroft M., Kubbutat M.H., Vousden K.H. 2000. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2:179–181 10.1038/35004057 [DOI] [PubMed] [Google Scholar]
- Lohrum M.A., Ludwig R.L., Kubbutat M.H., Hanlon M., Vousden K.H. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 3:577–587 10.1016/S1535-6108(03)00134-X [DOI] [PubMed] [Google Scholar]
- Maguire M., Nield P.C., Devling T., Jenkins R.E., Park B.K., Polański R., Vlatković N., Boyd M.T. 2008. MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Res. 68:3232–3242 10.1158/0008-5472.CAN-07-5271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko N.D., Moll U.M. 2007. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 6:1718–1723 10.4161/cc.6.14.4503 [DOI] [PubMed] [Google Scholar]
- Marechal V., Elenbaas B., Piette J., Nicolas J.-C., Levine A.J. 1994. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell. Biol. 14:7414–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine J.C., Lozano G. 2010. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 17:93–102 10.1038/cdd.2009.68 [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I. 2005. Cellular stress and nucleolar function. Cell Cycle. 4:1036–1038 10.4161/cc.4.8.1925 [DOI] [PubMed] [Google Scholar]
- Mekhail K., Khacho M., Carrigan A., Hache R.R., Gunaratnam L., Lee S. 2005. Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 170:733–744 10.1083/jcb.200506030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro E., Hoang T.Q., Jobart-Malfait A., Hernandez-Verdun D. 2008. In nucleoli, the steady state of nucleolar proteins is leptomycin B-sensitive. Biol. Cell. 100:303–313 10.1042/BC20070117 [DOI] [PubMed] [Google Scholar]
- O’Hagan H.M., Ljungman M. 2004. Nuclear accumulation of p53 following inhibition of transcription is not due to diminished levels of MDM2. Oncogene. 23:5505–5512 10.1038/sj.onc.1207709 [DOI] [PubMed] [Google Scholar]
- Olson M.O. 2004. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004:pe10 10.1126/stke.2242004pe10 [DOI] [PubMed] [Google Scholar]
- Olson M.O., Dundr M. 2005. The moving parts of the nucleolus. Histochem. Cell Biol. 123:203–216 10.1007/s00418-005-0754-9 [DOI] [PubMed] [Google Scholar]
- Rubbi C.P., Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22:6068–6077 10.1093/emboj/cdg579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina A.A., Ilyinskaya G.V., Rubtsova S.N., Agapova L.S., Chumakov P.M., Kopnin B.P. 1998. Activation of p53-mediated cell cycle checkpoint in response to micronuclei formation. J. Cell Sci. 111:977–984 [DOI] [PubMed] [Google Scholar]
- Sherr C.J., Weber J.D. 2000. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 10:94–99 10.1016/S0959-437X(99)00038-6 [DOI] [PubMed] [Google Scholar]
- Sleeman J. 2007. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J. Cell Sci. 120:1540–1550 10.1242/jcs.001529 [DOI] [PubMed] [Google Scholar]
- Sleeman J.E., Ajuh P., Lamond A.I. 2001. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 114:4407–4419 [DOI] [PubMed] [Google Scholar]
- Stavreva D.A., Kawasaki M., Dundr M., Koberna K., Müller W.G., Tsujimura-Takahashi T., Komatsu W., Hayano T., Isobe T., Raska I., et al. 2006. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol. Cell. Biol. 26:5131–5145 10.1128/MCB.02227-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A., Liu G., Mirsaliotis A., Xirodimas D.P. 2009. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 10:1132–1139 10.1038/embor.2009.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Levine A.J. 1999a. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl. Acad. Sci. USA. 96:3077–3080 10.1073/pnas.96.6.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Levine A.J. 1999b. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA. 96:6937–6941 10.1073/pnas.96.12.6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. 2005. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168:179–184 10.1083/jcb.200409053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Orian A., Grim J.E., Eisenman R.N., Clurman B.E. 2004. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 14:1852–1857 10.1016/j.cub.2004.09.083 [DOI] [PubMed] [Google Scholar]
- Xirodimas D.P. 2008. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 36:802–806 10.1042/BST0360802 [DOI] [PubMed] [Google Scholar]
- Xirodimas D., Saville M.K., Edling C., Lane D.P., Laín S. 2001. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 20:4972–4983 10.1038/sj.onc.1204656 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wolf G.W., Bhat K., Jin A., Allio T., Burkhart W.A., Xiong Y. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23:8902–8912 10.1128/MCB.23.23.8902-8912.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]