RNAi screens in Drosophila and human cells for novel actin regulators revealed conserved roles for proteins involved in nuclear actin export, RNA splicing, and ubiquitination.
Abstract
Although a large number of actin-binding proteins and their regulators have been identified through classical approaches, gaps in our knowledge remain. Here, we used genome-wide RNA interference as a systematic method to define metazoan actin regulators based on visual phenotype. Using comparative screens in cultured Drosophila and human cells, we generated phenotypic profiles for annotated actin regulators together with proteins bearing predicted actin-binding domains. These phenotypic clusters for the known metazoan “actinome” were used to identify putative new core actin regulators, together with a number of genes with conserved but poorly studied roles in the regulation of the actin cytoskeleton, several of which we studied in detail. This work suggests that although our search for new components of the core actin machinery is nearing saturation, regulation at the level of nuclear actin export, RNA splicing, ubiquitination, and other upstream processes remains an important but unexplored frontier of actin biology.
Complete screen data
Drosophila:
https://doi.org/10.1083/jcb.201103168.dv
Human actinome I:
https://doi.org/10.1083/jcb.201103168.dv
Human actinome II:
Introduction
The ability of animal cells to change their shape is essential for diverse processes from cell division to tissue remodeling during development, homeostasis, and disease (Rungger-Brändle and Gabbiani, 1983; Lecuit and Lenne, 2007; St Johnston and Ahringer, 2010). Moreover, for specialized cell types such as blood cells and neurons (Tahirovic and Bradke, 2009; Diez-Silva et al., 2010), dynamic form plays a critical role in cellular physiology. In all cases, forces generated by cortical actin filament dynamics and by the ATP-dependent movement of myosin motors along filaments play key roles in reshaping cells (Pollard, 2007). Generating a form appropriate to function therefore depends on local remodeling of the actin–myosin network driven by signals from the environment as well as the complement of actin accessory proteins expressed by a given cell.
The core actin cytoskeletal regulators are surprisingly well conserved across diverse species from yeast to humans. These include actin itself; two conserved filament nucleation pathways, one mediated by formins and the other by Arp2/3; regulators of filament dynamics such as profilin, capping protein, and cofilin; and upstream regulators, such as Ste20 family kinases and small Rho GTPases (Cvrcková et al., 2004; Rohn and Baum, 2010). Much of our knowledge about actin regulation in metazoan organisms relies on extrapolations from work done in yeast and on data from biochemical studies in a variety of systems. Furthermore, metazoan genomes encode a large number of conserved genes of unknown function that contain protein domains known to bind to or regulate actin. In this study our goal was to extend this work by using RNAi screening to better define a conserved metazoan “actinome” based upon gene function.
In recent years, with the development of high-throughput RNAi screening in cell culture, it has become possible to search in an unbiased fashion for new players involved in a variety of cell biological processes (Echeverri and Perrimon, 2006). Previous screens have used RNAi to define regulators of Drosophila cell shape (Kiger et al., 2003) and to identify novel components of the SCAR complex (Kunda et al., 2003; Rogers et al., 2003), whereas other groups used RNAi together with automated computational approaches to screen for clusters of actin regulators (Bakal et al., 2007). More recently, Fuchs et al. (2010) applied genome-wide RNAi screening and automated image analysis to survey genes regulating the shape of human HeLa cells, whereas D’Ambrosio and Vale (2010) used an automated analysis in a genome-wide screen to study cell spreading in Drosophila S2 cells. Although automating the image analysis speeds up annotation, minimizes user bias, and generates quantitative data, the trained human eye is still superior when searching for novel and subtle phenotypes. Indeed, it remains a mainstay for many types of screen (Eggert et al., 2004; Sönnichsen et al., 2005; Schnorrer et al., 2010).
Here, to identify a core set of actin regulators, we performed a visual genome-wide RNAi screen in Drosophila S2R+ cells, and a more focused screen in human HeLa cells. By comparing the orthologous human and fly RNAi datasets, we were able to eliminate genes from our analysis with cell type– or species-specific functions and to limit the number of genes identified with an indirect effect on the actin cytoskeleton. We then followed up a subset of the hits. This analysis identified a set of novel, conserved regulators of the actin cytoskeleton, including components of the Skp1-Cul1-F-box-protein (SCF) E3 ubiquitin ligase complex, the spliceosome and genes affecting the formation of actin filaments in the nucleus. The data suggest that although few previously uncharacterized core actin-binding proteins remain to be identified, understanding the complete picture of upstream regulation of actin cytoskeletal dynamics remains an important challenge. We further believe that this simple cross-species approach can be used as a simple, cheap, and effective way to screen for conserved regulators of a wide number of cell biological processes.
Results
A genome-wide Drosophila RNAi screen for cell morphology
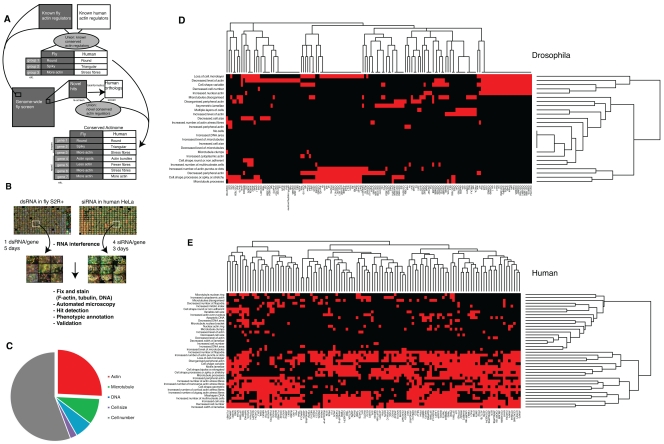
RNA interference enables systematic loss-of-function screens across a genome (Mohr et al., 2010). Our goal was to use parallel cell-based RNAi screens in fly and human cell culture to gain a more comprehensive picture of metazoan actin regulators and their phenotypes. To do so, we first performed a genome-wide, high-content RNAi screen in the hemocyte-derived adherent Drosophila cell line S2R+ (Fig. 1, A and B; Table S1; Yanagawa et al., 1998), whose read-out was a visual inspection of images of fixed cells stained with α-tubulin, F-actin, and DNA (see Materials and methods for details). After filtering and annotating hits with a controlled vocabulary, we identified a number of broad phenotypic categories (Fig. 1, C and D; see Table S2 for all hit annotations). The largest of these was the “viability” cluster, where gene silencing resulted in a considerable reduction in cell number. These genes were eliminated from the morphological analysis, except where evidence was available that interacting proteins displayed similar phenotypes (as for the SCF complex, described later).
Figure 1.
Screen overview. (A) A genome-wide morphology RNAi screen was performed in Drosophila S2R+ cells and in a subset of human HeLa cells for genes corresponding to a comprehensive set of all known human actin regulatory genes and genes predicted to play a role based on their domain structure. Comparing the two screens we were able to cluster the hits into morphological groups and arrive at a shortlist of conserved known and new actin regulatory genes. (B) A flowchart of the methodology. (C) Breakdown of all Drosophila hits by dominant phenotype; many hits fell into multiple categories, but these have only been accounted for in one category in this graph (see Table S2 for details). (D) Fly and (E) human hits clustered as a heat map; red indicates a hit in the specified category.
As expected, many hits in this visual screen were previously identified as having a reduced cell area as a result of growth and/or adhesion defects in an automated image analysis of the same dataset (Jani and Schöck, 2007; Sims et al., 2009). A cluster of dsRNAs induced a multinucleate phenotype associated with cytokinesis defects (Echard et al., 2004; Eggert et al., 2004). Of these, 17 were hits in previous studies, such as Rho (Rho1), Myosin II/MHC (zipper), and Anillin (scraps). Another cluster of dsRNAs induced various microtubule phenotypes, including various tubulin genes, the known microtubule regulator Tao-1 (Liu et al., 2010), dynein heavy chain (Dhc64C; Rasmusson et al., 1994), and nine eukaryotic initiation factor genes (Table S2).
The remaining 143 dsRNAs induced defects in the actin cytoskeleton—the focus of this study (Table S2). Of these, 22 were already known to be involved in the regulation of actin filament dynamics. These included representatives of the core conserved actin machinery previously described, including several actins (these are 95% identical in Drosophila), Profilin (chic), Capping protein (cpa and cpb subunits), Cofilin (twinstar), and three members of the Arp2/3 complex. Other known actin regulators identified as hits in our screen included the Rho family GTPases Rac1, Rac2, and Cdc42, along with all known members of the SCAR complex (SCAR, Abi, Hem, Sra-1) except the HSPC300 subunit (Kunda et al., 2003; Rogers et al., 2003), which all share a spiky cell phenotype (Table S2). Significantly, this Rac/SCAR-like spiky phenotypic cluster included another ten dsRNAs targeting nine genes not previously linked to Rac/SCAR signaling. Furthermore, a further set of genes had spiky morphology after RNAi silencing similar to that of SCAR, although with a range of additional subtle differences that led us to assign them to a distinct category. This alternative spiky cluster included clathrin heavy chain (Chc), which was recently characterized as having a role in SCAR-mediated lamellipodia formation independent from its role in vesicle trafficking (Gautier et al., 2011).
Our set of 143 putative actin regulators also contained two large clusters characterized by altered levels of actin filaments (Table S2). The “high phalloidin” cluster included actin-capping protein (cpa and cpb), whose knockdown is known to lead to an increase in F-actin (Kiger et al., 2003; Rogers et al., 2003), whereas the “low phalloidin” class included several actin genes (Act42A, Act5C, Act87E) and Profilin (chic). However, because the overall intensity of actin staining was variable across plates and was affected by cell density, we chose not to include these two clusters in our further analysis. Instead, in addition to the spiky cluster, we focused on dsRNAs giving rise to an assortment of rarer actin-related phenotypes (category “other actin”), including an increase in intracellular actin structures such as stress fibers or cytoplasmic speckles and/or changes in the level or organization of peripheral actin. This set included Pak3, which we previously characterized as an actin regulator (Asano et al., 2009), and the WH2 motif containing adenylate cyclase-associated protein Capulet/Act up (Capt; Baum et al., 2000; Benlali et al., 2000). In addition, a cluster of 29 genes was characterized by a novel phenotype in which actin filaments accumulated as a bar or cable-like structure within the nucleus.
Next, we used hierarchical clustering tools within the FLIGHT database to reveal hits with similar phenotypes forming part of the same interaction network or protein complex. This analysis revealed a prominent cluster with spiky morphology containing slmb (supernumerary limbs), lin19, and Roc1b, all of which are members of the same well-characterized complex, the SCF family of E3 ubiquitin ligases. Roc1b is a RING domain–containing protein that facilitates transfer of charged ubiquitin from the E2 ligase; Lin19 is a member of the cullin scaffold family, whereas Slmb is a member of the F-box family of proteins, which select protein targets for ubiquitination by the SCF complex (Jiang and Struhl, 1998; Deshaies, 1999; Bocca et al., 2001; Noureddine et al., 2002; Ou et al., 2002; Donaldson et al., 2004; Reynolds et al., 2008). We inspected the original screen images to determine whether other genes in the complex were identified by phenotype. It was clear from this analysis that, despite having a reduced cell number, Cul-4 and Roc1a manifested the same spiky, Rac/SCAR-like phenotype, so these genes were annotated accordingly. In contrast, dsRNAs targeting other complex members including Cul-2, Cul-3, Cul-5, and SkpA were indistinguishable from controls.
Based upon this analysis of the screen data, we selected a subset of genes for further validation: (1) 38 genes with strong actin-related phenotypes (spiky or “other” annotations), which included novel genes along with core known genes; and (2) 8 representative genes with a nuclear actin phenotype (Table I). To exclude the possibility of sequence-specific off-target effects (Perrimon and Mathey-Prevot, 2007), we retested each of our chosen hits and their interactors using a second independent dsRNA (Table S2). From group I, 36/38 genes were confirmed as hits, including additional Arp2/3 genes and the extra SCF genes, and 6/8 group II genes were successfully validated (Table I). These included genes with a known function in the export of actin monomers from the nucleus, namely chic and Exp6 (Stüven et al., 2003).
Table I.
Final list of genes with actin phenotypes conserved between Drosophila and mammalian cells
| FBgn | Gene | Phenotype group | Known actin regulator | Validated | Function | Mammalian orthologue(s) | HeLa actinome hit(s) | Mouse nuclear hit(s) |
| FBgn0015610 | Caf1 | High actin | No | Yes | Chromatin remodeling | RBBP4 | No | N/A |
| FBgn0034577 | cpa | High actin | Yes | Yes | F-actin capping | CAPZA1, CAPZA2, CAPZA3 | CAPZA1, CAPZA3 | N/A |
| FBgn0011570 | cpb | High actin | Yes | Yes | F-actin capping | CAPZB | CAPZB | N/A |
| FBgn0000042 | Act5C | Low actin | Yes | Yes | Actin filament formation | ACTG1 | ACTG1 | N/A |
| FBgn0000308 | chic | Low actin | Yes | Yes | G-actin binding | PFN1, PFN2, PFN3, PFN4 | PFN1, PFN4 (PFN3 not screened) | N/A |
| FBgn0011785 | BRWD3 | Other actin | No | Yes | WD40 domain protein | BRWD1,BRWD3, PHIP | No | N/A |
| FBgn0028388 | Capt | Other actin | Yes | Yes | G-actin binding | CAP1, CAP2 | No | N/A |
| FBgn0035586 | CG10671 | Other actin | No | No | Endoplasmic reticulum | FITM1, FITM2 | No | N/A |
| FBgn0039205 | CG13623 | Other actin | No | Yes | Mitochondrial | ISCA2 (HBLD1) | ISCA2 | N/A |
| FBgn0001491 | L(1)10Bb | Other actin | No | Yes | GPCR signaling | BUD31 (G10) | BUD31 | N/A |
| FBgn0044826 | Pak3 | Other actin | Yes | Yes | Rac GTPase signaling | PAK3 | No | N/A |
| FBgn0021967 | Pdsw | Other actin | No | No | Mitochondial electron transport chain | NDUFB10 | No | N/A |
| FBgn0011726 | tsr | Other actin | Yes | Yes | F-actin severing | CFL1, CFL2, DSTN | CFL1, DSTN | N/A |
| FBgn0052138 | CG32138 | Spiky | Moderately | Yes | Uncharacterized formin family member | FMNL1, FMNL2, FMNL3 | FMNL1 | N/A |
| FBgn0002183 | dre4 | Spiky | No | Yes | DNA repair | SUPT16H | SUPT16H | N/A |
| FBgn0015509 | lin19 | Spiky | No | Yes | Ubiquitin-dependent protein degradation | CUL1 | Not screened | N/A |
| FBgn0040291 | Roc1b | Spiky | No | Yes | Ubiquitin-dependent protein degradation | RBX1 | RBX1 | N/A |
| FBgn0003415 | skd | Spiky | No | Yes | Mediator complex | MED13, MED13L | No | N/A |
| FBgn0005411 | U2af50 | Spiky | No | Yes | RNA splicing | U2AF2 | U2AF2 | N/A |
| FBgn0020510 | Abi | Spiky (SCAR-like) | Yes | Yes | SCAR complex | ABI1, ABI2 | No | N/A |
| FBgn0031781 | Arc-p20a | Spiky (SCAR-like) | Yes | Yes | ARP2/3 complex | ARPC4 | ARPC4 | N/A |
| FBgn0011742 | Arp14D | Spiky (SCAR-like) | Yes | Yes | ARP2/3 complex | ACTR2 | No | N/A |
| FBgn0011744 | Arp66B | Spiky (SCAR-like) | Yes | Yes | ARP2/3 complex | ACTR3 | No | N/A |
| FBgn0039754 | CG9747 | Spiky (SCAR-like) | No | Yes | Fatty acid desaturase | SCD, SCD5, TRMT2A, TRMT2B | SCD (only one screened) | N/A |
| FBgn0011771 | Hem | Spiky (SCAR-like) | Yes | Yes | SCAR complex | NCKAP1 | NCKAP1 | N/A |
| FBgn0031437 | p16-arc | Spiky (SCAR-like) | Yes | Yes | ARP2/3 complex | ARPC5 | No | N/A |
| FBgn0010333 | Rac1 | Spiky (SCAR-like) | Yes | Yes | Rho family GTPase | RAC1 | RAC1 | N/A |
| FBgn0014011 | Rac2 | Spiky (SCAR-like) | Yes | Yes | Rho family GTPase | RAC2 | No | N/A |
| FBgn0041781 | SCAR | Spiky (SCAR-like) | Yes | Yes | SCAR complex | WASF1, WASF2, WASF3 | WASF3 | N/A |
| FBgn0016984 | Sktl | Spiky (SCAR-like) | Yes | Yes | Lipid kinase | PIPK51A, PIPK51B, PIPK51C | PIP5K1C | N/A |
| FBgn0023423 | slmb | Spiky (SCAR-like) | No | Yes | Ubiquitin-dependent protein degradation | BTRC, FBXW11 | Not screened | N/A |
| FBgn0038320 | Sra-1 | Spiky (SCAR-like) | Yes | Yes | SCAR complex | CYFIP1, CYFIP2 | CYFIP1 | N/A |
| FBgn0032859 | Arc-p34 | Spiky (SCAR-like)a | Yes | Yes | ARP2/3 complex | ARPC2 | ARPC2 | N/A |
| FBgn0038369 | Arpc3a | Spiky (SCAR-like)a | Yes | Yes | ARP2/3 complex | ARPC3A | No | N/A |
| FBgn0065032 | Arpc3b | Spiky (SCAR-like)a | Yes | Yes | ARP2/3 complex | ARPC3B | No | N/A |
| FBgn0001961 | Sop2 | Spiky (SCAR-like) | Yes | Yes | ARP2/3 complex | ARPC1A, ARPC1B | ARPC1A | N/A |
| FBgn0033260 | Cul-4 | Spiky (SCAR-like)b | No | Yes | ubiquitin-dependent protein degradation | CUL4A, CUL4B | Not screened | N/A |
| FBgn0025638 | Roc1a | Spiky (SCAR-like)b | No | Yes | ubiquitin-dependent protein degradation | RBX1 | RBX1 | N/A |
| FBgn0022213 | Cas | Nuclear actin | No | Yes | Nuclear transport | Cse1l | N/A | No |
| FBgn0030121 | CG17446 | Nuclear actin | No | Yes | Transcription | Cxxc1 | N/A | No |
| FBgn0031492 | CG3542 | Nuclear actin | No | No | Splicing | Prpf40a, Prpf40b | N/A | N/A |
| FBgn0037093 | CG7597 | Nuclear actin | No | Yes | Splicing (kinase) | Crkrs, Cdc2l5 | N/A | Cdc2l5 (Crkrs not tested) |
| FBgn0001337 | Exp6 | Nuclear actin | Yes | Yes | Nuclear transport | Xpo6 | N/A | Yes |
| FBgn0037657 | hyx | Nuclear actin | No | Yes | Transcription | Cdc73 | N/A | Yes |
| FBgn0016696 | Pitslre | Nuclear actin | No | No | Splicing (kinase) | Cdc2l1 | N/A | N/A |
| FBgn0003449 | snf | Nuclear actin | No | Yes | Splicing | Snrpa, Snrpb2 | N/A | Too toxic to assess |
Spiky (SCAR-like) annotation added after re-assessment with a longer RNAi period. See text for details.
Spiky (SCAR-like) annotation added after network analysis and reassessment. See text for details.
Identification of a conserved set of actin regulators
This analysis in Drosophila cells identified phenotypic profiles for the entire set of known actin regulators along with poorly characterized or unknown genes bearing actin-binding domains, and also identified a set of potentially novel actin regulators. To determine which of these genes perform conserved functions across species and cell types, we wanted to expand the analysis to a mammalian system. As a framework for the analysis of functional conservation of putative novel actin regulators, we therefore generated phenotypic profiles for the entire set of known mammalian actin regulators for comparison. To do so, we performed a duplicate siRNA screen of 516 known or predicted actin and Rho-GTPase family regulators (the “actinome”; Table S3) in human HeLa cells (Fig. 1, A and B). In addition, we tested the functions of representative close human orthologues of the set of putative novel actin regulators identified in the fly screen (Table I). 116 genes from this set were annotated as hits (Table S4), based upon having a reproducible phenotype with ≥2 individual siRNAs.
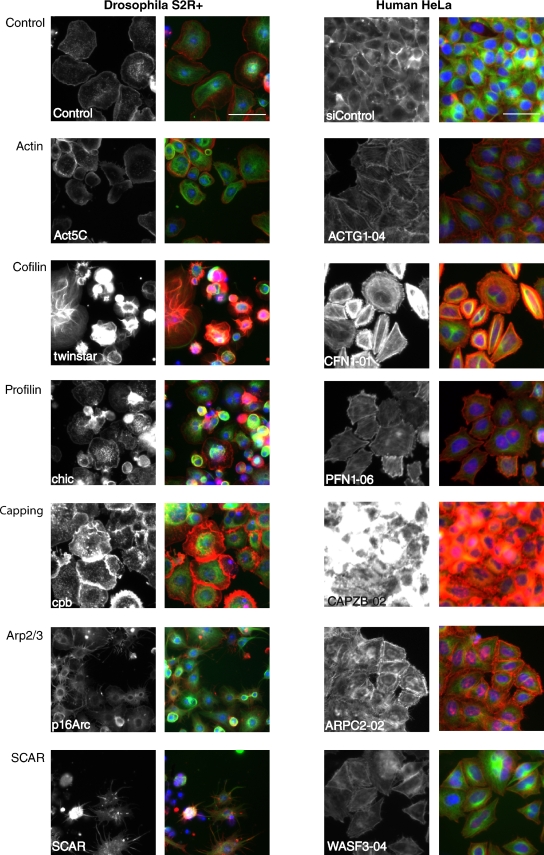
Comparison of the results of the S2R+ and HeLa screens revealed several hits among orthologous actin regulators (see Fig. 2 for representative images), including capping proteins A and B, profilin, cofilin, actin, Arp2/3, SCAR, nonmuscle myosin II, Rac, Rho, and Cdc42. When comparing fly versus human phenotypes, some fly/human orthologous pairs had similar morphological attributes; for example, both twinstar and CFL-1 depletion caused increased peripheral F-actin and multinucleated cells; and both cpb and CAPZB silencing induced a dramatic increase in F-actin levels (Fig. 2). In contrast, others gene pairs manifested different phenotypes (e.g., Act5C/ACTG1 and SCAR/WASF3; Fig. 2). These differences are likely due to cell- and species-specific differences in the interaction networks controlling cell shape.
Figure 2.
Images of representative conserved core fly/human gene pairs. Grayscale panels, F-actin; color panels, red is F-actin, green is α-tubulin, blue is DNA. In the case of human siRNAs, the representative siRNA ID number is indicated in the label. Bar, 50 µm.
We then inspected phenotypes for mammalian homologues of our novel validated Drosophila hits (Table I; see Table S4 for individual annotations and Fig. 1 E for their phenotypic clustering). For this analysis we used human siRNAs in HeLa cells, with the exception of the small cluster manifesting actin bars in the nucleus, for which we depleted the mouse orthologues in murine R3A4 cells. This analysis identified eight human orthologues of the fly hits together with two genes not previously implicated in nuclear actin regulation (Table I; discussed in detail later).
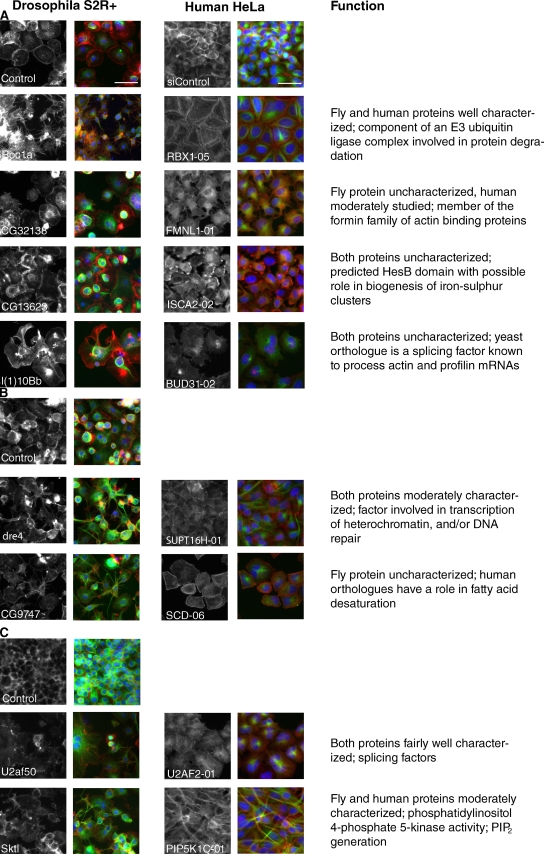
The eight pairs of conserved fly/human hits represent a diverse range of predicted cellular processes (Fig. 3, A–C). We recovered only one pair, CG32138/FMNL1, among the many uncharacterized genes predicted by domain structure to be involved in actin regulation. Drosophila CG32138 is a previously uncharacterized member of the formin family of actin-nucleating proteins (Goode and Eck, 2007) whose depletion was associated with an actin phenotype marked by a failure to spread, broken and disorganized lamellipodia, and multiple cytoplasmic actin cables, primarily positioned above the nucleus. Likewise, knockdown of FMNL1, one of the three human orthologues of CG32138, which has only recently been characterized (Han et al., 2009; Mersich et al., 2010; Mason et al., 2011), also led to an accumulation of actin filaments in the apical cell body and lamellipodia with a serrated appearance.
Figure 3.
Images of conserved novel fly/human gene knockdown pairs. Color and siRNA labeling as in Fig. 2. The fly gene phenotypes manifested best under different conditions: (A) fixed after respreading on concanavalin A–coated surface; (B) fixed after respreading on serum-coated surfaces; or (C) fixed after continuous growth under RNAi, no respreading. Bar, 50 µm. On the right is a brief description of the known or predicted function of each set of genes.
Two of the fly/human hit pairs, l(1)10Bb/BUD31 and CG13623/ISCA2, were more unexpected. Fly l(1)10Bb RNAi caused a particularly striking phenotype on knockdown that was unique in our screen: extremely large actin clumps and broken, distorted lamellae. Depletion of its sole human orthologue, the highly conserved BUD31, also led to large actin clumps and disorganized lamellipodia. The Saccharomyces cerevisiae homologue of this gene (also called BUD31) has been shown to be part of a splicing complex (Masciadri et al., 2004), and has been implicated in the splicing of actin and profilin transcripts. This effect is, however, unlikely to be a general consequence of aberrant RNA splicing because this striking phenotype is not shared by other splicing factors in fly cells (Table I) or in yeast (Masciadri et al., 2004). Even less is known about CG13623 and its human orthologue ISCA2 (HBLD1), which are entirely uncharacterized proteins whose main distinguishing feature is the presence of a hesB/yadR/yfhF domain, which in bacteria is thought to play a role in the biogenesis of iron–sulfur clusters for electron transfer processes (Cózar-Castellano et al., 2004). In our experiments, depletion of CG13623 led to asymmetrical lamellipodia, clumps of actin, and occasionally transverse stress fibers. Similarly, with the human orthologue ISCA2, knockdown led to polarized, peripheral clumps of actin in a flattened, roughly geometric shape, with the frequent occurrence of a thick actin bar at cell–cell junctions.
The five remaining gene pairs have been studied in other contexts. Skittles functions in phosphatidylinositol 4,5-bisphosphate production, and Xu et al. (2010) have demonstrated a role for the human orthologue PIP5K1C (phosphatidylinositol-4-phosphate 5-kinase type-1, also known as PIP5K1-gamma) in neutrophil polarity and adhesion. Depletion of these genes in both human and fly cells in our screen resulted in an elongated cell shape and an increase in actin puncta, which is consistent with the important role of PIP2 in the regulation of a large number of actin-binding proteins (Yin and Janmey, 2003).
The human gene U2AF2 (also known as U2AF65) encodes the large subunit of the U2AF heterodimeric complex, which is involved in the initial steps of spliceosome assembly on preRNA (Mollet et al., 2006). In Drosophila, the orthologous large subunit U2AF50 has been additionally implicated in the nuclear export of intronless mRNAs (Blanchette et al., 2004). In our screen, depletion of these two genes in both species causes disorganized actin and multinucleated cells, perhaps indicative of a defect in cytokinesis.
The FACT complex (Belotserkovskaya et al., 2003) is a heterodimeric complex involved in the modulation of chromatin assembly to influence gene expression, including Hox genes (Shimojima et al., 2003). One of its subunits, SPT16, was an actin hit in this category. Depletion of the Drosophila dre4/dSPT16 gene in fly cells or its human orthologue, SupT16H (SPT16) in HeLa cells caused an elongated cell shape and changes in peripheral actin levels in both cell types.
CG9747 is an unstudied fly gene, but one of its human orthologues, SCD (SCD1; stearoyl-CoA desaturase [delta-9-desaturase]), is an enzyme involved in the formation of saturated fatty acids (Igal, 2010). In our screen, depletion of CG9747 caused a spiky cell shape and increased actin puncta in S2R+ cells, whereas in HeLa cells, SCD knockdown led to an increase in actin stress fibers, a variable cell shape ranging from elongated to geometric, and an increase in multinucleated cells.
The final gene pair in this category, Roc1a/RBX1, which we studied in more detail, is described fully in the next section.
The SCF ubiquitin ligase pathway plays a role in cell shape
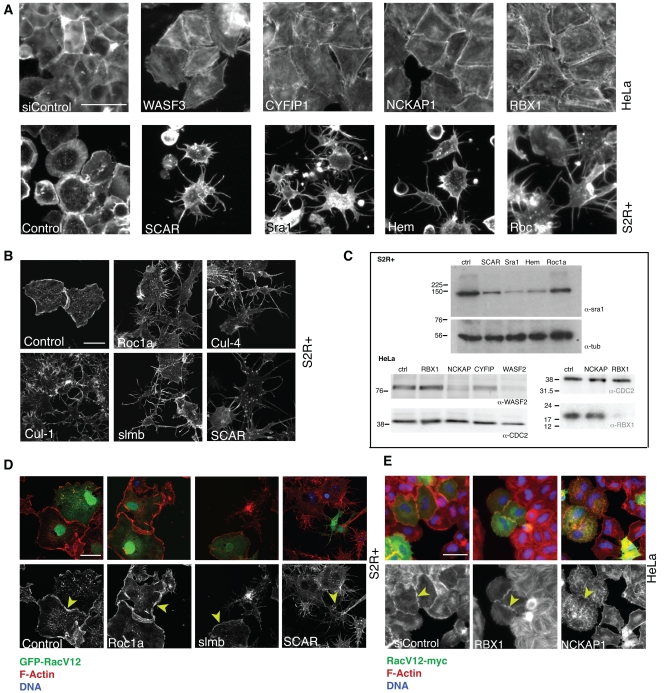
Components of the Rac/SCAR pathway, including its upstream regulators, Rac and Cdc42, the SCAR complex itself, and its downstream target, the Arp2/3 complex, which drives actin nucleation, were previously identified as having a common “spiky” RNAi phenotype in fly cells resulting from the loss of lamellipodia (Kunda et al., 2003; Rogers et al., 2003; see Fig. 4 A). RNAi-mediated depletion of the orthologous gene products from HeLa cells leads to a related phenotype, in which cells lose lamellipodia, take on a large geometrical shape, and accumulate stress fibers (Fig. 4 A; Innocenti et al., 2004; Derivery et al., 2008). This readout provided us with a useful assay for uncovering novel conserved components of this signaling pathway, and our parallel RNAi screening approach revealed several conserved genes were whose knockdown recapitulated the Rac/SCAR phenotype in both systems. This included five members of the SCF complex.
Figure 4.
The SCF ubiquitin ligase complex is involved in actin regulation. For all grayscale panels, staining is for F-actin. RBX1 and roc1a phenocopy the SCAR complex in human (A, top row) and fly (A, bottom row) cells. Bar, 50 µm. (B) Depletion of various SCF members phenocopies SCAR knockdown in fly cells. Bar, 20 µm. (C) roc1a/RBX1 depletion does not affect SCAR complex stability as measured by Sra1 or WASF2 protein levels in Western blot, respectively in fly cells (top) or human cells (bottom left), in the same situation when depleting other SCAR complex members does. Equal loading is indicated by tubulin staining of the same gels. The RBX1 knockdown in parallel cultures causes significant depletion (bottom right), with loading shown by staining for CDC2. (D) RacV12 overexpression (as marked by GFP, green; F-actin is red in top panels and grayscale in the bottom; blue is DAPI) rescues spiky phenotype of roc1a and slmb but not that of scar. Bar, 50 µm. (E) RacV12 overexpression (as marked by myc-tag, green; same staining for rest as in D) rescues RBX1 knockdown but not that of NCKAP1. Bar, 50 µm.
The core SCF complex recruits its substrate via an F-box receptor protein. Although our data suggested that slmb served that function, we wanted to inspect the phenotypes of all other F-box genes in our fly screening data. Drosophila has 37 members with domains characteristic of the F-box group (Ho et al. [2006] have reported 31), but slmb was the only one with a spiky phenotype. Although it is certainly possible that not all of these genes were efficiently depleted in the screen, these observations are consistent with Slmb being the sole or major F-box protein involved in the SCF-mediated actin phenotype in our system. Having validated these hits using independent RNAi reagents (Fig. 4 B shows representative examples), we chose to focus on a single component of this complex, Roc1a/RBX1, for the follow up analysis because silencing of this gene led to a robust loss of lamellipodia in S2R+ and HeLa cells (Fig. 4 A).
The stability of SCAR complex components (consisting in Drosophila of SCAR, Abi, Hem, Sra-1, and HSPC300) has previously been shown to be coordinately regulated by proteasome-dependent proteolysis, so that if one member of the complex is depleted by RNAi, the other components are also degraded (Kunda et al., 2003; Rogers et al., 2003; Derivery et al., 2008). To determine whether Roc1a/RBX1 is involved in this process, we therefore silenced Roc1a in S2R+ cells and then tested for the presence of Sra-1 protein by Western blot, as a marker for whole SCAR complex stability. Although knockdown of Sra1, SCAR, or Hem led to decreased levels of Sra-1 protein as expected, levels of Sra-1 remained unchanged in Roc1a RNAi cells (Fig. 4 C, top), under conditions in which nearly all cells manifested the spiky phenotype. Similarly, in human cells, the stability of the SCAR homologue WASF2 was unaffected by depleting RBX1, whereas knockdown of the Sra1 homologue CYFIP2 or the Hem homologue NCKAP1 led to degradation of WASF2 protein in HeLa cells (Fig. 4 C, bottom left), even though RBX1 levels were dramatically reduced after RNAi (Fig. 4 C, bottom right; the results of five replicate experiments in human and fly cells are quantified in Fig. S1 A). These results suggest that although the SCF complex regulates the stability of many targets and appears to regulate the formation of lamellipodial actin, it is unlikely to interfere directly with the stability of SCAR complex components.
Two recent papers have reported that the FERM domain containing tumor-suppressor protein Merlin/NF2 interacts indirectly with the RBX1/Cul4 complex (Huang and Chen, 2008; Li et al., 2010). Moreover, other work has shown a link between Merlin and Rac (Shaw et al., 2001; Sherman and Gutmann, 2001; Xiao et al., 2002; Okada et al., 2005; Morrison et al., 2007). These reports led us to investigate the role of Merlin in our system. Western blots on HeLa cells depleted for RBX1 showed no significant increase in the amount of Merlin protein (Fig. S1 B), in agreement with Huang and Chen (2008). Moreover, we were unable to detect a morphological or actin phenotype in S2R+ cells treated with independent dsRNAs targeting Drosophila Merlin (original fly screening data, and validated in Fig. S1 C), ruling out Merlin as the key link between the SCF complex and Rac.
Finally, to determine whether we could place Roc1a genetically upstream or downstream of Rac1, we tested the effect of introducing activated RacV12-GFP into S2R+ cells in which the expression of Roc1a and slmb as representative SCF members had been silenced using RNAi, compared with knockdown of SCAR as a representative SCAR complex member. In control cells, activated RacV12 causes a characteristic phenotype, namely a very spread cell with high levels of lamellipodial actin. As shown in Fig. 4 D, although RacV12 was unable to rescue the spiky phenotype of SCAR RNAi, Roc1a-depleted and slmb-depleted cells positive for RacV12-GFP developed prominent lamellipodia. Similarly, in HeLa cells, an activated RacV12-myc construct was able to rescue RBX1 knockdown, but not that of the downstream SCAR complex member NCKAP1 (Fig. 4 E). These data suggest that Rac can activate lamellipodial formation independently of Roc1a/RBX1; i.e., that Roc1a/RBX1 acts genetically upstream of Rac function. As a second test of this hypothesis, we performed double knockdown of roc1a in S2R+ cells with Pak3, a gene whose knockdown phenotype resembles the expression of constitutively active Rac (Asano et al., 2009). Again this condition resulted in a partial rescue of the spiky phenotype (Fig. S1 D; note that Pak3 protein levels were unaffected by Roc1a silencing, as shown in Fig. S1 E). Taken together, these data suggest that Roc1a protein exerts its effects upstream of Rac in a Pak3-independent manner.
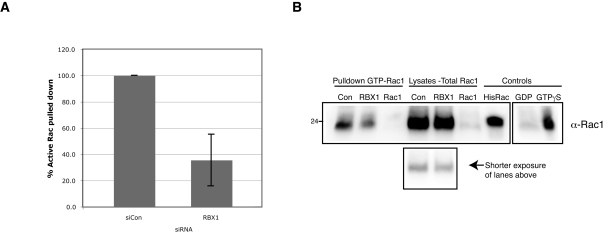
To further confirm this genetic interaction between RBX1 and the Rac pathway, we wanted to investigate whether endogenous activated Rac1 was reduced after depletion of RBX1. First, we used siRNA to deplete either Rac1 or RBX1 in HeLa cells. We then replated cells to stimulate Rac activity during respreading over the course of 5 h, and performed a pull-down assay using beads conjugated to the PBD domain of PAK, which specifically binds to the GTP-bound active form of Rac1. These experiments showed that levels of activated Rac1 were reduced to 36 ± 19% compared with overall Rac1 levels after RBX1 silencing (quantified in Fig. 5 A, with a representative Western blot shown in Fig. 5 B). Second, RBX1-depleted HeLa cells were stained with an antibody that targets the active, GTP-bound form of Rac1 (Haralalka et al., 2011). In this experiment, the high levels of active Rac1 seen in the cytoplasm and at the edges of spreading control cells were dramatically reduced after both RBX1 and Rac1 depletion (Fig. S1 F). Taken together, these experiments support the hypothesis that RBX1 acts upstream of Rac1.
Figure 5.
RBX1 depletion reduces active, GTP-bound Rac1. PAK-PBD bead pull-down of HeLa cell lysates depleted for control, RBX1, or Rac1 by siRNA, compared with the corresponding cellular lysate, probed with α-Rac1. (A) The mean of three independent experiments, normalized to control levels, is shown. Error bars show standard deviation; B shows a representative blot. The reaction control samples, preloaded with GTPγS and GDP (rightmost two lanes), were run in parallel but on a separate gel and membrane, as indicated.
Hyx/Cdc73 and CG7597/Cdc2l5 knockdown lead to excess actin in the nucleus
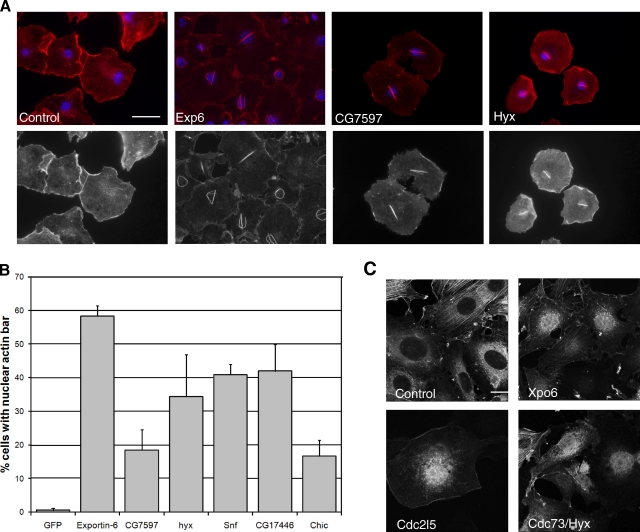
As mentioned previously, among the hits causing defects in the organization of the actin cytoskeleton, a subset displayed a prominent phalloidin-stainable bar in the nucleus of S2R+ cells (Fig. 6 A), whereas fewer than 1% of control cells did so (Fig. 6 B). The identification of this phenotypic class was surprising given that F-actin in the nucleus in other systems has traditionally not been visible using the phalloidin stain (Vartiainen, 2008). However, Exp6 and chic were identified among this set, both of which were previously shown to be required for the nuclear export of globular actin (Fig. 6 A and B; Stüven et al., 2003), suggesting a mechanism by which these nuclear filaments might form. This result led us to follow up the analysis of the nuclear actin phenotypic cluster in both fly and mammalian cells.
Figure 6.
Novel genes regulate actin in the nucleus. (A) Images showing actin bars in the nucleus of S2R+ after gene depletion. Bar, 10 µm (B) Quantitation of bars of experiment represented in A. (C) Depletion of key genes induces actin accumulation in mammalian cells. Red is F-actin, blue is DNA, and green is GFP-actin. Bar, 20 µm.
From the set of 28 dsRNAs that initially gave rise to a nuclear actin phenotype, we chose to follow up 8 representatives, and reproducibly confirmed 6 of these as strong hits in S2R+ cells using two independent dsRNAs, including Exp-6 (Table I; note chic was primarily categorized due to its more dominant phenotype, “decreased actin”, but it also validated as a seventh nuclear actin gene). Notably, several of the hits we had initially identified were components of the spliceosome, including snf (sans fille), which is an integral component of U1 and U2 small nuclear ribonucleoprotein particles (Cline et al., 1999). Snf was chosen as the representative example of the spliceosome complex for further analysis. In addition, depletion of the transcriptional regulator hyx/cdc73, which is a component of the Paf transcription complex (Shi et al., 1996), or of CG7597, which is the orthologue of mammalian Cdc2l5 known to regulate alternative splicing (Even et al., 2006), also produced a similar nuclear actin phenotype, as did the nuclear export gene Cas/CSE1 segregation protein and CG17446.
When we tested the mouse orthologues of these Drosophila hits (Table I) by siRNA knockdown in a GFP-actin mouse fibroblast line R3A4, which are more amenable than HeLa cells to studying nuclear export (unpublished data), we found that the depletion of the Exp6 orthologue Xpo6 led to the clear accumulation of nuclear GFP-actin, as expected (Fig. 6 C). In contrast, we found that depletion of the Cas orthologue Cse1l and the CG17446 orthologue Cxxc1 did not produce a nuclear actin phenotype by this test, and depletion of the snf orthologue Snrpb2 was too toxic to assess the phenotype. However, silencing of the hyx orthologue Cdc73 and the CG7597 orthologue Cdc2l5 resulted in a similar nuclear accumulation of GFP-actin (Fig. 6 C). These results suggest that, even though we were unable to see phalloidin-stained nuclear actin bars in these cells, the pathways regulating nuclear actin levels are at least partially conserved through evolution.
Cdc73, also known as parafibromin, is in humans encoded by the HRPT2 gene, mutations of which cause the hyperparathyroidism-jaw tumor syndrome (Carpten et al., 2002). As a component of the Paf complex, Cdc73 is involved in many transcription-related processes, including communication with transcription factors, regulation of histone modification, and recruitment of premRNA processing factors (Jaehning, 2010). Accordingly, cells from Cdc73 knockout mice display altered gene expression profiles (Wang et al., 2008). Although Cdc73 is mainly a nuclear protein (Bradley et al., 2007), it has also been found in the cytoplasm, where it may interact with the actin cross-linking proteins actinin-2 and actinin-3 (Agarwal et al., 2008). Cdc2l5 belongs to a subfamily of Cdc2-related kinases (Marqués et al., 2000), and is also called the cyclin-dependent kinase 13 (Cdk13) due to its ability to bind to L-type cyclins (Chen et al., 2007). It has been linked to alternative splicing (Even et al., 2006), and binds, for example, to the HIV-1 Tat-protein to regulate viral mRNA splicing (Berro et al., 2008). It remains to be determined how these genes and the spliceosome function to regulate levels of nuclear actin.
In summary, we have used a genome-scale RNAi screen in fly cells and a secondary screen in mammalian cells to define a conserved actinome based upon phenotype. This includes a number of new conserved actin regulators and implicates several core molecular processes in the regulation of the actin cytoskeleton. This simple strategy could be applied to enhance our understanding of a wide range of cell biological processes.
Discussion
Many actin regulators have been characterized over the years in a variety of organisms, including cell- and species-specific regulators, and a set of proteins that has subsequently proven to be functionally conserved. We were interested in using a systematic approach to define a core “actinome” based upon phenotype in cultured cells from two different lineages derived from two disparate animal species, Drosophila and mammals. This comparative screening strategy yielded four broad categories of hits: (1) the expected suite of proteins with well-known biochemical functions related to actin (e.g., Arp2/3 complex, SCAR, Rac); (2) poorly studied proteins with predicted biochemical functions related to actin (e.g., CG32138/FMNL1, Sktl/PIP5KC); (3) proteins with known biochemical functions but little previous relationship to actin or its regulation (e.g., the ubiquitin ligase component Roc1a/RBX1, the cyclin-dependent kinase Hyx/Cdc73); and (4) genes encoding proteins with no known biochemical function but with a conserved actin phenotype in our screen (e.g., CG13623/ISCA2).
Our analysis failed to identify good candidates for novel proteins that alter actin filament dynamics through direct binding to actin aside from CG32138/FMNL1, whose actin-related functions have been recently described (Katoh and Katoh, 2003; Perdigoto et al., 2008; Han et al., 2009; Fabian et al., 2010; Mersich et al., 2010; Mason et al., 2011). Instead, our hits fell into functional categories likely to act upstream of actin, such as specific splicing factors, nuclear export pathways, and proteasomal regulators. This highlights the importance of studying the regulation of actin dynamics in a wider cell-biological context (Pollard and Cooper, 2009), to which this and other unbiased functional screens can make an important contribution. Intriguingly, several of these proved to be small genes (e.g., l(1)10Bb, CG13623, and Roc1a), which are often missed in classical genetics screens. Conversely, the vast majority of unstudied genes with predicted actin-binding domains but no previously identified phenotype did not have an actin phenotype in our study. Some of these genes may simply have been missed due to inefficient silencing; however, it is likely that many of these proteins were effectively depleted by our reagents but were not morphological hits because they may have evolved more subtle tissue-specific actin-related functions, or because they may simply act as scaffolds to tether other processes to the actin cytoskeleton.
Among the basic cell processes upstream of actin regulation revealed by our screen, ubiquitination mediated by the SCF complex emerged as a conserved regulator upstream of Rac-mediated actin polymerization. Our phenotypic analysis of fly screen data suggests that Slmb/BTRC/β-TrCP is the main F-box receptor required for this activity. Slmb is known to regulate transcription and cyclin-dependent kinases (Frescas and Pagano, 2008), but thus far no known roles for SCF/Slmb suggest a link to cell shape and the actin cytoskeleton. It is possible, for example, that SCF/Slmb targets a Rac inhibitor for degradation, although we could detect no relevant phenotype in any of the known Rac inhibitors present in our screens. Further studies will be required to define the pathway in more detail.
In addition, the screen identified a set of dsRNAs that gave rise to a previously undescribed phenotype, the accumulation of phalloidin-stained nuclear actin bars. It has become increasingly obvious that actin has many important functions in the cell nucleus, especially in the process of gene expression. For example, nuclear actin has been implicated in the regulation of transcription factor activity, in transcription by all three RNA polymerases, in chromatin remodeling, and in premRNA processing (Skarp and Vartiainen, 2010). Despite these essential roles, very little is known about how nuclear actin is regulated. In our genome-wide fly screen, we identified several factors whose depletion caused accumulation of F-actin in the nucleus. These proteins are therefore candidates for regulators of nuclear actin export or proteins that limit inappropriate actin polymerization within the nucleus. Both processes may be functionally significant, as recent studies have suggested that modulation of nuclear actin levels appears to correlate with cellular quiescence (Spencer et al., 2011), and functional studies point to a requirement for polymerized actin within the nucleus for this role (McDonald et al., 2006; Ye et al., 2008). How nuclear actin levels are regulated in cells remains poorly understood, although increased actin in the nucleus has been detected upon the induction of different cellular stresses (Vartiainen, 2008).
Nuclear export of actin is mediated by the importin-β family member Exportin-6, which seems to use the small actin-binding protein profilin as a cofactor (Stüven et al., 2003). In our experiments in both fly and mammalian cells, depletion of either Exportin-6 or profilin resulted in the nuclear actin phenotype, demonstrating that our screen was sensitive enough to recover known factors involved in this process. The polymerization status of actin in the nucleus is still somewhat unclear, but it is thought that the majority of nuclear actin would be monomeric (McDonald et al., 2006). Indeed, phalloidin does not generally stain the nucleus, and we did not observe phalloidin-stainable bars in mammalian cells. Nevertheless, several studies have suggested a role for F-actin in the nucleus (Miyamoto et al., 2011) and S2R+ cells may therefore represent a good system in which to study this phenomenon.
Of note, many of the nuclear actin hits from Drosophila cells are components of the spliceosome. Although the spliceosome component we chose to validate in mouse cells, Snrpb2, could not be analyzed due to cell death, Cdc2l5, which produced the phenotype in both systems, has also been linked to splicing (Even et al., 2006). How defects in spliceosome function result in the nuclear actin phenotype is presently unclear, but it may be due to altered expression of proteins required for nuclear export of actin, or to a general stress response in the cell, as this has been shown to increase nuclear actin levels (Welch and Suhan, 1985). Alternatively, actin itself has been implicated in premRNA processing, and binds directly to several hnRNP proteins (Percipalle et al., 2002) that play a key role in this process. Perhaps disruption of the splicing process releases actin from these factors, resulting in uncontrolled actin polymerization within the nucleus. Future studies will reveal at which stage the identified factors impinge on nuclear actin to regulate its nuclear export and/or polymerization properties.
In summary, our species-comparative approach, as exemplified by the Rac/SCAR cluster and the actin-in-the-nucleus cluster, revealed that entire functional modules can be recapitulated across species, and that such “phenoprinting” can be used as a powerful shortcut to find novel actin regulators. Even when the precise nature or appearance of phenotype itself is not always conserved between species (e.g., a spiky appearance in S2R+ cells versus a triangular, stress-fiber appearance in HeLa cells; or the phalloidin stainable bar in S2R+ versus actin accumulation in mouse cells), consistent phenoprints within a species can still be used to pinpoint conserved functional modules. By making our data freely available, we expect that they can be further mined by the community for functional pathways tailored to individual laboratory interests, and as such should serve as an important resource for the future.
Materials and methods
Recombinant DNA cloning
We constructed pMT-RacV12 from a preexisting pUAS-RacV12 plasmid template using PCR with oligo Rac1(NT_BP) forward (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGATGCAGGCGATCAAGTGCGTCG-3′) and Rac1(NT_BP) reverse (5′-GGGACCACTTTGTACAAGAAAGCTGGGTCTTAGAGCAGGGCGCACTTGCGC-3′). The PCR amplimer was gel purified (QIAGEN), then a BP Clonase (Invitrogen) reaction was performed with the pDoner201 vector and purified RacV12 PCR product. pDoner-RacV12 was used for an LR reaction (Invitrogen) with the pMT-NT-GFP-destination vector (Liu et al., 2010) to create the final construct. The sequence as well as the activating V12 point mutation was confirmed by sequence analysis.
Cell culture
HeLa-Kyoto human carcinoma cells were cultured at 37°C in a humidified incubator under 5% CO2 in 9-cm dishes in DME (Invitrogen) supplemented with 10% fetal bovine serum (FBS, PAA) and antibiotics (50 U/ml penicillin and 50 µg/ml streptomycin; Invitrogen). Adherent S2R+ Drosophila cells were cultured at 24°C in a humidified incubator in T25 flasks in Shields and Sang M3 insect medium (Sigma-Aldrich) supplemented with FBS and antibiotics as for HeLa cells. A tetracyclin-inducible GFP-actin–expressing mouse fibroblast NIH3T3 cell line (R3A4) was created using the T-REX system (Invitrogen) according to the manufacturer’s instructions. R3A4 cells were maintained in DME supplemented with 10% FBS, GlutaMAX (Invitrogen) and antibiotics, as for HeLa cells, at 37°C and 5% CO2. Blasticidin and Zeocin (InvivoGen) were added during passaging but omitted during transfection.
RNAi reagent design and synthesis
The genome-wide screen was performed at the Drosophila RNAi Screening Center (Harvard Medical School, Boston, MA) using a library of 21,306 distinct dsRNAs (See Table S1 for primer details) targeting 12,061 protein-coding genes. Because the library targets 88% of the genome, we would expect this dataset to contain phenotypic information for the vast majority of known actin regulators, along with phenotypes for previously uncharacterized actin regulators. For validation experiments, PCR primers were based on a genome-wide library purchased from Invitrogen/Ambion, the DRSC validation library (http://flyrnai.org), or designed de novo using SnapDragon (http://flyrnai.org/snapdragon_doc1.html; see Table S2 for details). Gene-specific amplicons (∼200–500 bp; average 400 bp) were amplified from genomic DNA by PCR using HotStart Taq polymerase (QIAGEN) with primer pairs synthesized to order (EuroGentec). RNA was synthesized using a MegaScript reverse transcription kit (Invitrogen), and purified using a Multiscreen HTS kit (Millipore) or NucAway spin column (Applied Biosystems), and annealed by heating at 65°C for 10 min then cooling slowly to room temperature. dsRNAs were quantified by NanoDrop spectrometry, and gel electrophoresis was used to confirm the integrity of the dsRNAs. dsRNAs were stored at −40°C until use.
For the human screens, we used a computational approach to nominate actin- and Rho GTPase–related genes to create the “actinome” library. The actinome library contained (1) genes with known actin cytoskeleton association; (2) genes with predicted actin-binding domains; and (3) Rho family GTPases, GAPs, and GEFs. We also determined the human orthologues of Drosophila actin morphology hits by comparing orthologue assignments from Homologene (http://www.ncbi.nlm.nih.gov/homologene), Inparanoid (http://inparanoid.sbc.su.se/cgi-bin/index.cgi), and Ensembl (http://www.ensembl.org/index.html). Synthetic siRNAs for all target genes were obtained from Thermo Fisher Scientific/Dharmacon (Table S3). For controls, we included multiple instances of the following four Thermo Fisher Scientific controls: siCONTROL (a scrambled negative-control siRNA sequence); INCENP and KIF11, which give strong and distinctive phenotypes; and siTOX, an siRNA reagent that induces cell death. All siRNAs were reconstituted in nuclease-free water in 1x buffer provided by the manufacturer. For the mouse cell siRNA experiments, we purchased siRNA from QIAGEN and Sigma-Aldrich (Table S3).
High-throughput RNAi screening
The genome-wide S2R+ cell screen was performed as described previously (Kiger et al., 2003). In brief, a semi-automated method was used to seed 10 µl of a trypsinized suspension of S2R+ cells at a concentration of 2 × 106 cells/ml in serum-free M3 medium into black, thin-bottomed 384-well tissue culture plates (Corning) already containing a 3-µl droplet of dsRNA (∼0.3 µg). After a 30-min incubation at room temperature, wells were supplemented with 30 µl of complete M3 medium. All manipulations were performed with an alcohol-sterilized WellMate liquid-handling robot inside a tissue culture hood. Plates were spun briefly, sealed with parafilm, and incubated in a 25°C humidified incubator for 5 d before being processed: using a semi-automated method with the WellMate robot, cells were fixed with freshly prepared 4% formaldehyde in PBS for 20 min at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 5 min, then blocked with 5% BSA in PBS for 30 min. Next, cells were stained with a mixture of FITC-conjugated anti–α-tubulin antibody (clone DM1A at 1:400; Sigma-Aldrich), TRITC-conjugated phalloidin (0.125 µg/ml; Sigma-Aldrich), and DAPI (1 µg/ml; Sigma-Aldrich) in a PBS solution containing 1% BSA for 1 h. After 4x washing with PBS, plates were sealed with adhesive foil and stored in PBS containing 0.1% sodium azide (Sigma-Aldrich). Images were acquired using a 20x objective lens (NA 0.45) on a modified Eclipse TE-2000E microscope (Nikon) equipped with a Prior Proscan motorized stage and MetaMorph software (Molecular Devices), autofocusing on the DAPI channel. Images were screened visually using the MetaMorph “Review Screen Data” application. The uploaded .tif files were converted from 16-bit to 8-bit but were otherwise unprocessed.
The fly cell screen was performed once, and three independent researchers visually inspected images captured from two sites per well to nominate potential hits with morphology phenotypes of any description. We filtered the set of nominated hits to exclude dsRNAs with (1) >1 predicted amplicon; (2) one or more CAN repeats (a stretch of a particular trinucleotide sequence [CAn] repeated five or more times, which is known to cause nonspecific effects (Echeverri et al., 2006)); (3) >5 predicted off-target effects (19 bp; Echeverri et al., 2006); and (4) no known protein-encoding gene target according to FlyBase version 5.23 (http://flybase.org; Table S1). Images from the remaining dsRNAs were then annotated using a controlled vocabulary to describe phenotypes affecting cell number, cell size, and all aspects of nuclear and cytoskeletal morphology. DsRNAs with weak or inconsistent phenotypes (defined as present in only one of the two sites) were excluded from this analysis. In inspecting this refined list of dsRNAs associated with morphological phenotypes (see hit annotation details in Table S2), we found that when multiple dsRNAs targeting the same gene were annotated as hits, a similar range of phenotypes was recorded, demonstrating the robustness of the method.
For HeLa cell screens, siRNAs were arrayed into the Corning screening plates at a final concentration of 0.5 µM (3 µl) using both individual siGENOME duplexes and SmartPools (Thermo Fisher Scientific) using a Biomek Fx robotic liquid-handling robot (Beckman Coulter), sealed with foil adhesives and stored at −40°C until use. Trypsinized HeLa cells (2,000 cells per well) without antibiotics were reverse transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions at a final volume of 80 µl (final siRNA concentration of 25 nM). All transfection manipulations were performed sterilely as described for S2R+ cells, and the plates were spun briefly to eliminate any bubbles and incubated at 37°C in a humidified incubator under 5% CO2 for 3 d before processing as for the fly screen. Each human gene was targeted using four independent siRNAs, each individually assessed in four separate wells, and, for genes in the actinome, with the addition of a fifth well containing those four siRNAs pooled together. The entire screen was performed in duplicate, and images (three sites per well) from both screens were loaded into FLIGHT. Images were annotated by two independent researchers blinded to well designations; although we assessed the same general phenotypic categories as with the fly screen, we used more extensive sub-descriptions to reflect the greater detail visible in these cells. Genes were considered hits if at least two independent siRNA constructs were identified as strong morphological hits with similar phenotypes in both screen replicates by both researchers.
Hierarchical clustering and network analysis
We performed hierarchical clustering of both Drosophila and human binary RNAi phenotype annotations using the correlation distance and Wards method using R (http://www.r-project.org). To identify related hits that formed part of the same complex or interaction group, we used the network analysis tool on the FLIGHT database, which queries interaction data from a large set of online databases including Reactome and BioGrid (Sims et al., 2006, 2010).
Morphological phenotype validation
For validation experiments, we delivered dsRNAs to S2R+ cells in a similar manner as the high-throughput protocol and incubated them for 6 d. This longer incubation ensured that all phenotypes had time to manifest. Under these conditions, for example, we also validated the remaining four Arp2/3 complex components (Arc-p34, Arpc3a, Arpc3b and Arc-p20) that were not identified as hits initially because these proteins have a low turnover (Kunda et al., 2003). Cells were then trypsinized and replated at a lower density into two black, thin-bottomed confocal-ready 384-well tissue culture plates (Greiner bio-one)—one whose wells had been precoated overnight at 37°C with FBS, the other coated with concanavalin A (Con A IV-S; Sigma-Aldrich), dissolved at 5 µg/ml in water. Cells were allowed to spread for 1–3 h and then fixed. We also fixed replicates in which the cells were not replated after RNAi. For RNAi experiments destined to be replated onto glass coverslips, we suspended cells in serum-free medium at a concentration of 2 × 106 cells/ml (100 µl droplet in a well of a 4-well plastic tissue culture plate) and mixed these with 3 µg of dsRNA corresponding to the gene of interest or, as a negative control, to dsRNA targeting LacZ or dsRed, whose sequences are not present in the fly genome. This mixture was incubated at 24°C for 30 min before addition of complete serum-containing medium (300 µl). Cells were grown for 3–7 d at 24°C, depending on the experiment, then replated onto FBS- or Con A–coated circular 13-mm-diam coverslips and allowed to spread before analysis (typically 1–3 h). In experiments where cells were not replated before analysis (e.g., validating the nuclear actin hits in S2R+ cells), cell seeding was slightly different: 50,000 cells in 200 µl directly onto coverslips in a 24-well plate, analyzed after 6 d.
For experiments requiring plasmid transfection, S2R+ cells were transfected in 4-well plates with 1 ug total of DNA using FugeneHD (Roche), following the manufacturer’s protocol. We used GFP-tagged RacV12 under the control of a metallothionein promoter (pMT), or as a negative control, pMT-Gal4. 18 h before processing the experiments, a copper sulphate solution was added to the medium (final concentration, 70 nM) to induce expression from the pMT promoter.
For human cell validation, our strategy was to repeat the screen twice and to use pools as well as individual siRNAs for depletion, so on average each gene was tested five different ways in two separate sessions. We relied upon independent siRNAs and not pools for validation because 75 genes of the 116 hits failed to display a phenotype when using the pool in a situation when two or more of its individual siRNA components had a positive, gene-consistent phenotype. This result was likely due in part to dosage effects, as we kept the overall concentration of siRNA constant for all conditions, meaning that on average, each single siRNA was present at four times the concentration of its pool counterpart.
Small-scale HeLa experiments were performed in 384-well plates or by scaling up appropriately into 8-well chamber slides (LabTek; Thermo Fisher Scientific) precoated with fibronectin (10 µg/µl in PBS; Sigma-Aldrich). We used Thermo Fisher Scientific siCONTROL as a negative control in all cases. When plasmid transfection of RNAi cultures was required, FugeneHD was used as for S2R+ cells together with a myc-tagged RacV12 construct (Hogan et al., 2009) or as a negative control, a histone-2B-RFP construct, 18 h before processing. For experiments with mouse R34A cells, 5,000 cells were seeded on coverslips in a 24-well plate overnight. Cells were transfected with 10 nM siRNA using Interferin siRNA transfection reagent (Polyplus). After transfection, cells were incubated at 37°C with 5% CO2 for 4 d before induction of GFP-actin expression with 1 µg/ml tetracycline for 24 h, and were then analyzed. A GFP-alone construct was used as a negative control.
For immunostaining, all cell types were fixed and permeabilized as described for screening (except we used 4% paraformaldehyde instead of formaldehyde for the nuclear DNA experiments). For some experiments, wells that had received tagged RacV12 constructs were stained with the same mix, except DM1A-stained cells were visualized using a goat anti–mouse immunoglobulin antibody conjugated to Alexa 647 (1:500; Invitrogen); GFP signal was bolstered using anti-GFP antibody (Invitrogen), and myc-tagged-RacV12 was visualized using α-Myc tag (Clone 4A6; Millipore). For experiments assessing the presence and localization of GTP-bound Rac1, we used the monoclonal antibody α-Active Rac1 (NewEast Biosciences) at a concentration of 1:200. For HeLa cells, slides were mounted in FluorSave Reagent (EMD) and imaged using a scanning confocal microscope (model SP5, Leica; either the 40x lens or the 63x oil lens, NA 1.25 and 1.4, respectively;). For mouse cells, we used a confocal microscope (TCS SP5 MP SMD FLIM; Leica) equipped with a 63x (NA 1.3) oil objective. For both Leica microscopes, LAS AF software (Leica) was used for image acquisition. For fly cell experiments in which we were assessing phalloidin-stainable bars in the nucleus, actin was visualized with Alexa Fluor 594–conjugated phalloidin (Invitrogen) instead of TRITC-phalloidin, slides were mounted with Moviol-DABCO (Sigma-Aldrich), and images were acquired using a microscope (AX70 Provis; Olympus) equipped with an Plan Apochromat 60x, NA 1.40 oil objective. The camera used was F-view II FW (Olympus), and images were acquired with the analySIS software (Olympus).
All images were processed according to good practice using ImageJ/FIJI or Photoshop; in some cases we altered the brightness and contrast of the entire image to improve the image. Color channels were merged using either ImageJ or Photoshop.
Western blot analysis and GTP-bound Rac1 pull-down assays
For Western blotting to inspect SCAR complex stability, we either lysed samples directly in 2x Laemmli buffer (Sigma-Aldrich) and disrupted them with a fine-gauge needle, or lysed them in ice-cold RIPA buffer (50 mM Hepes, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS, pH 7.7) to which protease inhibitor cocktail (Sigma-Aldrich) had been added, incubated and agitated the samples for 10 min on ice, centrifuged them to harvest protein-containing supernatants, then made them up to 1x Laemmli buffer. All samples were heated at 99°C for 10 min before loading onto SDS-PAGE gels ranging from 15% for small proteins to 8% for large ones; or NuPAGE 4–12% Bis/Tris gradient gels (Invitrogen) for the Rac pull-down assay (see the next paragraph). We then transferred proteins to Immobilon-P (Millipore) membrane by Western blotting. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline/0.05% Triton X-100 (TBS-T) for 1 h. All primary antibody incubations were at room temperature for several hours or at 4°C overnight in TBS-T (1:1,500 for the α-dSra1 antibody (a gift from Alexis Gautreau, LEBS Centre National de la Recherche Scientifique, Gif-sur-Yvette, Paris, France); 1:1,000 for the human NF2/Merlin antibody (B12; Santa Cruz Biotechnology, Inc.); 1:1,500 for the WAVE2/WASF2 antibody (Gautreau et al., 2004); 1:2,000 for the dPak3 antibody (Asano et al., 2009); 1:2,000 for the α-tubulin antibody (DM1A; Sigma-Aldrich); and 1:2,000 for the anti-cdc2 antibody (anti-PSTAIR; Sigma-Aldrich), and then washed five times with TBS-T. We next incubated membranes with the appropriate HRP-conjugated secondary antibodies (1:1,000; Dako) in TBS-T for 1 h, and washed them as before, followed by an ECL procedure and detection on either Hyperfilm EC or an ImageQuant LAS4000 (solutions, film, and apparatus all from GE Healthcare). When reprobing was required, we stripped membranes in a hot solution of 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris, pH 6.8 (50°C) for 30 min with agitation, then washed them 2x 10 min in TBS-T until they were odorless. We always tested membranes with ECL to confirm absence of signal before rewashing and reblocking as normal. Protein bands were quantified on unsaturated exposures using the ImageQuantTL software according to the manufacturer’s instructions.
For assessing the activity state of Rac1 biochemically in HeLa cells, we scaled up the siRNA transfection to 9-cm plates (2.6 × 106 cells 18 μL Lipofectamine 2000 and 12 µl of siRNA [stock: 20 µM] in a total of 4 ml). The media was changed the next day, and 3 d after transfection with siRNA, when cultures were less than 50% confluent, we trypsinized the cells and replated them to stimulate Rac1 activity. At 5 h after replating, when the cells were actively spreading, we performed the pull-down assay using the Rac1 Activation Assay Biochem kit (Cytoskeleton) according to the manufacturer’s instructions. The samples were run on NuPAGE gradient gels, Western blotted, and probed with the kit’s monoclonal antibody to Rac1. Along with the washed beads and samples of total cell-clarified lysates, we also ran a purified His-Rac protein as a control for the Western blot, and each replicate assay included a GTPγS-loaded positive control, and a GDP-loaded negative control for the pull-down. Active Rac levels were normalized for total basal Rac1 protein levels and quantified as in the previous paragraph; we confirmed that the levels of total Rac1 were not altered by the siCONTROL or RBX1 siRNA treatments.
Online supplemental material
Fig. S1 shows additional cell biological and biochemical observations. Table S1 provides details of the 21,306 fly dsRNAs. Table S2 provides annotations for all fly hits (worksheet 1) and dsRNA fly validation primer details (worksheet 2). Table S3 provides all mammalian siRNAs and their details. Table S4 provides the annotation for all human hits. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103168/DC1.
Acknowledgments
We thank the Actin RNAi Consortium, members of our laboratory for helpful discussions, and Jonathan Lees for additional bioinformatical advice. We thank Alexis Gautreau for helpful discussions and for providing anti-dSra1 and anti–human WASF2 antiserum; Helen Morrison for the human NF2/Merlin antibody; Catherine Hogan for the myc antibody and human RacV12 construct; Rob de Bruin for the CDC2 antibody; and Anne Ridley, Francisco Vega, Harry Mellor, Shruti Haralalka, and Heike Laman for helpful discussions.
J.L. Rohn was funded by a fellowship from the Wellcome Trust. M.K. Vartiainen was funded by Academy of Finland, University of Helsinki Research Funds, and Sigrid Juselius foundation. J. Dopie was funded by a fellowship from the Helsinki Graduate Program in Biotechnology and Molecular Biology. N. Perrimon is an investigator of the Howard Hughes Medical Institute. B. Baum was funded by the Royal Society, the EMBO YIP program, UCL, and Cancer Research UK.
Footnotes
Abbreviations used in this paper:
- SCF
- Skp1-Cul1-F-box-protein
- Slmb
- supernumerary limbs
References
- Agarwal S.K., Simonds W.F., Marx S.J. 2008. The parafibromin tumor suppressor protein interacts with actin-binding proteins actinin-2 and actinin-3. Mol. Cancer. 7:65 10.1186/1476-4598-7-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y., Jiménez-Dalmaroni A., Liverpool T.B., Marchetti M.C., Giomi L., Kiger A., Duke T., Baum B. 2009. Pak3 inhibits local actin filament formation to regulate global cell polarity. HFSP J. 3:194–203. 10.2976/1.3100548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C., Aach J., Church G., Perrimon N. 2007. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 316:1753–1756. 10.1126/science.1140324 [DOI] [PubMed] [Google Scholar]
- Baum B., Li W., Perrimon N. 2000. A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10:964–973. 10.1016/S0960-9822(00)00640-0 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science. 301:1090–1093. 10.1126/science.1085703 [DOI] [PubMed] [Google Scholar]
- Benlali A., Draskovic I., Hazelett D.J., Treisman J.E. 2000. act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell. 101:271–281. 10.1016/S0092-8674(00)80837-5 [DOI] [PubMed] [Google Scholar]
- Berro R., Pedati C., Kehn-Hall K., Wu W., Klase Z., Even Y., Genevière A.M., Ammosova T., Nekhai S., Kashanchi F. 2008. CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J. Virol. 82:7155–7166. 10.1128/JVI.02543-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Labourier E., Green R.E., Brenner S.E., Rio D.C. 2004. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 14:775–786. 10.1016/j.molcel.2004.06.012 [DOI] [PubMed] [Google Scholar]
- Bocca S.N., Muzzopappa M., Silberstein S., Wappner P. 2001. Occurrence of a putative SCF ubiquitin ligase complex in Drosophila. Biochem. Biophys. Res. Commun. 286:357–364. 10.1006/bbrc.2001.5394 [DOI] [PubMed] [Google Scholar]
- Bradley K.J., Bowl M.R., Williams S.E., Ahmad B.N., Partridge C.J., Patmanidi A.L., Kennedy A.M., Loh N.Y., Thakker R.V. 2007. Parafibromin is a nuclear protein with a functional monopartite nuclear localization signal. Oncogene. 26:1213–1221. 10.1038/sj.onc.1209893 [DOI] [PubMed] [Google Scholar]
- Carpten J.D., Robbins C.M., Villablanca A., Forsberg L., Presciuttini S., Bailey-Wilson J., Simonds W.F., Gillanders E.M., Kennedy A.M., Chen J.D., et al. 2002. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 32:676–680. 10.1038/ng1048 [DOI] [PubMed] [Google Scholar]
- Chen H.H., Wong Y.H., Geneviere A.M., Fann M.J. 2007. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem. Biophys. Res. Commun. 354:735–740. 10.1016/j.bbrc.2007.01.049 [DOI] [PubMed] [Google Scholar]
- Cline T.W., Rudner D.Z., Barbash D.A., Bell M., Vutien R. 1999. Functioning of the Drosophila integral U1/U2 protein Snf independent of U1 and U2 small nuclear ribonucleoprotein particles is revealed by snf(+) gene dose effects. Proc. Natl. Acad. Sci. USA. 96:14451–14458. 10.1073/pnas.96.25.14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cózar-Castellano I., del Valle Machargo M., Trujillo E., Arteaga M.F., González T., Martín-Vasallo P., Avila J. 2004. hIscA: a protein implicated in the biogenesis of iron-sulfur clusters. Biochim. Biophys. Acta. 1700:179–188. [DOI] [PubMed] [Google Scholar]
- Cvrcková F., Rivero F., Bavlnka B. 2004. Evolutionarily conserved modules in actin nucleation: lessons from Dictyostelium discoideum and plants. Review article. Protoplasma. 224:15–31. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio M.V., Vale R.D. 2010. A whole genome RNAi screen of Drosophila S2 cell spreading performed using automated computational image analysis. J. Cell Biol. 191:471–478. 10.1083/jcb.201003135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Fink J., Martin D., Houdusse A., Piel M., Stradal T.E., Louvard D., Gautreau A. 2008. Free Brick1 is a trimeric precursor in the assembly of a functional wave complex. PLoS ONE. 3:e2462 10.1371/journal.pone.0002462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435–467. 10.1146/annurev.cellbio.15.1.435 [DOI] [PubMed] [Google Scholar]
- Diez-Silva M., Dao M., Han J., Lim C.T., Suresh S. 2010. Shape and biomechanical characteristics of human red blood cells in health and disease. MRS Bull. 35:382–388. 10.1557/mrs2010.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson T.D., Noureddine M.A., Reynolds P.J., Bradford W., Duronio R.J. 2004. Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases. Mol. Biol. Cell. 15:4892–4903. 10.1091/mbc.E04-03-0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Hickson G.R., Foley E., O’Farrell P.H. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14:1685–1693. 10.1016/j.cub.2004.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Perrimon N. 2006. High-throughput RNAi screening in cultured cells: a user’s guide. Nat. Rev. Genet. 7:373–384. 10.1038/nrg1836 [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., et al. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 3:777–779. 10.1038/nmeth1006-777 [DOI] [PubMed] [Google Scholar]
- Eggert U.S., Kiger A.A., Richter C., Perlman Z.E., Perrimon N., Mitchison T.J., Field C.M. 2004. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2:e379 10.1371/journal.pbio.0020379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even Y., Durieux S., Escande M.L., Lozano J.C., Peaucellier G., Weil D., Genevière A.M. 2006. CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo. J. Cell. Biochem. 99:890–904. 10.1002/jcb.20986 [DOI] [PubMed] [Google Scholar]
- Fabian L., Wei H.C., Rollins J., Noguchi T., Blankenship J.T., Bellamkonda K., Polevoy G., Gervais L., Guichet A., Fuller M.T., Brill J.A. 2010. Phosphatidylinositol 4,5-bisphosphate directs spermatid cell polarity and exocyst localization in Drosophila. Mol. Biol. Cell. 21:1546–1555. 10.1091/mbc.E09-07-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D., Pagano M. 2008. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer. 8:438–449. 10.1038/nrc2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs F., Pau G., Kranz D., Sklyar O., Budjan C., Steinbrink S., Horn T., Pedal A., Huber W., Boutros M. 2010. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol. Syst. Biol. 6:370 10.1038/msb.2010.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J.J., Lomakina M.E., Bouslama-Oueghlani L., Derivery E., Beilinson H., Faigle W., Loew D., Louvard D., Echard A., Alexandrova A.Y., et al. 2011. Clathrin is required for Scar/Wave-mediated lamellipodium formation. J. Cell Sci. In press. [DOI] [PubMed] [Google Scholar]
- Gautreau A., Ho H.Y., Li J., Steen H., Gygi S.P., Kirschner M.W. 2004. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA. 101:4379–4383. 10.1073/pnas.0400628101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Eck M.J. 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76:593–627. 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- Han Y., Eppinger E., Schuster I.G., Weigand L.U., Liang X., Kremmer E., Peschel C., Krackhardt A.M. 2009. Formin-like 1 (FMNL1) is regulated by N-terminal myristoylation and induces polarized membrane blebbing. J. Biol. Chem. 284:33409–33417. 10.1074/jbc.M109.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S., Shelton C., Cartwright H.N., Katzfey E., Janzen E., Abmayr S.M. 2011. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 138:1551–1562. 10.1242/dev.057653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M.S., Tsai P.I., Chien C.T. 2006. F-box proteins: the key to protein degradation. J. Biomed. Sci. 13:181–191. 10.1007/s11373-005-9058-2 [DOI] [PubMed] [Google Scholar]
- Hogan C., Dupré-Crochet S., Norman M., Kajita M., Zimmermann C., Pelling A.E., Piddini E., Baena-López L.A., Vincent J.P., Itoh Y., et al. 2009. Characterization of the interface between normal and transformed epithelial cells. Nat. Cell Biol. 11:460–467. 10.1038/ncb1853 [DOI] [PubMed] [Google Scholar]
- Huang J., Chen J. 2008. VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene. 27:4056–4064. 10.1038/onc.2008.44 [DOI] [PubMed] [Google Scholar]
- Igal R.A. 2010. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 31:1509–1515. 10.1093/carcin/bgq131 [DOI] [PubMed] [Google Scholar]
- Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L.B., Steffen A., Stradal T.E., Di Fiore P.P., Carlier M.F., Scita G. 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6:319–327. 10.1038/ncb1105 [DOI] [PubMed] [Google Scholar]
- Jaehning J.A. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta. 1799:379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani K., Schöck F. 2007. Zasp is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 179:1583–1597. 10.1083/jcb.200707045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Struhl G. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 391:493–496. 10.1038/35154 [DOI] [PubMed] [Google Scholar]
- Katoh M., Katoh M. 2003. Identification and characterization of human FMNL1, FMNL2 and FMNL3 genes in silico. Int. J. Oncol. 22:1161–1168. [PubMed] [Google Scholar]
- Kiger A.A., Baum B., Jones S., Jones M.R., Coulson A., Echeverri C., Perrimon N. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2:27 10.1186/1475-4924-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., Baum B. 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13:1867–1875. 10.1016/j.cub.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Lecuit T., Lenne P.F. 2007. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8:633–644. 10.1038/nrm2222 [DOI] [PubMed] [Google Scholar]
- Li W., You L., Cooper J., Schiavon G., Pepe-Caprio A., Zhou L., Ishii R., Giovannini M., Hanemann C.O., Long S.B., et al. 2010. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 140:477–490. 10.1016/j.cell.2010.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Rohn J.L., Picone R., Kunda P., Baum B. 2010. Tao-1 is a negative regulator of microtubule plus-end growth. J. Cell Sci. 123:2708–2716. 10.1242/jcs.068726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqués F., Moreau J.L., Peaucellier G., Lozano J.C., Schatt P., Picard A., Callebaut I., Perret E., Genevière A.M. 2000. A new subfamily of high molecular mass CDC2-related kinases with PITAI/VRE motifs. Biochem. Biophys. Res. Commun. 279:832–837. 10.1006/bbrc.2000.4042 [DOI] [PubMed] [Google Scholar]
- Masciadri B., Areces L.B., Carpinelli P., Foiani M., Draetta G., Fiore F. 2004. Characterization of the BUD31 gene of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 320:1342–1350. 10.1016/j.bbrc.2004.05.228 [DOI] [PubMed] [Google Scholar]
- Mason F.M., Heimsath E.G., Higgs H.N., Soderling S.H. 2011. Bi-modal regulation of a formin by srGAP2. J. Biol. Chem. 286:6577–6586. 10.1074/jbc.M110.190397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Carrero G., Andrin C., de Vries G., Hendzel M.J. 2006. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J. Cell Biol. 172:541–552. 10.1083/jcb.200507101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersich A.T., Miller M.R., Chkourko H., Blystone S.D. 2010. The formin FRL1 (FMNL1) is an essential component of macrophage podosomes. Cytoskeleton (Hoboken). 67:573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Pasque V., Jullien J., Gurdon J.B. 2011. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 25:946–958. 10.1101/gad.615211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S., Bakal C., Perrimon N. 2010. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 79:37–64. 10.1146/annurev-biochem-060408-092949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet I., Barbosa-Morais N.L., Andrade J., Carmo-Fonseca M. 2006. Diversity of human U2AF splicing factors. FEBS J. 273:4807–4816. 10.1111/j.1742-4658.2006.05502.x [DOI] [PubMed] [Google Scholar]
- Morrison H., Sperka T., Manent J., Giovannini M., Ponta H., Herrlich P. 2007. Merlin/neurofibromatosis type 2 suppresses growth by inhibiting the activation of Ras and Rac. Cancer Res. 67:520–527. 10.1158/0008-5472.CAN-06-1608 [DOI] [PubMed] [Google Scholar]
- Noureddine M.A., Donaldson T.D., Thacker S.A., Duronio R.J. 2002. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev. Cell. 2:757–770. 10.1016/S1534-5807(02)00164-8 [DOI] [PubMed] [Google Scholar]
- Okada T., Lopez-Lago M., Giancotti F.G. 2005. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J. Cell Biol. 171:361–371. 10.1083/jcb.200503165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C.Y., Lin Y.F., Chen Y.J., Chien C.T. 2002. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16:2403–2414. 10.1101/gad.1011402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P., Jonsson A., Nashchekin D., Karlsson C., Bergman T., Guialis A., Daneholt B. 2002. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 30:1725–1734. 10.1093/nar/30.8.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto C.N., Gervais L., Overstreet E., Fischer J., Guichet A., Schweisguth F. 2008. Overexpression of partner of numb induces asymmetric distribution of the PI4P 5-Kinase Skittles in mitotic sensory organ precursor cells in Drosophila. PLoS ONE. 3:e3072 10.1371/journal.pone.0003072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., Mathey-Prevot B. 2007. Matter arising: off-targets and genome-scale RNAi screens in Drosophila. Fly (Austin). 1:1–5. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36:451–477. 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. 2009. Actin, a central player in cell shape and movement. Science. 326:1208–1212. 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson K., Serr M., Gepner J., Gibbons I., Hays T.S. 1994. A family of dynein genes in Drosophila melanogaster. Mol. Biol. Cell. 5:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P.J., Simms J.R., Duronio R.J. 2008. Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping. PLoS ONE. 3:e2918 10.1371/journal.pone.0002918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Wiedemann U., Stuurman N., Vale R.D. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088. 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn J.L., Baum B. 2010. Actin and cellular architecture at a glance. J. Cell Sci. 123:155–158. 10.1242/jcs.049759 [DOI] [PubMed] [Google Scholar]
- Rungger-Brändle E., Gabbiani G. 1983. The role of cytoskeletal and cytocontractile elements in pathologic processes. Am. J. Pathol. 110:361–392. [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Schönbauer C., Langer C.C., Dietzl G., Novatchkova M., Schernhuber K., Fellner M., Azaryan A., Radolf M., Stark A., et al. 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 464:287–291. 10.1038/nature08799 [DOI] [PubMed] [Google Scholar]
- Shaw R.J., Paez J.G., Curto M., Yaktine A., Pruitt W.M., Saotome I., O’Bryan J.P., Gupta V., Ratner N., Der C.J., et al. 2001. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev. Cell. 1:63–72. 10.1016/S1534-5807(01)00009-0 [DOI] [PubMed] [Google Scholar]
- Sherman L.S., Gutmann D.H. 2001. Merlin: hanging tumor suppression on the Rac. Trends Cell Biol. 11:442–444. [DOI] [PubMed] [Google Scholar]
- Shi X., Finkelstein A., Wolf A.J., Wade P.A., Burton Z.F., Jaehning J.A. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima T., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S. 2003. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 17:1605–1616. 10.1101/gad.1086803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Bursteinas B., Gao Q., Zvelebil M., Baum B. 2006. FLIGHT: database and tools for the integration and cross-correlation of large-scale RNAi phenotypic datasets. Nucleic Acids Res. 34(Database issue):D479–D483. 10.1093/nar/gkj038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Duchek P., Baum B. 2009. PDGF/VEGF signaling controls cell size in Drosophila. Genome Biol. 10:R20 10.1186/gb-2009-10-2-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D., Bursteinas B., Jain E., Gao Q., Baum B., Zvelebil M. 2010. The FLIGHT Drosophila RNAi database: 2010 update. Fly (Austin). 4:344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarp K.P., Vartiainen M.K. 2010. Actin on DNA-an ancient and dynamic relationship. Cytoskeleton (Hoboken). 67:487–495. [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E., et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- Spencer V.A., Costes S., Inman J.L., Xu R., Chen J., Hendzel M.J., Bissell M.J. 2011. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J. Cell Sci. 124:123–132. 10.1242/jcs.073197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Ahringer J. 2010. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 141:757–774. 10.1016/j.cell.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Stüven T., Hartmann E., Görlich D. 2003. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 22:5928–5940. 10.1093/emboj/cdg565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S., Bradke F. 2009. Neuronal polarity. Cold Spring Harb. Perspect. Biol. 1:a001644 10.1101/cshperspect.a001644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M.K. 2008. Nuclear actin dynamics—from form to function. FEBS Lett. 582:2033–2040. 10.1016/j.febslet.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Wang P., Bowl M.R., Bender S., Peng J., Farber L., Chen J., Ali A., Zhang Z., Alberts A.S., Thakker R.V., et al. 2008. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol. Cell. Biol. 28:2930–2940. 10.1128/MCB.00654-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch W.J., Suhan J.P. 1985. Morphological study of the mammalian stress response: characterization of changes in cytoplasmic organelles, cytoskeleton, and nucleoli, and appearance of intranuclear actin filaments in rat fibroblasts after heat-shock treatment. J. Cell Biol. 101:1198–1211. 10.1083/jcb.101.4.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G.H., Beeser A., Chernoff J., Testa J.R. 2002. p21-activated kinase links Rac/Cdc42 signaling to merlin. J. Biol. Chem. 277:883–886. 10.1074/jbc.C100553200 [DOI] [PubMed] [Google Scholar]
- Xu W., Wang P., Petri B., Zhang Y., Tang W., Sun L., Kress H., Mann T., Shi Y., Kubes P., Wu D. 2010. Integrin-induced PIP5K1C kinase polarization regulates neutrophil polarization, directionality, and in vivo infiltration. Immunity. 33:340–350. 10.1016/j.immuni.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa S., Lee J.S., Ishimoto A. 1998. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J. Biol. Chem. 273:32353–32359. 10.1074/jbc.273.48.32353 [DOI] [PubMed] [Google Scholar]
- Ye J., Zhao J., Hoffmann-Rohrer U., Grummt I. 2008. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 22:322–330. 10.1101/gad.455908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.L., Janmey P.A. 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65:761–789. 10.1146/annurev.physiol.65.092101.142517 [DOI] [PubMed] [Google Scholar]