Abstract
The retinoblastoma suppressor pRB belongs to the family of so-called pocket proteins, which also includes p107 and p130. These proteins may functionally overlap in cell cycle control and tumor suppression. We have generated an isogenic set of embryonic stem (ES) cell lines carrying single or compound loss-of-function mutations in the Rb gene family, including a cell line completely devoid of all three pocket proteins. None of the knockout combinations affected the growth characteristics of ES cells; however, concomitant ablation of all three pocket proteins strongly impaired their differentiation capacity. For the generated genotypes, primary mouse embryonic fibroblasts (MEFs) also were obtained. While inactivation of Rb alone did not alleviate the senescence response of MEFs, pRB/p107-deficient MEFs, after having adapted to in vitro culturing, continued to proliferate at modest rate. Additional ablation of p130 rendered MEFs completely insensitive to senescence-inducing signals and strongly increased their proliferation rate. Although triple-knockout MEFs retained anchorage dependence, they lacked proper G1 control and showed increased cell turnover under growth-inhibiting conditions.
Keywords: Rb, p107, p130, pocket proteins, cell cycle control, cell turnover
Loss of function of the retinoblastoma suppressor gene, RB, is a common event in the development of many tumor types in human, including hereditary retinoblastoma and sporadic lung, breast, and bladder carcinomas (Harbour et al. 1988; Lee et al. 1988; Horowitz et al. 1990). Numerous biochemical studies have placed pRB at the heart of the cellular machinery that controls passage from G1 into S phase of the cell cycle (Weinberg 1995). pRB can exist in hyper- and hypophosphorylated forms, the latter binding to and inhibiting a class of transcription factors, collectively known as E2 factors (E2F), whose activity is required for the transcription of genes that are essential for DNA synthesis (Bernards 1997; Dyson 1998; Nevins 1998). Phosphorylation of pRB by cyclin-dependent kinases (CDK) leads to dissociation of the pRB/E2F complex, releasing the transcriptional activity of E2Fs. CDK activity is positively regulated by heterodimerization with cyclins but also negatively regulated by cyclin-dependent-kinase inhibitors (CKIs). pRB function can thus be described as a cell cycle switch: cyclin D1-stimulated kinase activity of CDK4 turns pRB -OFF- through phosphorylation; this releases E2F activity and promotes G1-S transition; the pRB -ON- state is favored by inhibition of CDK4 activity by the CKI p16INK4A, which promotes cell cycle arrest. The importance of this G1-S control pathway is underscored by the finding that in the majority of human cancers, genetic alterations have been found that favor the pRB -OFF- state. These include genetic inactivation of RB in retinoblastoma and many other cancers; cyclin D1 overexpression in carcinomas of the breast, oesophagus, and head and neck; CDK4 amplification or mutational activation in melanomas; and abrogation of p16INK4A activity in melanomas, pancreatic, and bladder carcinomas (Hall and Peters 1996; Sherr 1996).
Although the G1-S control pathway described above provided a framework for understanding the tumor-suppressor function of pRB, cell cycle control in mammalian cells is a far more complex circuitry of different, at least partially overlapping and interacting pathways that are regulated by positive and negative feedback mechanisms and can be modulated at numerous levels. Thus, pRB shares its E2F-regulating activity with two homologs, p107 and p130 (for review, see Mulligan and Jacks 1998; Nevins 1998; Lipinski and Jacks 1999). The three proteins share extensive structural homology, most notably located in two regions, A and B, which together form the so-called pocket domain responsible for binding to E2Fs. These domains also form the binding site for many viral oncoproteins, including adenovirus E1A, simian virus 40 large-T antigen, and human papillomavirus E7, all of which abrogate interactions with E2F (DeCaprio et al. 1988; Whyte et al. 1988; Dyson et al. 1989). Also outside the A/B domain, p107 and p130 share extensive homology, whereas corresponding pRB regions and even the pRB spacer region that separates the A and B subdomains appear to be unique. The closer resemblance between p107 and p130 is underscored by a number of observations suggesting that the two proteins can functionally substitute for each other. p107 and p130 almost exclusively interact with E2F4,5, although they do so at consecutive stages of the cell cycle: Complexes of p130 and E2F4,5 predominate in quiescent or differentiated cells; p107/E2F4,5 complexes are only detected in cycling cells. In contrast, pRb has strongest affinity for E2F1,2,3, although it can also bind E2F4 (for review, see Dyson 1998). Rather than merely sequestering E2Fs, pocket-protein/E2F complexes appear to be actively involved in transcriptional repression (Zhang et al. 1999). While no differences in expression of E2F target genes were found on ablation of p107 or p130 alone, combined loss in p107−/−p130−/− primary mouse embryonic fibroblasts (MEFs) resulted in derepression of a number of genes, including B-myb, Cdc2, E2f1, Ts, Rrm2, and Cyclin A2. Expression of these genes was normal in Rb−/− MEFs, but in these cells, Cyclin E and p107 were slightly up-regulated (Herrera et al. 1996; Hurford et al. 1997). Thus, p107 and p130 appeared to be redundant in regulating a specific set of E2F-responsive genes that is not subject to pRb/E2F-mediated repression. However, overexpression of each of the pocket proteins in vitro can cause cell cycle arrest, an activity that can be relieved through phosphorylation by cyclinD/CDK4,6. Full inactivation of pRB also requires cyclinE/CDK2, which adds an additional level of regulation to the pRB pathway. Thus, the three pocket proteins may to some extent be redundant in blocking progression of cells through the cell cycle. In resting T lymphocytes, the main pocket protein/E2F complex is p130/E2F4. However, in the absence of p130, E2F4 was mainly found complexed to p107 or, when this protein was also absent, to pRb (Mulligan et al. 1998). Apparently, in this system pocket proteins can compensate for each other in regulating E2F4 activity, providing a rationale for their shared activities in blocking cell cycle progression.
Specific and redundant functions of pocket proteins also became manifest in mice carrying single or compound disruptions in the Rb gene family (Mulligan and Jacks 1998). Complete inactivation of Rb appeared not to be compatible with normal development (Clarke et al. 1992; Jacks et al. 1992; Lee et al. 1992). Rb−/− embryos died at midgestation, presenting with extensive apoptosis in the central nervous system and eye lens and defects in erythropoiesis and myogenesis. These defects could at least partially be attributed to increased free E2F1 activity, inducing p53-dependent apoptosis (Morgenbesser et al. 1994; Tsai et al. 1998). In contrast, genetic ablation of either p107 or p130 did not show an overt phenotype (Cobrinik et al. 1996; Lee et al. 1996). However, combination of p107 and p130 loss-of-function mutations caused excessive chondrocyte proliferation leading to malformation of the long bones and ribs. Respiratory problems associated with this defect led to rapid neonatal death. Remarkably, a dramatic phenotype was observed when loss-of-function mutations in either p107 or p130 were crossed into the BALB/c background (LeCouter et al. 1998a,b): p107 deficiency caused impaired growth of newborns, while embryonic fibroblasts displayed a twofold reduction in doubling time; p130-deficient embryos died at midgestation and showed marked apoptosis in the central nervous system. BALB/c mice were found to carry a point mutation in the p16Ink4a gene (Zhang et al. 1998). These observations suggest that p107 and p130 can functionally substitute for each other, but only in the presence of a pathway that requires wild-type p16 and most likely involves pRB. Conversely, p107 appeared to attenuate the consequences of pRB deficiency in development as p107/pRB-deficient embryos showed a reduced life span with respect to embryos lacking only pRB (Lee et al. 1996). p107 activity also appeared to modulate the tumor suppressor function of pRB. In contrast to humans, where hemizygosity for the retinoblastoma suppressor gene RB highly predisposes to retinoblastoma, Rb+/− mice developed tumors originating from the intermediate lobe of the pituitary gland while the retina remained unaffected (Hu et al. 1994; Robanus- Maandag et al. 1994). No predisposition to tumorigenesis was found to be associated with p107 or p130 deficiency (Cobrinik et al. 1996; Lee et al. 1996). However, concomitant inactivation of Rb and p107 in chimeric mice gave rise to retinoblastoma development, indicating that p107 can act as a tumor suppressor in the context of pRb deficiency (Robanus-Maandag et al. 1998).
Primary mouse embryonic fibroblasts represent a rather well-defined cell type that has been widely used to identify the consequences of gene ablation for cell cycle control. Although pRB deficiency or combined p107/p130 deficiency caused a slight acceleration of S-phase entry on restimulation of serum-starved cells, the overall growth characteristics of single- or double-knockout MEFs did not deviate from those of wild-type MEFs (Cobrinik et al. 1996; Herrera et al. 1996). In particular, the single- and double-knockout genotypes tested thus far did not affect the growth arrest response of cells to a number of growth-inhibiting conditions such as serum deprivation, contact inhibition, and prolonged passaging. This again suggests functional redundacy within the pocket-protein family. To test this directly, we have now generated MEFs that are completely devoid of all three pocket proteins. Our results identify the pocket proteins as critical mediators of senescence and of the response of cells to a variety of growth-restricting conditions.
Results
Inactivation of Rb, p107, and p130 in ES cells
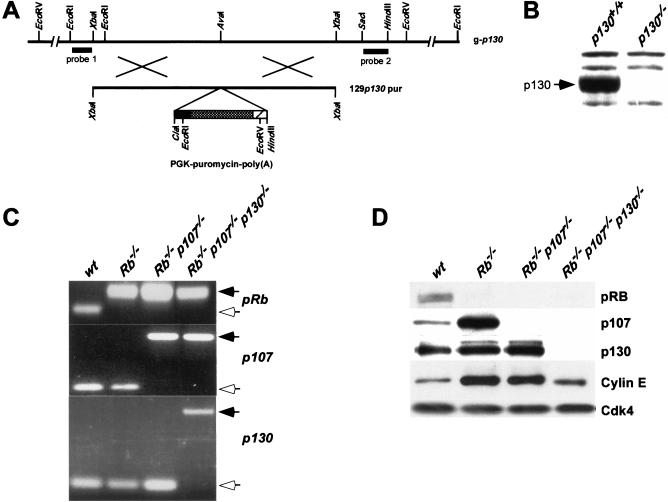
An isogenic set of wild-type (wt), Rb−/−, Rb−/−p107−/−, and Rb−/−p107−/−p130−/− embryonic stem (ES) cell lines was generated by consecutive rounds of homologous recombination. The generation of Rb−/− and Rb−/−p107−/− ES cell lines has been described before (Te Riele et al. 1992; Robanus-Maandag et al. 1998). To inactivate p130, we generated a 129p130-pur targeting vector (Fig. 1A). Rb−/−p107−/−p130−/− cells were obtained in two ways: First, p130 was inactivated in Rb−/−p107−/− ES cells and Rb−/−p107−/−p130+/− cells were cultured at high puromycin concentrations to select for inactivation of the remaining p130 wild-type allele; second, both p130 alleles were inactivated in p107−/− ES cells and then Rb was inactivated using the targeting vectors 129Rb-hyg and 129Rb-his. Figure 1B shows that p130 protein was not detectable in brain tissue obtained from a p130−/− mouse carrying the same disruption in p130, indicating that the p130− allele can be considered a null allele. Thus, Rb−/− p107−/−p130−/− cells are completely devoid of pocket proteins and will hereafter be designated as triple-knockout (TKO) cells. Each genotype was derived in duplicate by independent rounds of gene targeting starting from wild-type cells. None of the generated genotypes in ES cells resulted in an obvious phenotype, indicating that the pocket proteins pRB, p107, and p130 are dispensable for normal ES cell growth.
Figure 1.
Generation of Rb−/−p107−/−p130−/− ES cells and MEFs. (A) Restriction map of the wild-type p130 allele around codon 405 at the AvaI site and 129p130-pur DNA targeting construct. Probes 1 and 2 detect modifications at p130. (B) Western blot analysis of p130 in lysates prepared from p130+/+ and p130−/− brain tissue. (C) PCR analysis of Rb, p107 and p130 of DNA isolated from passage 1 wt, Rb−/−, Rb−/−p107−/− and TKO MEFs. PCR products resulting from wild-type and knockout alleles of each gene are indicated by open and solid arrows, respectively. (D) Western blot analysis of pRB, p107, p130, and Cyclin E in lysates prepared from indicated MEFs. Cdk4 served as a loading control.
Limited differentiation capacity of Rb−/−p107−/−p130−/− ES cells in teratocarcinomas
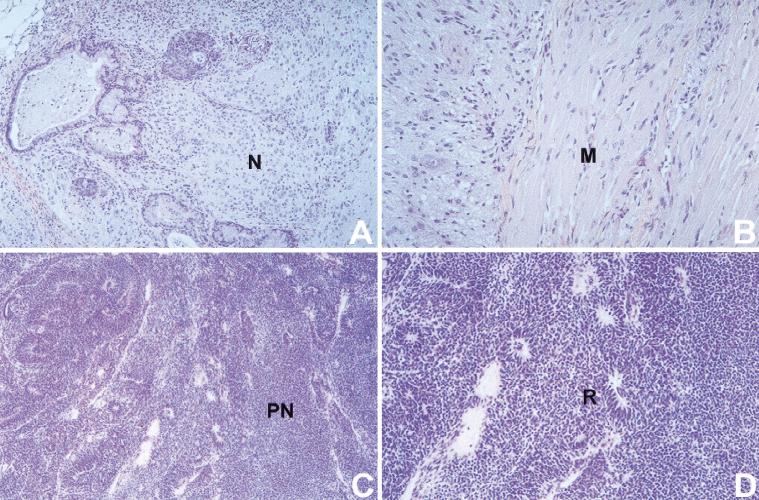
To study whether pocket proteins become essential during differentiation of ES cells, we injected wt, Rb−/−, Rb−/−p107−/−, and Rb−/−p107−/−p130−/− ES cells subcutaneously into nude mice. In this environment, ES cells form teratocarcinoma-like tumors in which a variety of differentiated cell types can be identified, including neuronal and muscle cells. Three weeks after injection the teratocarcinomas were removed and histologically examined. Tumors derived from wt, Rb−/−, and Rb−/−p107−/− ES cells were highly similar with respect to their strongly heterogeneous appearance with extensive neuronal differentiation (Fig. 2A) and striated muscle (Fig. 2B). Extensive neuronal differentiation was confirmed by staining with the neuron-specific antibodies anti-glial fibrillary acidic protein (GFAP) and anti-neuron specific enolase (NSE; not shown). In contrast, TKO ES cells formed a rather homogeneous tumor mass, without any muscle cell differentiation (Fig. 2C). Rosette-like structures and positive staining with a p75LNGFR antibody (Zorick and Lemke 1996) indicated that TKO tumors predominantly consisted of neuroblast-like cells (Fig. 2D; not shown). Furthermore, immunohistochemistry using antibodies directed against the proliferation marker Ki-67 showed that the fraction of proliferating cells in TKO tumors was 15-fold higher than in teratocarcinomas of the other genotypes. These data show that in this system, ablation of the Rb gene family abrogates neuronal and muscular differentiation.
Figure 2.
Limited differentiation in TKO teratocarcinomas. Histological sections of teratocarcinomas were stained with hematoxylin-eosin. (A,B) Advanced neuronal (N) and muscle (M) differentiation in teratocarcinomas generated with wt ES cells. Similar results were obtained with Rb−/− and Rb−/−p107−/− ES cells. (C,D) Cell-dense TKO teratocarcinomas lack muscle differentiation and exist of primitive, relatively undifferentiated neuronal cells (PN) with rosette-like structures (R). Magnification (A,C) objective 10×, (B,D) objective 20×.
Pocket-protein-deficient MEFs obtained from chimeric embryos
Primary MEFs have been instrumental in studying the consequences of gene ablation for cell cycle control. We have derived an isogenic set of MEFs from chimeric embryos generated by blastocyst injections of mutant ES cells. In these experiments, we used ES cells that also contained a neomycin resistance gene under the control of the Rosa26 promoter (Friedrich and Soriano 1991; J.-H. Dannenberg and H. te Riele, in prep.). Brief culturing in the presence of G418 provided a uniform method to obtain MEF preparations that were exclusively ES-cell derived. This was verified by genotyping these cells by PCR for the presence of the wild-type and knockout alleles of Rb, p107, and p130 (Fig. 1C). The presence or absence of pocket proteins was further confirmed by Western blot analyses (Fig. 1D). We were able to generate MEF cultures with the following genotypes: wt, Rb−/−, Rb−/− p107−/−, and somewhat surprisingly, Rb−/−p107−/−p130−/−.
Pocket proteins are essential for G1 control
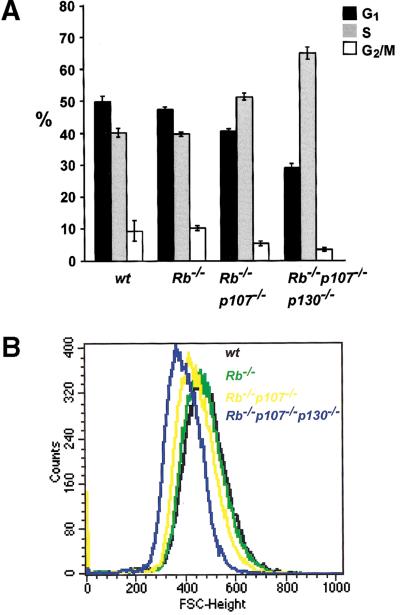
Proliferating MEF cultures of early passage were labeled with the thymidine analogue BrdU and analyzed by FACS. Figure 3A shows that concomitant inactivation of Rb, p107, and p130 strongly reduced the percentage of cells in G1. This decrease in G1 fraction was accompanied by an increase of the S-phase population, with TKO MEF cultures having up to 65% S-phase cells. In Rb−/− MEFs, only a minor decrease in G1 population was observed, whereas disruption of both Rb and p107 gave an intermediate phenotype. Together with the strongly reduced population-doubling time of TKO cells (see below; Fig. 4), these observations suggest that successive ablation of pRb, p107, and p130 reduces the length of the G1 phase of the cell cycle.
Figure 3.
Deregulated G1-control in TKO MEFs. (A) Percentages of G1, S, and G2/M phases of the cell cycle in proliferating wild-type (wt) and pocket-protein-deficient MEFs. Results are indicated as average values of three independent experiments. Error bars indicate standard deviations of the mean. (B) FSC-H histogram showing cell-size analysis of MEFs deficient for Rb, p107, and p130 compared with wt counterparts. The forward scatter, FSC-H, is indicative for the cell diameter, and a shift to the left, relative to the wt MEFs, represents smaller cells.
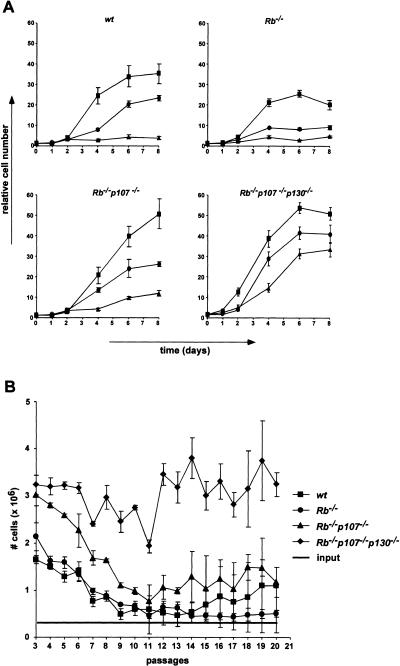
Figure 4.
Growth characteristics of pocket-protein-deficient MEFs. (A) Growth curves of wt, Rb−/−, Rb−/−p107−/−, and TKO MEFs at passages 3 (squares), 5 (circles), and 7 (triangles). The figure shows a representative of three independent experiments, each performed in triplicate. (B) Cell proliferation on a 3T9-based protocol of indicated MEFs for 20 passages. At 3.5-d intervals, the total numbers of cells of three independent cultures of each genotype were determined before redilution of the cells to 3 × 105 per six-well plate (input). The experiment was performed twice. Error bars indicate standard deviations of the mean.
G1 shortening has previously been correlated to a decrease in cell size in several other experimental settings, such as overexpression of cyclin E (Ohtsubo and Roberts 1993) and concomitant inactivation of Rb and p21 (Brugarolas et al. 1998). We therefore determined the size of wild-type and mutant MEFs. Proliferating cultures of early passage were fixed, stained with propidiumiodide (PI), and analyzed by FACS (Fig. 3B). Relative cell sizes closely correlated to the proportion of cells in G1: TKO<Rb−/−p107−/−<Rb−/− = wt. This again indicates that ablation of pocket proteins reduces the length of G1.
It is of interest to note that in cyclin-E-overexpressing or Rb/p21-deficient cells, G1 shortening has been correlated with increased cyclin E levels or elevated CDK2 kinase activity. However, whereas Rb−/− and Rb−/− p107−/− MEFs showed increased levels of cyclin E, TKO MEFs expressed cyclin E at wild-type levels (Fig. 1D). Thus, shortening of G1 is not obligatorily accompanied by elevated levels of cyclin E.
Rb−/−p107−/−p130−/− MEFs are immortal
Wild-type MEFs have a limited growth capacity on prolonged passaging: Their growth rate gradually decreases until they finally completely arrest with an enlarged, flattened morphologoly and 2n DNA content, a state that is referred to as replicative senescence (Todaro and Green 1963; Campisi 1997). The involvement of the Rb gene family in senescence was assessed in two ways. First, we determined the growth rates of wt, Rb−/−, Rb−/− p107−/−, and TKO MEFs at increasing passages. Figure 4A shows that the growth rates of wt and Rb−/− MEFs gradually decreased on prolonged passaging and that these cells entered senescence around passage seven. Rb−/− p107−/− MEFs showed a fivefold reduction in growth rate; however, they had not completely ceased proliferating at passage seven (Fig. 4A) and clearly had not adopted a senescence-like morphology (not shown). TKO MEFs retained the strongest proliferative capacity at later passages, their growth rate being reduced less than twofold. Second, the long-term proliferative capacity of MEFs was assessed following a 3T9-based protocol (Fig. 4B). Wild-type, Rb−/−, and Rb−/−p107−/− MEFs showed a decline in proliferation rate up to passage 11. While the Rb−/− cells completely stopped growing in this experiment, wt cells ultimately escaped from senescence as a result of a genetic alteration (see also below). Rb−/− p107−/− MEFs, although initially subject to growth inhibition, did not enter senescence but established a constant proliferation rate of one to two population doublings every 3 d. Also, TKO cells initially showed some decline in growth rate. However, this decline was marginal, and the cells continued to proliferate at a rate that was about threefold higher than the double knockouts. These results demonstrate that inactivation of Rb alone does not alleviate the senescence response of MEFs; concomitant inactivation of Rb and p107, although not completely alleviating growth inhibition on in vitro culturing, confers a continuing proliferation capacity, albeit at a modest rate, whereas ablation of all Rb-gene family members renders cells virtually insensitive to growth inhibition on in vitro culturing and strongly increases their proliferation rate.
Immortal TKO MEFs retain an intact p19ARF/p53 pathway
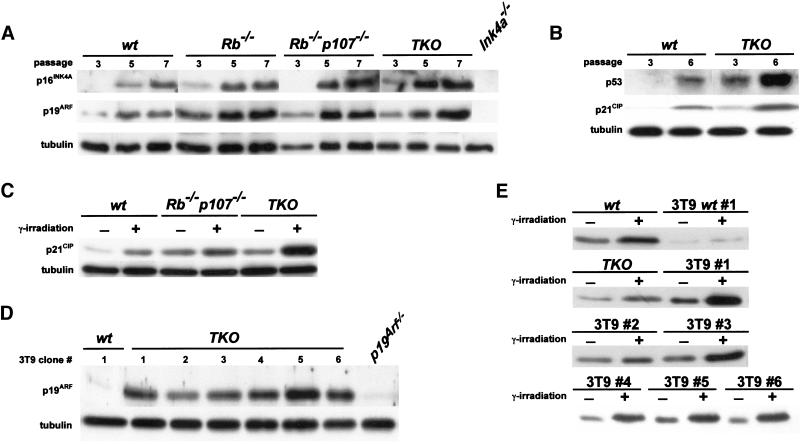
It has previously been hypothesized that replicative senescence results from the p53-dependent growth inhibitory effect of p19ARF, which gradually accumulates on prolonged passaging (Quelle et al. 1995; Zindy et al. 1997). Thus, inactivation of Ink4a, p19Arf, or p53 has been shown to result in immortalization of MEFs (Serrano et al. 1996, 1997; Kamijo et al. 1997). Furthermore, spontaneous immortalization of MEFs was accompanied by either mutation of p53 (Harvey and Levine 1991; Rittling and Denhardt 1992) or deletion of the Ink4a locus (Kamb et al. 1994; Nobori et al. 1994; Zindy et al. 1998), ablating both p16INK4A and p19ARF (Kamijo et al. 1997). We therefore tested the functionality of this pathway in immortal TKO MEFs. Figure 5A shows that p16Ink4a and p19Arf were properly expressed in all genotypes including TKO and tended to increase on prolonged passaging. Also, p53 function was induced as evidenced by increased protein levels and accumulation of p21CIP (Fig. 5B). Furthermore, when TKO MEFs were γ irradiated, they showed a strong induction of p21Cip, as in wt and Rb−/−p107−/− MEFs, again indicating that the cells contained functional p53 (Fig. 5C). These observations indicate that complete loss of function of the Rb gene family renders MEFs resistant toward p19ARF/p53-induced senescence and predicts that TKO MEFs at passage numbers where wild-type MEFs spontaneously escape from senescence can grow without mutations in p53 or deletions of the Ink4a locus. Indeed, we found that in six independent clones that were grown on a 3T9-based protocol for >20 passages, p19ARF was still expressed, in contrast to a wt 3T9 clone that had lost p19ARF expression, probably because of a deletion of the Ink4a locus (Fig. 5D). Finally, all six TKO 3T9 clones showed increased p21Cip expression on γ irradiation, as in wt and TKO MEFs of early passage, indicating that p53 also remained functional in these clones (Fig. 5E).
Figure 5.
Intact p19ARF/p53 pathway in immortal TKO MEFs. (A) Indicated MEFs were passaged according to a 3T9 protocol, and cell pellets were collected at passages 3, 5, and 7. Cell lysates were analyzed for p16INK4A, p19ARF and tubulin levels by immunoblotting. Ink4a−/− MEFs served as a negative control. (B) Expression of p53 and p21CIP in wt and TKO MEFs at passages 3 and 6. (C) Expression analysis of p21CIP in control and γ-irradiated early passage wt, Rb−/−p107−/− and TKO MEFs showing induction of p21Cip in all genotypes 24 h after irradiation. (D) Expression analysis of p19ARF in six independent TKO 3T9 clones (1–6). All clones expressed p19Arf, whereas a wild-type 3T9 clone (3T9wt, 1) had lost expression of this protein. p19Arf−/− MEFs served as a negative control. (E) Induction of p21Cip in γ-irradiated wt (1) and TKO (1–6) 3T9 MEF cell lines compared to γ-irradiated early passage wt and TKO MEFs.
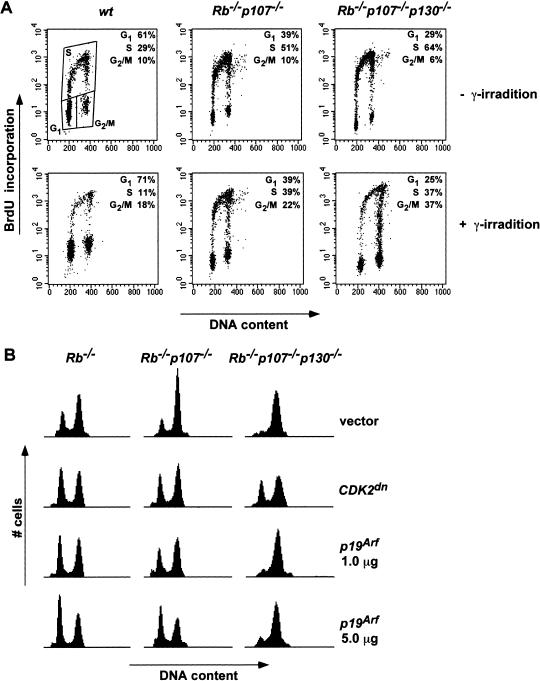
p53 can induce both a pRB-dependent G1 and a pRB-independent G2/M arrest on γ irradiation (Agarwal et al. 1995; Brugarolas et al. 1995; Harrington et al. 1998). Figure 6A shows that a γ-irradiation-induced G1 arrest was indeed abrogated in Rb−/−p107−/− and TKO MEFs, while the G2/M checkpoint was still intact. This again indicates that in Rb−/−p107−/− and TKO cells, p53 remained functional and that pocket proteins are dispensable for radiation-induced G2/M arrest.
Figure 6.
Defective G1 checkpoint in TKO MEFs on irradiation and ectopic p19Arf expression. (A) γ-irradiation induced a G2/M arrest in Rb−/−p107−/− and TKO MEFs. Percentages of G1, S, and G2/M phases of the cell cycle in nonirradiated and irradiated (5.5 Gy) MEFs were measured by FITC-PI FACS analysis and plotted as FL3-A (X-axis)–FL1-H (Y-axis) dot plots. (B) Ectopic expression of p19Arf induces a pocket-protein-dependent G1 arrest. FACS analysis of GFP-positive Rb−/−, Rb−/−p107−/− and TKO MEFs expressing either vector plasmid, a dominant-negative variant of CDK2, or p19Arf (1.0 or 5.0 μg) together with a histone H2B-GFP expression construct in a 10 : 1 ratio. Representative results of three independent experiments are shown.
Finally, we asked whether MEFs deficient for the Rb-gene family are still sensitive to G1 arrest following ectopic expression of p19ARF. Cells were electroporated and analyzed by FACS. First, we found that introduction of a CDK2 dominant-negative protein increased the G1 population to the same extent in Rb−/−, Rb−/−p107−/−, and TKO MEFs (Fig. 6B). Ectopic expression of p19ARF induced a similar G1 arrest in cells lacking pRB or both pRB and p107. In contrast, in TKO MEFs, G1 arrest was only marginally induced by p19ARF compared to the effect of CDK2dn (Fig. 6B). Together with the observation that ablation of the Rb gene family causes immortalization despite normal levels of INK4A proteins and functional p53, this result indicates that pocket proteins are essential mediators of the senescence response to p19ARF.
Inactivation of pocket proteins leads to increased cell turnover under growth-inhibiting conditions
TKO cells are largely refractory to growth inhibition by p19ARF. We therefore examined the behavior of these cells under a number of other growth-inhibiting conditions.
Growth factor depletion
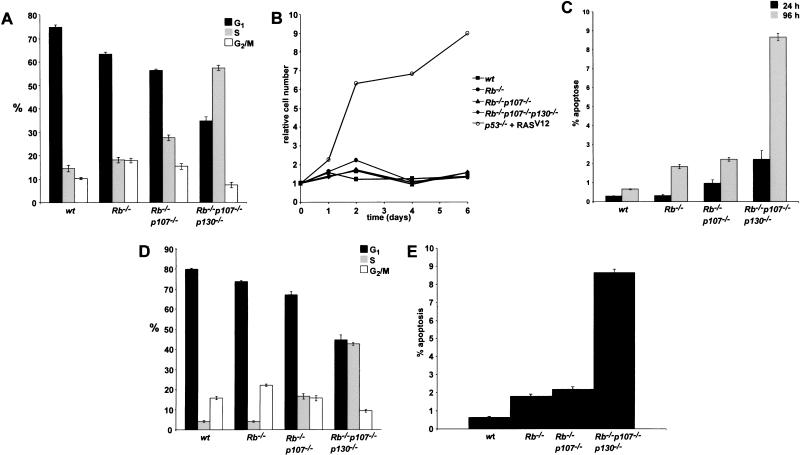
MEFs were plated at low density, supplied with 0.1% serum, and analyzed by BrdU-FACS. Wt and Rb−/− cells strongly arrested as evidenced by the accumulation of cells in G1 and a block in DNA synthesis (Fig. 7A). The response of Rb−/−p107−/− MEFs was less pronounced, although the level of DNA synthesis was still significantly decreased compared to 10% serum (see also Fig. 3A). In contrast, TKO MEFs continued to incorporate BrdU almost at the same rate as under high serum conditions. Suprisingly, under these conditions, no increase in cell number was observed on prolonged culturing (Fig. 7B). The appearance of floating cells in the medium at 24 h after the application of low serum suggested that cells became apoptotic. Indeed, PI-FACS analysis revealed a significant sub- G1 population in TKO MEFs at 24 h after low serum addition, increasing up to 10% of the population after 4 d (Fig. 7C). Again, Rb−/−p107−/− showed an intermediate phenotype. We did not observe an increased population of multinucleated MEFs, indicating that continuous BrdU incorporation did not result from endoreduplication (data not shown).
Figure 7.
Increased cell turnover in TKO MEFs under growth-inhibiting conditions. (A) Distribution of G1, S, and G2/M phases of indicated MEFs at 0.1% FCS. (B) Growth curves of indicated MEFs at 0.1% FCS. (C) Percentage of apoptotic cells in indicated MEF cultures at 0.1% FCS, 24 h (black), and 96 h (grey) after growth-factor depletion. Percentages were obtained by determing the sub-G1 population in a FL3A-log histogram. (D) Distribution of G1, S, and G2/M phases of indicated contact-inhibited MEFs. (E) Percentage apoptotic cells in contact-inhibited MEF cultures. Average values of three independent experiments are indicated in percentages. Error bars indicate standard deviations from the mean (A,C,D,E).
Contact inhibition
Another extracellular signal that provokes a G0/G1 arrest in normal fibroblasts is cell–cell contact at confluency (Nilausen and Green 1965). This phenomenon, known as contact inhibition, was shown to be accompanied by increased p27KIP1 and decreased cyclin D1 levels (Polyak et al. 1994; St. Croix et al. 1998). Furthermore, hypophosphorylated isoforms of pRb accumulated, suggesting that pRB is essential for G1 arrest. To study the role of pocket proteins in contact inhibition, MEFs were grown to confluency and cultured for an additional 4 d. DNA synthesis was measured by BrdU-FACS. Again, deletion of all pocket proteins revealed a severe defect in establishing a G0/G1 arrest (Fig. 7D). Instead, TKO MEFs continued to incorporate BrdU and to generate apoptotic cells (Fig. 7E).
Nonadherent conditions
Anchorage independance is a characteristic of most oncogenically transformed cell types and was attributed to constitutive cyclin E- (Fang et al. 1996) or A- (Guadagno et al. 1993) dependent CDK2 activity. In nonmalignant cells, growth arrest under nonadherent conditions was associated with loss of cyclin E-CDK2 activity (Fang et al. 1996) and underphosphorylation of pRB and p107 (Schulze et al. 1996). However, we found that TKO MEFs, although giving very tiny colonies containing up to 20 cells, were not capable of sustained proliferation in soft agar (Table 1). This indicates that ablation of pocket-protein activity is not sufficient for growth under nonadherent conditions and suggests that loss of CDK2 activity can confer a pocket-protein-independent growth arrest. This view is consistent with our observation that TKO cells can still be blocked by ablation of CDK2 activity (Fig. 6B). The anchorage dependence of TKO MEFs is consistent with their inability to grow under the skin of nude mice (not shown), demonstrating that these cells are not transformed.
Table 1.
Immortal TKO MEFs do not grow anchorage independently
| Genotype MEFs
|
Exp I
|
Exp II
|
Exp III
|
|---|---|---|---|
| wt | 0 | 0 | 0 |
| Rb−/− | 0 | 0 | 0 |
| RB−/−p107−/− | 0 | 0 | 0 |
| Rb−/−p107−/−p130−/− | 0a | 0 | 0 |
| p53−/− + RASV12 | 229 | 238 | 273 |
Number of foci of indicated early passage MEFs appearing after 2 wk in soft agar.
A small percentage of the MEFs developed into tiny colonies (up to 20 cells) but did not sustain growth.
In conclusion, complete ablation of the Rb gene family does not allow anchorage-independent growth; however, it severely compromises cell cycle arrest in response to growth-factor depletion and contact inhibition. Under these growth-inhibiting conditions, pocket-protein-deficient cells undergo increased cell turnover.
Discussion
We have generated an ES cell line carrying inactivating disruptions in all three retinoblastoma gene family members and found that this strongly limited their differentiating capacity. In MEFs, complete ablation of the pocket-protein family severely compromised G1 control. This was most strikingly manifested by the finding that triple-knockout MEFs were immortal and sustained continuous proliferation and apoptosis under growth-restricting conditions.
Differentiation
ES cells have a very high proliferation rate (doubling time of 7–8 h), and in a proliferating population, ∼70% of the cells were found in S phase (Savatier et al. 1994). Savatier et al. (1996) made the surprising observation that undifferentiated ES cells do not contain detectable cyclin D1/CDK4 activity and are insensitive to growth inhibition by p16INK4A. Furthermore, ES cells do not express p130 (LeCouter et al. 1996), whereas pRB and p107 could be readily detected (Robanus-Maandag et al. 1998). Apparently, in ES cells the antiproliferative effects of pRB and p107 are neutralized, possibly by the presence of an E1A-like activity (E1A-LA), as was postulated before (Murray et al. 1991). The dramatic increase in cyclin D1/CDK4 activity on induction of differentiation likely reflects the pRB/p107/p130 control pathways becoming active. Our observations are fully consistent with this view: Ablation of the pRB family did not affect the growth potential of ES cells but severely compromised their differentiating potential. Thus, pocket proteins are required for differentiation of ES cells. Moreover, as both pRB/p107-deficient ES cells (this work) and p130-deficient ES cells (not shown) showed normal muscular and neuronal differentiation in teratocarcinomas, p130 can compensate for the absence of pRB and p107.
Despite their limited differentiation capacity when placed under the skin of nude mice, TKO ES cells injected into blastocysts did not show unrestricted proliferaton but, instead, were capable of contributing to embryonic development at least until day 15 of gestation, allowing the derivation of TKO MEFs. This observation may suggest that differentiation into certain cell types, for example, fibroblasts, does not require pocket-protein function. More intriguing is the possibility that differentiation of TKO cells in chimeric embryos is directed and controlled by surrounding wild-type cells. In either case, during development, the cells have apparently acquired characteristics that, unlike their embryonic stem cell progenitors, prevent them from growing anchorage independently or under the skin of nude mice.
Immortalization
In both human and mouse primary fibroblasts, prolonged culturing generates a growth-inhibiting signal that ultimately leads to a state of replicative senescence reflected by an enlarged, flattened morphology and the absence of DNA synthesis (Hayflick and Moorhead 1961; Todaro and Green 1963; Campisi 1997). This type of growth arrest is accompanied by gradually increasing levels of the CDK2/4 inhibitors, p21CIP and p16INK4A, the cell cycle inhibitor p19ARF, and p53 (Lloyd et al. 1997; Palmero et al. 1997; Zindy et al. 1997, 1998). p16INK4A and p19ARF are encoded by one genetic locus, Ink4a, whereby p19ARF is expressed from an alternative reading frame (Quelle et al. 1995). Whereas p16INK4A was shown to act upstream of pRB to promote cell cycle arrest (Serrano et al. 1993), p19ARF can physically interact with p53 and/or MDM2, thereby antagonizing the function of MDM2 and stabilizing p53 (Kamijo et al. 1998; Pomerantz et al. 1998; Zhang et al. 1998). Spontaneous immortalization of MEFs is usually accompanied by either deletion of the Ink4a locus (Kamb et al. 1994; Nobori et al. 1994; Kamijo et al. 1997; Zindy et al. 1998) or loss of p53 function (Harvey and Levine 1991; Rittling and Denhardt 1992). Furthermore, Ink4a−/− and p53−/− MEFs are immortal. These observations suggest that induction of INK4A proteins and p53 is causally related to induction of senescence. However, mice and MEFs lacking only p19ARF showed a phenotype indistinguishable from that resulting from disruption of the Ink4a locus abrogating both p16INK4A and p19ARF. Although the consequences of ablation of p16INK4A alone are unknown, this suggests that the p19ARF/p53 rather than the p16INK4A/pRB pathway is responsible for inducing senescence.
We now show that MEFs deficient for pRB, p107, and p130 are immortal, cannot be arrested by ectopic p19ARF expression, and do not sustain deletions of the Ink4a locus or mutations in p53 on prolonged passaging. Also, pRB/p107-deficient MEFs can be considered immortal: Although they respond to supraphysiological levels of p19ARF by a G1 arrest, after having adapted to in vitro culturing, they sustained a moderate but constant proliferation rate on prolonged passaging. These observations indicate that both p19ARF-induced senescence and cell cycle arrest require pRB family members. It therefore remains plausible that both products of the Ink4a locus are implicated in senescence induction, p16INK4A not being sufficient but facilitating the activation of pocket proteins by p19ARF (Carnero et al. 2000). How p19ARF signals to the pRB family remains unclear. The immortal phenotype of p53−/− MEFs suggests this pathway to be p53 dependent. However, p19ARF-p53-induced senescence is not implemented via p21CIP-induced inhibition of cyclin E/CDK2, as p21−/− and p21−/−p27−/− MEFs still undergo senescence and/or are responsive to overexpression of p19ARF (Pantoja and Serrano 1999; Groth et al. 2000). Furthermore, our data show that TKO MEFs are not blocked in G1 by p19ARF overexpression, although they can still be blocked by inhibition of CDK2 activity on expression of a dominant-negatively acting CDK2 mutant. A picture thus emerges wherein p19ARF induces senescence via the pRB family but independent of p21CIP-mediated inhibition of cyclin E/CDK2 activity. Whether the link between p19ARF and the Rb gene family is p53 dependent remains obscure. In contrast to one report (Kamijo et al. 1997), another showed that p19ARF could induce a cell cycle arrest in p53−/− MEFs, which can be relieved by overexpression of E2F-1 or by blocking p16INK4A function (Carnero et al. 2000). These data suggest that p19ARF can target the pRB pathway independent of p53. The only known target of p19ARF is MDM2, which can bind p53 but also pRB (Xiao et al. 1995) and may, thus, be a candidate to mediate the p19ARF/pRB connection.
The role of pocket proteins in oncogenic transformation
Expression of so-called transforming oncogenes such as oncogenic RAS (Palmero et al. 1998) and v-Abl (Radfar et al. 1998) strongly increases the level of p19ARF. This activates a p53-dependent checkpoint that protects cells from abnormal mitogenic signaling by inducing replicative arrest. MEFs lacking p19ARF or p53 function are no longer susceptible to this type of protection; they escape from replicative senescence and become oncogenically transformed (Harvey and Levine 1991; Harvey et al. 1993; Kamijo et al. 1997). Overexpression of another class of oncogenes such as MYC, adenovirus E1A, and E2F-1 in MEFs has been shown to enhance proliferation but also apoptosis (Lowe and Ruley 1993; Hermeking and Eick 1994; Wagner et al. 1994; Qin et al. 1994; Querido et al. 1997). Apoptosis is strongly enhanced by depriving cells of extracellular survival factors (Evan et al. 1992; Lowe and Ruley 1993) and is dependent on a functional p19ARF-p53 pathway that does not involve p21CIP function (Lowe et al. 1994; Wagner et al. 1994; Bates et al. 1998; De Stanchina et al. 1998; Zindy et al. 1998). The behavior of MYC- or E1A-expressing cells is highly reminiscent of that of triple-knockout cells: Pocket-protein ablation increased the proliferative capacity of cells but also their sensitivity to apoptosis: in particular, under conditions of low serum or high density. The picture that now emerges is that p19ARF acts as a sensor of abnormal or conflicting mitogenic signaling, and activates a p53-dependent response that can either cause cell cycle arrest or sensitize cells to apotosis. The behavior of triple-knockout cells indicates that this decision depends on pocket-protein functions. In their presence, cells arrest; in their absence, for example, by genetic ablation, sequestration by E1A, or inhibition following overexpression of MYC (Berns et al. 2000; Lasorella et al. 2000), cells become immortal but also highly sensitive to apoptosis.
Indeed, we observed that culturing of triple-knockout MEFs under growth-restricting conditions readily induced apoptosis. At the same time, however, continuing cell proliferation was observed. Thus, under certain growth-restricting conditions, loss of pocket-protein function allows continuous cell turnover without net increase in cell number. We envisage that loss of pocket-protein function in vivo may also lead to increased cell turnover, which would strongly facilitate the acquisition of additional oncogenic events. Several observations lend support to this scenario. For example, conditional inactivation of Rb in the mouse pituitary gland resulted in increased cell turnover. Tumors appeared to arise from cells in which apoptosis was blocked, presumably through an additional mutation (Vooijs et al. 1997). Also, inactivation of the pRB family members in the mouse brain epithelium by the large T variant T121 induced aberrant proliferation and p53-dependent apoptosis. Inactivation of p53 in this setting resulted in a dramatic reduction in apoptosis (Yin et al. 1997). We speculate that redundancy of pocket proteins in cell cycle control is cell-type specific. For example, in the mouse pituitary gland, p107 and p130 may not be expressed or functional in compensating for loss of pRB, whereas in the mouse retina both pRB and p107 are active and, therefore, loss of both proteins is required to sustain prolonged cell turnover ultimately leading to retinoblastoma development (Robanus-Maandag et al. 1998).
In conclusion, we have identified three characteristics associated with loss of pocket-protein function that may provide a rationale for its frequent involvement in human cancer. These are reduced differentiation capacity, loss of the senescence response, and increased cell turnover under growth-restricting conditions. We expect that a careful study of the fate of ES cells carrying single or compound loss-of-function mutations in the Rb gene family in chimeric mice will provide further insight into the role of pocket protein ablation in tumorigenesis.
Materials and methods
Generation of ES cells deficient for pRb, p107, and p130
Knockout alleles of pRb, p107, and p130 were generated in 129Ola (E14/IB10) embryonic stem cells. The targeting vectors 129Rb-hyg, 129Rb-his, and 129p107-IRESβgeo have been described (te Riele et al. 1992; Robanus-Maandag et al. 1998). To inactivate p130, we generated the targeting construct 129p130-pur, in which a PGK-puromycin cassette was introduced into codon 405 residing in the A-domain of the pocket region (LeCouter et al. 1996). On electroporation and selection at 1.8 μg/mL of puromycin, homologous recombinants were obtained with a frequency of 80%. The remaining allele of p130 was inactivated by selection at high puromycin concentrations (14 μg/mL). p130−/− mice were generated using chimeric mice which gave germ-line transmission.
Generation of MEFs deficient for pRb, p107, and p130
Chimeric embryos were generated by injection of mutant ES cells into a 3.5-d-old C57Bl/6 blastocyst. Embryos we isolated 12 d after implantation and put into culture. Briefly, embryonic tissue without organs was washed in PBS, minced and incubated in 100 μL trypsin/EDTA overnight on ice and then 30 min at 37°C with an additional 100 μL of trypsin/EDTA. Mutant MEFs were selected for 48 h with 800 μg/mL G418. These cells were designated passage 1. Nonchimeric MEF cultures, serving as a control, all died within 48 h of selection.
ES cell culture, generation and characterization of teratocarcinomas
ES cells were cultured as described (Robanus-Maandag et al. 1998). Nude mice were injected subcutaneously with 106 ES cells. Tumors were removed 4 wk after injection, fixed in phosphate-buffered formalin, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin according to standard procedures. For immunohistochemical characterization, rehydrated sections were stained with antibodies against glial fibrillary acidic protein (GFAP, DAKO), low-affinity nerve growth factor receptor (p75LNGFR, Chemicon International), neuron specific enolase (NSE, DAKO), and proliferation marker Ki-67 (MIB-5, Immunotech). The fraction of proliferating cells was determined by counting Ki-67-positive cells in six different areas in a teratocarcinoma section at a 160× magnification.
Culturing of MEFs, growth curves, 3T9 protocol
MEFs were cultured in GMEM (GIBCO) supplemented with 10% fetal calf serum, 1 mM nonessential amino acids, 1 mM sodium pyruvate, and 0.1 mM β-mercaptoethanol. For growth curves, MEFs were seeded at 2.5 × 104 cells per 12-well plate, in triplicate. At various time points, cells were washed with PBS, fixed for 5 min in 4% formaldehyde, and stained with 0.1% crystal violet (Sigma) in distilled water for 30 min. After washing the cells two times with water, they were allowed to dry. Staining was retrieved from the cells by adding 1 mL 10% acetic acid per well. Optical density was measured of 100 μL retrieved staining at 590 nm. Values were normalized to the optical density at day 0.
A 3T9 protocol was performed by seeding 3 × 105 cells per 6-well plate, in triplicate. Cultures were incubated at 37°C for 3.5 d, and the total amount of cells was determined using a cell counter (Casy 1) followed by replating 3 × 105 cells. This protocol was repeated 20 times.
Electroporation of MEFs
MEFs were grown in 10-cm dishes to subconfluency. One to 2 × 106 cells were resuspended in 100 μL electroporation buffer (2 mM Hepes at pH 7.2, 15 mM K2HPO4/KH2PO4, 250 mM mannitol, and 1 mM MgCl2) at 37°C. The cell suspension was mixed with plasmid DNA (histone H2B-GFP expression plasmid [Kanda et al. 1998], together with either a dominant-negative variant of CDK2 [Van den Heuvel and Harlow 1993] or pcDNA-p19Arf in a 1 : 10 ratio) and transferred to an electroporation cuvette (0.1 cm, Biorad). After 5 min of incubation at room temperature, the cells were electroporated with Gene Pulser II/Gene Pulser II RF module apparatus (Biorad). Five minutes later, electroporation cells were taken up in 8 mL 37°C medium and seeded into a 10-cm dish. To measure effects on G1, electroporated cells were blocked in G2/M with nocodazole (0.5 μg/mL) 32 h after electroporation for 16 h. Cells were trypsinized and resuspended in 250 μL 0.1% Triton X-100 and 50 μg/mL propidium iodide in PBS. For each sample, 10.000 GFP-expressing cells were collected by FACS (FACscan, Becton Dickinson) and analyzed for their cell cycle distribution with the CellQuest program (Becton Dickinson).
FACS analysis
Cell cycle distribution of MEFs was determined by incubating MEFs with 5-Bromodeoxyuridine (0.3 μg/mL) plus 5-Fluoro-5-deoxyuridine (0.03 μg/mL) for 5 h. Cells were collected and resuspended in 70% alcohol at 4°C, stained with DAKO mouse anti-BrdU antibody, and prepared for FACS analysis as described in Brugarolas et al. (1995).
For cell size analysis, cultures were trypsinized, fixed in 70% alcohol at 4°C, stained with propidium iodide, and analyzed by FACS as described in Brugarolas et al. (1998).
Protein analysis
Protein levels were determined by Western blot analyses following established protocols. Antibodies against pRB (C-15), p107 (C-18), p130 (C-20), p16INK4A(M-156), CDK4 (C-22), cyclin E (M-20), p21CIP (C-19), and p53 (FL-393) were obtained from Santa Cruz; the p19ARF antibody (R562) was obtained from Abcam. Goat antirabbit and goat antimouse secondary antibodies were from Biosource.
Irradiation of MEFs
After 5 × 105 cells were plated onto 10-cm dishes, tghey were irradiated the next day with 20 Gy of γ-radiation. Cell lysates were prepared after 24 h and analyzed on Western blots.
For cell cycle analysis, cells were irradiated with 5.5 Gy. After 14 h, cells were incubated with BrdU/FdU for 4 h and analyzed by FACS.
Soft agar assay
In GMEM containing 10% serum, 3 × 104 cells were resuspended in 2 mL 0.4% low–melting point agarose (Sigma) and seeded, in duplicate, into six-well plates coated with 1% low–melting point agarose in GMEM containing 10% serum. The number of foci was scored after 2 wk.
Acknowledgments
We thank Karin van 't Wout and Karin van Veen-Buurman for blastocyst injections and injections of ES cells into nude mice; Els Robanus-Maandag and Marleen Dekker for providing Rb−/− ES cells and targeting vectors; Kees de Goeij, Dennis Hoogervorst, and Jurjen Bulthuis for histotechnical assistance; Martin van der Valk for histological examination of teratocarcinomas; Thijn Brummelkamp for pcDNA-p19ARF expression plasmid; Charles Sherr and Frédérique Zindy for p19Arf−/− MEFs; Jos Jonkers for p53−/− MEFs; Ronald DePinho for Ink4a−/− MEFs; Eric Noteboom and Anita Pfauth for help with FACS analyses; and Reuven Agami, Daniel Peeper, and René Bernards for helpful discussions and critically reading of the manuscript. This work was supported by the Netherlands Cancer Foundation through project grant NKI 95–956 to H.t.R. (J.-H.D., A.v.R., L.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hriele@nki.nl; FAX 31-20-512-2086.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.847700.
References
- Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Bernards R. E2F: A nodal point in cell cycle regulation. Bichim Biophys Acta. 1997;1333:33–40. doi: 10.1016/s0304-419x(97)00027-9. [DOI] [PubMed] [Google Scholar]
- Berns K, Martins C, Dannenberg J-H, Berns A, te Riele H, Bernards R. p27kip-independent cell cycle regulation by MYC. Oncogene. 2000;19:4822–4827. doi: 10.1038/sj.onc.1203879. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Bronson RT, Jacks T. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. The biology of replicative senescence. Eur J Cancer. 1997;33:703–709. doi: 10.1016/S0959-8049(96)00058-5. [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM, Beach DH. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Robanus Maandag E, Van Roon M, Van der Lugt NMT, Van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Cobrinik D, Lee M-H, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes & Dev. 1996;10:1633–16344. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- De Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes & Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–936. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters C, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fang F, Orend G, Watanabe N, Hunter T, Ruoslahti E. Dependence of cyclin E-CDK2 kinase activity on cell anchorage. Science. 1996;271:499–502. doi: 10.1126/science.271.5248.499. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embyonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes & Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Groth A, Weber JD, Willumsen BM, Sherr CJ, Roussel MF. Oncogenic Ras induces p19ARF and growth arrest in mouse embryo fibroblasts lacking p21Cip1 and p27Kip1 without activating cyclin D-dependent kinases. J Biol Chem. 2000;275:27473–27480. doi: 10.1074/jbc.M003417200. [DOI] [PubMed] [Google Scholar]
- Guadagno TM, Ohtsubo M, Roberts JM, Assoian RK. A link between cyclin A expression and adhesion-dependent cell cycle progression. Science. 1993;262:1572–1575. doi: 10.1126/science.8248807. [DOI] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Lai S-L, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988;241:353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DM, Levine AJ. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine fibroblasts. Genes & Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated of p53-deficient mice. Oncogene. 1993;8:2547–2467. [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz JM, Park S-H, Bogenmann E, Cheng J-C, Yandell DW, Kaye FJ, Minna JD, Drya TP, Weinberg RA. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Gutsmann A, Herbert DC, Bradley A, Lee W-H, Lee EY-HP. Heterozygous Rb-1Δ20/+ mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene. 1994;9:1021–1027. [PubMed] [Google Scholar]
- Hurford RK, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes & Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppresion at the mouse INK4a locus mediated by the alternative reading frame product p19arf. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci. 1998;95:8292–8287. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- LeCouter JE, Whyte PF, Rudnicki MA. Cloning and expression of the Rb-related mouse p130 mRNA. Oncogene. 1996;12:1433–1440. [PubMed] [Google Scholar]
- LeCouter J, Kablar B, Hardy WR, Ying C, Megeney LA, May LL, Rudnicki MA. Strain-dependent myeloid hyperplasia, growth deficiency and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol Cell Biol. 1998a;18:7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCouter J, Kablar B, Whyte PFM, Ying C, Rudnicki MA. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development. 1998b;125:4669–4679. doi: 10.1242/dev.125.23.4669. [DOI] [PubMed] [Google Scholar]
- Lee EY-HP, To H, Shew J-Y, Bookstein R, Scully P, Lee W-H. Inactivation of the retinoblastoma susceptibility gene in human breast cancers. Science. 1988;241:218–221. doi: 10.1126/science.3388033. [DOI] [PubMed] [Google Scholar]
- Lee EY-HP, Chang C-Y, Hu N, Wang Y-C, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lee M-H, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Targeted disruption of p107: Functional overlap between p107 and Rb. Genes & Dev. 1996;10:1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- Lloyd AC, Obermuller F, Staddon S, Barth CF, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes & Dev. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes & Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Jacks T, Housman DE, Ruley HE. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci. 1994;91:2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Mulligan G, Jacks T. The retinoblastoma gene family: Cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- Mulligan GJ, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: Evidence for functional compensation by p107 and pRB. Mol Cell Biol. 1998;18:206–220. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EJ, Stott D, Rigby PWJ. Sequences and factors required for the F9 embryonal carcinoma stem cell E1a-like activty. Mol Cell Biol. 1991;11:5534–5540. doi: 10.1128/mcb.11.11.5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Diff. 1998;9:585–593. [PubMed] [Google Scholar]
- Nilausen K, Green H. Reversible arrest of growth in G1 of an established cell fibroblast line (3T3) Exp Cell Res. 1965;40:166–168. doi: 10.1016/0014-4827(65)90306-x. [DOI] [PubMed] [Google Scholar]
- Nobori T, Miura K, Wu DJ, Lois A, Tkabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Roberts JM. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Palmero I, McConnell B, Parry D, Brookes S, Hara E, Bates S, Jat P, Peters G. Accumulation of p16INK4a in mouse fibroblasts as a function of replicative senescence and not of retinoblastoma gene status. Oncogene. 1997;15:495–503. doi: 10.1038/sj.onc.1201212. [DOI] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Pantoja C, Serrano M. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene. 1999;18:4974–4982. doi: 10.1038/sj.onc.1202880. [DOI] [PubMed] [Google Scholar]
- Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip, a cyclin-cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes & Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Qin X-Q, Livingston DM, Kaelin WG, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Querido E, Teodoro JG, Branton PE. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radfar A, Unnikrishnan I, Lee HW, DePinho RA, Rosenberg N. p19(Arf) induces p53-dependent apoptosis during abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci. 1998;95:13194–13197. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittling SR, Denhardt DT. p53 mutations in spontaneously immortalized 3T12 but not 3T3 mouse embryo cells. Oncogene. 1992;7:935–942. [PubMed] [Google Scholar]
- Robanus-Maandag EC, Van der Valk M, Vlaar M, Feltkamp C, O'Brien J, Van Roon M, Van der Lugt N, Berns A, te Riele H. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 1994;13:4260–4268. doi: 10.1002/j.1460-2075.1994.tb06746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robanus-Maandag E, Dekker M, Van der Valk M, Carrozza M-L, Jeanny J-C, Dannenberg J-H, Berns A, te Riele H. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes & Dev. 1998;12:1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;18:309–322. [PubMed] [Google Scholar]
- Schulze A, Zerfass-Thome K, Bergès J, Middendorp S, Jansen-Dürr P, Henglein B. Anchorage-dependent transcription of the cyclin A gene. Mol Cell Biol. 1996;16:4632–4638. doi: 10.1128/mcb.16.9.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, Mila EM, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16ink4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- St. Croix B, Sheehan C, Rak JW, Florenes VA, Slingerland JM, Kerbel RS. E-cadherin-dependent growth suppression is mediated by the cyclin-dependent kinase inhibitor p27(KIP1) J Cell Biol. 1998;142:557–571. doi: 10.1083/jcb.142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Riele H, Robanus-Maandag E, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropiate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- Van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- Vooijs M, van der Valk M, te Riele H, Berns A. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene. 1998;17:1–12. doi: 10.1038/sj.onc.1202169. [DOI] [PubMed] [Google Scholar]
- Wagner AJ, Kokontis JM, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes & Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Association between an oncogene and an anti-oncogene: The adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, Livingston DM. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- Yin C, Knudson CM, Korsmeyers JS, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Postigo AA, Dean DC. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4A, TGFβ, and contact inhibiton. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ramsay ES, Mock B. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle D, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorick TS, Lemke G. Schwann cell differentiation. Curr Opin Cell Biol. 1996;8:870–876. doi: 10.1016/s0955-0674(96)80090-1. [DOI] [PubMed] [Google Scholar]