Abstract
Growth factor activity is localized within the natural extracellular matrix (ECM) by specific non-covalent interactions with core ECM biomolecules, such as proteins and proteoglycans. Recently, these interactions have inspired us and others to develop synthetic biomaterials that can non-covalently regulate growth factor activity for tissue engineering applications. For example, biomaterials covalently or non-covalently modified with heparin glycosaminoglycans can augment growth factor release strategies. In addition, recent studies demonstrate that biomaterials modified with heparin-binding peptides can sequester cell-secreted heparin proteoglycans and, in turn, sequester growth factors and regulate stem cell behavior. Another set of studies show that modular versions of growth factor molecules can be designed to interact with specific components of natural and synthetic ECMs, including collagen and hydroxyapatite. In addition, layer-by-layer assemblies of GAGs and other natural polyelectrolytes retain growth factors at a cell-material interface via specific non-covalent interactions. This review will detail the various bioinspired strategies being used to non-covalently localize growth factor activity within biomaterials, and will highlight in vivo examples of the efficacy of these materials to promote tissue regeneration.
Keywords: Biomimetics, Drug Delivery, Hydrogels, Tissue Engineering, Monolayers
1. Introduction
A primary goal of tissue engineering is to repair or replace tissues that have been damaged due to disease or injury. Unlike bone, skin, and peripheral nerves, which demonstrate repair after injury,[1–4] most tissues of the human body are incapable of restoring proper function to extensively damaged tissue. Instead, significant tissue damage is commonly associated with secretion of excess extracellular matrix components by myofibroblasts, a process known as fibrosis.[5–9] This results in the formation of hard scar tissue that adversely affects organ function. Traditionally, organ and tissue transplantation have been successful in replacing or restoring function to extensively damaged tissues. However, organ transplantation is only an effective therapy for certain tissue types, and the number of individuals requiring a transplant is often significantly greater than the number of organs or tissues available for transplantation. In light of these limitations, the field of tissue engineering emerged with a focus on restoring proper physiological function to damaged organs by limiting scar formation and directing the growth of new, native tissue.[10, 11]
Since its inception, the field of tissue engineering has progressed toward developing materials that controllably deliver cells and biomolecules to sites of tissue damage.[12] In particular, a wide variety of naturally-derived macromolecules, synthetic polymers, minerals, or combinations thereof have been used as “carriers” to localize cells and biomolecules at the site of tissue damage, and their applicability in this regard has been extensively reviewed elsewhere.[13–21] More recently, however, it has been widely proposed that biomaterials engineered to specifically regulate biomolecule activity may, in turn, control cell behavior during re-growth of functional tissue. As such, recent approaches have moved beyond materials that serve as relatively inert cell and biomolecule “carriers” toward materials that actively regulate biomolecule activity via specific, non-covalent interactions. This review will highlight biomaterials that regulate the activity of growth factors, a class of soluble signaling biomolecules, by mimicking non-covalent interactions commonly observed within the natural extracellular matrix (ECM).
Due to the well-defined role of growth factors in tissue development (reviewed in[22]), tissue engineering approaches often rely on biomaterials that deliver growth factors. For example, numerous methods have encapsulated a growth factor within a material (e.g. plastic microspheres or highly hydrated polymeric ‘hydrogels’) and allowed it to diffuse out from the material and into the surrounding tissue.[18, 23–28] Although successful at delivering biologically active growth factors in vivo, these approaches are typically complicated by rapid, burst release that provides relatively short-term control over growth factor availability. Moreover, growth factor localization is often limited by the propensity of these molecules to transport through tissue. Other efforts have eliminated the transport component by covalently linking a growth factor onto a biomaterial.[29–33] However, covalent linkage may directly influence growth factor activity by changing protein conformation or by masking the active sites required for growth factor binding to cell surface receptors. Covalent immobilization also inhibits internalization of growth factor-receptor complexes, which, in turn, may influence the response of a cell.[34–37]
Synthetic polymer hydrogels (e.g. poly(ethylene glycol), poly(acrylic acid), and poly(NIPAAM)) and naturally-derived polymer hydrogels (e.g. collagen, gelatin, alginate, and chitosan) have emerged as common biomaterials to deliver growth factors, as they mimic the highly hydrated, macromolecular network structure of the natural ECM.[38] Recently, numerous studies have incorporated functional epitopes derived from the solid-phase of the ECM, such as cell adhesion ligands, into the solid-phase of hydrogel-based biomaterials to further mimic the natural ECM.[39] Aside from cell adhesion ligands, however, biological macromolecules within the ECM also present epitopes that bind to growth factors to limit diffusion and, in turn, localize growth factor activity. Incorporation of growth factor-binding sites into synthetic biomaterials has recently emerged as a promising strategy to non-covalently localize growth factor activity. Additionally, engineering growth factors or growth factor-mimicking peptides to include sites that bind to the natural ECM has emerged as a useful strategy to localize growth factor activity within natural biomaterials. This review will detail the recent development of biomaterials that localize growth factor activity by mimicking non-covalent interactions observed in the native ECM. Additionally, we will highlight studies that have used biomaterials that mimic non-covalent growth factor-ECM interactions in tissue engineering strategies in vivo.
2. ECM components in synthetic biomaterials to localize growth factor activity
The ECM is a self-assembled network of diverse biomolecules that organizes cells, provides mechanical support, and presents cell anchorage sites.[40] For example, specific subunits within ECM proteins, such as the tri-peptide Arg-Gly-Asp (RGD), mediate cell adhesion to the ECM,[41–45] while proteoglycans (PGs) are involved in cell-ECM[46–48] and cell-cell adhesion.[49–51] In addition to their role in cell adhesion, PGs also influence growth factor activity. Several specific examples from fundamental biology demonstrate the role of PGs. For example, non-covalent interactions between chondroitin sulfate PGs and the growth factor midkine are required for midkine-mediated macrophage migration.[52] Additionally, the glycosaminoglycans (GAGs) heparin and heparan sulfate mediate the dimerization of FGFs, such as acidic FGF[53] and basic FGF[54], which is integral to FGF receptor activation. FGF-heparin interactions also influence FGF binding specificity to various FGF receptors, as high concentrations of heparin promote preferential binding of FGF-4 with FGF receptor-1, while low heparin concentrations favor FGF-4 binding with FGF receptor-2.[55] Conversely, some growth factor-PG interactions can also down-regulate growth factor activity. The core protein of the PG perlecan binds to FGF-18 and inhibits its mitogenic effect on growth plate chondrocytes.[56] Interactions between platelet-derived growth factor (PDGF) and chondroitin sulfate inhibit phosphorylation of PDGF-receptor-β and down-regulate fibroblast proliferation.[57] Additionally, binding of bone morphogenetic protein-2 (BMP-2) to heparan sulfate down-regulates osteogenesis by C2C12 cells.[58] Taken together, these examples show that PGs and their GAG subunits can non-covalently bind to growth factors and either up-regulate or down-regulate their effects. In view of the importance of PGs in fundamental biology, recent studies have relied on diverse covalent and non-covalent methods to incorporate GAGs and PGs into synthetic biomaterials, which we highlight in the following sections.
2.1 Covalent incorporation of GAGs and PGs into biomaterials
Numerous covalent mechanisms have been used to conjugate GAGs onto various reactive functional groups presented by synthetic biomaterials. This section will highlight covalent chemistries that are commonly used to conjugate GAGs onto biomaterials, which are schematically represented in Figure 1A, and the use of these GAG-modified biomaterials in tissue engineering applications.
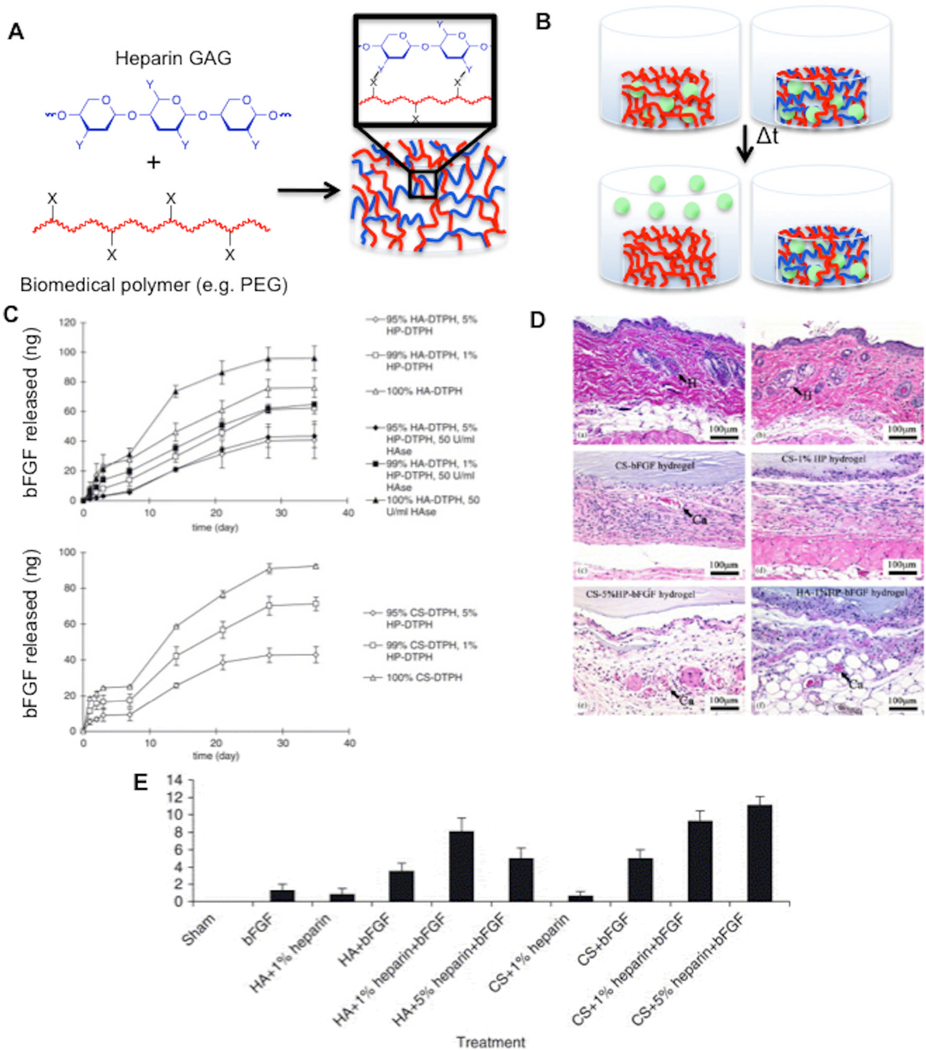
Figure 1. Figure 1: Heparin covalently incorporated into hydrogels controls heparin-binding growth factor release and enhances tissue regeneration in vivo.
Schematic representations of (A) covalent incorporation of heparin GAGs into hydrogels comprised of biomedical polymers (Y is often COOH, OH, SH, acrylate, or methacrylate, X is often NH2, COOH, acrylate, or methacrylate) and (B) augmented release of heparin-binding growth factors ( ) from heparin-modified hydrogels in vitro. C) bFGF release profiles from hyaluronic acid-PEG-heparin and chondroitin sulfate-PEG-heparin hydrogels. Hematoxylin and eosin staining (D) and neovascularization index (E) of skin and subcutaneous tissue at implant site 14 days after sham, bolus FGF-2 injection, implantation of hyaluronic acid-PEG-heparin, chondroitin sulfate-PEG-heparin, or control hydrogels. Reproduced with permission from[81] 2005 Elsevier.
) from heparin-modified hydrogels in vitro. C) bFGF release profiles from hyaluronic acid-PEG-heparin and chondroitin sulfate-PEG-heparin hydrogels. Hematoxylin and eosin staining (D) and neovascularization index (E) of skin and subcutaneous tissue at implant site 14 days after sham, bolus FGF-2 injection, implantation of hyaluronic acid-PEG-heparin, chondroitin sulfate-PEG-heparin, or control hydrogels. Reproduced with permission from[81] 2005 Elsevier.
2.1.1 Photo-crosslinkable GAGs
Photo-polymerization of synthetic polymer chains into covalently cross-linked hydrogel networks is a common biomaterial design strategy due to the efficiency of the crosslinking reactions under physiological conditions and the ability to encapsulate viable cells directly into the hydrogel.[59, 60] In light of these benefits, Anseth and co-workers prepared heparin GAGs bearing photo-crosslinkable vinyl moieties and covalently incorporated them into poly(ethylene glycol) (PEG) hydrogels during photo-crosslinking.[61] In turn, they used these materials to characterize the influence of heparin on multiple cell types cultured on or within PEG hydrogels. For example, they characterized the influence of heparin-modified PEG hydrogels on the inhibition of interstitial cell myofibroblast activation by FGF-2.[62] Cells cultured on heparin-modified PEG hydrogels showed decreased activation of mitogen-activated protein kinase (MAPK), a protein that is part of the FGF-2 signaling cascade,[63] when compared to cells cultured on PEG hydrogels lacking heparin. Furthermore, heparin-modified PEG hydrogels increased interstital cell expression of alpha smooth muscle actin, a key marker of myofibroblast activation of valvular interstital cells.[64] Together, these results demonstrated that heparin GAGs covalently incorporated into a biomaterial regulate heparin-binding growth factor activity which, in turn, influences cell behavior.
In a second set of studies, Anseth and co-workers characterized the influence of heparin-modified PEG hydrogels on human mesenchymal stem cell behavior.[61, 65, 66] Heparin incorporated into the hydrogel promoted hMSC adhesion and also provided long-term release of FGF-2 (∼5 weeks), which enhanced hMSC proliferation.[61] Moreover, heparin-modified PEG hydrogels enhanced cellular production of alkaline phosphatase, as well as up-regulation of osteopontin and collagen I gene expression - all markers that indicate osteogenic differentiation.[67] When cells were cultured in osteogenic differentiation medium that was first passed over a heparin column to deplete heparin-binding proteins and then supplemented with fibronectin, BMP-2, or both, a similar increase in osteogenic markers was observed, indicating that differentiation was dependent on fibronectin and BMP-2 (a heparin-binding ECM protein and growth factor, respectively).[65] An additional set of studies demonstrated that BMP-2 secreted by MSCs in response to fluvastatin was also retained within heparin-presenting PEG hydrogels.[66] Together, these results demonstrate that heparin-modified PEG hydrogels amplify the local activity of cell-secreted growth factors, analogous to the heparin-mediated regulation of growth factor activity observed in natural ECMs, and described in the introduction section above.
2.1.2 Other covalent conjugation mechanisms
A number of additional covalent chemistries that proceed efficiently under physiological conditions are commonly used to form hydrogels. For example, the Michael-type addition reaction between thiol groups and α,β-unsaturated carbonyls (e.g. acrylates, methacrylates, and maleimides) has been used to crosslink multi-arm polymer chains into hydrogel networks.[68] Scatena and co-workers modified heparin GAGs to bear thiol groups that react with bi-functional PEG diacrylate to form a hydrogel.[69] These PEG-heparin hydrogels were then used to deliver osteoprotegerin, which is a heparin-binding growth factor that influences osteoclast differentiation[70] and also promotes angiogenesis, the growth of new blood vessels from a pre-existing vascular supply.[71] Osteoprotegerin was retained within the hydrogel for up to 500 hours in vitro. After subcutaneous implantation in mice, osteoprotegerin was maintained at the implant site for 2 weeks, and a nearly two-fold increase in vascular density surrounded the implant when compared to control PEG implants lacking heparin GAGs.
Kiick and co-workers also relied on the Michael-type addition reaction to covalently conjugate heparin GAGs to PEG hydrogels. They modified low molecular weight heparin GAGs with malemide moieties that can react with thiol-terminated multi-arm PEG chains to form hydrogel networks.[72–74] The FGF-2 release rate from heparin-modified PEG hydrogels was significantly slower than the release rate from control PEG hydrogels lacking heparin. The FGF-2 release rate also correlated with hydrogel erosion when heparin was incorporated into degradable PEG hydrogels.[72]
Although these examples demonstrate facile incorporation of a single GAG within a PEG hydrogel, natural ECMs typically include multiple PGs and GAGs. In particular, the ECM often includes members of the three PG ‘families’: the large aggregating chondroitin sulfate proteoglycans (e.g. lecticans), the small leucine-rich chondroitin sulfate proteoglycans, and the heparan sulfate proteoglycans, which are decorated with various, biochemically-distinct GAGs (e.g. heparan sulfate, chondroitin sulfate, dermatan sulfate, and keratin sulfate),[75, 76] as well as hyaluronan, the only GAG lacking a protein core.[77] Hyaluronic acid and chondroitin sulfate GAGs are of particular interest for tissue engineering applications, as they are important components of natural tissue development processes.[78–80] Recently, Prestwich and co-workers prepared heparin, hyaluronic acid, and chondroitin sulfate GAGs modified with thiol groups that can react with PEG diacrylate to form hydrogels.[81] Mixtures of thiol-modified heparin and hyaluronic acid or thiol-modified heparin and chondroitin sulfate reacted rapidly with PEG diacrylate to form hydrogels. FGF-2 release from heparin-modified PEG-hyaluronic acid and PEG-chondroitin sulfate hydrogels was significantly slower than FGF-2 release from heparin-free hydrogels (Fig. 1C). The FGF-2 released from the heparin-modified hydrogels was bioactive, as it stimulated NIH 3T3 fibroblast proliferation in vitro. Importantly, FGF-2-loaded heparin-modified hydrogels significantly increased blood vessel in-growth after subcutaneous implantation when compared to FGF-2-loaded heparin-free hydrogels (Fig. 1D–E). Taken together, these results indicate that multiple, distinct GAGs covalently incorporated into biomaterials enhance blood vessel growth, likely by enhancing growth factor activity in vivo.
Although the Michael-type addition reaction allows for GAG conjugation to biomaterials, GAGs typically do not contain the thiol or α,β-unsaturated carbonyl moieties required for the reaction to proceed.[82] So, immobilization of GAGs via Michael-type addition requires chemical modification of the GAG chain. Instead of relying on GAG chemical modification, many groups have conjugated GAGs onto biomaterials through covalent mechanisms that rely on reactive groups already present on native GAGs. For example, Mizushima and co-workers used carbodiimide condensation to crosslink carboxylate moieties in heparin to carboxylate moieties in alginate via an ethylenediamine linker.[83] Heparin conjugated to alginate hydrogels significantly decreased FGF-2 release rate in vitro, and heparin-modified alginate gels loaded with FGF-2 significantly increased angiogenesis in the dorsal area of the rat two weeks after implantation. In another set of studies, Kuo and co-workers used carbodiimide condensation to conjugate heparin to chitosan-alginate polyelectrolyte complexes used as drug delivery vehicles.[84, 85] The amount of FGF-2 bound to heparin-modified chitosan-alginate complexes was dependent on the amount of heparin present, and the FGF-2 release rate was significantly slower than the release rate from heparin-free complexes. Additionally, the released FGF-2 promoted human foreskin fibroblast proliferation during in vitro culture. Alginate and chitosan are of significant interest for tissue engineering applications, as they are naturally-derived, bio-compatible polysaccharides that are not susceptible to enzymatic degradation and readily crosslink to form hydrogels in the presence of multivalent ions.[86], [87, 88] These examples demonstrate that polysaccharide biomaterials modified with heparin localize growth factor activity in vitro and in vivo, and suggests that modifying other natural polysaccharides with heparin may be of interest for tissue engineering applications.
Taken together, the multiple examples presented here demonstrate that heparin GAGs covalently incorporated into biomaterials significantly decrease the release rate of heparin-binding growth factors. The heparin-growth factor interactions within these biomaterials also help to maintain long-term growth factor bioactivity. Growth factor bioactivity typically decreases rapidly in protease-rich tissues and fluids, so maintaining long-term bioactivity is an important goal.[89, 90] Implanting heparin-modified biomaterials has led to enhanced blood vessel growth in multiple studies. Therefore, materials that localize and maintain growth factor bioactivity may favor regeneration of complex tissue architectures that require a functional blood supply.
2.2 Non-covalent incorporation of GAGs and PGs into biomaterials
Although covalent chemistries are useful for introducing GAGs into synthetic biomaterials, natural PGs instead integrate into the ECM through non-covalent interactions with various proteins.[91, 92] Recent studies have mimicked these non-covalent interactions to introduce GAGs or PGs into natural and synthetic biomaterials, which we highlight in the following sections and schematically represent in Figure 2A.
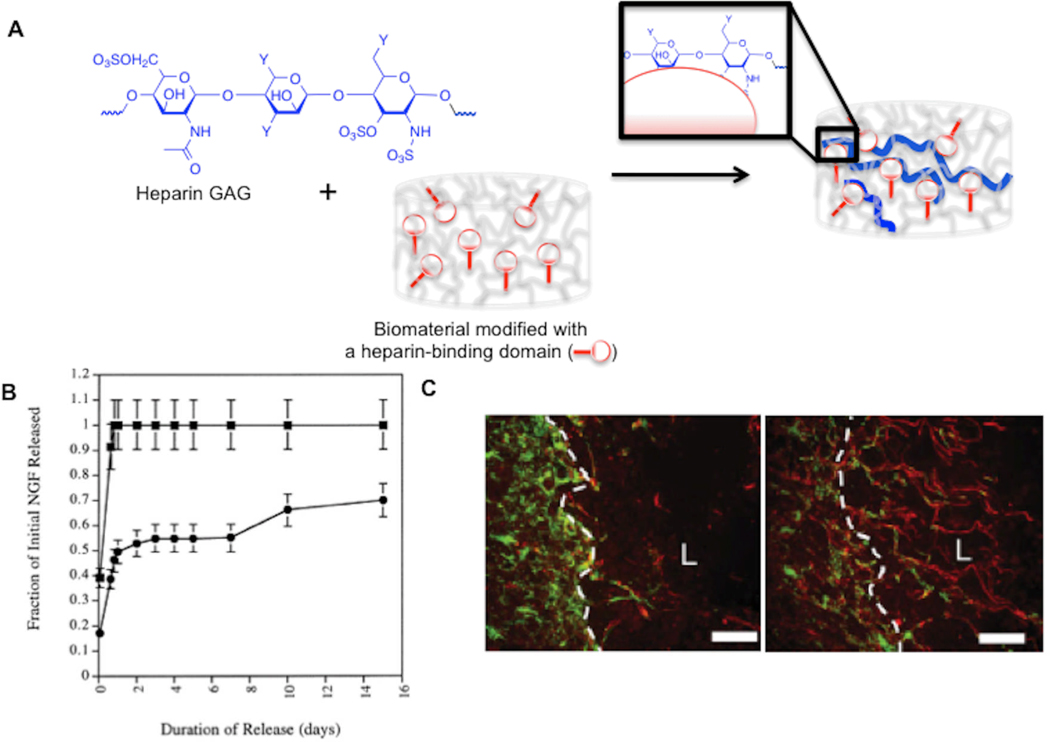
Figure 2. Heparin non-covalently incorporated into hydrogels controls heparin-binding growth factor release and enhances tissue regeneration in vivo.
(A) Schematic representation of non-covalent binding of heparin GAGs within a hydrogel biomaterial presenting a heparin-binding domain. (B) Release profile of nerve growth factor from unmodified (■) and heparin-modified (●) fibrin matrices. Reproduced with permission from[96] 2000 Elsevier. (C) Heparin-modified fibrin matrices loaded with neurotrophin-3 promote extension of neural processes (red) across the glial scar border (white) at a spinal cord lesion site. Reproduced with permission from[100] Wiley Interscience 2009.
2.2.1 Non-covalent incorporation of GAGs into biomaterials
GAGs and PGs are localized within the natural ECM through specific non-covalent interactions with natural ECM components. There are several examples from nature that illustrate this point. For example, the ECM glycoprotein fibronectin contains a heparin-binding domain within domain III-13,[93] and the ECM glycoprotein laminin binds to heparin, heparan sulfate, dermatan sulfate, and chondroitin sulfate.[94] A number of recent studies have mimicked non-covalent PG incorporation into the ECM to incorporate GAGs and PGs into biomaterials. Sakiyama-Elbert and co-workers covalently linked a peptide derived from the heparin-binding domain of antithrombin to the backbone of fibrin hydrogels, which in turn bound to heparin GAGs that were present during hydrogel crosslinking.[95] Importantly, these heparin-binding fibrin hydrogels significantly decreased the release rate of heparin-binding proteins, including nerve growth factor,[96] (Fig. 2B), glial-derived neurotrophic factor,[97] platelet-derived growth factor-BB,[98] and neurotrophin-3.[99, 100] The utility of these hydrogels in tissue engineering was demonstrated in four separate examples, summarized as follows: 1) heparin-binding fibrin hydrogels loaded with FGF-2 significantly enhanced neurite outgrowth from explanted chick dorsal root ganglia;[95] 2) heparin-modified fibrin matrices loaded with neurotrophin-3 promoted neural process extension across a glial scar border at a spinal cord lesion site;[100] (Fig. 2C) 3) nerve growth factor delivery from heparin-binding fibrin hydrogels enhanced peripheral nerve regeneration in a rat sciatic nerve defect;[101] and 4) platelet-derived growth factor BB released from heparin-binding fibrin matrices improved gliding properties of sutured tendons in a canine model.[102]
Stupp and co-workers used non-covalent interactions to introduce heparin GAGs onto self-assembling peptide amphiphile biomaterials. In particular, they engineered peptide amphiphiles to include a heparin-binding site that is displayed on the surface of peptide nanofibers.[103] Heparin-bound nanofibers loaded with vascular endothelial growth factor (VEGF) or FGF-2 significantly enhanced blood vessel growth within the rat cornea when compared to nanofibers lacking a heparin-binding domain. A second study demonstrated that heparin-bound peptide nanofibers loaded with VEGF and FGF-2 significantly enhanced islet cell engraftment in diabetic mice and, in turn, resulted in a significantly higher level of normoglycemia.[104] Therefore, natural and synthetic biomaterials engineered to mimic natural GAG-ECM interactions can enhance growth factor activity and improve tissue regeneration, similar to the aforementioned biomaterials (section 2.1) that were covalently modified with heparin GAGs.
2.2.2 Non-covalent incorporation of recombinant PGs into a biomaterial
Heparin GAGs are not the only component of PGs that bind to growth factors. For example, the core protein of the PG perlecan binds to FGF-18 and thereby decreases proliferation of growth plate chondrocytes.[56] Thus, biomaterials that non-covalently bind to PGs, rather than GAGs alone, are of significant interest for tissue engineering applications. Farach-Carson and co-workers recently demonstrated that a PG synthesized using recombinant DNA technology (a “recombinant PG”) could bind to a collagen-based hydrogel and mediate growth factor binding. Specifically, recombinant perlecan domain I, which is the chondroitin sulfate and heparan sulfate bearing domain, bound to collagen I hydrogels in a heparan sulfate-dependent manner, and bound to collagen II hydrogels in a heparan sulfate- and chondroitin sulfate-dependent manner.[105, 106] In turn, the recombinant perlecan domain I mediated FGF-2 binding to collagen I matrices and BMP-2 binding to collagen II matrices. The bound FGF-2 significantly enhanced the proliferation of MG63 osteoblasts and human bone marrow stromal cells. The bound BMP-2 enhanced C3H10T1/2 cell chondrogenic differentiation on collagen II fibrils or within poly(lactic acid) matrices modified with collagen II fibrils. This body of work demonstrates that a synthetic polymer and a natural ECM protein mixed into a composite biomaterial can mimic natural ECM-PG interactions and enhance growth factor activity. Mixing synthetic polymers and natural proteins into composite biomaterials may provide a simple method to prepare ECM-mimicking biomaterials without requiring chemical modification to the protein or polymer.
2.2.3 Biomaterials that sequester natural, cell-secreted PGs
During natural tissue development and wound healing, numerous cell types secrete PGs that integrate into the ECM and regulate cell function. For example, heparin PGs secreted by mast cells increase endothelial cell migration during angiogenesis,[107] presumably by causing competitive release of pro-angiogenic growth factors bound to heparan sulfates in the natural ECM.[108] Additionally, cartilage-specific proteoglycan expression is negligible in the early stages of chick wing bud development, but it is then significantly increased at sites of early cartilage differentiation.[109] Moreover, human mesenchymal stem cells (hMSCs) produce PGs that are primarily located inside the cell during culture in maintenance medium, but secrete PGs throughout the extracellular environment during culture in osteogenic induction medium.[110] Recently, we and others proposed that biomaterials modified with heparin-binding sites could sequester cell-secreted PGs that, in turn, direct cell behavior in a manner that is analogous to cell-secreted PGs within the natural ECM. For example, Thomson and coworkers covalently immobilized FGF-2, a heparin-binding growth factor, onto self-assembled monolayers and demonstrated that immobilized FGF-2 sequesters PGs present in mouse embryonic feeder (MEF) layer conditioned medium. The sequestered PGs mediated human embryonic stem cell adhesion onto the substrate.[111]
We recently developed chemically well-defined cell culture substrates that sequester cell-secreted PGs via a short peptide derived from a natural heparin-binding domain. Specifically, self-assembled monolayers (SAMs) modified with a peptide derived from the heparin-binding domain of FGF-2 specifically sequestered heparin PGs from stem cell growth medium. When this heparin-binding peptide was immobilized alongside a fibronectin-derived cell adhesion peptide, Arg-Gly-Asp-Ser-Pro (RGDSP), the two peptides worked synergistically to promote hMSC adhesion and spreading in a heparin-dependent manner,[112] similar to native cell adhesion mediated by the integrin- and heparin-binding domains of fibronectin.[46]
We also used these substrates to characterize the influence of sequestered serum-borne PGs on cell proliferation. SAMs modified with a heparin-binding peptide enhanced FGF-2-mediated endothelial cell proliferation. Cell proliferation increased as a function of the heparin-binding peptide density on the SAM. Additionally, the sequestered PGs enhanced FGF-mediated hMSC proliferation while maintaining hMSC phenotype.[113] These results were observed during culture in standard stem cell culture medium supplemented only with 10% fetal bovine serum (FBS). Strikingly, the increase in stem cell proliferation via PG sequestering was equivalent to the increase caused by adding FGF-2, which has previously been shown to promote hMSC proliferation.[114] This result indicates that sequestering cell-secreted heparin PGs is sufficient to enhance hMSC proliferation without adding growth factors to the medium, and suggests that biomaterials engineered to sequester cell-secreted biomolecules may allow for optimized stem cell expansion without the need for expensive supplements, such as recombinant growth factors.
Serum-borne PGs sequestered by a substrate can also enhance hMSC differentiation in the absence of recombinant growth factor supplements.[113] Specifically, hMSCs cultured on heparin-binding SAMs in osteogenic induction medium showed significantly increased alkaline phosphatase activity when compared to cells cultured on SAMs unable to sequester heparin. Interestingly, the increase in alkaline phosphatase activity was dependent on BMP-2, a heparin-binding growth factor, as cells cultured on heparin-binding SAMs in medium supplemented with a BMP receptor inhibitor showed no increase in alkaline phosphatase activity. Collectively, our results demonstrate that by simply changing the culture medium formulation, heparin-sequestering biomaterials can up-regulate different signaling pathways (e.g. FGFR-mediated or BMPR-mediated) and, in turn, amplify different stem cell behaviors, such as proliferation or osteogenic differentiation. These observations suggest that the influence of heparin-binding biomaterials on stem cell behavior in vivo may depend not only on the heparin-binding growth factors secreted by the implanted stem cell type, but also on heparin-binding growth factors secreted by cells adjacent to the implant site. Thus, further in vitro studies using heparin-binding biomaterials to characterize the influence of different heparin-binding growth factors, either alone or in combination, on hMSC behavior may be useful for predicting cell response to the highly variable and circumstantial heparin-binding growth factor composition likely to be present in vivo. It is noteworthy that sequestered PGs that present diverse GAG chains with varying sulfation profiles may be capable of binding a broader range of growth factors than heparin alone, leading to new strategies to control growth factor activity.
It is also possible to locally influence stem cell behavior by spatially patterning heparin-binding peptides. We recently developed a method to spatially pattern peptides within an otherwise bio-inert SAM.[115] In particular, poly(dimethyl siloxane) (PDMS) microchannels placed onto a SAM substrate allow for injection of aqueous solutions of peptide within spatially defined locations on a substrate, which, in turn, spatially controls peptide conjugation to the SAM. Using this method, we demonstrated that a spatially patterned heparin-binding peptide locally increases hMSC proliferation in standard cell culture medium.[113] Interestingly, the degree of increased proliferation observed in patterned regions was similar to that observed on SAMs presenting a heparin PG-binding peptide over the entire SAM surface. Therefore, the influence of bound heparin PGs on hMSC behavior is dependent on spatial co-localization of cells and heparin PGs. Spatially patterning sites of PG binding onto a biomaterial may provide a mechanism to spatially control diverse hMSC processes by localizing the activity of cell-secreted molecules, analogous to the natural ECM.
2.3 Engineered peptides that mimic GAGs and PGs
In light of the importance of GAGs and PGs in tissue development, recent efforts have also developed synthetic peptides that mimic GAGs. Synthetic peptides bearing sulfate groups on various amino acid side-chains have been synthesized as an analog to sulfated heparin PGs. For example, Hubbell and co-workers screened a library of sulfated tetrapeptides against the heparin-binding growth factor VEGF, and identified a sequence SY(SO(3))DY(SO(3)) that bound to VEGF with an affinity 100-fold stronger than suramin, a heparin mimic.[116] Kiick and co-workers prepared synthetic sulfated peptides that bound with specific, high-affinity to multiple heparin-binding peptides, as well as the heparin-binding growth factor VEGF.[117] Although not explicitly demonstrated to date, these results suggest that incorporating these peptides into biomaterials may provide an alternate mechanism to non-covalently bind growth factors.
2.4 Summary
The numerous covalent and non-covalent mechanisms described herein highlight the diversity of strategies to introduce GAGs or PGs into synthetic and natural biomaterials. While many of these examples rely on materials modified with xenogeneic GAGs or recombinant PGs to localize recombinant growth factor activity, recent work has shown that biomaterials presenting a heparin-binding peptide can sequester native, cell-secreted heparin PGs and, in turn, locally enhance cell-secreted growth factor activity. Importantly, the multiple demonstrations that GAG- and PG-modified biomaterials enhance cell proliferation and differentiation in vitro and enhance tissue regeneration in vivo represent an extensive toolkit for emerging tissue engineering approaches.
3. Growth factors engineered to bind components of the natural ECM
In typical tissue engineering applications, biomaterials are introduced at the site of tissue damage or disease to promote functional tissue regeneration. Since an ECM composed of natural biomolecules exists within the tissue adjacent to an implant site, matrices comprised of ECM-derived proteins, such as collagen and gelatin, have emerged as promising biomaterials to allow for implant assimilation into the surrounding tissue.[118] Additionally, mineral-based biomaterials have been widely used in bone tissue engineering applications as a mimic of the mineralized ECM of hard tissues.[119] As such, there has been a recent interest in strategies to enhance growth factor activity within collagen- and mineral-based biomaterials. This section highlights engineered growth factors that non-covalently bind to native ECM components.
3.1 Growth factor fusion proteins with collagen-binding domains
The ECM protein collagen is the most abundant protein in humans. Recently, Tabata and co-workers demonstrated that FGF-2 interacts with collagen sponges and, in turn, is released in a sustained manner in vitro and in vivo.[120] When subcutaneously implanted in mice, FGF-2-loaded collagen sponges enhanced blood vessel growth, and implantation in an ischemic hindlimb resulted in significantly increased blood flow.
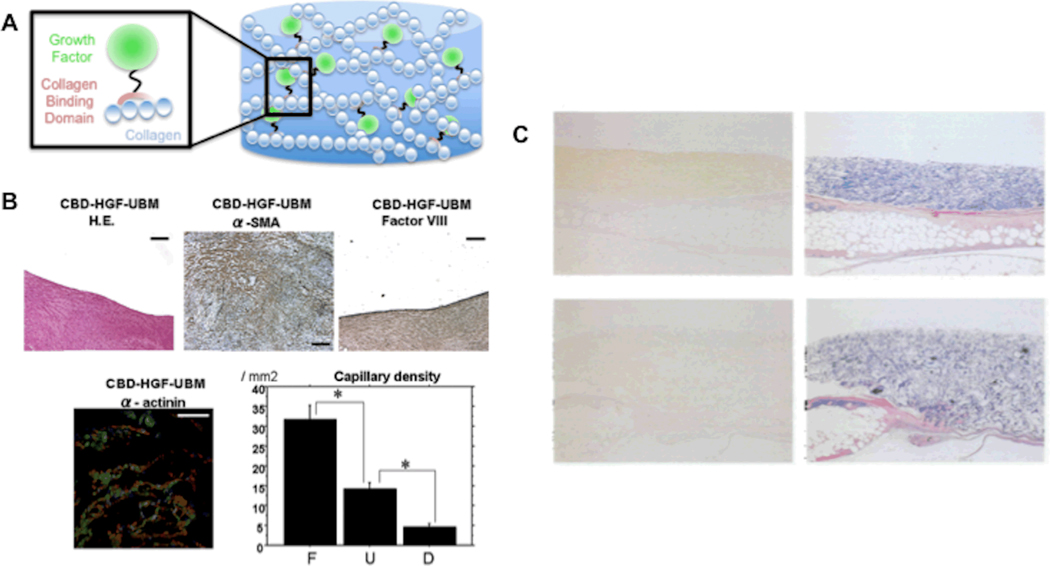
Unlike FGF-2, however, most growth factors of interest for tissue engineering applications lack collagen-binding domains.[121] As such, numerous groups have engineered growth factors to include domains that bind to members of the collagen protein family, which are schematically represented in figure 3A. For example, Zenati and co-workers prepared a fusion protein of hepatocyte growth factor (HGF) and the collagen-binding domain of fibronectin.[122] Collagen-rich cardiac patches loaded with collagen-binding HGF significantly improved cardiac function (e.g. linear local shortening and mean electrical activity), and also significantly increased the number of cells expressing the cardiac muscle markers α-smooth muscle actin and α-actinin when implanted into a surgically created defect within the porcine right ventricular wall ENREF 124 (Fig. 3B).
Figure 3. Engineered collagen-binding growth factors enhance tissue regeneration in vivo.
A) Schematic representation of collagen-binding domain-growth fusion proteins bound to collagenous matrices. B) Implantation site of collagen-binding hepatocyte growth factor loaded patches stained positively for endothelial cells and cardiomyocytes at 60 days. The density of capillaries at the patch implant site is also significantly greater than the density at control or Dacron patch site implantation. Reproduced with permission from[122] 2008 Elsevier. C) Day 4 post-implantation anti-EGF (left) and azan staining (right) of collagen sponges loaded with collagen-binding epidermal growth factor (top) or epidermal growth factor (bottom) (sponges were implanted on day 3 after epidermal wounding in diabetic mice). Reproduced with permission from[123] 2001 Oxford.
In a different study, Kitajima and co-workers prepared a fusion of epidermal growth factor (EGF) with the collagen-binding domain of fibronectin.[123] NRK49F rat kidney cells proliferated more robustly on collagen substrates exposed to collagen-binding EGF compared to those exposed to native EGF. Importantly, this result indicates that collagen-binding EGF is retained in an active form within collagen matrices. Collagen sponges loaded with collagen-binding EGF also significantly enhanced epidermal wound healing of diabetic mice, an animal model of impaired wound healing, when compared to collagen sponges loaded with native EGF (Fig. 3C).
In a third study, Dai and co-workers prepared a fusion of VEGF with a collagen type 1-binding heptapeptide.[124] Collagen-binding VEGF specifically bound to type 1 collagen and enhanced human umbilical vein endothelial cell proliferation in vitro. Collagen matrices loaded with collagen-binding VEGF were highly vascularized 14 days after subcutaneous implantation in a rat model. Additionally, collagen-binding VEGF loaded matrices delivered to a rat cardiac infarct border zone after myocardial infarction improved myocardial function, as measured by echocardiography and hemodynamic analysis.
Together, these results demonstrate that the activity of growth factors engineered to contain a collagen-binding domain is localized to collagen-based materials in vitro and in vivo. Formation of multiple tissue types, including myocardium and epidermis, with this approach suggests a widespread potential to use this method in tissue engineering strategies related to other collagen-rich tissues and biomaterials.
3.2. Mimicking growth factor binding to ECM proteins
Numerous ECM proteins regulate growth factor activity and, unlike the ubiquitous GAG-growth factor interactions discussed in section 2 above, these interactions often have well-defined binding specificity. For example, VEGF binds to fibronectin in a pH-dependent manner,[125] and fibronectin-bound VEGF enhances endothelial cell proliferation and migration.[126] VEGF-mediated differentiation of endothelial progenitor cells (EPCs) into endothelial colonies is also enhanced by fibronectin when compared to ECM proteins that do not bind VEGF, such as collagen I, collagen IV, or vitronectin.[127] Hepatocyte growth factor bound to fibronectin or vitronectin up-regulates HGF-mediated endothelial cell migration when compared to free HGF.[128] Connective tissue growth factor (CTGF) bound to fibronectin mediates oval cell adhesion and migration, as well as oval cell activation during liver regeneration.[129] Insulin-like growth factor-2 (IGF-II) bound to vitronectin enhances protein synthesis and cell migration of skin keratinocytes, but only enhances protein synthesis of corneal epithelial cells.[130]. The latter example nicely demonstrates that growth factor-ECM interactions can cause different cell types to respond differently to the same growth factor.
Interestingly, ECM proteins can also inhibit growth factor activity via specific binding. For example, latent complexes of various TGF-β isoforms and accessory proteins are stored in the ECM via interactions with ECM proteins, such as fibronectin and fibrillins.[131, 132] In addition, von Willebrand factor type C (VWC) domains, which are present in chordin and other ECM proteins, inhibit BMP-2 activity by binding to the receptor-binding domain of the growth factor.[133] Interestingly, chordin inhibition increases osteogenic differentiation of mesenchymal stem cells,[134] which suggests that chordin or VWC domains may be useful to regulate stem cell fate within biomaterials.
These examples of natural ECM proteins as regulators of growth factor activity may be applicable also to synthetic biomaterials. Recently, Anseth and co-workers modified PEG hydrogel matrices with a peptide that binds to tumor necrosis factor-α (TNF-α).[135] TNF-α bound to the TNF-α-binding peptide in a dose-dependent manner, leading to decreased TNF-α-mediated apoptosis of multiple cell types (PC12 cells, hMSCs, and pancreatic islets) encapsulated within the hydrogel. Although this study only provides one example of modifying a synthetic biomaterial with a peptide that binds to a target growth factor, the multitude of growth factor binding sites present within different ECM proteins suggests that this approach may provide a level of control that is unattainable with heparin PG- or GAG-modified biomaterials.
3.3. Growth factors engineered to bind to mineral-based biomaterials
The ECM of hard tissues (e.g. bone and teeth) is characterized by a collagen-rich organic phase and a phase of inorganic calcium-phosphate (Ca-P) minerals, principally hydroxyapatite. Numerous proteins present within hard tissue ECMs, such as bone sialoprotein, osteopontin, and osteocalcin, demonstrate high affinity binding to hydroxyapatite minerals via amino acids with phosphate- or carboxylate-terminated side chains.[136, 137] However, most growth factors involved in hard tissue development lack a specific mineral-binding domain. Recently, we and others have localized growth factor activity to mineralized ECMs and mineral-based biomaterials by preparing modular growth factors that contain a growth factor unit and mineral-binding unit (schematically represented in figure 4A). We highlight these engineered, mineral-binding growth factors and their application in tissue engineering in the following sections.
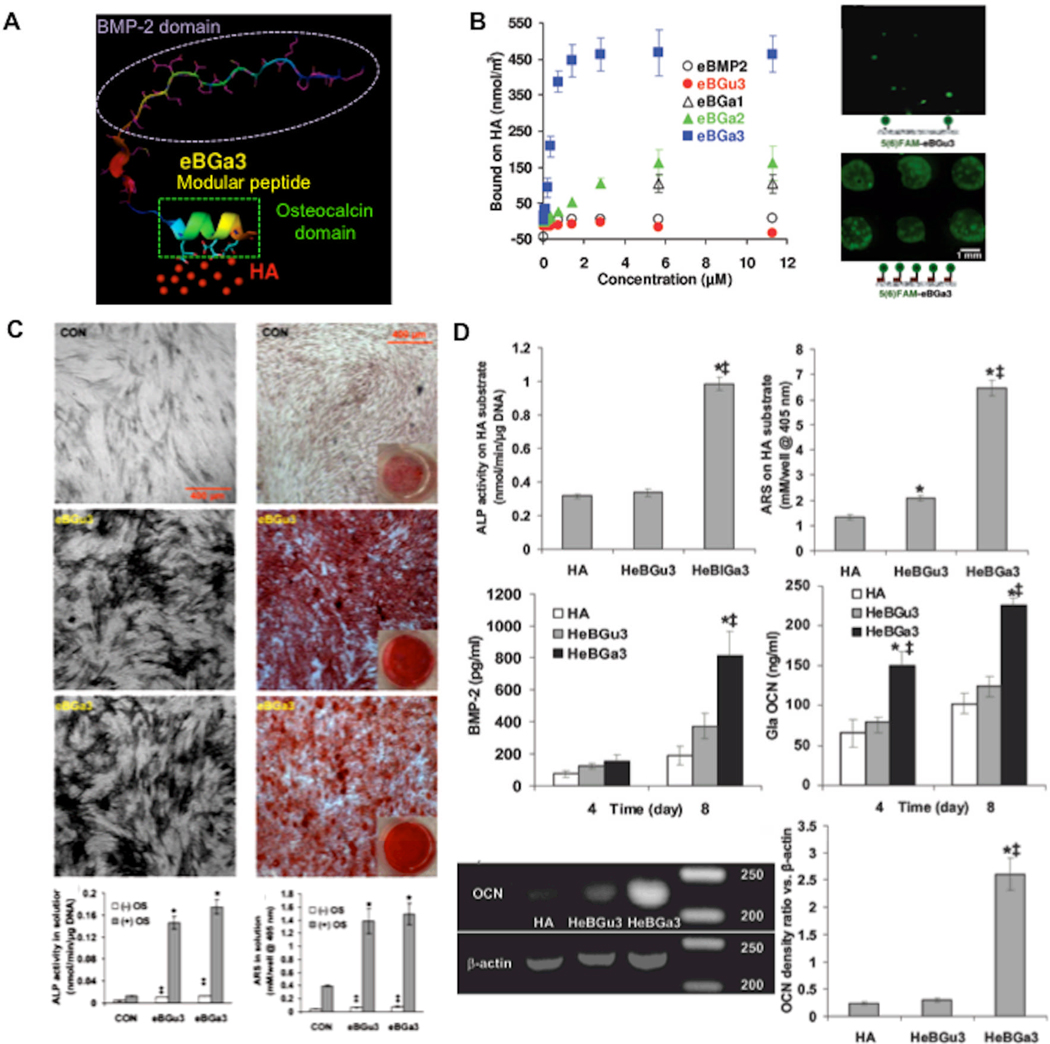
Figure 4. Modular peptide containing a mineral-binding sequence and a sequence derived from bone morphogenetic protein-2 bind to mineral-based biomaterials and enhance hMSC osteogenic differentiation.
A) Schematic representation of modular BMP-2 peptide bound to hydroxyapatite (HA). B) Fluorescent photomicrograph and binding curve demonstrating controllable binding of modular BMP-2 peptides to HA. (C) Both the BMP-2 peptide alone (left) and the modular, mineral-binding BMP-2 peptide (right) enhanced hMSC osteogenic differentiation when added in solution, indicating that the mineral-binding sequence did not interfere with biological activity (differentiation demonstrated by enhanced alkaline phosphatase staining and alizarin red staining for mineralized tissue). (D) The modular BMP-2 peptide up-regulated osteogenic differentiation when bound to a hydroxyapatite biomaterial, here indicated by upregulated mRNA expression of osteogenic markers. Reproduced with permission from[143] 2009 Wiley Interscience.
3.3.1 GAGs, proteins, and peptides engineered to bind to mineral-based materials
Uludag and co-workers proposed that incorporating a mineral-binding bisphosphonate moiety into non-mineral binding heparin would allow for heparin binding to mineralized matrices. The bisphosphonate-heparin affinity for hydroxyapatite depended on the bisphosphonate density on the GAG chain,[138] and heparin-bisphosphonate promoted FGF-2 and BMP-2 binding to hydroxyapatite in a heparin dose-dependent manner. Interestingly, Uludag and co-workers also demonstrated that bisphosphonate-modified albumin bound to hydroxyapatite, and its release rate from hydroxyapatite was significantly decreased when compared to native albumin.[139] When injected into the medullary cavity of an ovariectomized rat tibia, the concentration of bisphosphonate-modified albumin was 12-fold greater than the concentration of bisphosphonate-free albumin after 3 days.[140] Although albumin is not a growth factor, this suggests that growth factor activity can be localized within mineralized ECMs via bisphosphonate-modification.
Recent efforts have also relied on the concept of recombinant ‘fusion proteins’ to create mineral-binding growth factors. Jang and co-workers demonstrated that FGF-2 fused to the mineral-binding domain of osteocalcin bound onto hydroxyapatite with significantly higher affinity than native FGF-2.[141] Osteoblast cells cultured in the presence of the osteocalcin-FGF-2 fusion protein demonstrated increased proliferation and differentiation when compared to cells cultured in the presence of native FGF-2.
We recently developed a class of modular peptides with a mineral-binding sequence and a growth factor-derived sequence using a standard solid-phase peptide synthesis approach.[142, 143] Specifically, we generated a series of modular peptides with mineral-binding sequences inspired by osteocalcin and a BMP-2-derived sequence previously shown to promote osteogenic differentiation and ectopic bone formation.[144] Hydroxyapatite binding affinity of these modular peptides was dependent on the number of carboxyglutamic acid residues included in the osteocalcin-inspired mineral-binding sequence (Fig. 4B). Hydroxyapatite-bound BMP-2 peptides increased hMSC differentiation into bone-forming cells, as measured by increased alkaline phosphatase activity, mineralized tissue formation, BMP-2 expression, and osteocalcin expression (Fig. 4C–D). Importantly, our observation that mineral-binding modular peptides bind to hydroxyapatite and enhance hMSC differentiation in vitro suggested that these modular peptides could be advantageous for bone tissue engineering applications.
In a second set of studies, we characterized the influence of mineral-binding BMP-2 peptides on bone-tendon healing in vivo. Interference screws, which are commonly used to join bone and tendon in close proximity for anterior cruciate ligament reconstruction,[145] often provide limited bone-tendon healing in the absence of additional biological factors, such as recombinant bone morphogenetic protein-2.[146] We “dip-coated” mineral binding BMP-2 peptides onto interference screws and characterized the influence of local BMP-2 activity on bone-tendon healing in a sheep bone-tendon healing model.[147] Screws coated with the modular BMP-2 peptide significantly increased mesenchymal precursor cell density and new bone formation at the bone-tendon healing site 6 weeks after surgery. This localized effect of the modular BMP-2 peptide suggests the potential of this approach to improve bone-tendon healing over a longer term. More recent studies in our group have shown that intra-operative “dip-coating” of hydroxyapatite-coated titanium implants dramatically improves new appositional bone formation at the implant-bone interface in a sheep model, suggesting that these modular peptides may be clinically translatable in multiple contexts.
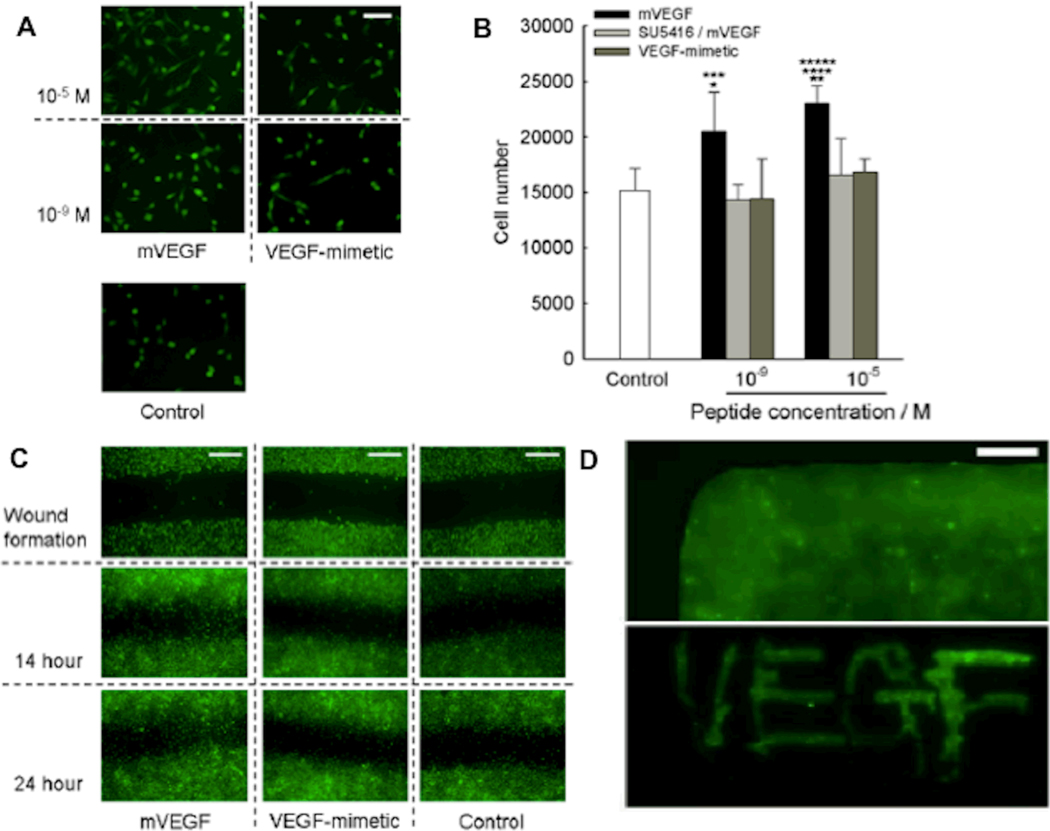
Up-regulation of stem cell differentiation into bone-forming cells is an important facet of bone regeneration. However, blood vessel growth is perhaps equally critical for bone tissue engineering, and there has been limited clinical success in promoting concerted bone and blood vessel regeneration to date.[148] VEGF is commonly used to promote blood vessel growth into biomaterials. Recently, Pedone and co-workers demonstrated that the ““VEGF-mimicking” peptide KLTWQELYQLKYKGI binds to VEGF receptors and promotes endothelial cell proliferation.[149] To localize VEGF activity within mineral-based biomaterials, we prepared a modular, mineral-binding version of the VEGF-mimicking peptide. The modular VEGF peptide bound strongly and rapidly to the surface of hydroxyapatite biomaterials, and promoted endothelial cell proliferation and migration (Fig. 5).[150] This demonstration that modular VEGF can promote endothelial cell functions involved in early blood vessel growth, coupled with the observation that modular BMP-2 promotes osteogenic differentiation, suggests that growth factor-inspired peptides bound to hydroxyapatite may work in concert to induce osteogenesis and angiogenesis. Importantly, these studies also showed that modular versions of VEGF and BMP-2 can be applied to biomaterials simply by “painting” or “dipping” procedures, which facilitate spatial control and intra-operative activation of biomaterials in clinical applications.
Figure 5. A modular peptide containing a mineral-binding sequence and a sequence that mimics the vascular endothelial growth factor active site enhances endothelial cell proliferation and migration during culture on mineral-based biomaterials.
Fluorescent photomicrographs (A) or cell number (B) of C166-GFP endothelial cells after 48 hour culture on hydroxyapatite slabs coated with a mineral binding VEGF peptide (mVEGF), mVEGF plus a VEGF signaling inhibitor (SU5416), or the VEGF peptide without a mineral-binding sequence (VEGF-mimetic). C) Fluorescent photomicrographs of C166-GFP endothelial cell migration in a “wound assay” on hydroxyapatite slabs coated with a mineral-binding VEGF peptide, a soluble VEGF peptide, or basal medium. D) Fluorescent photomicrographs demonstrating that modular mineral-binding peptides can be dip-coated (top) or ‘painted’, as demonstrated by writing VEGF on slab with fluorescent mVEGF peptide (bottom), onto mineral-based biomaterials. Reproduced with permission from[150] Wiley Interscience 2010.
3.4 Summary
Importantly, the strategies described in this section allow for growth factor activity to be localized on protein- and mineral-based biomaterials, whose chemical composition is often difficult to modify without altering bulk material properties or biochemical function. The range of in vivo tissue engineering examples also show that combining natural biomaterials with engineered growth factors is a widely applicable strategy.
4.1 Non-covalent polyelectrolyte assemblies
GAGs are anionic polymers of carboxylated and sulfated saccharides that can associate into highly ordered structures via ionic interactions with polycationic polymers. Polyelectrolyte assemblies play an important role in tissue mechanics and as biophysical barriers in the ECM. For example, the glycocalyx, which is a polyelectrolyte assembly of primarily heparan sulfate and hyaluronan that is present on the surface of most eukaryotic cells, plays an important role in platelet-endothelial cell adhesion and as a barrier in the blood vessel lumen.[151] The aggrecan aggregate, on the other hand, is an assembly formed between the PG aggrecan, a binding protein, and hyaluronan, that is found within the ECM of articular cartilage and the intervertebral disk. The aggrecan aggregate organizes other ECM components (e.g collagen fibrils), modulates transport of GAG-binding proteins (e.g. growth factors), and confers the cartilage ECM with a relatively high compressive modulus by maintaining high water content.[152] Together, these natural polyelectrolyte assemblies provide an interesting physical model to design ECM-mimicking biomaterials for tissue engineering applications.
Layer-by-layer assembly, a process of forming materials by sequentially adsorbing oppositely charged polymers onto a substrate, provides a method to mimic the polyelectrolyte assemblies within natural ECMs. For example, heparin (an anionic GAG) and chitosan (a cationic polysaccharide) can be sequentially adsorbed onto a variety of materials, including poly(ethylene terepthalate),[153] titanium,[154] stainless steel,[155] polycaprolactone,[156] and poly(L-lactide),[157] resulting in ordered polyelectrolyte layers. The layer-by-layer process controls the polyelectrolyte assembly’s physical properties, such as thickness and mechanics, by simply varying the number of oppositely charged polymer adsoption steps. To date, polyelectrolyte assemblies have been extensively used as biomaterials to carry cells or deliver DNA/RNA, and their application in this regard has been extensively reviewed elsewhere.[158] Layer-by-layer assemblies can also be used to deliver biologically active proteins, including growth factors.
In an early example of polyelectrolyte assemblies for protein delivery, Schaaf and co-workers demonstrated that protein A incorporated into poly(lysine) and poly(glutamate) assemblies is presented to macrophages in an active conformation, and the rate of protein A presentation can be controlled by changing the thickness of the adsorbed layers or by varying the molar ratio of poly-l-lysine and poly-l-glutamate (i.e. enzymatically degradable amino acid polymers) with poly-d-lysine and poly-d-glutamate (i.e. non-degradable amino acid polymers) polymers in the assembly.[159] Although protein A is not a growth factor, and neither poly(lysine) nor poly(glutamate) are ECM-derived polyelectrolytes, this initial demonstration suggested that a similar approach may be used to deliver growth factors from ECM-derived GAG assemblies as well. Toward that end, Lavalle and co-workers demonstrated that photoreceptor cell adhesion and differentiation on poly-l-lysine and chondroitin sulfate assemblies are improved by adsorbing basic fibroblast growth factor, which non-covalently binds to chondroitin sulfate GAGs.[160] More recently, Picart and colleagues showed that poly(L-lysine)/hyaluronan assemblies loaded with recombinant BMP-2 induce myoblast differentiation into osteoblasts.[161] Interestingly, the bioactivity of BMP-2 was not due to film degradation or protein release, as only ∼3% of the total protein included in the film was released into culture medium. Instead, the bioactivity of BMP-2 within these films was likely related to the specific interaction of BMP-2 with hyaluronan GAGs, which mimics the influence of the same GAGs on BMP-2 within natural ECMs.
4.2 Summary
Together, these studies demonstrate that layer-by-layer polyelectrolyte films, which are inspired by the polyelectrolyte assemblies in natural ECMs, can non-covalently localize growth factor activity for in vitro cell culture applications. Although not demonstrated to date, these studies suggest that polyelectrolyte assemblies containing ECM-derived GAGs may be advantageous for in vivo tissue engineering therapies.
5. Conclusions
A key component of many tissue engineering approaches involves growth factor delivery to sites of damaged or diseased tissue. Within the natural ECM, PGs and proteins influence growth factor activity in diverse ways. Inspired by the natural ECM, several investigators have developed biomaterials that localize growth factor activity. GAGs and PGs in synthetic hydrogels decrease the rate of heparin-binding growth factor release and enhance blood vessel growth within soft tissues. Growth factors engineered to include collagen-binding domains are localized to collagen-rich matrices, where they improve myocardial repair after myocardial infarction and enhance epidermal wound healing. Additionally, growth factors and peptide-based growth factor mimics engineered to include a mineral-binding domain promote cell proliferation and differentiation on mineral-based biomaterials. Layer-by-layer assemblies of GAGs and other natural polyelectrolytes non-covalently retain growth factors at a cell-material interface. Together, these examples highlight the potential of bioinspired, non-covalent interactions to regulate growth factor activity for diverse tissue engineering applications.
Throughout this review, we also highlight examples of biomaterials that mimic nature’s sequestering interactions to control native growth factor activity. The examples introduce a new paradigm, in which growth factors are not necessarily released from a biomaterial, but instead a biomaterial is designed to bind to growth factors that are already present in standard cell culture or in vivo. One particular advantage of this approach relates to temporal dynamics. Biomaterials engineered to adapt to temporal fluctuations in growth factor expression, as well as the proteins and proteoglycans that regulate their activity, may ultimately mimic natural ECMs and control cell behavior over time. Recently, we and others developed biomaterials that specifically bind to and sequester cell-secreted biomolecules, and the equilibrium nature of these interactions suggests that the type and density of sequestered biomolecules will vary as a function of cell behavior. Thus, biomaterials engineered to mimic the various sequestering interactions in natural ECMs may provide ECM mimics that dynamically regulate tissue development and regeneration.
Acknowledgements
This work was supported by the National Institutes of Health (R01HL093282 and R01EY017367).
Biographies

Murphy is an Associate Professor of Biomedical Engineering, Pharmacology, and Orthopedics/Rehabilitation at the University of Wisconsin, where he has been since 2004. He received his Ph.D. in Biomedical Engineering from the University of Michigan in 2002, and was a postdoctoral fellow in Chemistry at the University of Chicago from 2002–2004. Murphy’s research interests focus on designing “bioinspired” materials that mimic and exploit biological systems. He has published over 50 manuscripts and filed 15 patents.

Hudalla is a postdoctoral fellow in Surgery and Chemistry at the University of Chicago, where he has been since 2010. He received his Ph.D. in Biomedical Engineering from the University of Wisconsin in 2010. Hudalla’s doctoral work focused on materials that use bio-inspired mechanisms to modulate stem cell behavior, while his current research interests focus on bio-inspired approaches to develop immunomodulatory materials.
References
- 1.Fawcett JW, Keynes RJ. Annu Rev Neurosci. 1990;13:43. doi: 10.1146/annurev.ne.13.030190.000355. [DOI] [PubMed] [Google Scholar]
- 2.Martin P. Science. 1997;276:75. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 3.McKibbin B. J Bone Joint Surg Br. 1978;60-B:150. doi: 10.1302/0301-620X.60B2.350882. [DOI] [PubMed] [Google Scholar]
- 4.Yannis IV. In: Scarless Wound Healing. Garg HG, Longaker MT, editors. Marcel Dekker; 2000. [Google Scholar]
- 5.Wynn TA. J Pathol. 2008;214:199. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diegelmann RF, Evans MC. Front Biosci. 2004;9:283. doi: 10.2741/1184. [DOI] [PubMed] [Google Scholar]
- 7.Mutsaers SE, Bishop JE, McGrouther G, Laurent GJ. Int J Biochem Cell Biol. 1997;29:5. doi: 10.1016/s1357-2725(96)00115-x. [DOI] [PubMed] [Google Scholar]
- 8.Weber KT. Semin Nephrol. 1997;17:467. [PubMed] [Google Scholar]
- 9.Desmouliere A, Chaponnier C, Gabbiani G. Wound Repair Regen. 2005;13:7. doi: 10.1111/j.1067-1927.2005.130102.x. [DOI] [PubMed] [Google Scholar]
- 10.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 11.Hubbell JA. Nature Biotech. 1995;13:565. doi: 10.1038/nbt0695-565. [DOI] [PubMed] [Google Scholar]
- 12.Smith IO, Ma PX. Sci. Techno. Adv. Mater. 2010;11:1. doi: 10.1088/1468-6996/11/1/014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badylak SF, Freytes DO, Gilbert TW. Acta Biomater. 2009;5:1. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Habraken WJ, Wolke JG, Jansen JA. Adv Drug Deliv Rev. 2007;59:234. doi: 10.1016/j.addr.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Lin CC, Anseth KS. Pharm Res. 2009;26:631. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto T, Okazaki M, Nakahira A, Sasaki J, Egusa H, Sohmura T. Curr Med Chem. 2007;14:2726. doi: 10.2174/092986707782023208. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt JJ, Rowley J, Kong HJ. J Biomed Mater Res A. 2008;87:1113. doi: 10.1002/jbm.a.32287. [DOI] [PubMed] [Google Scholar]
- 18.Tessmar JK, Gopferich AM. Adv Drug Deliv Rev. 2007;59:274. doi: 10.1016/j.addr.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Vinatier C, Guicheux J, Daculsi G, Layrolle P, Weiss P. Biomed Mater Eng. 2006;16:S107. [PubMed] [Google Scholar]
- 20.Yang C, Hillas PJ, Baez JA, Nokelainen M, Balan J, Tang J, Spiro R, Polarek JW. BioDrugs. 2004;18:103. doi: 10.2165/00063030-200418020-00004. [DOI] [PubMed] [Google Scholar]
- 21.Yoshikawa H, Myoui A. J Artif Organs. 2005;8:131. doi: 10.1007/s10047-005-0292-1. [DOI] [PubMed] [Google Scholar]
- 22.Unsicker K, Krieglstein K. In: in 1–2. Wiley VCH, editor. 2006. [Google Scholar]
- 23.Sokolsky-Papkov M, Agashi K, Olaye A, Shakesheff K, Domb AJ. Adv Drug Deliv Rev. 2007;59:187. doi: 10.1016/j.addr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Lee SH, Shin H. Adv Drug Deliv Rev. 2007;59:339. doi: 10.1016/j.addr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Chung HJ, Park TG. Adv Drug Deliv Rev. 2007;59:249. doi: 10.1016/j.addr.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Silva AK, Richard C, Bessodes M, Scherman D, Merten OW. Biomacromolecules. 2009;10:9. doi: 10.1021/bm801103c. [DOI] [PubMed] [Google Scholar]
- 27.Varde NK, Pack DW. Expert Opin Biol Ther. 2004;4:35. doi: 10.1517/14712598.4.1.35. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Huang G. Crit Rev Ther Drug Carrier Syst. 2009;26:29. doi: 10.1615/critrevtherdrugcarriersyst.v26.i1.20. [DOI] [PubMed] [Google Scholar]
- 29.Mann BK, Schmedlen RH, West JL. Biomaterials. 2001;22:439. doi: 10.1016/s0142-9612(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 30.Kuhl PR, Griffith-Cima LG. Nat Med. 1996;2:1022. doi: 10.1038/nm0996-1022. [DOI] [PubMed] [Google Scholar]
- 31.DeLong SA, Moon JJ, West JL. Biomaterials. 2005;26:3227. doi: 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 32.Bentz H, Schroeder JA, Estridge TD. J Biomed Mater Res. 1998;39:539. doi: 10.1002/(sici)1097-4636(19980315)39:4<539::aid-jbm6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 33.Backer MV, Patel V, Jehning BT, Claffey KP, Backer JM. Biomaterials. 2006;27:5452. doi: 10.1016/j.biomaterials.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Hsia E, Richardson TP, Nugent MA. J Cell Biochem. 2003;88:1214. doi: 10.1002/jcb.10470. [DOI] [PubMed] [Google Scholar]
- 35.Richardson TP, Trinkaus-Randall V, Nugent MA. J Cell Sci. 2001;114:1613. doi: 10.1242/jcs.114.9.1613. [DOI] [PubMed] [Google Scholar]
- 36.Sperinde GV, Nugent MA. Biochemistry. 1998;37:13153. doi: 10.1021/bi980600z. [DOI] [PubMed] [Google Scholar]
- 37.Sperinde GV, Nugent MA. Biochemistry. 2000;39:3788. doi: 10.1021/bi992243d. [DOI] [PubMed] [Google Scholar]
- 38.Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin H, Jo S, Mikos AG. Biomaterials. 2003;24:4353. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 40.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. in 2002, 1090. [Google Scholar]
- 41.Hayman EG, Pierschbacher MD, Ruoslahti E. J Cell Biol. 1985;100:1948. doi: 10.1083/jcb.100.6.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pytela R, Pierschbacher MD, Ginsberg MH, Plow EF, Ruoslahti E. Science. 1986;231:1559. doi: 10.1126/science.2420006. [DOI] [PubMed] [Google Scholar]
- 43.Gardner JM, Hynes RO. Cell. 1985;42:439. doi: 10.1016/0092-8674(85)90101-1. [DOI] [PubMed] [Google Scholar]
- 44.Horwitz A, Duggan K, Greggs R, Decker C, Buck C. J Cell Biol. 1985;101:2134. doi: 10.1083/jcb.101.6.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dedhar S, Ruoslahti E, Pierschbacher MD. J Cell Biol. 1987;104:585. doi: 10.1083/jcb.104.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saoncella S, Echtermeyer F, Denhez F, Nowlen JK, Mosher DF, Robinson SD, Hynes RO, Goetinck PF. Proc Natl Acad Sci U S A. 1999;96:2805. doi: 10.1073/pnas.96.6.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McQuade KJ, Beauvais DM, Burbach BJ, Rapraeger AC. J Cell Sci. 2006;119:2445. doi: 10.1242/jcs.02970. [DOI] [PubMed] [Google Scholar]
- 48.Hozumi K, Suzuki N, Nielsen PK, Nomizu M, Yamada Y. J Biol Chem. 2006;281:32929. doi: 10.1074/jbc.M605708200. [DOI] [PubMed] [Google Scholar]
- 49.Cole GJ, Loewy A, Glaser L. Nature. 1986;320:445. doi: 10.1038/320445a0. [DOI] [PubMed] [Google Scholar]
- 50.Celie JW, Keuning ED, Beelen RH, Drager AM, Zweegman S, Kessler FL, Soininen R, van den Born J. J Biol Chem. 2005;280:26965. doi: 10.1074/jbc.M502188200. [DOI] [PubMed] [Google Scholar]
- 51.Luo J, Kato M, Wang H, Bernfield M, Bischoff J. J Cell Biochem. 2001;80:522. [PubMed] [Google Scholar]
- 52.Hayashi K, Kadomatsu K, Muramatsu T. Glycoconj J. 2001;18:401. doi: 10.1023/a:1014864131288. [DOI] [PubMed] [Google Scholar]
- 53.Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury JE, Pinchasi D, Huang J, Jaye M, Crumley G, Schlessinger J, Lax I. Cell. 1994;79:1015. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 54.Ornitz DM, Yayon A, Flanagan JG, Svahn CM, Levi E, Leder P. Mol Cell Biol. 1992;12:240. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aviezer D, Safran M, Yayon A. Biochem Biophys Res Commun. 1999;263:621. doi: 10.1006/bbrc.1999.1434. [DOI] [PubMed] [Google Scholar]
- 56.Smith SM, West LA, Hassell JR. Arch Biochem Biophys. 2007;468:244. doi: 10.1016/j.abb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fthenou E, Zafiropoulos A, Katonis P, Tsatsakis A, Karamanos NK, Tzanakakis GN. J Cell Biochem. 2008;103:1866. doi: 10.1002/jcb.21570. [DOI] [PubMed] [Google Scholar]
- 58.Jiao X, Billings PC, O’Connell MP, Kaplan FS, Shore EM, Glaser DL. J Biol Chem. 2007;282:1080. doi: 10.1074/jbc.M513414200. [DOI] [PubMed] [Google Scholar]
- 59.Hill RS, Cruise GM, Hager SR, Lamberti FV, Yu X, Garufis CL, Yu Y, Mundwiler KE, Cole JF, Hubbell JA, Hegre OD, Scharp DW. Ann N Y Acad Sci. 1997;831:332. doi: 10.1111/j.1749-6632.1997.tb52208.x. [DOI] [PubMed] [Google Scholar]
- 60.Bryant SJ, Nuttelman CR, Anseth KS. J Biomater Sci Polym Ed. 2000;11:439. doi: 10.1163/156856200743805. [DOI] [PubMed] [Google Scholar]
- 61.Benoit DS, Anseth KS. Acta Biomater. 2005;1:461. doi: 10.1016/j.actbio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Cushing MC, Liao JT, Jaeggli MP, Anseth KS. Biomaterials. 2007;28:3378. doi: 10.1016/j.biomaterials.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Tsang M, Dawid IB. Sci STKE. 2004;2004:e17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- 64.Skalli O, Schurch W, Seemayer T, Lagace R, Montandon D, Pittet B, Gabbiani G. Lab Invest. 1989;60:275. [PubMed] [Google Scholar]
- 65.Benoit DS, Durney AR, Anseth KS. Biomaterials. 2007;28:66. doi: 10.1016/j.biomaterials.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 66.Benoit DS, Collins SD, Anseth KS. Adv Funct Mater. 2007;17:2085. doi: 10.1002/adfm.200700012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lian JB, Stein GS. Iowa Orthop J. 1995;15:118. [PMC free article] [PubMed] [Google Scholar]
- 68.Elbert DL, Pratt AB, Lutolf MP, Halstenberg S, Hubbell JA. J Control Release. 2001;76:11. doi: 10.1016/s0168-3659(01)00398-4. [DOI] [PubMed] [Google Scholar]
- 69.McGonigle JS, Tae G, Stayton PS, Hoffman AS, Scatena M. J Biomater Sci Polym Ed. 2008;19:1021. doi: 10.1163/156856208784909381. [DOI] [PubMed] [Google Scholar]
- 70.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Cell. 1998;93:165. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 71.Cross SS, Yang Z, Brown NJ, Balasubramanian SP, Evans CA, Woodward JK, Neville-Webbe HL, Lippitt JM, Reed MW, Coleman RE, Holen I. Int J Cancer. 2006;118:1901. doi: 10.1002/ijc.21606. [DOI] [PubMed] [Google Scholar]
- 72.Yamaguchi N, Kiick KL. Biomacromolecules. 2005;6:1921. doi: 10.1021/bm050003+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, Furst EM, Kiick KL. J Control Release. 2006;114:130. doi: 10.1016/j.jconrel.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nie T, Baldwin A, Yamaguchi N, Kiick KL. J Control Release. 2007;122:287. doi: 10.1016/j.jconrel.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wight TN, Heinegard DK, Hascall VC. Cell Biology of the Extracellular Matrix. Plenum Press; 1991. p. 45. [Google Scholar]
- 76.Bosman FT, Stamenkovic I. J Pathol. 2003;200:423. doi: 10.1002/path.1437. [DOI] [PubMed] [Google Scholar]
- 77.Fraser JR, Laurent TC, Laurent UB. J Intern Med. 1997;242:27. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 78.Toole BP. Semin Cell Dev Biol. 2001;12:79. doi: 10.1006/scdb.2000.0244. [DOI] [PubMed] [Google Scholar]
- 79.Maeda N, Fukazawa N, Ishii M. Front Biosci. 15:626. doi: 10.2741/3637. [DOI] [PubMed] [Google Scholar]
- 80.Shepard JB, Krug HA, LaFoon BA, Hoffman S, Capehart AA. Int J Biol Sci. 2007;3:380. doi: 10.7150/ijbs.3.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai S, Liu Y, Zheng Shu X, Prestwich GD. Biomaterials. 2005;26:6054. doi: 10.1016/j.biomaterials.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 82.Esko JD, Kimata K, Lindahl U. Essentials of Glycobiology. Cold Spring Harbor; 2009. p. 229. [Google Scholar]
- 83.Tanihara M, Suzuki Y, Yamamoto E, Noguchi A, Mizushima Y. J Biomed Mater Res. 2001;56:216. doi: 10.1002/1097-4636(200108)56:2<216::aid-jbm1086>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 84.Xu Y, Zhan C, Fan L, Wang L, Zheng H. Int J Pharm. 2007;336:329. doi: 10.1016/j.ijpharm.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 85.Babister JC, Tare RS, Green DW, Inglis S, Mann S, Oreffo RO. Biomaterials. 2008;29:58. doi: 10.1016/j.biomaterials.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 86.Augst AD, Kong HJ, Mooney DJ. Macromol Biosci. 2006;6:623. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 87.Berger J, Reist M, Mayer JM, Felt O, Gurny R. Eur J Pharm Biopharm. 2004;57:35. doi: 10.1016/s0939-6411(03)00160-7. [DOI] [PubMed] [Google Scholar]
- 88.Berger J, Reist M, Mayer JM, Felt O, Peppas NA, Gurny R. Eur J Pharm Biopharm. 2004;57:19. doi: 10.1016/s0939-6411(03)00161-9. [DOI] [PubMed] [Google Scholar]
- 89.Huang Z, Ni C, Chu Y, Wang S, Yang S, Wu X, Wang X, Li X, Feng W, Lin S. J Biotechnol. 2009;142:242. doi: 10.1016/j.jbiotec.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 90.Ryou H-W, Lee J-W, Yoon KA, Park E-S, Chi S-C. Arch Pharm Res. 1997;20:34. doi: 10.1007/BF02974039. [DOI] [PubMed] [Google Scholar]
- 91.Yamaguchi Y. Cell Mol Life Sci. 2000;57:276. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yurchenco PD, Amenta PS, Patton BL. Matrix Biol. 2004;22:521. doi: 10.1016/j.matbio.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 93.Ingham KC, Brew SA, Migliorini MM, Busby TF. Biochemistry. 1993;32:12548. doi: 10.1021/bi00097a035. [DOI] [PubMed] [Google Scholar]
- 94.Del Rosso M, Cappelletti R, Viti M, Vannucchi S, Chiarugi V. Biochem J. 1981;199:699. doi: 10.1042/bj1990699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakiyama-Elbert SE, Hubbell JA. J Control Release. 2000;65:389. doi: 10.1016/s0168-3659(99)00221-7. [DOI] [PubMed] [Google Scholar]
- 96.Sakiyama-Elbert SE, Hubbell JA. J Control Release. 2000;69:149. doi: 10.1016/s0168-3659(00)00296-0. [DOI] [PubMed] [Google Scholar]
- 97.Wood MD, Borschel GH, Sakiyama-Elbert SE. J Biomed Mater Res A. 2009;89:909. doi: 10.1002/jbm.a.32043. [DOI] [PubMed] [Google Scholar]
- 98.Thomopoulos S, Zaegel M, Das R, Harwood FL, Silva MJ, Amiel D, Sakiyama-Elbert S, Gelberman RH. J Orthop Res. 2007;25:1358. doi: 10.1002/jor.20444. [DOI] [PubMed] [Google Scholar]
- 99.Taylor SJ, McDonald JW, 3rd, Sakiyama-Elbert SE. J Control Release. 2004;98:281. doi: 10.1016/j.jconrel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 100.Johnson PJ, Parker SR, Sakiyama-Elbert SE. Biotechnol Bioeng. 2009;104:1207. doi: 10.1002/bit.22476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee AC, Yu VM, Lowe JB, 3rd, Brenner MJ, Hunter DA, Mackinnon SE, Sakiyama-Elbert SE. Exp Neurol. 2003;184:295. doi: 10.1016/s0014-4886(03)00258-9. [DOI] [PubMed] [Google Scholar]
- 102.Thomopoulos S, Das R, Silva MJ, Sakiyama-Elbert S, Harwood FL, Zampiakis E, Kim HM, Amiel D, Gelberman RH. J Orthop Res. 2009;27:1209. doi: 10.1002/jor.20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rajangam K, Behanna HA, Hui MJ, Han X, Hulvat JF, Lomasney JW, Stupp SI. Nano Lett. 2006;6:2086. doi: 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- 104.Stendahl JC, Wang LJ, Chow LW, Kaufman DB, Stupp SI. Transplantation. 2008;86:478. doi: 10.1097/TP.0b013e3181806d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang W, Gomes RR, Brown AJ, Burdett AR, Alicknavitch M, Farach-Carson MC, Carson DD. Tissue Eng. 2006;12:2009. doi: 10.1089/ten.2006.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang WD, Gomes RR, Jr, Alicknavitch M, Farach-Carson MC, Carson DD. Tissue Eng. 2005;11:76. doi: 10.1089/ten.2005.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Azizkhan RG, Azizkhan JC, Zetter BR, Folkman J. J Exp Med. 1980;152:931. doi: 10.1084/jem.152.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bashkin P, Doctrow S, Klagsbrun M, Svahn CM, Folkman J, Vlodavsky I. Biochemistry. 1989;28:1737. doi: 10.1021/bi00430a047. [DOI] [PubMed] [Google Scholar]
- 109.Mallein-Gerin F, Kosher RA, Upholt WB, Tanzer ML. Dev Biol. 1988;126:337. doi: 10.1016/0012-1606(88)90144-3. [DOI] [PubMed] [Google Scholar]
- 110.Manton KJ, Leong DF, Cool SM, Nurcombe V. Stem Cells. 2007;25:2845. doi: 10.1634/stemcells.2007-0065. [DOI] [PubMed] [Google Scholar]
- 111.Levenstein ME, Berggren WT, Lee JE, Conard KR, Llanas RA, Wagner RJ, Smith LM, Thomson JA. Stem Cells. 2008;26:3099. doi: 10.1634/stemcells.2007-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hudalla GA, Murphy WL. Langmuir. 2010;26:6449. doi: 10.1021/la1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hudalla GA, Kouris NA, Koepsel JT, Ogle BM, Murphy WL. Nature Chemical Biology. 2011 doi: 10.1039/c1ib00021g. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Biochem Biophys Res Commun. 2001;288:413. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- 115.Koepsel JT, Murphy WL. Langmuir. 2009;25:12825. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maynard HD, Hubbell JA. Acta Biomater. 2005;1:451. doi: 10.1016/j.actbio.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 117.Kim SH, Kiick KL. Peptides. 2007;28:2125. doi: 10.1016/j.peptides.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hunt NC, Grover LM. Biotechnol Lett. 2010 doi: 10.1007/s10529-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 119.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Chem Rev. 2008;108:4754. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kanematsu A, Marui A, Yamamoto S, Ozeki M, Hirano Y, Yamamoto M, Ogawa O, Komeda M, Tabata Y. J Control Release. 2004;99:281. doi: 10.1016/j.jconrel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 121.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. J Biol Chem. 2002;277:4223. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 122.Ota T, Gilbert TW, Schwartzman D, McTiernan CF, Kitajima T, Ito Y, Sawa Y, Badylak SF, Zenati MA. J Thorac Cardiovasc Surg. 2008;136:1309. doi: 10.1016/j.jtcvs.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ishikawa T, Terai H, Kitajima T. J Biochem. 2001;129:627. doi: 10.1093/oxfordjournals.jbchem.a002900. [DOI] [PubMed] [Google Scholar]
- 124.Zhang J, Ding L, Zhao Y, Sun W, Chen B, Lin H, Wang X, Zhang L, Xu B, Dai J. Circulation. 2009;119:1776. doi: 10.1161/CIRCULATIONAHA.108.800565. [DOI] [PubMed] [Google Scholar]
- 125.Goerges AL, Nugent MA. J Biol Chem. 2004;279:2307. doi: 10.1074/jbc.M308482200. [DOI] [PubMed] [Google Scholar]
- 126.Wijelath ES, Rahman S, Namekata M, Murray J, Nishimura T, Mostafavi-Pour Z, Patel Y, Suda Y, Humphries MJ, Sobel M. Circ Res. 2006;99:853. doi: 10.1161/01.RES.0000246849.17887.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wijelath ES, Rahman S, Murray J, Patel Y, Savidge G, Sobel M. J Vasc Surg. 2004;39:655. doi: 10.1016/j.jvs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 128.Rahman S, Patel Y, Murray J, Patel KV, Sumathipala R, Sobel M, Wijelath ES. BMC Cell Biol. 2005;6:8. doi: 10.1186/1471-2121-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pi L, Ding X, Jorgensen M, Pan JJ, Oh SH, Pintilie D, Brown A, Song WY, Petersen BE. Hepatology. 2008;47:996. doi: 10.1002/hep.22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hyde C, Hollier B, Anderson A, Harkin D, Upton Z. J Invest Dermatol. 2004;122:1198. doi: 10.1111/j.0022-202X.2004.22527.x. [DOI] [PubMed] [Google Scholar]
- 131.Chaudhry SS, Cain SA, Morgan A, Dallas SL, Shuttleworth CA, Kielty CM. J Cell Biol. 2007;176:355. doi: 10.1083/jcb.200608167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dallas SL, Sivakumar P, Jones CJ, Chen Q, Peters DM, Mosher DF, Humphries MJ, Kielty CM. J Biol Chem. 2005;280:18871. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
- 133.Zhang JL, Huang Y, Qiu LY, Nickel J, Sebald W. J Biol Chem. 2007;282:20002. doi: 10.1074/jbc.M700456200. [DOI] [PubMed] [Google Scholar]
- 134.Kwong FN, Richardson SM, Evans CH. Arthritis Res Ther. 2008;10:R65. doi: 10.1186/ar2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin CC, Metters AT, Anseth KS. Biomaterials. 2009;30:4907. doi: 10.1016/j.biomaterials.2009.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Goldberg HA, Warner KJ, Li MC, Hunter GK. Connect Tissue Res. 2001;42:25. doi: 10.3109/03008200109014246. [DOI] [PubMed] [Google Scholar]
- 137.Dowd TL, Rosen JF, Li L, Gundberg CM. Biochemistry. 2003;42:7769. doi: 10.1021/bi034470s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gittens SA, Bagnall K, Matyas JR, Lobenberg R, Uludag H. J Control Release. 2004;98:255. doi: 10.1016/j.jconrel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 139.Uludag H, Kousinioris N, Gao T, Kantoci D. Biotechnol Prog. 2000;16:258. doi: 10.1021/bp990154m. [DOI] [PubMed] [Google Scholar]
- 140.Uludag H, Gao T, Wohl GR, Kantoci D, Zernicke RF. Biotechnol Prog. 2000;16:1115. doi: 10.1021/bp000066y. [DOI] [PubMed] [Google Scholar]
- 141.Jeon E, Jang JH. Protein Pept Lett. 2009;16:664. doi: 10.2174/092986609788490267. [DOI] [PubMed] [Google Scholar]
- 142.Lee JS, Lee JS, Murphy WL. Acta Biomater. 2010;6:21. doi: 10.1016/j.actbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee JS, Lee JS, Wagoner-Johnson A, Murphy WL. Angew Chem Int Ed Engl. 2009;48:6266. doi: 10.1002/anie.200901618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saito A, Suzuki Y, Ogata S, Ohtsuki C, Tanihara M. Biochim Biophys Acta. 2003;1651:60. doi: 10.1016/s1570-9639(03)00235-8. [DOI] [PubMed] [Google Scholar]
- 145.Harvey A, Thomas NP, Amis AA. J Bone Joint Surg Br. 2005;87:593. doi: 10.1302/0301-620X.87B5.15803. [DOI] [PubMed] [Google Scholar]
- 146.Rodeo SA, Suzuki K, Deng XH, Wozney J, Warren RF. Am J Sports Med. 1999;27:476. doi: 10.1177/03635465990270041201. [DOI] [PubMed] [Google Scholar]
- 147.Lu Y, Markel MD, Nemke B, Lee JS, Graf BK, Murphy WL. Arthroscopy. 2009;25:1427. doi: 10.1016/j.arthro.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Santos MI, Reis RL. Macromol Biosci. 2010;10:12. doi: 10.1002/mabi.200900107. [DOI] [PubMed] [Google Scholar]
- 149.D’Andrea LD, Iacarrino G, Fattorusso R, Sorriento D, Carranante C, Capasso D, Trimarco B, Pedone C. Proc Natl Acad Sci. 2005;102:14215. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee JS, Wagoner Johnson AJ, Murphy WL. Adv Mater. 2010 doi: 10.1002/adma.201002970. [DOI] [PubMed] [Google Scholar]
- 151.Weinbaum S, Tarbell JM, Damiano ER. Annu Rev Biomed Eng. 2007;9:121. doi: 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- 152.Roughley PJ. Eur Cell Mater. 2006;12:92. doi: 10.22203/ecm.v012a11. [DOI] [PubMed] [Google Scholar]
- 153.Fu J, Ji J, Yuan W, Shen J. Biomaterials. 2005;26:6684. doi: 10.1016/j.biomaterials.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 154.Ruan Q, Zhu Y, Li F, Xiao J, Zeng Y, Xu F. J Coll Int Sci. 2009;333:725. doi: 10.1016/j.jcis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 155.Meng S, Liu Z, Shen L, Guo Z, Chou LL, Zhong W, Du Q, Ge J. Biomaterials. 2009;301:2276. doi: 10.1016/j.biomaterials.2008.12.075. [DOI] [PubMed] [Google Scholar]
- 156.Liu L, Guo S, Chang J, Ning C, Dong C, Yan D. J Biomed Mater Res B. 2008;87:244. doi: 10.1002/jbm.b.31103. [DOI] [PubMed] [Google Scholar]
- 157.Liu ZM, Gu Q, Xu ZK, Groth T. Macromol Biosci. 2010;10:1043. doi: 10.1002/mabi.201000086. [DOI] [PubMed] [Google Scholar]
- 158.Boudou T, Crouzier T, Ren K, Blin G, Picart C. Adv Mater. 2010;22:441. doi: 10.1002/adma.200901327. [DOI] [PubMed] [Google Scholar]
- 159.Benkirane-Jessel N, Lavalle P, Hubsch E, Holl V, Senger B, Haikel Y, Voegel J-C, Ogier J, Schaaf P. Adv Func Mater. 2005;15:648. [Google Scholar]
- 160.Tezcaner A, Hicks D, Boulmedais F, Sahel J, Schaaf P, Voegel J-C, Lavalle P. Biomacromolecules. 2006;7:86. doi: 10.1021/bm0505134. [DOI] [PubMed] [Google Scholar]
- 161.Crouzier T, Ren K, Nicolas C, Roy C, Picart C. Small. 2009;5:598. doi: 10.1002/smll.200800804. [DOI] [PubMed] [Google Scholar]