Abstract
Cells respond to genotoxic insults by triggering a DNA damage checkpoint surveillance mechanism and by activating repair pathways. Recent findings indicate that the two processes are more related than originally thought. Here we discuss the mechanisms involved in responding to UV-induced lesions in different phases of the cell cycle and summarize the most recent data in a model where Nucleotide Excision Repair (NER) and exonucleolytic activities act in sequence leading to checkpoint activation in non replicating cells. The critical trigger is likely represented by problematic intermediates that cannot be completely or efficiently repaired by NER. In S phase cells, on the other hand, the replicative polymerases, blocked by bulky UV lesions, re-initiate DNA synthesis downstream of the lesions, leaving behind a ssDNA tract. If these gaps are not rapidly refilled, checkpoint kinases will be activated.
Keywords: UV irradiation, DNA damage checkpoint, DNA repair
1. Introduction
Cellular DNA is constantly threatened by genotoxic events arising from cellular metabolisms (e.g., free oxygen radicals, replication errors) and induced by environmental factors (e.g., ionizing and UV radiations, chemicals). To prevent the effect of endogenous and exogenous mutagenic agents and to maintain genome integrity, cells have evolved a complex response to DNA damage (DDR), which includes repair mechanisms and regulatory circuits. A key role in this response is played by signaling pathways that we will refer to as DNA damage checkpoints, surveillance mechanisms responsible for the coordination of cell cycle progression, DNA replication, transcription with DNA repair and apoptosis. Checkpoint activation temporarily halts or delays cell cycle progression, possibly providing the cell with enough time to remove DNA lesions before these are converted in secondary and more dangerous lesions (e.g., replication through a single strand gap would generate a double strand break). The checkpoints also actively stimulate the repair processes [1–9] and, in higher eukaryotes, trigger the apoptotic response, if damage cannot be dealt with successfully [10–12].
2. DNA damage checkpoint
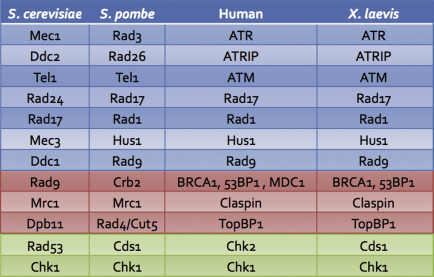
The importance of the DNA damage checkpoint in the maintenance of genomic stability is underlined by the existence of many syndromes linked to mutations in checkpoint genes, causing increased cancer proneness or other clinical symptoms, especially neurological defects [13,14]; it is thus not surprising that these pathways are extremely conserved throughout evolution (Table 1).
Table 1.
Checkpoint functions are evolutionarily conserved. The table shows the correspondence between various checkpoint factors in different organisms. The upstream factors are in blue, mediators are in pink and downstream effectors are in green.
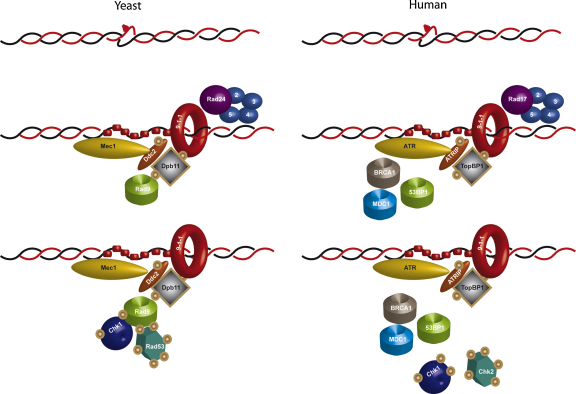
The DNA damage checkpoint response consists of a signal transduction cascade mainly based on phosphorylation events; the mechanistic details of the pathway have been recently discussed elsewhere [15,16], and will be just briefly summarized here to give a schematic picture to the reader. The first signaling event is carried out by the apical checkpoint kinases and is triggered after DNA damage detection. Two complexes are independently recruited at the lesion sites [17]: the human ATR/ATRIP or Saccharomyces cerevisiae Mec1/Ddc2 complex, and the 9-1-1 checkpoint clamp complex, composed of Rad9-Rad1-Hus1 in human or their orthologue subunits Rad17-Mec3-Ddc1 in yeast. The co-localization of these complexes is sufficient to trigger at least a partial checkpoint signaling even in the absence of actual DNA damage [18]. In S. cerevisiae the Mec1 apical kinase can be activated both by the Ddc1 subunit of the checkpoint clamp and by the adaptor protein Dpb11 which is recruited at the lesion through interaction with Ddc1 [19–23]. In human cells, the 9-1-1 complex is not able to directly activate ATR, but it is needed to recruit TopBP1 (the Dpb11 orthologue) which, in turn, stimulates ATR activity [24]. The apical kinases phosphorylate checkpoint mediators or adaptors, which are held close to the lesion by the interaction with post-translationally modified histone residues and with other checkpoint factors [25]. The mediators amplify the signaling cascade providing a platform to recruit effector kinases close to the apical kinases, and facilitating their activation. In budding yeast, Mec1 activates both Rad53 and Chk1 [26], while in human cells Chk2 is activated by ATM and Chk1 by ATR [27]. The prototype of checkpoint mediators is S. cerevisiae Rad9, which, once phosphorylated by the apical kinase, recruits Rad53 at the damage site allowing its phosphorylation by Mec1. Oligomerization of Rad9 seems to be critical to provide a scaffold for Rad53 binding, leading to a local increase in Rad53 molecules and stimulating its auto-phosphorylation; this event is responsible for full Rad53 activation [28,29]. Chk1 activation also requires Rad9, but the mechanism through which this mediator facilitates Chk1 phosphorylation by Mec1 is still poorly understood [30]. In human cells, the identity of the functional Rad9 orthologue is still debated: multiple candidates exist – i.e., MDC1 (mediator of DNA-damage checkpoint 1), 53BP1 (p53-binding protein 1) and BRCA1 (breast cancer 1 early-onset) – all characterized by the presence of tandem BRCT (BRCA1 C-terminal repeat) domains. Since these three proteins are involved in checkpoint signal transduction and each of them seems to carry out, separately and sometimes redundantly, some of Rad9 functions, they may be all considered as Rad9 orthologues [31]. Finally, effector kinases are responsible for the phosphorylation of a great number of targets, including cell cycle machinery factors and key proteins important for replication and repair [32,33].
The checkpoint response can act in at least three different phases of the cell cycle: in G1, to prevent chromosomes with problematic lesions from entering S phase, in S phase to control their replication, and in G2 (or M in some organisms) to avoid loss of genetic information due to mitotic segregation of severely damaged chromosomes. The general scheme of the checkpoint cascade is similar in all three cases, but significant differences can be found, depending on the nature of the DNA lesion and on the cell cycle phase in which the damage is detected [13,34–36]. Furthermore, in human cells the two apical kinases seem to be partly specialized in the response to different classes of DNA damaging agents. In fact, ATM (Ataxia Telangiectasia Mutated) is activated by double-strand breaks (DSBs) caused, for example, by ionizing radiation (IR), while ATR (ATM and Rad3-Related) is activated by ssDNA coated with the RPA heterotrimeric complex and mainly triggers checkpoint activation after UV irradiation or replication-stress. This specialization is possibly imputable to the different networks of physical interactions that these kinases participate to, and that are responsible for their recruitment at the sites of lesion [37–40]. The situation is somewhat complicated by the finding that ATR can be also recruited to DSBs and this binding depends upon ATM [41,42]. This separation of tasks is not found in budding yeast, where Mec1 (the ATR homologue) is the main player of checkpoint activation after all kind of DNA lesions, while Tel1 (the ATM homologue) is especially devoted to telomere maintenance. The redundant role of Tel1 in the DNA damage checkpoint is uncovered only in the absence of Mec1 [15,43].
In this review, we will focus our attention on UV-induced lesions and we will discuss the reciprocal interactions between NER, post replication repair (PRR), and the checkpoint pathway. In particular, we will discuss how NER plays a role in the activation of the checkpoint response after UV treatment, and how checkpoint kinases contribute to modulating the actual repair events.
Given the variety of DNA lesions the cell has to deal with, it was hypothesized that the first responders, among checkpoint factors, had to be recruited to a common DNA intermediate, which was later identified as long regions of ssDNA covered by RPA [44]. While ATRIP and Ddc2 directly bind RPA-covered ssDNA [44], loading of the 9-1-1 complex requires the activity of an RFC-like complex, that places it at the junction between dsDNA and 5′ ssDNA [45]. In the case of a single DSB, a large amount of evidence indicates that recombination factors are first recruited at the DSB, and then the 5′ ends of the DSB are processed through the concerted action of several proteins, including helicases and nucleases. This action generates long ssDNA tails that, on one hand will recruit checkpoint factors, and on the other will initiate repair through homologous recombination mechanisms [46]. On the other hand, extensive resection is not required when a DSB is repaired through a Non-Homologous End Joining (NHEJ) process [47]. For a long time, it was unclear how UV irradiation, which causes bulky lesions on DNA (mainly cyclobutane pyrimidine dimers (CPD) and 6,4 photoproducts (6-4PP)) responsible for inducing a distortion of the DNA helix [48], triggers the same checkpoint response in the absence of any DSB (Fig. 1).
Fig. 1.
The DNA damage checkpoint cascade. The DNA damage checkpoint is triggered by a ssDNA region. The left side of the figure reports the checkpoint cascade in budding yeast. RPA-covered ssDNA recruits the Mec1-Ddc2 and the 9-1-1 complexes. Phosphorylated Ddc1 interacts with Dpb11 which recruits the Rad9 mediator. Rad53 and Chk1 kinases are activated upon binding to oligomeric Rad9 and then leave chromatin to find their own targets. The right side of the figure summarizes the same signaling cascade in human cells.
3. Nucleotide excision repair
UV-induced DNA lesions are mainly removed through NER that efficiently identifies 6-4PPs and more slowly takes care of CPDs. The lesion recognition mechanism of NER depends upon the physical location of the lesion, with TC-NER acting on lesions that block transcription, and GG-NER taking care of the rest of the genome [48].
NER has been reconstituted in vitro and the mechanism is discussed elsewhere in this issue. Briefly, once the lesion has been recognized a pre-incision complex is assembled at the damage site. Endonucleolytic incision 5′ and 3′ to the lesion produces a short gap containing ssDNA covered by RPA, which is then refilled by DNA polymerase activities.
How long this ssDNA tract is exposed for, before a DNA polymerase refills the gap is not clear, but most reports seem to agree that the refilling is extremely rapid and tightly coordinated with the incision process [49,50] (see review by Fagbemi et al. in this issue of DNA Repair).
Since UV lesions are bulky and block the progression of replicative DNA polymerases, when the replication forks collide with UV-induced lesions during the S phase of the cell cycle re-priming events may take place downstream of the lesions, leaving ssDNA gaps behind the fork. Such structures have been detected by electron microscopy on replicating UV-damaged DNA and are likely responsible for the rapid and sensitive response observed in UV-irradiated S phase cells [51]. Outside of S phase and in non cycling cells the situation is quite different. Recent analysis showed that UV lesions themselves cannot activate the checkpoint and NER plays a major role in triggering the checkpoint response, although contrasting results have also been reported [52–57].
4. NER and DNA damage checkpoint
The tight relationship between NER and the checkpoint response started to become clear when, in budding yeast, a rad14 mutant, which is defective in assembling a competent pre-incision complex, was identified in a screen for mutations specifically inactivating the DNA damage checkpoint in response to UV irradiation, while leaving intact the DSB-induced checkpoint [55]. Furthermore, a direct interaction between Rad14 and the 9-1-1 checkpoint complex was reported, albeit its physiological significance has not been fully addressed. This work also showed that, in non cycling cells, any NER mutation affecting the incision event caused a deficient checkpoint activity, demonstrating that UV lesions per se are not sufficient to trigger the apical checkpoint kinase, and that their processing by a repair mechanism is necessary for recruiting the Mec1/Ddc2 and the 9-1-1 complexes to damaged chromosomes and for a prompt checkpoint response [55]. Such results are also consistent with the finding that UV irradiation in G1 of a cycling rad14Δ strain results in a strong arrest at the beginning of S phase [58], accompanied by the accumulation of replication-dependent ssDNA regions [51]. Altogether, it was suggested that a NER intermediate, possibly the ssDNA gapped structure generated by the double incision event may be responsible for recruiting and activating checkpoint factors. In cycling NER-deficient cells, UV irradiation would not cause a G1 delay or G2/M arrest and replication of the damaged template would lead to the accumulation of ssDNA regions resulting in Mec1 activation.
Extension of this kind of analysis to human cells derived from XP patients confirmed that lack of NER-dependent processing prevented UV-induced checkpoint activity in non cycling fibroblasts, revealing that XP cells are not only deficient in repairing UV lesions, but they are also deficient in the G1 and G2/M UV-induced checkpoint [56,57]. Interestingly, while XPC cells, defective in GG-NER, exhibit a checkpoint failure, cells obtained from Cockayne syndrome patients, which are defective in TC-NER, are instead able to activate the checkpoint, possibly thanks to the activity of GG-NER. Intriguingly, there seems to be a correlation between the capacity of these cells to properly control G1 and G2/M transitions after UV, their genomic instability and the proneness of XP and CS patients to develop tumors [56]. In budding yeast, the analysis of mutants specifically defective in the TC-NER or in the GG-NER branches of NER, revealed that activity of either one of the sub-pathways was sufficient to trigger a checkpoint response [55]. Interestingly, although UV-induced photoproducts and DSBs are processed by different DNA repair pathways and trigger signaling responses controlled by distinct apical kinases (see above) they eventually generate the same epigenetic mark involving H2A ubiquitination [59].
The model suggesting that gapped NER intermediates are responsible for checkpoint activation in UV-irradiated cells poses a few problems: (a) the gaps are very short (∼30 nt); (b) repair synthesis is very rapid, so the gaps are virtually absent; (c) it is not clear what would be the advantage of activating the checkpoint and arresting cell cycle progression, once the damage is practically repaired.
Recent work shed light on these problems, showing that normal NER-intermediates are not directly responsible for activating checkpoint kinases. In order to achieve a full and prompt checkpoint activity after UV irradiation in non cycling yeast cells, NER is necessary but not sufficient: in fact, the nuclease activity of Exo1 is also required [60,61]. Exo1 belongs to the Rad2 family of nucleases and has multiple cellular roles (see [62] for a review). This work shows that UV irradiation causes the accumulation, in yeast chromosomes, of long ssDNA regions that are dependent upon NER and Exo1 and correlate with Mec1 kinase activation. Preventing completion of repair synthesis by genetic or chemical means strongly increases accumulation of ssDNA and checkpoint activation, in agreement with a previous report [63]. The frequency of these large ssDNA gaps is much lower than the expected frequency of UV damages, suggesting that only a minor fraction of lesions undergo Exo1-dependent processing. Intriguingly, this mechanism is conserved also in human cells (Sertic et al., in preparation). These results suggest that the ∼30 nt long ssDNA gaps produced by NER can be refilled by DNA polymerases or extended by Exo1; given that polymerases refill a DNA gap at a rate of about 3700 nt/min and Exo1 excises DNA at 160 nt/min, most UV lesions are normally rapidly repaired by NER. This is consistent with the observation that very low UV doses do not seem to activate the G1 checkpoint [61,64,65]; if NER can rapidly and effectively deal with a low number of lesions, there would be no point in triggering a checkpoint response. If for any reason the repair synthesis step is perturbed, Exo1 may have a kinetic opportunity to process the NER gap, generating a long ssDNA region, which recruits checkpoint factors and triggers the signaling cascade (Fig. 2). This situation may arise, for example, at higher UV doses, in case repair synthesis factors become limiting or if the refilling polymerase encounters an insurmountable block. In these conditions, repair may not be completed and the extension of the ssDNA region may meet two purposes: activating the checkpoint response and channeling the problematic lesion to a different repair pathway (e.g., recombination) [66–68].
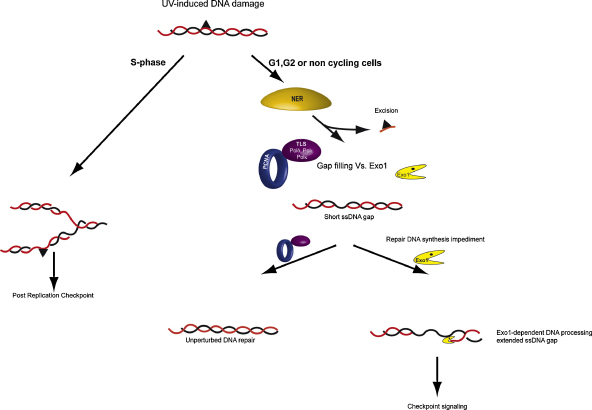
Fig. 2.
UV-induced checkpoint response. In cells that are not replicating their genome (i.e., G1, G2 or non-cycling cells), NER removes UV lesions efficiently and DNA polymerases (i.e., pol δ, pol ɛ, TLS polymerases) begin the refilling process. If the repair reaction is impeded after the excision step, a competition between the refilling polymerases and Exo1 nuclease can take place. Problematic refilling (e.g., closely opposing lesions) allows Exo1 to further process the gapped intermediate generating long ssDNA gaps which recruit checkpoint factors and trigger the signaling. At low UV-doses G1 and G2 cells do not accumulate large ssDNA gaps since UV lesions can be efficiently removed by NER. If the damages are still present when the cell enters S phase, the replicative polymerase will be blocked by the bulky lesion and will reinitiate DNA synthesis further downstream, leaving ssDNA gaps behind the replication fork. These gaps can be refilled by post replication repair and trigger a post-replication checkpoint.
Interestingly, translesion DNA polymerase activities (TLS) seem to counteract the generation of the UV-induced checkpoint signal [61,69]. Moreover, an unexpected role for TLS polymerases in NER was described in human cells, where DNA polymerase κ was found to be responsible for approximately 30% of the unscheduled DNA synthesis detected in UV-irradiated cells [70,71]. The actual role of TLS polymerases in NER is not completely understood and it will be interesting to determine if their activity is limited to particular regions of the genome and/or to particular configurations, such as the presence of a lesion in the template strand that may interfere with the refilling step of NER, as previously suggested in bacteria [72].
5. Closely opposing lesions
The possibility that Exo1-depedent processing may be facilitated by a polymerase blocking lesion in the template is intriguing. One instance where this might happen is when two UV lesions, one on each DNA strand, are generated in a limited region; this configuration has been defined “closely opposing UV lesions” [73–75]. Probability calculations would predict that the frequency of closely opposing lesions increases with the square of the UV dose and the chance of generating such situation in a yeast chromosome is expected to be very low. On the other hand, UV lesion formation has a strong sequence bias, and actual measurements on irradiated DNA proved that approximately 1% of all UV-induced lesions are configured as closely opposing lesions [76–78]. When NER encounters two closely spaced lesions, one on each strand, a major problem arises. NER can only process one damage at a time because the lesion needs to be in a double-stranded configuration [79]. Incision and removal of the first UV-induced dimer leaves to the refilling polymerase a gap containing a lesion in the template strand. DNA polymerase δ or ɛ, which normally take care of repair synthesis, cannot replicate past the template lesion and stall, strongly resembling a blocked replication fork. During S phase, such situation would be bypassed via Post Replication Repair (PRR), which entails TLS polymerases and/or template switching mechanisms. Interestingly, it was recently shown that DNA polymerase κ directly participates to NER repair synthesis in human cells [70,71] and that loss of TLS activity greatly potentiates the checkpoint response to UV irradiation in yeast G1 cells [61], suggesting that closely opposing UV lesions may indeed be at least partly responsible for checkpoint activation. These particular lesions may also contribute to explain the observation that both in wild-type yeast and bacteria cells most of the UV-induced mutagenesis depends upon a functional NER and takes place in G1 cells, while in the absence of NER the mutagenesis is S-phase specific [80,81]. Moreover, a role for TLS in G1-irradiated cells is also supported by the finding that G1 synchronized cultures of yeast mutants lacking TLS polymerases are more sensitive to UV light than asynchronous cultures, while this is not the case for strains that are TLS proficient [61].
6. Replicating UV damaged DNA
As mentioned above, in NER deficient cells, the lack of lesion removal coupled to the failure to activate the G1 DNA damage checkpoint in response to UV irradiation allows a large amount of DNA lesions to go into S phase. Here, there is no need for lesion processing to generate the checkpoint activating structures since ssDNA regions are generated by the stalling of replication forks at the polymerase blocking lesions. Blocked polymerase can leave the lesion and PCNA behind and re-initiate downstream of the lesion via a re-priming mechanism, generating numerous ssDNA gaps behind the forks, which are likely responsible for the strong activation of Mec1 during S phase in UV-irradiated cells [51,82–85]. Consistently, even at low UV doses (5 J/m2), NER deficient yeast cells exhibit a strong cell cycle arrest at the beginning of S phase, due to Mec1 DNA damage checkpoint activation [58]. An active checkpoint leads to Rev1 phosphorylation [86,87], possibly increasing TLS activity and progressively reducing the amount of RPA-covered ssDNA, thus promoting the switch off of the checkpoint itself.
In a wild type background, elegant time lapse experiments showed that after an acute low dose of UVC light (5 J/m2) yeast cells do not delay cell cycle progression until they proceed through S phase; for this reason this response was called post-replication checkpoint [64]. With low levels of UV-induced lesions the NER mechanism is very efficient and rapidly takes care of most lesions, so that neither the G1 or the G2 checkpoints are activated. The few lesions that are encountered by replicating polymerases in these conditions, on the other hand, block the replication fork and trigger a checkpoint response; this response may be detected in late S phase, when most replicons have completed duplication and the left-over ssDNA gaps need to be refilled (Fig. 2). An important point in this regard was made by irradiating budding yeast cells with very low (0.18 J m−2 min−1) chronic UV dose (CLUV): the only pathway necessary and sufficient to ensure cell survival was found to be the RAD5-dependent branch of PRR [65]. Indeed, NER deficient cells (rad14Δ) and DNA damage checkpoint deficient cells (mec1Δ) are not particularly sensitive to the CLUV treatment, contrary to the rad18Δ and rad5Δ cells, deficient in PRR [65], which irreversibly activate Mec1 DNA damage checkpoint and die in the G2 phase. In fact, S phase can be completed in the absence of PRR, but the gapped replicated DNA needs to be refilled by PRR in G2 in order to warrant cell survival [66,67].
In mammalian cells the situation is more complex, because NER acts also in S phase, where GG-NER is enhanced [88] and is stimulated by active ATR [89,90]. Indeed, ATR-deficient Seckel syndrome fibroblasts exhibit attenuation of S phase specific GG-NER and a similar effect has been detected in XPV skin fibroblasts, deficient in pol η [91]. Thus, in human cells both ATR-dependent DNA damage checkpoint and TLS influence S phase-specific GG-NER: defects in ATR or pol η may cause the abnormal persistence of ssDNA gaps opposite a template lesion and this would inhibit DNA adducts excision by GG-NER.
7. Recruitment of checkpoint factors by NER
In the last few years evidence has emerged that the role of NER in checkpoint activation is not limited to the generation of the ssDNA signal but, in addition, NER proteins seem to be involved in directly recruiting checkpoint factors to the proximity of DNA lesions.
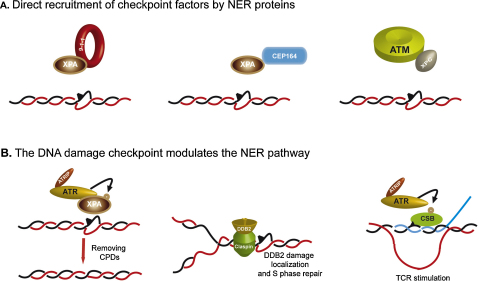
In S. cerevisiae, a physical interaction was identified between Rad14 and both Ddc1 and Mec3, two subunits of the 9-1-1 checkpoint complex. Although the physiological relevance of this interaction has not been directly investigated further, association of Ddc1 and Ddc2 to UV-damaged chromosomes is lost in a rad14Δ strain, suggesting that the 9-1-1 complex may be initially recruited at the sites of DNA lesions by directly interacting with the key NER factor Rad14 [55].
Additional observations indicate that this mechanism is most likely conserved in higher eukaryotes. In human G1 cells the recruitment of the 9-1-1 complex onto damaged DNA is dependent on XPA and XPC proteins [92]; analogously, Cep164, a checkpoint mediator protein in the ATR signaling pathway required for Chk1 phosphorylation after UV damage, was shown to be recruited to CPD sites in a NER-dependent manner, through UV-induced physical interaction with XPA [93]. In addition, a role for NER in the activation of ATM after cisplatin treatment was discovered. Immunoprecipitation experiments revealed a physical interaction between ATM and NER factors and this association is required for ATM recruitment to DNA [94] (Fig. 3).
Fig. 3.
Crosstalks between NER and checkpoint factors. A two-way functional interaction exists between the checkpoint machinery and the NER apparatus: some examples (discussed in the text) are shown. (A) NER factors recruit checkpoint proteins to damaged chromosomes, thus facilitating the activation of the signaling cascade. (B) DNA damage checkpoint factors modulate NER activity allowing for efficient repair of the lesions.
Combining the notion that processing of lesions by repair machineries is a pre-requisite for checkpoint activation outside of S phase, with the observation that checkpoint factors interact with repair proteins depicts a model where the repair machinery, which is specialized for direct lesion recognition, increases the local concentration of checkpoint sensors on damaged chromosomal regions facilitating a robust checkpoint response.
Interestingly, a NER-independent mechanism for activating checkpoint kinases seems to exist in non cycling cells. If NER-deficient yeast cells are blocked in the G1 phase of the cell cycle, UV irradiated and held in non dividing conditions indefinitely, a delayed activation of the Mec1-dependent pathway has been reported [95,96]. Recent evidence indicates that UV-induced signaling may proceed via NER-independent mechanisms also in non-dividing mammalian cells likely through generation of DNA strand breaks [97].
8. NER modulation by checkpoint proteins
The interplay between NER and the DNA damage checkpoint is even more complex; in fact, while NER is involved in checkpoint activation, the checkpoint pathway actively stimulates NER by modulating cellular levels, localization and activity of NER factors, through transcriptional regulation, direct protein–protein interactions and post-translational modifications (Fig. 3).
The S. cerevisiae Rad9 gene product plays a role in the repair of both UV-damaged strands of an actively transcribed gene; this effect on NER is likely indirect and probably occurs through up-regulation of some NER genes (i.e., RAD2, RAD7, RAD16 and RAD23), consistently with a previously reported Rad9-dependent stimulation of NER genes transcription [98]. Rad9 does not seem to be required for the repair of non-transcribed regions, suggesting that it is acting only when repair is coupled to transcription [8]. Along similar lines, Rad26, a NER factor critical for TC-NER, is a direct target of Mec1 kinase and its phosphorylation enhances TC-NER, possibly by stimulating its ATPase activity or by modulating its interaction with other TC-NER proteins [99]. Notably, CSB (the human orthologue of Rad26) was identified in a screen for putative ATM/ATR substrates [33], pointing to a conservation of this regulatory mechanism through evolution. Other NER factors were found in the same screening, namely XPA, XPC, RPA1 and RAD23B, but further characterization will be required to prove the significance of the interaction between ATM/ATR and the NER factors identified in the screen. Another example on the possible feedback of the checkpoint on NER is the regulation of XPA by ATR. It has been found that XPA nuclear import and its stable accumulation at nuclear foci after UV irradiation is dependent upon ATR. ATR also seems to be responsible for XPA phosphorylation after UV radiations, although this modification is not required for XPA foci formation [100–102]. Further studies will be required to firmly establish the role of ATR in the modulation of the XPA function. Finally, an S-phase specific role for the DNA damage checkpoint in regulating GG-NER was recently suggested. In particular, ATR inhibition was shown to specifically abrogate NER in S-phase, while the repair rate was unaffected in G1 and G2/M cells [89]. Another report revealed a role for the replication checkpoint mediator Claspin in regulating the DDB2 subunit of the UV-DDB factor, which is involved in the initial steps of GG-NER [103]. DDB2 is localized at the UV-induced DNA lesions, where its ubiquitination and subsequent degradation seems to control XPC recruitment and damage recognition, thus triggering the NER process [104]. Claspin knockdown affects DDB2 recruitment at damage sites and its subsequent ubiquitin-mediated degradation; in agreement with this observation, it has been shown that Claspin and DDB2 physically interact and their association is greatly enhanced upon UV irradiation [103].
9. Summary and perspectives
The complex interplay between NER and DNA damage checkpoints is not an isolate case in the DDR landscape, since in recent years a large cluster of papers highlighted the reciprocal interdependence of the checkpoint pathways and virtually all other known repair systems. It seems that, as a general rule, a two-way functional interaction exists between the checkpoint machinery and the repair apparatus. On one side, repair factors help to recruit checkpoint proteins at the damage sites onto DNA and, by modifying the primary lesions to RPA-covered ssDNA, trigger the checkpoint cascade; on the other side, once activated the checkpoint pathway stimulates the repair process mainly through direct protein–protein interactions or post-translational modifications. One of the key factors at the interface between checkpoint and repair is the 9-1-1 checkpoint clamp, and its involvement in such processes has been recently discussed [25].
The checkpoint response may be seen as a process that signals the cell that something that should have been working properly, has instead some problem; the cell can thus deploy a set of measures to attempt to solve the problems and avoid further complications.
The data obtained with low acute UV doses suggest that if the lesions are not frequent enough to interfere with G1 or G2 processes, the cell has no way (or need) to acknowledge their presence and activate the checkpoint. Indeed, NER can easily keep these lesions under control. When these damage-containing chromosomes are replicated, though, the DNA polymerases scanning the genome will eventually detect them, and re-initiate DNA synthesis further downstream leaving behind ssDNA gaps. Since the region hosting a polymerase-blocking lesion can be almost completely replicated by an incoming fork starting from an adjacent origin, at low levels of lesions the gaps will accumulate and activate the checkpoint kinases toward the end of S phase. This event has a clear relevance since checkpoint mutants will die in this situation, and only after checkpoint activation the gaps are refilled by PRR and the lesions are actually removed from the chromosomes [61]. It has to be noted that a checkpoint response can be triggered in G1 cells, even at these low UV doses, if something interferes with completion of NER. Indeed, alterations in the refilling step of repair will sensitize G1 cells more than S phase cells [58].
At higher UV doses (>20 J/m2), cells promptly respond also in non replicating conditions, consistently with the increased probability of repair problems arising. Repair DNA synthesis, in these situations, could be affected by the low level of dNTPs, by the formation of closely opposing lesions, by limiting level of particular factors in saturating conditions and by the higher possibility that lesions are generated in “difficult to repair” chromosomal locations. If the refilling reaction is problematic, nucleases like Exo1 have a greater chance to process the NER intermediates and elicit a checkpoint response [58].
What is surprising is what happens with chronic low UV doses (CLUV), which are supposed to best mimic sunlight exposure. Experiments performed in yeast cells have suggested that in CLUV conditions no checkpoint is activated, not even during S phase; indeed, checkpoint deficient strains do not exhibit sensitivity to CLUV treatment [62]. Even more surprisingly, NER is not important in these conditions, since NER-deficient cells are also not sensitive to CLUV. The possibility to extend these findings beyond yeast cells remains to be determined, indeed they seem to contrast with the situation observed in XP patients, who are deficient in NER and clearly hypersensitive to sunlight. In the future the actual events happening with sunlight exposure will need to be investigated.
NER is the most versatile repair system and eliminates a wide repertoire of DNA lesions, among which are UV-induced CPD and 6-4PP, that represent the main determinants in solar mutagenesis and skin cancer [105,106]. The importance of the findings summarized here may thus expand further than the problems related to exposure to UV light.
It is expected that genome-wide analysis of protein–protein interaction networks provided by high throughput screenings will progressively increase the number of known physical interactions between checkpoint proteins and repair factors, thus strengthening and expanding the model describing the functional connections between these two key genome stability pathways.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Funding
Work in the authors’ lab is supported by grants from AIRC, Fondazione Cariplo, MIUR and EU FP6 IP DNA Repair.
Acknowledgments
The authors apologize for the many interesting papers that they were not able to discuss or acknowledge. FP6 IP DNA Repair. F.L. is supported by Fondazione Adriano Buzzati-Traverso.
Contributor Information
Paolo Plevani, Email: paolo.plevani@unimi.it.
Marco Muzi-Falconi, Email: marco.muzifalconi@unimi.it.
References
- 1.Zhao X., Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao R., Zhang Z. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashkirov V.I., King J.S. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahnesorg P., Jackson S.P. The non-homologous end-joining protein Nej1p is a target of the DNA damage checkpoint. DNA Repair (Amst) 2007;6:190–201. doi: 10.1016/j.dnarep.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flott S., Alabert C. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell. Biol. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin I., Ngo H.P. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou W.C., Wang H.C. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 2008;27:3140–3150. doi: 10.1038/emboj.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Moghrabi N.M., Al-Sharif I.S., Aboussekhra A. The RAD9-dependent gene trans-activation is required for excision repair of active genes but not for repair of non-transcribed DNA. Mutat. Res. 2009;663:60–68. doi: 10.1016/j.mrfmmm.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe S.W., Schmitt E.M. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y., Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 12.Hirao A., Kong Y.Y. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 13.Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 14.Kerzendorfer C., O’Driscoll M. Human DNA damage response and repair deficiency syndromes: linking genomic instability and cell cycle checkpoint proficiency. DNA Repair (Amst) 2009;8:1139–1152. doi: 10.1016/j.dnarep.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Putnam C.D., Jaehnig E.J., Kolodner R.D. Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair (Amst) 2009;8:974–982. doi: 10.1016/j.dnarep.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancar A., Lindsey-Boltz L.A. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 17.Melo J.A., Cohen J., Toczyski D.P. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–3221. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonilla C.Y., Melo J.A., Toczyski D.P. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol. Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majka J., Niedziela-Majka A., Burgers P.M. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Elledge S.J. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puddu F., Granata M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol. Cell. Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navadgi-Patil V.M., Burgers P.M. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mordes D.A., Nam E.A., Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navadgi-Patil V.M., Burgers P.M. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 2009;8:996–1003. doi: 10.1016/j.dnarep.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazzaro F., Giannattasio M. Checkpoint mechanisms at the intersection between DNA damage and repair. DNA Repair (Amst) 2009;8:1055–1067. doi: 10.1016/j.dnarep.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez Y., Bachant J. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 27.Bartek J., Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 28.Sweeney F.D., Yang F. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert C.S., Green C.M., Lowndes N.F. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 30.Blankley R.T., Lydall D. A domain of Rad9 specifically required for activation of Chk1 in budding yeast. J. Cell Sci. 2004;117:601–608. doi: 10.1242/jcs.00907. [DOI] [PubMed] [Google Scholar]
- 31.FitzGerald J.E., Grenon M., Lowndes N.F. 53BP1: function and mechanisms of focal recruitment. Biochem. Soc. Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 32.Smolka M.B., Albuquerque C.P. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka S., Ballif B.A. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 34.Siede W., Friedberg A.S., Friedberg E.C. Rad9-dependent-G(1) arrest defines a second checkpoint for damaged DNA in the cell-cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulovich A.G., Hartwell L.H. A checkpoint regulates the rate of progression through S-phase in Saccharomyces cerevisiae in response to DNA-damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 36.Weinert T.A., Hartwell L.H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 37.Khanna K.K., Lavin M.F. Ionizing-radiation and UV induction of p53 protein by different pathways in ataxia–telangiectasia cells. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 38.Tibbetts R.S., Brumbaugh K.M. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canman C.E., Lim D.S. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 40.Lovejoy C.A., Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jazayeri A., Falck J. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 42.Adams K.E., Medhurst A.L. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrow D.M., Tagle D.A. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene, MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 44.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 45.Zou L., Liu D., Elledge S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison J.C., Haber J.E. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 47.Weterings E., Chen D.J. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 48.Friedberg E.C., Walker G.C., Siede W. ASM Press; Washington, D.C.: 2006. DNA repair and mutagenesis. [Google Scholar]
- 49.Staresincic L., Fagbemi A.F. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luijsterburg M.S., von Bornstaedt G. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopes M., Foiani M., Sogo J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Jiang G., Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol. Cell. Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bomgarden R.D., Lupardus P.J. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Driscoll M., Ruiz-Perez V.L. A splicing mutation affecting expression of ataxia–telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 55.Giannattasio M., Lazzaro F. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marini F., Nardo T. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marti T.M., Hefner E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 2006 doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neecke H., Lucchini G., Longhese M.P. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marteijn J.A., Bekker-Jensen S. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009;186:835–847. doi: 10.1083/jcb.200902150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakada D., Hirano Y., Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannattasio M., Follonier C. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Tran P.T., Erdeniz N. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 63.Matsumoto M., Yaginuma K. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 64.Callegari A.J., Kelly T.J. UV irradiation induces a postreplication DNA damage checkpoint. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15877–15882. doi: 10.1073/pnas.0607343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hishida T., Kubota Y. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature. 2008;457:612–615. doi: 10.1038/nature07580. [DOI] [PubMed] [Google Scholar]
- 66.Karras G.I., Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 67.Daigaku Y., Davies A.A., Ulrich H.D. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stergiou L., Eberhard R. NER and HR pathways act sequentially to promote UV-C-induced germ cell apoptosis in Caenorhabditis elegans. Cell Death Differ. 2011;18:897–906. doi: 10.1038/cdd.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Callegari A.J., Clark E. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8219–8224. doi: 10.1073/pnas.1003449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogi T., Lehmann A.R. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 71.Ogi T., Limsirichaikul S. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 72.Bridges B.A., Brown G.M. Mutagenic DNA repair in Escherichia coli. XXI. A stable SOS-inducing signal persisting after excision repair of ultraviolet damage. Mutat. Res. 1992;270:135–144. doi: 10.1016/0027-5107(92)90124-k. [DOI] [PubMed] [Google Scholar]
- 73.Minton K., Friedberg E.C. Letter: evidence for clustering of pyrimidine dimers on opposite strands of U.V.-irradiated bacteriophage DNA. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1974;26:81–85. doi: 10.1080/09553007414550981. [DOI] [PubMed] [Google Scholar]
- 74.Lam L.H., Reynolds R.J. Bifilar enzyme-sensitive sites in ultraviolet-irradiated DNA are indicative of closely opposed cyclobutyl pyrimidine dimers. Biophys. J. 1986;50:307–317. doi: 10.1016/S0006-3495(86)83464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sage E., Cramb E., Glickman B.W. The distribution of UV damage in the lacI gene of Escherichia coli – correlation with mutation spectrum. Mutat. Res. 1992;269:285–299. doi: 10.1016/0027-5107(92)90211-j. [DOI] [PubMed] [Google Scholar]
- 76.Lam L.H., Reynolds R.J. A sensitive, enzymatic assay for the detection of closely opposed cyclobutyl pyrimidine dimers induced in human diploid fibroblasts. Mutat. Res. 1986;166:187–198. doi: 10.1016/0167-8817(86)90017-9. [DOI] [PubMed] [Google Scholar]
- 77.Lam L.H., Reynolds R.J. Repair of closely opposed cyclobutyl pyrimidine dimers in UV-sensitive human diploid fibroblasts. Mutat. Res. 1986;166:199–205. doi: 10.1016/0167-8817(86)90018-0. [DOI] [PubMed] [Google Scholar]
- 78.Lam L.H., Reynolds R.J. DNA sequence dependence of closely opposed cyclobutyl pyrimidine dimers induced by UV radiation. Mutat. Res. 1987;178:167–176. doi: 10.1016/0027-5107(87)90266-1. [DOI] [PubMed] [Google Scholar]
- 79.Svoboda D.L., Smith C.A. Effect of sequence, adduct type, and opposing lesions on the binding and repair of ultraviolet photodamage by DNA photolyase and (A)BC excinuclease. J. Biol. Chem. 1993;268:10694–10700. [PubMed] [Google Scholar]
- 80.Eckardt F., Teh S.J., Haynes R.H. Heteroduplex repair as an intermediate step of UV mutagenesis in yeast. Genetics. 1980;95:63–80. doi: 10.1093/genetics/95.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bridges B.A., Mottershead R. RecA+-dependent mutagenesis occurring before DNA replication in UV- and -γ irradiated Escherichia coli. Mutat. Res. 1971;13:1–8. doi: 10.1016/0027-5107(71)90120-5. [DOI] [PubMed] [Google Scholar]
- 82.Rupp W.D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 83.Lehmann A.R. Post-replication repair of DNA in ultraviolet-irradiated mammalian cells. No gaps in DNA synthesized late after ultraviolet irradiation. Eur. J. Biochem. 1972;31:438–445. doi: 10.1111/j.1432-1033.1972.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 84.di Caprio L., Cox B.S. DNA synthesis in UV-irradiated yeast. Mutat. Res. 1981;82:69–85. doi: 10.1016/0027-5107(81)90139-1. [DOI] [PubMed] [Google Scholar]
- 85.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 86.Pagès V., Santa Maria S.R. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 2009;23:1438–1449. doi: 10.1101/gad.1793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sabbioneda S., Bortolomai I. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair (Amst) 2007;6:121–127. doi: 10.1016/j.dnarep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 88.Gospodinov A., Anachkova B. Cells synchronized in S phase show increased rate of repair of UV damaged plasmids. FEBS Lett. 2004;572:99–102. doi: 10.1016/j.febslet.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 89.Auclair Y., Rouget R. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17896–17901. doi: 10.1073/pnas.0801585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Auclair Y., Rouget R., Drobetsky E.A. ATR kinase as master regulator of nucleotide excision repair during S phase of the cell cycle. Cell Cycle. 2009;8:1865–1871. doi: 10.4161/cc.8.12.8800. [DOI] [PubMed] [Google Scholar]
- 91.Auclair Y., Rouget R. Requirement for functional DNA polymerase eta in genome-wide repair of UV-induced DNA damage during S phase. DNA Repair (Amst) 2010;9:754–764. doi: 10.1016/j.dnarep.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 92.Warmerdam D.O., Freire R. Cell cycle-dependent processing of DNA lesions controls localization of Rad9 to sites of genotoxic stress. Cell Cycle. 2009;8:1765–1774. doi: 10.4161/cc.8.11.8721. [DOI] [PubMed] [Google Scholar]
- 93.Pan Y.R., Lee E.Y. UV-dependent interaction between Cep164 and XPA mediates localization of Cep164 at sites of DNA damage and UV sensitivity. Cell Cycle. 2009;8:655–664. doi: 10.4161/cc.8.4.7844. [DOI] [PubMed] [Google Scholar]
- 94.Colton S.L., Xu X.S. The involvement of ataxia–telangiectasia mutated protein activation in nucleotide excision repair-facilitated cell survival with cisplatin treatment. J. Biol. Chem. 2006;281:27117–27125. doi: 10.1074/jbc.M602826200. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H., Taylor J., Siede W. Checkpoint arrest signaling in response to UV damage is independent of nucleotide excision repair in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:9382–9387. doi: 10.1074/jbc.M300061200. [DOI] [PubMed] [Google Scholar]
- 96.Giannattasio M., Lazzaro F. DNA decay and limited Rad53 activation after liquid holding of UV-treated nucleotide excision repair deficient S. cerevisiae cells. DNA Repair (Amst) 2004;3:1591–1599. doi: 10.1016/j.dnarep.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 97.Vrouwe M.G., Pines A. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 2011;124:435–446. doi: 10.1242/jcs.075325. [DOI] [PubMed] [Google Scholar]
- 98.Al-Moghrabi N.M., Al-Sharif I.S., Aboussekhra A. The Saccharomyces cerevisiae RAD9 cell cycle checkpoint gene is required for optimal repair of UV-induced pyrimidine dimers in both G(1) and G(2)/M phases of the cell cycle. Nucleic Acids Res. 2001;29:2020–2025. doi: 10.1093/nar/29.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taschner M., Harreman M. A role for checkpoint kinase-dependent Rad26 phosphorylation in transcription-coupled DNA repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010;30:436–446. doi: 10.1128/MCB.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu X., Shell S.M. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006;66:2997–3005. doi: 10.1158/0008-5472.CAN-05-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shell S.M., Li Z. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J. Biol. Chem. 2009;284:24213–24222. doi: 10.1074/jbc.M109.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu X., Shell S.M. ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation. Oncogene. 2007;26:757–764. doi: 10.1038/sj.onc.1209828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Praetorius-Ibba M., Wang Q.E. Role of Claspin in regulation of nucleotide excision repair factor DDB2. DNA Repair (Amst) 2007;6:578–587. doi: 10.1016/j.dnarep.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 104.El-Mahdy M.A., Zhu Q. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 105.Brash D.E., Rudolph J.A. A role for sunlight in skin-cancer – UV-induced p53 mutations in squamous-cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tornaletti S., Pfeifer G.P. Slow repair of pyrimidine dimers at p53 mutation hotspots in skin-cancer. Science. 1994;263:1436–1438. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]