Abstract
Background
Protein kinase C-β2 (PKCβ2) is a splice-variant of the PRKCB1 gene and belongs to a family of serine/threonine-specific kinases that are predominantly activated by diacylglycerol, calcium, and phorbol ester. Cellular functions associated with PKCβ2 activation include transformation, proliferation, and inhibition of apoptosis. Enzastaurin (LY317615) is an oral, selective, potent inhibitor of the PKCβ2 kinase. Preclinical activity for this agent was predominantly reported in lymphoma, glioblastoma, and colorectal cancer. In patients with advanced non–small-cell lung cancer (NSCLC) whose previous therapy had failed, 13% of patients had disease control for 6 months with single-agent therapy.
Patients and Methods
We investigated whether biologically relevant variants of PRKCB1 exist in lung cancer cell lines in the context of enzastaurin-induced proliferation and kinase inhibition, using exon sequencing, immunoblotting, and cytotoxicity assays in NSCLC and small-cell lung cancer (SCLC) cell lines.
Results
We discovered a total of 6 single-nucleotide variants, but only 1 resulted in an amino acid substitution (T40I). This substitution was not located in the kinase domain of PKCβ2 and did not affect enzastaurin’s antiproliferative or phosphorylation-inhibitory activity. We found enzastaurin to be equally active in NSCLC and SCLC cell lines, with values of the 50% inhibitory concentration in a range of 0.05-0.2 μM.
Conclusion
The inhibition of phosphorylation of PKCβ2 and the downstream molecules glycogen synthase kinase-3β, S6RP, Akt, and forkhead transcription factor was evident in the same concentration range, which suggests the premise that the determination of phosphorylation levels of these molecules in human tissue specimens may be a useful pharmacodynamic parameter for in vivo target inhibition by enzastaurin.
Keywords: Akt, Drug activity, LY317615, Predictive marker, Sequence variants
Introduction
The protein kinase C (PKC) family of proteins is characterized by serine/threonine-specific kinase activity, triggered predominantly by diacylglycerol and calcium and by the tumor-promoting agent phorbol ester. They are encoded by 9 different genes (α, β, γ, δ, ε, ζ, η, θ, and ι) located on distinct chromosomal segments, and isoforms have been described as a result of alternative splicing. Their function is the phosphorylation of proteins involved in a variety of signal-transduction pathways, and several different family members display tissue-specific expression profiles.1
The PRKCB1 gene, located on chromosome segment 16p11.2, comprises 18 exons and encodes PKCβ, which occurs in 2 isoforms, β1 and β2. The isoforms are a result of the differential use of exons 17 and 18 through alternative splicing of the mRNA. Isoform β1 (NP_997700), which uses exon 18, has a shorter C-terminus than isoform β2 (NP_002729), which uses exon 17 (NCBI Gene Database, identification number 5579). Cellular functions associated with PKCβ activation include transformation, proliferation, inhibition of apoptosis,2-4 and activation of Akt (protein kinase B, PKB) pathways.5,6 The activation of Akt, which can be triggered by a loss of phosphatase and tensin homologue (PTEN), is associated with poor prognoses in non–small-cell lung cancer (NSCLC).7,8
Enzastaurin (LY317615) is an oral serine/threonine kinase inhibitor that targets the PKC and Akt pathways, and inhibits the phosphorylation of glycogen synthase kinase-3β (GSK-3β) and S6 ribosomal protein (S6RP).6 The antitumor activity of enzastaurin was indicated in experimental models of several human malignancies.6 In a phase II single-agent trial of oral enzastaurin (500 mg per day) in 55 patients with advanced-stage NSCLC whose previous chemotherapy had failed, a 6-month progression-free survival rate of 13% was reported.9
For a better definition of the group of patients who may benefit from enzastaurin, we studied the PRKCB1 gene for sequence variations, assessed these variations’ potential effects on enzastaurin efficacy, and investigated the relationship between the inhibition of PKCβ pathway phosphorylation and proliferation in lung cancer cell lines.
Patients and Methods
Cell Culture
Sixteen NSCLC cell lines and 12 small-cell lung cancer (SCLC) lines were obtained from their original sources or the American Type Culture Collection (Bethesda, MD). They were maintained in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and antibiotics, and their authenticity was confirmed by DNA fingerprinting or isoenzyme patterns. They were free of mycoplasma contamination.
Gene Sequencing
For sequencing of the human PRKCB1 gene in cell lines, genomic DNA was extracted using resin-exchange chromatography. The complete genomic sequence, as reported in GenBank accession number NC_000016, was used as a reference. Exons 2-18 were amplified using intronic primer pairs and sequenced in both directions. Exon 1 required two primer pairs for a complete sequence analysis. The primer sequences are listed in Table 1. Sequencing was performed on an ABI Prism 377 DNA Sequencer, using BigDye Terminator Cycle Sequencing (Applied Biosystems, Inc; Carlsbad, CA). Results were compared with the reference sequence, using Sequencher software (version 3.0, Gene Codes Corp; Ann Arbor, MI) and BLAST software.
Table 1.
Primers Used for Sequencing of PRKCB1 Gene in Lung Cancer
| Exon | Forward Primer | Reverse Primer | Amplicon Size |
|---|---|---|---|
| 1 | GCGTGCAGAATGACCAATGGGATGGA | GGAGCCGGAGCCCGAGAGG | 594 |
| 1 | CAGCAGCTGGGCGAGTGACA | CTCCTAGTCAGGTGGAGCAAAG | 657 |
| 2 | CTGCCTGACATACACCTCCTTC | CACCCTCTCTTCCCACAAATAG | 111 |
| 3 | CCTCCTCTCCGCTTTCCTTCT | CACAAGGCAGCATTGACTGT | 192 |
| 4 | GTTCTTGGTGAGTTCTCCAGTG | GAATGTATGGCAGACCTGGAC | 303 |
| 5 | CTGTCTCAAAGGAGGGAAACAG | CTCCCCTTCTCAATCACACAG | 304 |
| 6 | CACAAGTTCTGCAGCTGTCACT | AGGTAGGGAAGGAAGGAAACAG | 346 |
| 7 | CTACCTCCAGGTCTTGTCCTCTT | CCAGACTTCACTTTGGGACTCT | 346 |
| 8 | GGCCTCCTTTTCATATGCTG | AGAAAGAAAGAGGAGGGAGCAG | 261 |
| 9 | CCTACAAGATCGTCCTCAAACC | CGTCCCATAGGAACAGCAAA | 283 |
| 10 | GAACACCAGCTGCTCATAACTG | GTGCTGGGTGCTGCTAAGATAG | 328 |
| 11 | CAGAAGCTACGGGATGCTAATG | CACCCTCAAAGGAACCACTAAC | 207 |
| 12 | GTCTCACTATGTTGCCCAGCTA | GTGACACAGTCCACCGAGATT | 592 |
| 13 | CCCACACAACACCTAACACAGT | GGTACATCACCGCAATGTGA | 286 |
| 14 | GAACTGCAAACCTCCCTGATAG | GGGGTAGGGAGGACTTCTCTTAT | 167 |
| 15 | CAAGCAGGGATTCTTCTCCTC | GGTCCTGGTCACAGCTTTAAGA | 183 |
| 16 | GGTCCTATTATCAGGTCCTCTCC | GCAGTCAGAGAGAATGGGCTACT | 167 |
| 17 | GGGAAGGGAATGGAGAAAAG | CCACAATAGCCGTTGAGCTT | 329a |
| 18 | GCGTATCTTGGTCCTGTGTCTT | ACCAGGAACATCAGCTTCTGAC | 553b |
PKCβ2 isoform.
PKCβ1 isoform.
Proliferation Assays
Cell proliferation was assessed using a luminescent cell-viability assay (CellTiter-Glo; Promega; Madison, WI). This method is based on the quantification of adenosine triphosphate (ATP) levels, which correlate with the number of viable cells in culture. Cell lines H23, H125, H322, A549, H69, H211, DMS76, and SW210.5 were plated at the appropriate density (3000 cells per well for NSCLC; 50,000 cells per well for SCLC) in 96-well opaque white plates (Matrix Technologies, Hudson, NH) for 24 hours. The next day, growth medium was replaced with RPMI-1640 containing 1% FBS, at 90 μL per well. Enzastaurin was dissolved in phosphate-buffered saline (PBS) containing 0.02% dimethyl sulfoxide (DMSO), and we added 10 μL to each well. The final enzastaurin concentrations were 0.0 μM, 0.001 μM, 0.01 μM, 0.05 μM, 0.1 μM, 1.0 μM, and 5.0 μM. Cells were incubated for 72 hours at 37°C. We then added 100 μL of CellTiter-Glo reagent to each well. Cells and reagents were mixed with an orbital shaker for 2 minutes to induce cell lysis and were incubated at room temperature for 10 minutes to stabilize the luminescent signal. Luminescence was measured in a Veritas™ Microplate Luminometer (Turner Biosystems; Sunnyvale, CA). Cell proliferation was calculated as percentage of relative luminescence units of treated vs. untreated cells. Error bars represent the standard error, which was calculated using statistical tools in Microsoft Excel. At least three independent experiments were performed.
Immunoblotting
The NSCLC (H23, H125, H322, and A549) and SCLC (H69, H211, DMS76, and SW210.5) cell lines were incubated with 1 μM enzastaurin in RPMI-1640 supplemented with 1% FBS for up to 4 hours in six-well plates at approximately 1,000,000 cells/well. They were harvested in protein lysis buffer (20 mmol/L Tris-HCl at pH 7.6, 150 mmol/L NaCl, 1 mmol/L ethylenediaminetetraacetate, 0.5% NP40, 1 mmol/L dithiothreitol, 5 μmol/L trichostatin A, 1 mmol/L sodium orthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L NaF, and complete protease inhibitors; Roche Applied Science; Penzberg, Germany). Lysates were centrifuged at 4°C for 15 minutes at 13,000 × g, and the supernatants were recovered. Protein extracts (50 μg) were fractionated through 10% Novex tris-glycine gels (InVitrogen; Carlsbad, CA), blotted onto pure nitrocellulose membranes (Bio-Rad, Hercules, CA), and probed for the targets PKCβ2, phospho-PKCβ2 (ser660), GSK3β, phosphor-GSK3β (Ser9), S6RP, phosphor-S6RP (Ser240/244), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with commercially available antisera (Cell Signaling, Beverly, MA; Santa Cruz, Inc, Santa Cruz, CA). For final protein detection, a goat anti-mouse or anti-rabbit immunoglobulin G (IgG) or rabbit anti-goat IgG horseradish peroxidase secondary antibody was used (Santa Cruz, Inc), together with SuperSignal West Pico chemiluminescent substrate (Pierce; Rockford, IL).
In-Cell Western Assay
For in-cell Western blot analysis, NSCLC (H23, H125, H322, and A549) and SCLC (H69, H211, DMS76, and SW210.5) cell lines were seeded in RPMI-1640 with 10% FBS at 20,000 cells/well in 96-well clear-bottom plates (B.D. Falcon; Franklin, NJ). After 24 hours, the medium was changed to RPMI-1640 with 1% FBS, at 180 μL per well. Enzastaurin was dissolved in PBS containing 0.02% DMSO, and we added 20 μL to each well. The final enzastaurin concentrations were 0.0 μM, 0.001 μM, 0.01 μM, 0.1 μM, 1.0 μM, and 5.0 μM. After 4 hours of incubation, the medium was completely aspirated, cells were fixed with 3.7% formaldehyde in PBS for 20 minutes, and cells were permeabilized with five 5-minute washes in PBS + 0.1% Triton-X 100. Li-Cor Odyssey blocking buffer (Li-Cor Biosciences; Lincoln, NB) was added to each well (150 μL) for 90 minutes at room temperature with moderate shaking on a rotator, followed by overnight incubation with primary antibodies phospho-PKCβ2 (Ser660) and PKCβ2, 1:50; phospho-GSK3β (Ser9) and GSK3β, 1:100; S6RP and phospho-S6RP (Ser240/244), 1:500; phospho-Akt (Thr308), phospho-Akt (Ser473), and Akt, 1:100; and phospho-forkhead transcription factor (FKHR) (Ser256) and FKHR, 1:100, in Li-Cor Odyssey blocking buffer. After five 5-minute washes in PBS + 0.1% Tween-20, detection was performed using two species-specific infrared fluorescent dye-conjugated secondary antibodies (IRDye 800CW-conjugated goat anti-rabbit IgG 1:800, and IRDye 680CW-conjugated goat anti-mouse IgG 1:200 in Li-Cor Odyssey blocking buffer + 0.2% Tween-20). After 1 hour of incubation and washing, targets were simultaneously visualized using the Odyssey Infrared Imaging Scanner (Li-Cor), with the 700-nm fluorophore emitting a red color and the 800-nm fluorophore emitting a green color. Relative fluorescent units for enzastaurin-treated samples were divided by vehicle controls to determine percent change in expression. Error bars represent standard error, which was calculated using statistical tools in Microsoft Excel. At least three independent experiments were performed.
Results
PRKCB1 Gene Sequencing
Within the coding regions of PKCβ1/2 (the last 173 nucleotides of exon 1, all 1690 nucleotides of exons 2-16, the first 153 nucleotides of exon 17 for PKCβ2, and the first 159 nucleotides of exon 18 for PKCβ1), we observed six single-nucleotide variations and no deletions or insertions in 28 lung-cancer cell lines. Five variants were silent alterations (C79A in 13 cases, C606T in one case, G702A in one case, T1191C in 12 cases, and C1770T in nine cases). Only one variant (C119T in exon 1), as found in two SCLC cell lines (H69 and SW210.5), resulted in an amino-acid substitution (threonine 40 to isoleucine; Table 2). No apparent sequence variations led to splice variants.
Table 2.
Sequence Variations in the PRKCB1 Gene in Lung Cancer Cell Lines
| Cell Line | Histology | PRKCB1 Coding Region Variants |
Exon Number | Amino Acid Change |
|---|---|---|---|---|
| H23 | NSCLC | G702A | 7 | – |
| – | – | C1770T | 16 | – |
| H125 | NSCLC | C79A | 1 | – |
| H226 | NSCLC | C79A | 1 | – |
| – | – | C606T | 6 | – |
| H290 | NSCLC | T1191C | 10 | – |
| H292 | NSCLC | C79A | 1 | – |
| H322 | NSCLC | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| H324 | NSCLC | C79A | 1 | – |
| H358 | NSCLC | – | – | – |
| H520 | NSCLC | – | – | – |
| H522 | NSCLC | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| H661 | NSCLC | – | – | – |
| EPLC-65H | NSCLC | C79A | 1 | – |
| LCLC-97TM1 | NSCLC | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| LCLC-103H | NSCLC | – | – | – |
| EPLC-272H | NSCLC | C79A | 1 | – |
| A549 | NSCLC | – | – | – |
| H69 | SCLC | C79A | 1 | – |
| – | – | C119T | 1 | T40I |
| H82 | SCLC | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| H146 | SCLC | – | – | – |
| H209 | SCLC | C79A | 1 | – |
| H211 | SCLC | T1191C | 10 | – |
| H417 | SCLC | – | – | – |
| 16HC | SCLC | C79A | 1 | – |
| – | – | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| 22H | SCLC | C79A | 1 | – |
| – | – | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| 24H | SCLC | C79A | 1 | – |
| – | – | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| 86M1 | SCLC | C79A | 1 | – |
| – | – | T1191C | 10 | – |
| – | – | C1770T | 16 | – |
| DMS76 | SCLC | T1191C | 10 | – |
| SW210.5 | SCLC | C79A | 1 | – |
| – | – | C119T | 1 | T40I |
| – | – | T1191C | 10 | – |
Abbreviations: NSCLC = non–small-cell lung caner; SCLC = small-cell lung cancer
Effect of T40I on Enzastaurin’s In Vitro Activity
We studied the effect of enzastaurin on the level of phosphorylation of PKCβ2 and the downstream molecules GSK3β and S6RP, and on proliferation in SCLC cell lines with (H69 and SW210.5) and without (H211 and DMS79) the T40I amino-acid substitution. In all four cell lines, maximal inhibition of PKCβ2 phosphorylation was achieved after 2 hours of exposure to 1 μM enzastaurin. The maximal inhibition of GSK3β and S6RP phosphorylation was achieved after 4 hours of exposure, and no clear differences among these four cell lines were evident. The 50% inhibitory concentration (IC50) for the inhibition of proliferation was 0.2 μM for H69, 0.07 μM for SW210.5, 0.05 μM for H211, and 0.08 μM for DMS79. These results suggest that the T40I substitution has no effect on enzastaurin’s in vitro efficacy.
Effect of Enzastaurin on NSCLC and SCLC Cell Lines
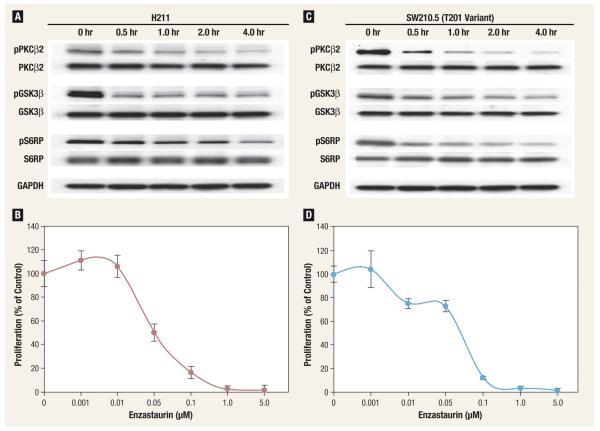
We assessed whether enzastaurin would have differential activity in NSCLC versus SCLC cell lines. In all four NSCLC cell lines studied, the maximal inhibition of PKCβ2, GSK3β, and S6RP phosphorylation was achieved after 2-4 hours of exposure to 1 μM enzastaurin. The IC50 concentrations for the inhibition of proliferation were 0.09 μM for H23, 0.08 μM for H125, 0.08 μM H322, and 0.07 μM for A549. These concentrations were similar to those observed in SCLC cell lines, suggesting that enzastaurin is equally effective in NSCLC and SCLC cell lines (Figure 1).
Figure 1. Phosphorylation (A and C) and Proliferation (B and D) Inhibition by Enzastaurin in 2 Small-Cell Lung Cancer Cell Lines.
Gene sequencing of PRKCB1 revealed the presence of a T40I variant in exon 1 in SW210.5; H211 presented no evidence of sequence variations.
Effect of Enzastaurin Concentration on Phosphorylation and Proliferation Inhibition
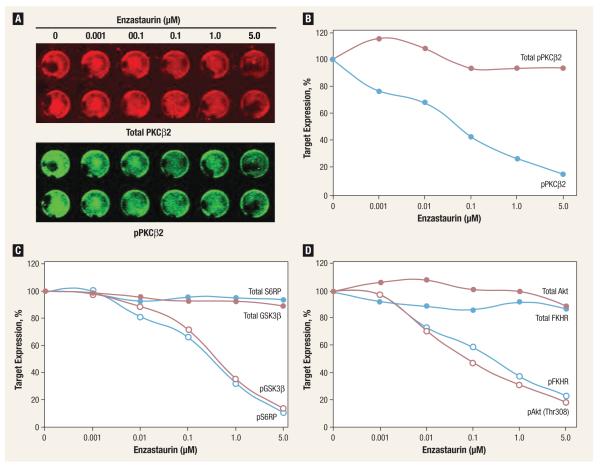
We assessed the relationship between inhibition of protein phosphorylation and proliferation, using different concentrations of enzastaurin. A clear concentration-dependent inhibition of PKCβ2, GSK3β, S6RP, Akt (Thr308), and FKHR phosphorylation occurred in cell line H23, with an IC50 of approximately 0.08 μM for PKCβ2. This concentration was similar to that required for a 50% inhibition of proliferation and suggests that the inhibition of PKCβ2 phosphorylation may be a useful surrogate marker of proliferation inhibition in lung cancer (Figure 2).
Figure 2. Concentration-Dependent Phosphorylation Inhibition of PKβ2 (A and B) and Downstream Targets GSK3~ (C, Red), S6RP (C, Blue), Akt (D, Red), and FKHR (D, Blue) After 1 Hour of Exposure to Enzastaurin in Non–Small-Cell Lung Cancer Cell Line H23.
Discussion
The discovery of tumor-promoting signal-transduction molecules and pathways invigorated the development of targeted agents, with hopes of reducing cancer-related morbidity and mortality. Because it is the leading cause of cancer deaths in the United States,10 lung cancer is one of the leading disease sites for this development. This is exemplified by the approval and implementation of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors as active therapeutic agents. The successful implementation of these agents into clinical practice was substantially facilitated by the identification of subgroups of patients with favorable efficacy characteristics. Most notable among these characteristics was the discovery of mutations in the EGFR gene that render tumors exquisitely sensitive to blockades of EGFR-mediated signal transduction.11-14
Many other signal-transduction pathways have been targeted for drug development, and novel inhibitors are undergoing clinical testing. Among these is enzastaurin, a selective and potent (IC50 of 6 nM) ATP-binding inhibitor of the serine/threonine kinase PKCβ2.15 The preclinical activity of this agent was predominantly reported in lymphoma, glioblastoma, and colorectal cancer.6,16 However, in patients with advanced NSCLC whose previous therapy had failed, the efficacy of this agent was marginal, with only 7 of 55 (13%) patients showing no disease progression 6 months after initiation of treatment.9 Moreover, two studies presented at the 2009 meeting of the American Society of Clinical Oncology did not show additional efficacy when enzastaurin was added to a combination of carboplatin and pemetrexed or carboplatin, pemetrexed, and bevacizumab as first-line therapy in patients with NSCLC.17,18
We sequenced the complete coding region of the PRKCB1 gene in 28 lung cancer cell lines and found only one sequence variation that resulted in an amino acid substitution (T40I) in exon 1. This variation was evident in two SCLC cell lines, in the conserved region 1, which is a cysteine-rich zinc-binding domain. However, position 40 has not been implicated in zinc, diacylglycerol, or phorbol ester binding (NP_002729.2). The T40I substitution did not result in an obvious alteration of enzastaurin on PKCβ2 pathway activation or proliferation inhibition. This lack of effect on efficacy is not surprising, because the catalytic domain of PKCβ2 encompasses amino acids 341-663, including the ATP-binding site. We found no evidence for PKCβ2 deletions, insertions, truncations, or other putatively activating mutations in the cell lines examined. Because we limited our sequencing efforts to 28 cell lines, we cannot rule out the existence of PKC mutations with a potentially meaningful biologic effect in a small subset of lung cancers. Examples of a low-frequency gene mutation with biologic and potentially clinical effects are found in the recently described EML4-Alk fusions, which have an estimated frequency of < 5% in NSCLC.19 However, the clinical experience with enzastaurin in lung cancer to date does not support the existence of a hypersensitive population of patients, a finding consistent with our PRKCB1 gene sequencing data.
Our results demonstrated the concentration-dependent proliferation inhibition and suppression of PKCβ2 kinase and pathway activity in NSCLC and SCLC cell lines with IC50 values in the 0.05–0.2-μM range, which are lower than the previously reported IC50 values in the 1.0-μM range in glioblastoma (U87MG), colorectal (HCT116), and prostate cancer (PC3) cell lines.6 These concentrations are also lower than those reported for lung cancer cell lines with IC50 values in the 1–10-μM range (A549; approximately 2-3 μM).20,21 This is most likely a result of differences in the reagents, cell densities, and exposure durations used for assessments of proliferation. In our studies, the drug concentrations required for the phosphorylation inhibition of PKCβ2 and the downstream molecules GSK3β, S6RP, Akt, and FKHR and proliferation inhibition are comparable (Table 3), which is consistent with previous data in colorectal cancer.6 Our IC50 concentrations were below the steady-state plasma enzastaurin concentrations reported in clinical trials.20
Table 3.
Enzastaurin Concentrations That Induced 50% Inhibition
| Cell Line | Proliferation, μM | pPKCβ2 (Ser660),μM |
pGSK3β (Ser9), μM |
pS6RP (Ser240/244), μM |
pAkt (Thr308), μM |
pAkt (Ser473), μM |
pFKHR (Ser256), μM |
|---|---|---|---|---|---|---|---|
| H69 | 0.20 | – | – | – | – | – | – |
| SW210.5 | 0.07 | – | – | – | – | – | – |
| H211 | 0.05 | – | – | – | – | – | – |
| DMS79 | 0.08 | – | – | – | – | – | – |
| H23 | 0.09 | 0.08 | 0.08 | 0.08 | 0.08 | > 5.00 | 0.30 |
| H125 | 0.08 | – | – | – | – | – | – |
| H322 | 0.08 | – | – | – | – | – | – |
| A549 | 0.07 | – | – | – | 0.20 | > 5.00 | 0.20 |
Our results suggest that a PRKCB1 gene variant (T40I) exists, and it does not affect enzastaurin’s antiproliferative or phosphorylation-inhibitory activity. In addition, enzastaurin appears to be equally active in NSCLC and SCLC cell lines, with IC50 values in the range of 0.05-0.2 μM. The inhibition of phosphorylation of PKCβ2 and the downstream molecules GSK3β, S6RP, Akt, and FKHR is evident in the same concentration range, which suggests that the determination of phosphorylation levels of these molecules in human tissue specimens may be a useful pharmacodynamic parameter for in vivo target inhibition by enzastaurin. In a limited number of pretreatment tumor samples from pancreatic cancer patients treated with enzastaurin and gemcitabine, no statistically significant association between drug efficacy and biomarker levels was evident.20 To our knowledge, a reduction in PKCβ2 phosphorylation has not been described in patients’ specimens during therapy. However, in xenograft-bearing mice receiving enzastaurin, a reduction in GSK3β phosphorylation was described.21 It is thus important to determine prospectively if a reduction in target phosphorylation by enzastaurin is a clinically useful predictive marker of therapeutic efficacy.
Conclusion
Apparently PKCβ2 does not possess sequence alterations that lead to enzastaurin hypersensitivity. The phosphorylation inhibition of PKCβ2, GSK3β, S6RP, Akt, and FKHR may constitute a useful pharmacodynamic parameter for in vivo target inhibition by enzastaurin.
Acknowledgments
This work was supported in part by grants U01-CA101222 and P50-CA119997 to G. B. from the National Cancer Institute.
Footnotes
Sang-Haak Lee and Tingan Chen contributed equally to this work.
Disclosures
Gerold Bepler has received research funding from Eli Lilly and Company. The remaining authors report no relevant potential conflicts of interest.
References
- 1.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7:281–94. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 2.Barr L, Campbell S, Baylin S. Protein kinase C-beta 2 inhibits cycling and decreases c-myc-induced apoptosis in small cell lung cancer cells. Cell Growth Differ. 1997;8:381–92. [PubMed] [Google Scholar]
- 3.Murray NR, Davidson LA, Chapkin RS, et al. Overexpression of protein kinase C-beta 2 induces colonic hyperproliferation and increased sensitivity to colon carcinogenesis. J Cell Biol. 1999;145:699–711. doi: 10.1083/jcb.145.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark A, West K, Blumberg P, et al. Altered protein kinase C (PKC) isoforms in non-small cell lung cancer cells: PKC-delta promotes cellular survival and chemotherapeutic resistance. Cancer Res. 2003;63:780–6. [PubMed] [Google Scholar]
- 5.Partovian C, Simons M. Regulation of protein kinase B/Akt activity and Ser473 phosphorylation by protein kinase C-alpha in endothelial cells. Cell Signal. 2004;16:951–7. doi: 10.1016/j.cellsig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Graff JR, McNulty AM, Hanna KR, et al. The protein kinase C-beta selective inhibitor, enzastaurin (LY317615.HCl), suppresses signaling through the AKT pathway, induces apoptosis, and suppresses growth of human colon cancer and glioblastoma xenografts. Cancer Res. 2005;65:7462–9. doi: 10.1158/0008-5472.CAN-05-0071. [DOI] [PubMed] [Google Scholar]
- 7.Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–85. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Tang J, He Q, Guo R, et al. Phosphorylated Akt overexpression and loss of PTEN expression in non-small cell lung cancer confers poor prognosis. Lung Cancer. 2006;51:181–91. doi: 10.1016/j.lungcan.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Oh Y, Herbst RS, Burris H, et al. Enzastaurin, and oral serine/threonine inhibitor, as second- or third-line therapy of non-small-cell lung cancer. J Clin Oncol. 2008;26:1135–41. doi: 10.1200/JCO.2007.14.3685. [DOI] [PubMed] [Google Scholar]
- 10.American Cancer Society [Accessed: March 25, 2010];Cancer Facts and Figures. 2009 Available at: http://www.cancer.org/downloads/STT/500809web.pdf.
- 11.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–68. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 14.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 15.Faul MM, Gillig JR, Jirousek MR, et al. Acyclic N-(azacycloalkyl)bisindolyl-maleimides: isozyme selective inhibitors of PKC-beta. Bioorg Med Chem Lett. 2003;13:1857–9. doi: 10.1016/s0960-894x(03)00286-5. [DOI] [PubMed] [Google Scholar]
- 16.Su TT, Guo B, Kawakami Y, et al. PKC-beta controls IkappaB kinase lipid raft recruitment and activation in response to BCR signaling. Nat Immunol. 2002;3:780–6. doi: 10.1038/ni823. [DOI] [PubMed] [Google Scholar]
- 17.Obasaju CK, Raju RN, Stinchcombe T, et al. Final results of a randomized phase II trial of pemetrexed and carboplatin with or without enzastaurin versus docetaxel and carboplatin as first-line treatment of patients with stage IIIB/IV non-small cell lung cancer. Proc Am Soc Clin Oncol. 2009;27:416s. doi: 10.1097/JTO.0b013e3181fd42eb. [DOI] [PubMed] [Google Scholar]
- 18.Casey EM, Harb W, Bradford D, et al. Randomized, double blind, multicenter, phase II study of pemetrexed, carboplatin, bevacizumab with enzastaurin or placebo in chemotherapy-naive patients with stage IIIB/IV non-small cell lung cancer: Hoosier Oncology Group (HOG) LUN06-116. Proc Am Soc Clin Oncol. 2009;27:415s. doi: 10.1097/JTO.0b013e3181ee820c. [DOI] [PubMed] [Google Scholar]
- 19.Mano H. Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci. 2008;99:2349–55. doi: 10.1111/j.1349-7006.2008.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards DA, Kuefler PR, Becerra C, et al. Gemcitabine plus enzastaurin or single-agent gemcitabine in locally advanced or metastatic pancreatic cancer: results of a phase II, randomized, noncomparative study. Invest New Drugs. 2009 doi: 10.1007/s10637-009-9307-8. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Bodo J, Durkin L, Hsi ED. Quantitative in situ detection of phosphoproteins in fixed tissues using quantum dot technology. J Histochem Cytochem. 2009;57:701–8. doi: 10.1369/jhc.2009.953547. [DOI] [PMC free article] [PubMed] [Google Scholar]