Abstract
In modern functional genomics registration techniques are used to construct reference gene expression patterns and create a spatiotemporal atlas of the expression of all the genes in a network. In this paper we present a software package called GCPReg, which can be used to register the expression patterns of segmentation genes in the early Drosophila embryo. The key task, which this package performs, is the extraction of spatially localized characteristic features of expression patterns. To facilitate this task, we have developed an easy-to-use interactive graphical interface. We describe GCPReg usage and demonstrate how this package can be applied to register gene expression patterns in wild type and mutants. GCPReg has been designed to operate on a UNIX platform and is freely available via the Internet at http://urchin.spbcas.ru/downloads/GCPReg/GCPReg.htm.
Keywords: image processing, confocal microscopy, quantitative gene expression data, spatial registration, segmentation genes
Introduction
The registration problem arises in biological imaging whenever images of biological molecules acquired from many individuals and in different experiments need to be matched. In modern functional genomics research registration techniques have two important applications. First, they are used to construct reference expression patterns, i.e., representative patterns of a given gene expression at a given time interval. Another application of these techniques is the creation of a spatiotemporal atlas of expression of all genes in a genetic network.
A particular case of the problem is registration of quantitative data on gene expression extracted from confocal images. Nowadays the confocal scanning microscopy of fluorescently tagged molecules is the most common method for acquisition of gene expression data at cellular resolution. Confocal images are of very high quality that makes it possible to extract high-precision quantitative information. However, currently available confocal microscopes permit scanning for expression of only a small number of genes at once. Thus to construct reference data or a spatiotemporal map of expression of all network genes it is necessary to combine data from many individuals scanned in different experiments, but natural variability precludes direct superposition of the data. To solve this problem the data need to be registered, i.e., brought to the one and the same coordinate system.
One common approach to registration of gene expression patterns is the point mapping technique. It is based on the extraction of “ground control points” (GCPs),1 which are a small number of characteristic features in each pattern, and application of a coordinate transformation to make the patterns coincide as closely as possible. The extraction of GCPs can be performed, for example, by spline approximation or wavelet decomposition techniques.
One of the best-studied gene regulatory networks in Drosophila controls the segmentation of the embryo. Drosophila segments are determined at the syncytial blastoderm stage, at which the embryo is an ellipsoidal shell of nuclei that are not surrounded by cell membranes. The future segmental pattern is produced by the spatiotemporal refinement in the expression of about 14 transcription factors, referred to as segmentation genes.2,3 Maternal coordinate genes (primarily bicoid and hunchback) provide asymmetric initial conditions to the rest of the segmentation system.4,5 Recently, methods for the acquisition of quantitative data on segmentation gene expression were introduced by two research groups.6–8
An important feature of the segmentation genetic network is its relative independence from genes controlling the dorsoventral (DV) patterning of the embryo.9,10 This makes it possible to consider the expression of segmentation genes in one dimension along the AP axis. Although such an approach was a simplification, its successful applications demonstrate the ability of the one-dimensional representation to reveal new interesting facts about dynamics of formation of segmentation gene expression domains.11–15
In this paper we present a new software tool called GCPReg. This tool implements the method which we have developed to register the one-dimensional expression patterns of segmentation genes.7 The method was successfully applied to register a large dataset on segmentation gene expression with high accuracy.7,16–19 In the already-formed late patterns of segmentation genes the standard deviations of the x coordinates of the extrema range within about 0.5–1% egg length and are small compared to the average size of a nucleus.7,17 The method consists of the following basic steps: (1) construction of the GCP template; (2) extraction of GCPs from each expression pattern; (3) application of an affine coordinate transformation to minimize the distance between the GCP template and corresponding GCPs in individual patterns; (4) update of the GCP template. The expression patterns of most segmentation genes are made up of several expression domains, each of which has at least one concentration maximum. It is natural to take the maxima and/or minima of one-dimensional gene expression pattern as GCPs for registration. Here we describe how to work with GCPReg and present several examples of its use.
GCPReg Usage
Construction of a new GCP template
The main function of GCPReg is to register the one-dimensional expression patterns of a segmentation gene against the set of GCPs. We define this set as a template. The package provides a set of ready-to-use GCP templates for nine segmentation genes knirps (kni), giant (gt), even-skipped (eve), fushi tarazu (ftz), hairy (h), runt (run), odd-skipped (odd), paired (prd) and sloppy-paired (slp) at cycle 14A. This cycle is subdivided into eight time intervals and the templates are separately computed for each time interval from 2 to 8.
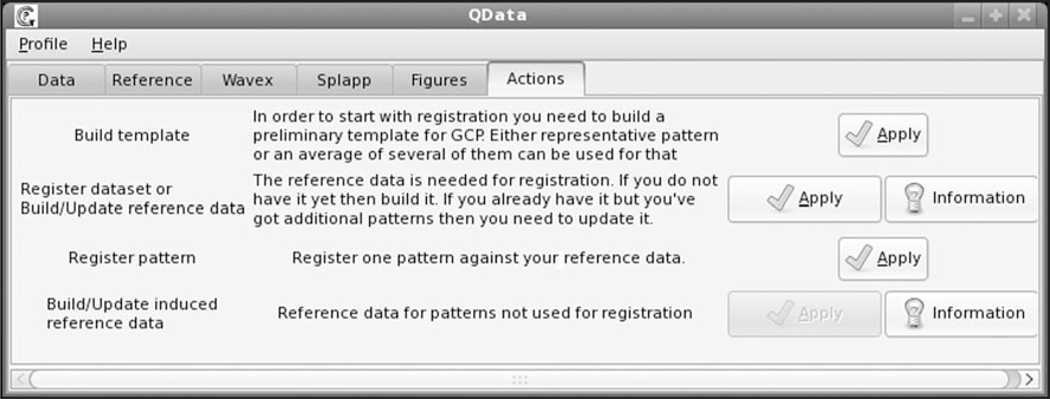
To construct a new template a user chooses the option “Build template” in the GCPReg Actions window presented in Figure 1. A new template can be created in three ways. The first method is recommended in case of expression patterns with well-resolved features. Then the user can just select a typical pattern and use the coordinates of its extrema as GCPs. The second mode is mostly applicable if there is a need to register poorly resolved patterns, from which it is not possible to extract well-defined GCPs. In this situation the user selects a set of individual patterns and applies GCPReg to average these patterns and extract GCPs from the averaged pattern. Finally the user can explicitly define the GCP template and save this information as a file in the working directory. The choice of characteristic features should be based on a thorough analysis of expression patterns of a given gene at a given time interval in the whole dataset. For better registration results it is recommended to choose as GCPs those characteristic features the position of which have the least variance at the given developmental time. The number of GCPs is an input parameter of the program.
Figure 1.
GCPReg interface.
A typical or averaged pattern used to extract GCPs is called a reference pattern and saved for the visualization of registration results. In case of user-defined GCPs the corresponding reference pattern may be optionally provided by the user and saved in the working directory.
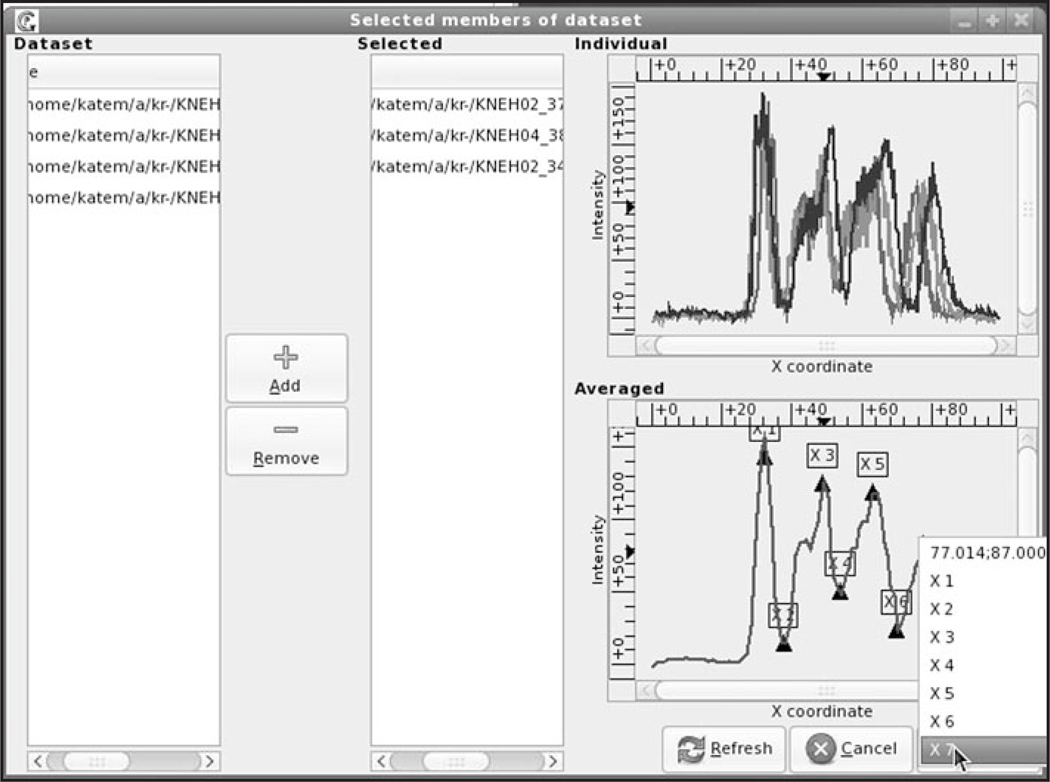
To facilitate the construction of the GCP template, an easy-to-use interactive graphical user interface (GUI) was developed (see Fig. 2). The user defines source directories containing unregistered data and selects either one or several files each containing quantitative data on the expression of a given gene at a given time interval. If several files were selected, the input patterns are automatically averaged and the resultant reference pattern is presented on the lower panel.
Figure 2.
GUI for extraction of the GCP template. The template is constructed for eve expression pattern in embryos mutant for Kr belonging to late cycle 14A (time class 6 according to our classification, see Materials and methods). The Dataset panel (left) shows the folders containing the dataset. Any number of files can be selected and moved to the Selected panel by pressing the Add button. The upper panel on the right (Individual) presents the graphs of selected patterns; while in the lower panel (Averaged) the average of these patterns is plotted. The pop-up menu shows cursor coordinates and the list of feasible GCPs.
To extract GCPs from the reference pattern the user places the mouse pointer on an extremum of the pattern diagram and clicks the right button. This opens the pop-up menu displaying the coordinates of the selected GCP and the list of feasible GCPs. Clicking the left mouse button on the highlighted GCP number automatically writes the x-coordinate of this point into the file. The procedure is repeated until all the points are extracted.
Feature extraction and registration
The method for registration of an individual expression pattern against the template is organized into two steps: Feature Extraction and Registration. Registration applies the affine coordinate transformation to minimize the distance between GCPs extracted by Feature Extraction methods and the GCP template. Two alternative feature extraction methods are implemented. The first one, the Spline method, approximates the expression pattern by a quadratic spline, finds positions of the spline extrema, and saves them as GCPs. The second method, FRDWT, implements the fast redundant dyadic wavelet transform20 to decompose the pattern into two sequences: low and high pass, and uses the zeros of high pass as GCPs.
A user can either choose the option Register pattern to register a single input pattern or select Register Dataset to register all input patterns one by one.
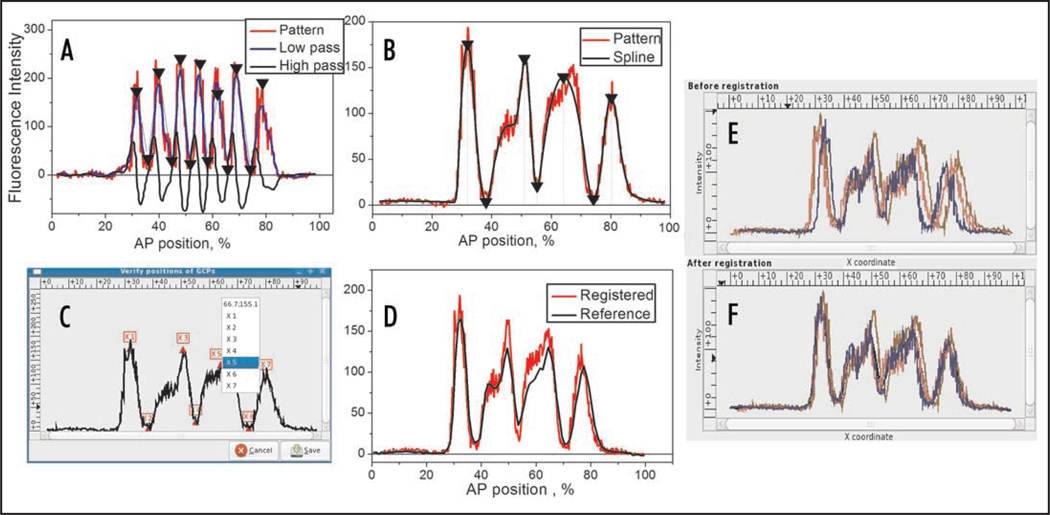
GCPReg operates in both automatic and interactive modes. In automatic mode the positions of extremal features are automatically found on a pattern by means of quadratic splines or FRDWT.7,17 These two methods provide very similar results in most cases, but it is generally recommended to apply spline approximation to broad expression domains, while for narrow sharp stripes the FRDWT method is preferable. To automatically extract extremal features and put them into correct correspondence with the template GCPs the user has to define a set of manually adjustable parameters such as the number of GCPs to be extracted, the allowable range of their positions, the minimal amplitude of the first domain (in order to distinguish it from small random peaks), etc. The correct choice of these parameters requires the thorough inspection of individual expression patterns. The feature extraction procedure is illustrated in Figure 3A and B.
Figure 3.
Feature extraction and registration. (A) The FRDWT decomposition of eve pattern in the wild-type embryo belonging to late cycle 14A (time class 6). The pattern is decomposed into two sequences, low pass and high pass, and GCPs are extracted as zeros of the high pass and marked on the pattern by arrows. (B) Spline approximation of eve pattern in a Kr− mutant embryo from the time class 5. GCPs are extracted as coordinates of the spline extrema (marked by arrows). (C) GUI for interactive correction of the automatically found GCPs. In this pattern the fifth GCP (third maximum) is misplaced and needs to be corrected (the corresponding GCP template is shown in Fig. 2). (D) Visualization of registration results: the registered pattern is superimposed on the reference pattern. (E and F) Screenshots of registration results: three eve patterns in Kr− embryos before (E) and after (F) registration. The expression pattern presented in (C and D) is the same as in (B).
The extracted GCPs are used to register a pattern against the GCP template by application of an affine coordinate transformation. By default the extraction of GCPs is fully automatic, but it often happens that due to individual peculiarities of a pattern (e.g., if the pattern is poorly resolved, noisy or spatially shifted) both Spline and FRDWT methods fail to automatically find the precise positions of extremal features. In such a situation GCPReg is run in interactive mode using the GUI. In this mode the user can interactively select the positions of all GCPs and correct erroneous GCPs that were automatically selected by either of the feature extraction methods. The GUI presents the pattern on which the automatically selected features are enumerated and marked as shown in Figure 3C. To correct the GCP position the user just places the mouse pointer on a new point of the diagram and clicks the right button to open the pop-up menu and chooses the GCP number. If the pattern is not yet completely formed and some of the characteristic features are missing, it is sufficient to only select well-defined features as GCPs. The smaller number of extracted GCPs does not preclude the pattern from registration with the GCP template.
There are two options for visual inspection of the registration results. First, the registration quality of a single pattern can be visualized by superposition of the registered pattern on the reference pattern (if available) as shown in Figure 3D. The second option is convenient to use if the set of patterns is registered. In this case the GUI presents two panels displaying the superimposed patterns before and after registration (see Fig. 3E and F).
The main input parameters of GCPReg are: the feature extraction method (splines or FRDWT), the GCP template, the run mode (automatic or interactive), the set of adjustable parameters, and the input dataset. The files with registered data are generated as output. These files contain the transformed coordinates of nuclei and mean fluorescence intensities of all gene products stained in the embryo.
If an embryo was stained for expression of several genes, registration of the data collected in one microscope channel results in registration of the data on expression of genes detected in other microscope channels. This kind of registration is called the induced registration as opposed to the direct registration described above. If there is a need to register expression patterns of all the genes stained in the embryo it is recommended to select the expression pattern with the most well-defined features for the direct registration.
Template update
A new GCP template is usually constructed using one or a few representative expression patterns, which may not contain full information about the localization of expression domains of a given gene. Thus it is recommended to update GCP templates from time to time as new data is being accumulated.
To update the GCP template and the reference pattern for a given gene and given time interval the user chooses the Build/Update Reference data option. On selection of this option GCPReg first checks, whether GCPs were extracted from all the data files in the directory and otherwise runs the feature extraction procedure in either automatic or interactive mode. Next, the GCP template and the reference pattern are updated by averaging all the GCP values and all the registered patterns correspondingly.
To build or update the reference patterns for genes scanned in other microscope channels the Build/Update induced reference data option is used. On selection of this option the number of microscope channel used to collect the data should be specified. Reference patterns are constructed by averaging corresponding patterns in the registered dataset.
GCPReg Usage Examples
Automated registration
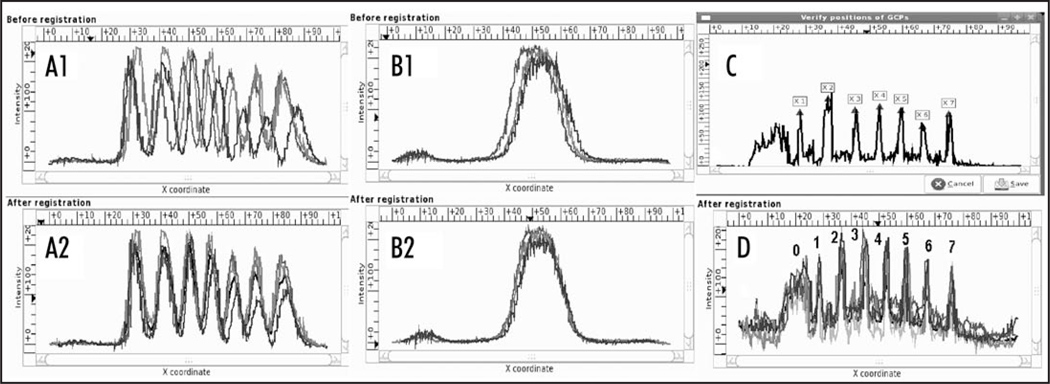
In late cycle 14A pair-rule genes form seven narrow expression domains (stripes) with well-localized characteristic features. Therefore the expression patterns of these genes can be registered in automatic mode with high precision. In such patterns the positions of all the stripe maxima and interstripe minima are taken as GCPs. The extraction of 13 extremal features, seven maxima and six minima, is done by either FRDWT or spline approximation. The result of feature extraction from a late eve pattern by the FRDWT method is shown in Figure 3A as an example. The results of registration of five eve expression patterns are presented in Figure 4A1 and A2. It is evident that the initial spatial variability of these patterns is reduced considerably after registration.
Figure 4.
GCPReg usage examples: Screenshots of registration results. (A) Automated registration: five expression patterns of the eve gene in embryos belonging to late cycle 14A (time class 6) before (A1) and after (A2) registration. (B) Induced registration: Three Kr expression patterns acquired in the same embryos as the eve patterns presented in (A1 and A2) and registered using these patterns. (C) Interactive registration: The GCP extraction from slp expression pattern in an embryo belonging to late cycle 14A. Maxima of stripes from 1 to 7 are only used as GCPs. (D) Results of the interactive registration of four late slp expression patterns in wild-type embryos from time class 8. Expression domains are enumerated from 0 to 7.
Interactive registration
In case of automatic registration of poorly resolved patterns some of the extracted GCPs may be mislocated or missed and such errors must be interactively corrected. Figure 3 illustrates how the interactive registration works by considering eve gene expression patterns in three Krüppel (Kr) null mutant embryos as an example. In these mutants the eve expression pattern differs greatly from that in wild type embryos, and hence a new GCP template has to be created. Besides, eve expression domains are not well-defined even at late times, thus it is necessary to use the interactive mode to control the GCP extraction. In this example the spline approximation method followed by manual correction was applied to extract seven GCPs, three minima and four maxima. The corresponding GCP template is shown in Figure 2. The results of the pattern registration are presented in Figure 3E and F.
Another example of the interactive registration is shown in Figure 4C and D. slp gene belongs to the pair-rule family and forms a classical pattern of seven transverse stripes. However, prior to gastrulation additional “segment-polarity-like” stripes appear between the primary slp pair-rule stripes and additional DV-dependent region of expression (stripe 0) forms in the presumptive head region. Due to high variability of these features the automatic registration of slp pattern is complicated and requires careful adjustment of program parameters. In these patterns only maxima of pair-rule-like stripes from 1 to 7 are taken as GCPs. It should be noted that FRDWT is the only recommended method for feature extraction in such situations.
Induced registration
The gap gene Kr forms a bell-shaped expression domain with only one well-localized extremal feature, which is obviously insufficient for the direct registration of patterns. The expression patterns of this gene are registered by induced registration using eve expression patterns acquired in the same embryos and presented in Figure 4A1 and A2. Figure 4B2 demonstrates that the registration of eve patterns brings about a decrease in spatial variability of Kr patterns and hence induces the registration of these patterns.
Discussion
We have designed the software tool GCPReg to register one-dimensional patterns of segmentation gene expression. The rationale behind this approach is the relative independence of segmentation gene expression from genes controlling the dorsoventral (DV) patterning of the embryo.9,10 This allows us to consider the expression of segmentation genes in one dimension along the AP axis, which is a level of representation suitable for answering many important biological questions.
Two other programs were developed to register gene expression patterns in early Drosophila embryo. The program of Sorzano et al.21 performs the elastic registration of segmentation gene expression patterns. This program deforms the coordinates non-linearly to make the images coincide as close as possible. After transformation all the registered images are brought to one and the same size. The method is very accurate, however it distorts the coordinates of nuclei and therefore cannot be used to register quantitative data on gene expression extracted from confocal images. The program developed by Fowlkes et al.22 registers three-dimensional quantitative gene expression data extracted from stacks of confocal images. This program warps the borders of expression domains using the regularized thin-plate splines in order to bring an individual pattern into alignment with a standard morphological template.
GCPReg has rich functionality. It allows a user to create GCP templates against new data to be registered, choose between two feature extraction methods, adjust program parameters according to pattern characteristics and correct manually the GCP positions using GUI. These options make it possible to use GCPReg for registration of any one-dimensional data on gene expression both at the RNA and protein levels and at a resolution of a single cell. The highest quality of registration can be achieved if expression domains are well resolved and have well-defined positional features. However, the possibility of interactively correcting the GCP positions allows the user to correctly extract characteristic features even from poorly resolved expression patterns, such as patterns of segmentation gene expression in wild type embryos at cleavage cycle 13 and early cleavage cycle 14A, in mutant embryos, or complicated segment-polarity-like pre-gastrulation patterns.
An especially convenient feature of GCPReg is the ability to update both GCP templates and reference patterns with the acquisition of new data. This option makes it possible to take full advantage of information about the localization of expression domains in the whole dataset, and thereby improves the registration quality.
GCPReg can be also used to register two-dimensional quantitative data on segmentation gene expression extracted from a whole 2D embryo image. As was shown in Kozlov et al.19 this data can be registered with good accuracy by applying the coordinate transformation estimated from corresponding one-dimensional data.
The friendly graphical user interface makes the GCPReg usage simple, functional and convenient. It facilitates the creation of templates and feature extraction. Rich means of data visualization enable the user to estimate both the accuracy of registration and the quality of the resulting reference data.
A limitation of the registration method is the requirement that the pattern has a sufficient number of detectable and well-localized characteristic features. However this limitation is not very restrictive, as it is enough to have only one such pattern, and all the rest will be simultaneously registered by induced registration.
Materials and Methods
Quantitative data on segmentation gene expression
To demonstrate how GCPReg works we use quantitative data on segmentation gene expression obtained in wild type and mutant embryos and stored in the FlyEx database (http://urchin.spbcas.ru/flyex/; http://flyex.ams.sunysb.edu/flyex/).
The quantitative gene expression data are acquired from images of gene expression patterns obtained by confocal scanning microscopy of fixed embryos immunostained for segmentation proteins. The immunostaining procedure was as described in refs. 6 and 23. The microscope at our disposal has four channels and thus allows to only scan four fluorescent labels at a time. Each embryo is scanned for the expression of three genes and histones to mark nuclei. Each gene is detected in a single channel. All the embryos belong to cleavage cycle 14A. The registration method was applied to 1,074 wild-type embryos immunostained to detect the expression of maternal genes bicoid (in 177 embryos) and caudal (in 110 embryos); gap genes Krüppel (in 286 embryos), knirps (121), giant (163), hunchback (258) and tailless (109) and pair-rule genes even-skipped (in 1,074 embryos), fushi tarazu (in 374 embryos), hairy (139), runt (72), odd-skipped (120), paired (121) and sloppy-paired (98), as well as to 63 Krüppel null mutants stained for expression of knirps (30), giant (31), hunchback (44) and even-skipped (34).
The quantitative data on gene expression is extracted through a data pipeline which consists of five steps: image segmentation, background removal, temporal characterization of an embryo, data registration and data averaging.17 The data are presented as a table containing nucleus number, x and y coordinates of its centroid measured in percent of the embryo length and width, as well as the averaged fluorescence intensities (relative expression levels) for each gene scanned in the embryo. One-dimensional data are extracted from the central 10% strip of an embryo along the AP axis.
Temporal classification of embryos makes it possible to reconstruct the temporal dynamics of gene expression from many embryos each fixed at a different stage of development. As cleavage cycle 14A is about 50 minutes long, it was divided into eight temporal equivalence classes based on thorough visual inspection of the expression pattern of the eve gene in individual embryos as described in references 7 and 24. Each class represents about 6.5 minutes of development. The registration method was applied to all the embryos belonging to time classes from 2 to 8. All the registered data are stored in the FlyEx database.
GCPReg technical information
The package is coded using a combination of shell and Perl scripts and applications in the C programming language. The external program gnuplot is used to draw graphs. Development was carried out on Fedora GNU/Linux Operating System for x86 with the gcc 4.3 compiler. The graphical user interface is based on gtk+. The package can be downloaded from http://urchin.spbcas.ru/downloads/GCPReg/GCPReg.html.
Acknowledgements
This work is supported by NIH grant RR07801, GAP award RUB1-1578 and RFBR grants 08-04-00712-a, 08-01-00315-a and 09-04-01590-a.
References
- 1.Brown LG. ACM computing surveys. 1992;24:16–19. [Google Scholar]
- 2.Akam M. The Molecular Basis for Metameric Pattern in the Drosophila Embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- 3.Ingham P. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- 4.Driever W, Nüsslein-Volhard C. A gradient of Bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 5.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the Hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 6.Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Development Genes and Evolution. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- 7.Myasnikova E, Samsonova A, Kozlov K, Samsonova M, Reinitz J. Registration of the Expression Patterns of Drosophila Segmentation Genes by Two Independent Methods”. Bioinformatics. 2001;17:3–12. doi: 10.1093/bioinformatics/17.1.3. [DOI] [PubMed] [Google Scholar]
- 8.Luengo-Hendriks CL, Keränen SV, Fowlkes CC, Simirenko L, Weber GH, DePace AH, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biology. 2006;7:123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll S, Winslow G, Twombly V, Scott M. Genes that control dorsoventral polarity affect gene expression along the anteroposterior axis of the Drosophila embryo. Development. 1987;99:327–332. doi: 10.1242/dev.99.3.327. [DOI] [PubMed] [Google Scholar]
- 10.Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holloway DM, Harrison LG, Spirov AV. Noise in the segmentation gene network of Drosophila with implications for mechanisms of body axis specification. In: Bezrukov SM, editor. Fuctuations and Noise in Biological, Biophysical and Biomedical Systems. Vol. 5110. 2003. pp. 180–191. [Google Scholar]
- 12.Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional Evolution of a cis-Regulatory Module. PLoS Biology. 2005;3:0588–0598. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yucel G, Small S. Morphogens: precise outputs from a variable gradient. Curr Biol. 2006;16:29–31. doi: 10.1016/j.cub.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov K, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- 15.Surkova S, Kosman D, Kozlov K, Manu, Myasnikova E, Samsonova A, et al. Characterization of the Drosophila segment determination morphome. Developmental Biology. 2008;313:844–862. doi: 10.1016/j.ydbio.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poustelnikova E, Pisarev A, Blagov M, Samsonova M, Reinitz J. A database for management of gene expression data in situ. Bioinformatics. 2004;20:2212–2221. doi: 10.1093/bioinformatics/bth222. [DOI] [PubMed] [Google Scholar]
- 17.Surkova S, Myasnikova E, Janssens H, Kozlov K, Samsonova A, Reinitz J, Samsonova M. Pipeline for acquisition of quantitative data on segmentation gene expression from confocal images. FLY. 2008;2:58–66. doi: 10.4161/fly.6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pisarev A, Poustelnikova E, Samsonova M, Reinitz J. FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution. Nucleic Acids Research. 2008;1:1–7. doi: 10.1093/nar/gkn717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlov K, Myasnikova E, Pisarev A, Samsonova M, Reinitz J. A method for two-dimensional registration and construction of the two-dimensional atlas of gene expression patterns in situ. In Silico Biology. 2002;2:125–141. [PubMed] [Google Scholar]
- 20.Unser M. A practical guide to the implementation of the wavelet transform. In: Aldroubi A, Unser M, editors. Wavelets in Medicine and Biology. Raton, Boston, London, New York, Washington D.C: CRC Press LLC; 1996. pp. 37–73. [Google Scholar]
- 21.Sorzano COS, Blagov M, Thevenaz P, Myasnikova E, Samsonova M, Unser M. Algorithm for Spline-Based Elastic Registration in Application to Confocal Images of Gene Expression. Pattern Recognition and Image Analysis. 2006;16:93–96. [Google Scholar]
- 22.Fowlkes C, Hendriks C, Keränen S, Weber G, Rübel O, Huang M, et al. A Quantitative Spatiotemporal Atlas of Gene Expression in the Drosophila Blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 23.Janssens H, Kosman D, Vanario-Alonso CE, Jaeger J, Samsonova M, Reinitz J. A high-throughput method for quantifying gene expression data from early Drosophila embryos. Development Genes and Evolution. 2005;215:374–381. doi: 10.1007/s00427-005-0484-y. [DOI] [PubMed] [Google Scholar]
- 24.Myasnikova E, Samsonova A, Samsonova M, Reinitz J. Support vector regression applied to the determination of the developmental age of a Drosophila embryo from its segmentation gene expression patterns. Bioinformatics. 2002;18:87–95. doi: 10.1093/bioinformatics/18.suppl_1.s87. [DOI] [PubMed] [Google Scholar]