Abstract
Cell adhesion molecule CD2 and its ligand CD58 provide good examples of protein-protein interactions in cells that participate in the immune response. To modulate the cell adhesion interaction, peptides were designed from the discontinuous epitopes of the β-strand region of CD2 protein. The two strands were linked by a peptide bond. β-strands in the peptides were nucleated by inserting a beta-sheet-inducing (D)-Pro-Pro sequence or a dibenzofuran (DBF)-turn mimetic with key amino acid sequences from CD2 protein that binds to CD58. The solution structures of the peptides (5–10) were studied by NMR and molecular dynamics simulations. The ability of these peptides to inhibit cell adhesion interaction was studied by E-rosetting and lymphocyte epithelial assays. Peptides 6 and 7 inhibit the cell adhesion activity with an IC50 value of 7 nM and 11 nM respectively, in lymphocyte-epithelial adhesion assay. NMR and molecular modeling results indicated that peptides 6 and 7 exhibited β-hairpin structure in solution.
Keywords: beta hairpin, CD2, CD58, cell adhesion, dibenzofuran, diproline sequence, immunomodulation, NMR
Introduction
Protein-protein interactions (PPI) play a crucial role in many biological processes, including transmembrane signal transduction, cell regulation, and the immune response 1–3. Analysis of 3D structures of protein complexes has suggested that the PPI surfaces are large, covering 700–1500 Å2, and the binding surfaces are relatively flat4. Some amino acid residues in the interface contribute substantially to the binding energy while others contribute only marginally to stabilize the protein complexes. Analyses of the PPI surfaces and the amino acid residues by several researchers have indicated that there are “hot spots” on the protein-protein interface area where major contributions to binding free energy are provided5. Protein-protein interactions can be modulated by small molecules and have implications in the treatment of diseases6. Cell adhesion molecule CD2 and its ligand CD58 provide good examples of protein-protein interactions in cells that participate in the immune response. CD2 is a 50 KDa protein that is involved in the early stages of immune response. It is expressed on the surface of T cells and natural killer (NK) cells in all mammalian species and binds to CD58 via the first portion of the extracellular domain. CD58 is also called leukocyte function-associated antigen-3 (LFA-3) in humans and CD48 in rodents 7–9. It is known that in the presence of CD2–CD58 interaction, T-cell receptors (TCR) on T cells recognize the correct peptide-major histocompatibility complex (pMHC) on antigen-presenting cells (APC) with 50- to 100-fold greater efficiency than in the absence of CD2–CD58 interaction 7. CD2–CD58 interaction affects expression of cell surface molecules, production of inflammatory cytokines, and T-cell antigen recognition. The CD2–CD58 pathway initiates strong antigen-independent cell adhesion, substantial expansion of naive T helper cells, and induction of large amounts of IFN-γ in memory cells 10, 11. The overall contribution of this pair of molecules is to enhance the T cell-antigen recognition. Monoclonal antibodies (mAbs) that block the CD2–CD58 interaction are being developed to treat autoimmune diseases. These mAbs inhibit T-cell adhesion or activation 12–15. Alefacept is a recombinant human CD58-Ig fusion protein that effectively binds to CD2 and prevents CD2 interaction with CD58 expressed on APC. Alefacept is a biologic that is approved for the treatment of chronic plaque psoriasis in adults 12, 16. However, therapeutic antibodies/fusion proteins often elicit significant side effects attributed to their immunogenicity, and are susceptible to enzymatic degradation. To circumvent these problems, one approach is to design short peptides or small molecular mimics of CD2 protein that will bind to critical areas in target proteins and, like antibodies, interfere with their activity. Peptides, although susceptible to enzymatic degradation, can be modified to protect the N-and C-termini from enzymatic degradation 17, 18. Our aim is to develop peptides that inhibit protein-protein interactions between CD2 and CD58 and utilize them as possible therapeutic agents for autoimmune diseases.
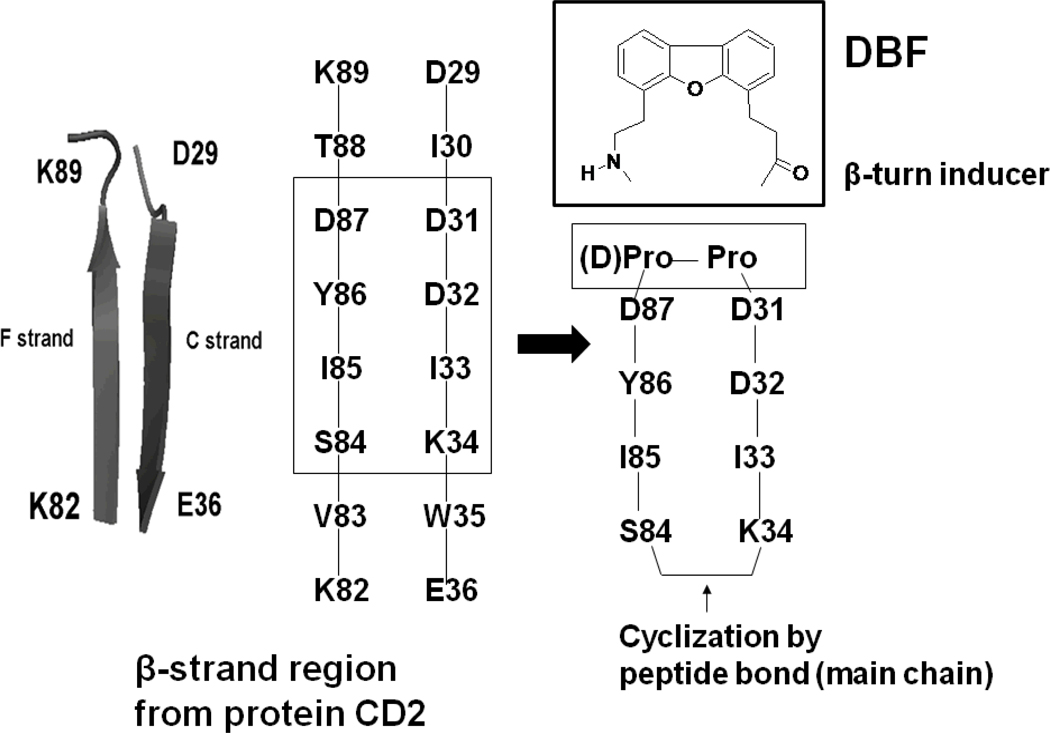
In our previous studies, we have shown that peptides designed from the β-strand region of CD2 protein block cell adhesion interactions between T cells and epithelial cells 19–22. Conformational constraints were introduced in the peptide by a Pro-Gly sequence and cyclization to generate β-hairpin structures from the strand sequences 20. In the present study, we wanted to introduce more rigid structures to nucleate β-hairpin structures in the peptide. A (D)-Pro-Pro sequence or a dibenzofuran moiety 23 was inserted to connect the F and C strands of CD2 protein between D31 and D87. The other end of the strand (K34-S84) was cyclized by main chain cyclization to acquire a stable peptide structure (Figure 1, Table 1). The use of (D)-Pro at the i+1 position of a β-turn facilitates the formation of type II" or I' turns. (D)-Pro-Pro sequences have been shown to be strong β-hairpin nucleators 24. The DBF moiety was also used to stabilize the β-hairpin structures23. Thus, peptides with (D)-Pro-Pro and DBF moiety were used to stabilize the sequences designed from F and C strands of CD2 protein. The designed peptides are shown in Table 1. The ability of the designed peptides to modulate the CD2–CD58 interaction was evaluated by using two cell adhesion assays. The structures of the peptides were studied by NMR and molecular dynamics simulations. Peptides 6 and 7 acquire a stable β-hairpin structure in solution. In the present study we describe the design of peptides that are novel in the sense that (D) Pro-Pro sequence and DBF were used to stabilize the structure and nucleate β-hairpin structures. Among the peptides designed in this series, peptides 6 and 7 inhibit cell adhesion activity in the nanomolar range. Furthermore, these peptides at nanomolar concentration can suppress antigen-specific immune response in vitro in cells isolated from humanized mice susceptible to inflammatory polyarthritis.
Figure 1.
Design of peptides from CD2 protein. F and C strands of CD2 protein from the crystal structures are shown with the sequences of amino acids that are important in binding to CD58 (rectangular box). The amino acids from the two strands of CD2 protein were used to design conformationally constrained peptides. One end of the strands was cyclized by introducing (D)-Pro-Pro in the peptide sequence. The other ends of the strands were joined by backbone cyclization.
Table 1.
Sequence of peptides used in the study
| Code | Sequence | Comments | Purity |
|---|---|---|---|
| 5 | Cyclo (1,8) I1Y2D3(D)P4P5D6D7K8 | (D)-Pro-Pro for β-hairpin, truncated form of 6 | >95 |
| 6 | Cyclo (1,10) S1I2Y3D4(D)P5P6D7D8I9K10 | (D)-Pro-Pro for β-turn, S1-D4 to D7-K10 residues from CD2 protein sequence | 95 |
| 7 | Cyclo(1,6)K1D2D3-DBF-D4Y5I6 | DBF for β-hairpin, sequence from protein CD2 used in anti-clockwise direction. Truncated form of 8 | >95 |
| 8 | Cyclo(1,8) K1I2D3D4-DBF-D5Y6I7S8 | DBF for β-hairpin, sequence from protein CD2 used in anti-clockwise direction | >95 |
| 9 | Cyclo(1,6) I1Y2D3-DBF-D4D5K6 | DBF for β-hairpin, sequence from protein CD2 used as it appears in protein. Truncated form of 10 | >95 |
| 10 | Cyclo(1,8) S1I2Y3D4-DBF-D5D6I7K8 | DBF for β-hairpin, sequence from protein used as it appears in protein | >95 |
| C | KGKTDAISVKAI-NH2 | Control peptide. Sequence from hot-spot region was reversed, Tyr replaced by Ala. | >95 |
Results
The design of the peptides was based on the structure of the CD2–CD58 complex 25, 26 as well as on our previous studies 19–22. The CD58 binding domain of CD2 consists of β-strands with charged residues. From our earlier report it is very clear that peptides designed from β-strands exhibit cell adhesion inhibition activity 20–21. In the present study, conformational constraints were introduced to stabilize the β-hairpin structure and to improve the cell adhesion inhibition activity of peptides. Designed peptides with conformational constraints are shown in Table 1. Linear control peptide was synthesized in the laboratory and cyclic peptides were custom synthesized from commercial sources. HPLC chromatograms of the peptides indicated that the peptides were ≥95% pure. HPLC chromatograms and high-resolution mass spectra of the peptides are available in Supporting Information.
Cell Adhesion Inhibition Activity
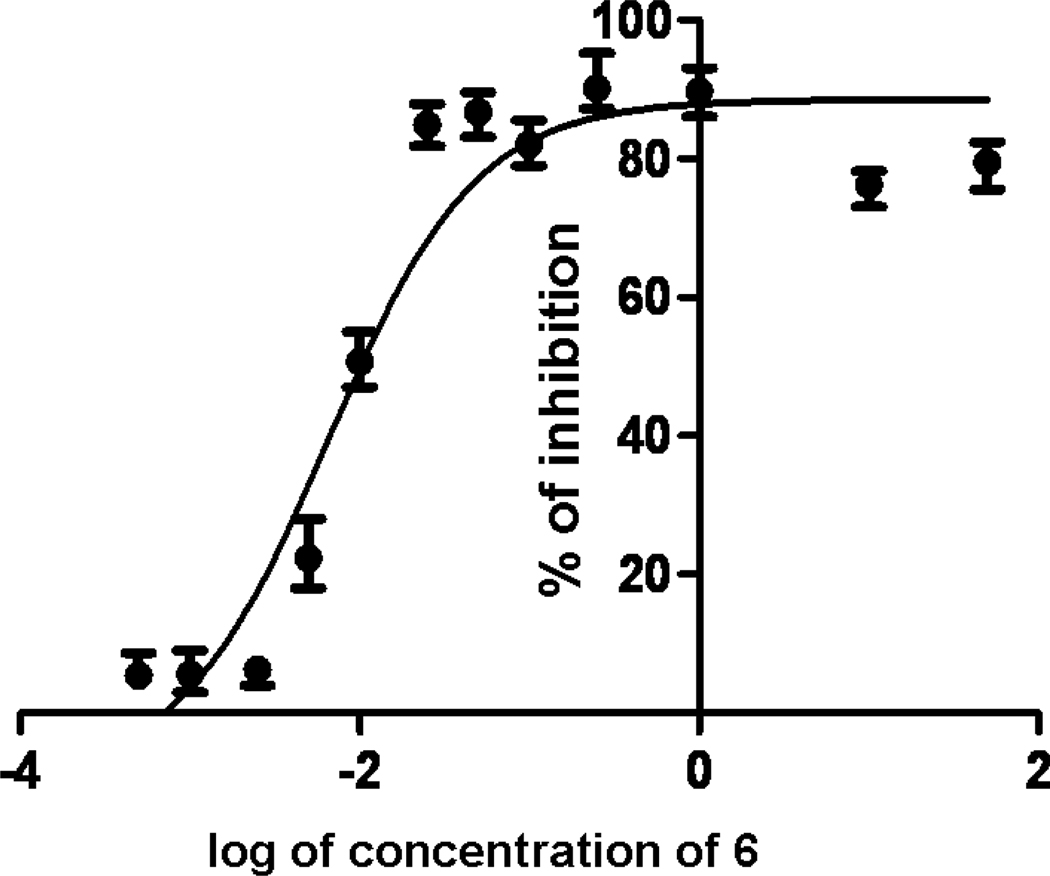
The ability of peptides to inhibit cell adhesion was evaluated using two cell adhesion assays, lypmphocyte-epithelial adhesion and E-rosetteing. Caco-2 cells are adherent cells that express CD58 protein and T cells are non-adherent cells that express CD2 protein. Caco-2 cells and T cells adhere to one another upon incubation. Thus, inhibition of cell adhesion interaction between T cells and Caco-2 cells (lypmphocyte-epithelial) can be used to evaluate the protein-protein interaction between CD2 and CD58. Similarly, interaction between sheep red blood cells (SRBC) and T cells can be used to evaluate the inhibitory activity of the peptides. SRBC express CD58 and Jurkat cells express CD2 protein. When these cells are incubated, they adhere to each other. Each Jurkat cell adheres to many sheep red-blood cells. Five or more SRBC adhering to a Jurkat cell is counted as positive E-rosetting 27. A representative dose-response curve for cell adhesion inhibition activity for peptide 6 is shown in Figure 2. Peptides 6 and 7 showed inhibition activity of nearly 80% in the concentration range of 0.1 µM or lower. IC50 values of inhibition of cell adhesion were calculated using graph-pad prism (GraphPad Software, La Jolla, CA) in a dose-response curve. The IC50 value of peptide 6 was 6.9 ± 0.4 nM while, for peptide 7 it was 11.1 ± 3.8 nM. Peptides 5, 8, 9, and 10 showed less than 20% cell adhesion inhibition in the lymphocyte epithelial assay. A control peptide exhibited nearly 18% inhibition activity. An antibody to CD58 adhesion domain inhibited cell adhesion nearly 100% at a concentration of ≤1 µM. Statistical analysis suggested that there is no difference in the activity of peptides 5, 8, 9, and 10 compared to the control peptide (p> 0.05) in the concentration range of 0.0005 to 150 µM. Since peptides 6 and 7 exhibited potential inhibitory activity of cell adhesion in lymphocyte epithelial assay, the inhibition activities of peptides 6 and 7 and control peptides were evaluated in E-rosetting assay. Inhibition activity of the peptide 6 in E-rosetting assay in the concentration range of 0.0005 to 50 µM is shown in Figure 3. For comparison inhibition activity of the control peptide is also shown in the figure. Peptides 6 and 7 showed inhibitory activity with IC50 values of 9.4 ± 0.3 nM and 25.7 ±1.5 nM respectively, in the E-rosetting assay.
Figure 2.
A representative dose-response curve for peptide 6 for inhibition of fluorescently labeled Jurkat cells and adherent Caco-2 cells. IC50 value was calculated based on four-parameter fit using GraphPad prism (see text for details).
Figure 3.
Inhibition of E-rosette formation by synthetic peptide 6 derived from CD2 protein. Peptides were added to AET-treated sheep red blood cells (SRBC) (expressing CD58 protein) first and Jurkat cells (expressing CD2 protein) labeled with BCECF was added later. Cells with five or more SRBC bound were counted as rosettes. Values are percent inhibition of peptide-treated cells and are expressed as the mean of three experimental values. Error bars represent standard error of the mean (SEM).
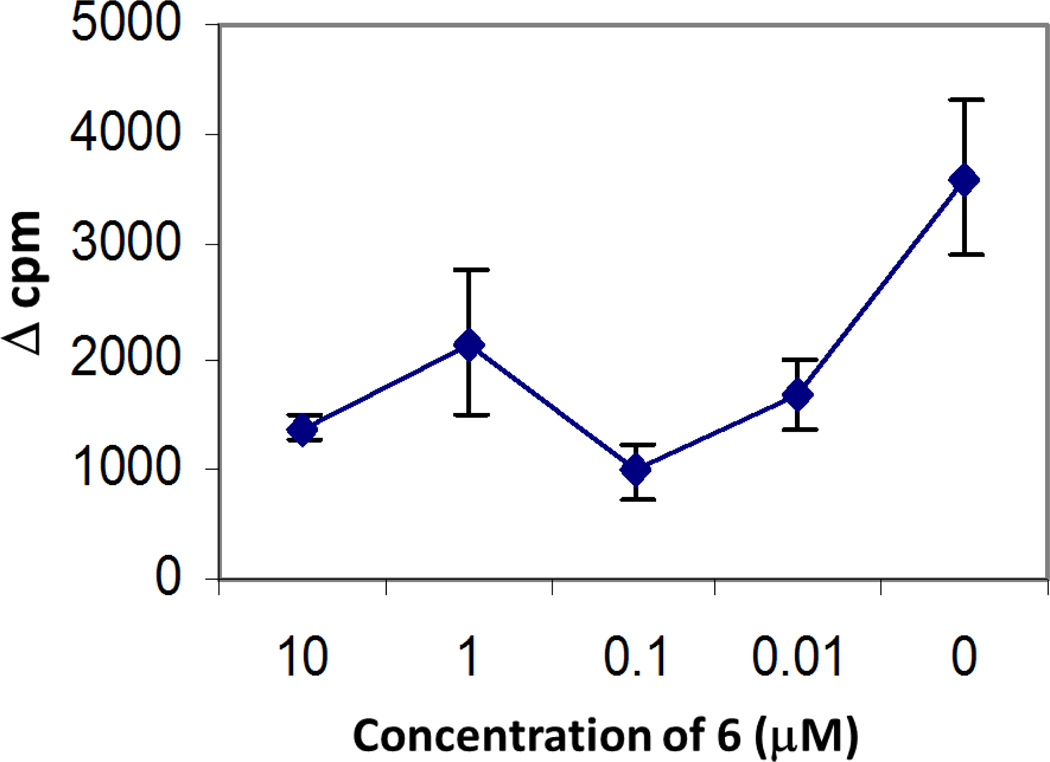
The ability of peptide 6 to suppress T cell response in vitro in response to a recall challenge with type collagen II (CII) was evaluated using lymph node cells (LNC) from CII-primed HLA-DQ8 transgenic mice. HLA-DQ8 mice develop cellular and humoral responses to CII that lead to development of arthritis following immunization with type II collagen. The arthritis-susceptible mouse model is relevant to humans28–30. The rationale for the experiment was that the peptides derived from CD2 should inhibit cell adhesion between T cells and antigen presenting cell, thus suppressing T cell response. Lymph node cells harvested from CII-primed mice were cultured in the presence or absence of CII and peptide 6. The T cell response monitored by tritium thymidine incorporation assay indicated that there was a dose-dependent suppression of T cell response to CII in vitro by peptide 6 (Figure 4).
Figure 4.
Dose-dependent suppression of T-cell response to type II collagen (CII) in vitro by peptide P6 in cells harvested from CII-primed from DQ8 mice, humanized transgenic mouse model of arthritis. In vitro T-cell response to CII was measured in the presence or absence of peptide 6 at four different concentrations. Change in counts per minute (cpm) was plotted to represent the T-cell proliferation. Delta cpm (Δcpm) was calculated by subtracting medium control from the test samples. The data show mean ±S.D. (N = 4).
CellTiter-Glo Assay for Peptide Cytotoxicity
The effect of the toxicity of peptides on Caco-2 cells and Jurkat cells was evaluated by a cell viability assay, CellTiter-Glo 31. This experiment provided confirmation that the observed biological activity (inhibition of adhesion) is not due to the cytotoxicity of peptides. Peptides were tested for their cytotoxicity for 2 h, which is the maximum exposure of peptides to cells during cell adhesion inhibition assays. Results indicated that Caco-2 cells and Jurkat cells showed more than 95% viability in the presence of peptides 6 and 7. Peptide 7 exhibited nearly 90% viability even at 200 µM concentration of peptide. For comparison, cells with 1% DMSO or <1% SDS and cells without peptide were incubated for 2 h were used. There was no statistical difference between the cells with peptides 6 and 7 (concentration range used: peptide 7, 200 to 0.025 µM; peptide 6, 200 to 0.01 µM) and cells without the peptide. Viability was calculated based on cells without peptides. The results clearly suggest that the peptides are not toxic to the cells studied.
Circular Dichroism
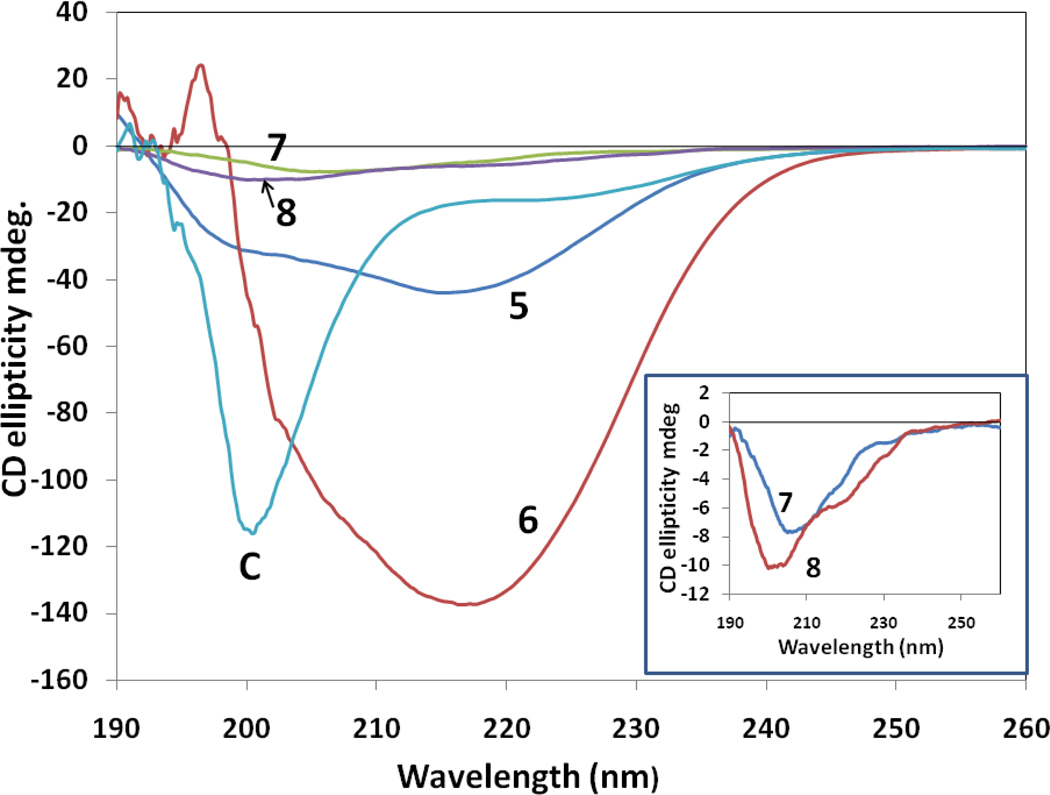
Circular dichroism spectra of peptides 5–8 in water are shown in Figure 5. Peptides 5 and 6 exhibited a negative CD band around 217 nm and a small positive band around 190 nm, suggesting a β-strand or β-hairpin type of structure of the peptide. Peptide 6 showed a very intense negative band around 217 nm, indicating a stable β-hairpin type of structure of the peptide in solution 32, 33. Peptide 8 showed a negative band around 205 nm and a negative shoulder around 217 nm, suggesting a possible β-hairpin type of structure of the peptide in solution34. Peptide 7 showed a negative band around 208 nm, indicating the possibility of a β-turn structure of the peptide in solution. A control peptide (labeled C) exhibited a negative band around 200 nm, which is suggestive of flexible or open structure of the peptide in solution34. The CD spectra of 9 and 10 exhibited a small negative band around 200 nm, indicative of flexible structure of peptides in solution. The spectrum of peptide 9 was noisy even after accumulation of 10 scans (Supporting Information), suggesting scattering of light due to possible aggregation of peptide in solution.
Figure 5.
Circular dichroism spectra of peptides 5–8 and control peptide. The legend for different peptides is shown in the figure. Peptide 6 exhibits stable secondary structure with a negative band around 217 nm indicative of β-strand structure, whereas the control peptide exhibits flexible structure as indicated by a negative CD band around 200 nm. Peptide concentration was 0.5 mM in water. Inset: Expanded regions of CD spectra of peptides 7 and 8 are shown.
NMR and structure calculation
To understand the structure-activity relationships of peptides 5–10, NMR, and NMR-restained molecular dynamics and energy-minimization studies were carried out. Peptides 5 and 6 were completely soluble in water at a concentration of 2.5 mg/mL (concentration at which NMR studies were carried out) and hence were studied in water. Peptides 7–10 with DBF moiety (Table 1) were not completely soluble in water at 2 mg/mL. Thus, they were studied in 100% DMSO-d6. To compare the results of NMR experiments for all the peptides presented in this study, peptides 5 and 6 were studied in 90% H2O/10% D2O as well as in 100% DMSO-d6. The NMR spectra of peptides in water and DMSO were not significantly different for peptides 5 and 6. The amide region of the 1D NMR spectra of the peptides showed dispersion of amide resonance chemical shifts for peptides 6 and 7 > 1.2 ppm 35. Two dimensional NMR spectral data of peptides 5–10 are available as Supporting Information. Based on the NMR data and NMR-restrained molecular dynamics simulations, the most probable structures of peptides 6–8 in solution are proposed. Three letter abbreviations are used for amino acids in peptides and single letter code for amino acid residues in protein in the text.
Peptide 6
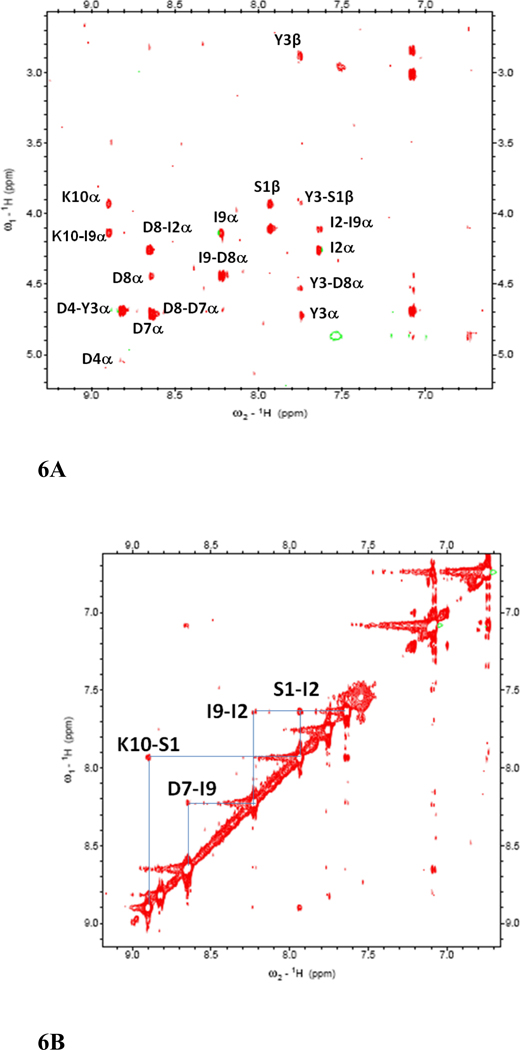
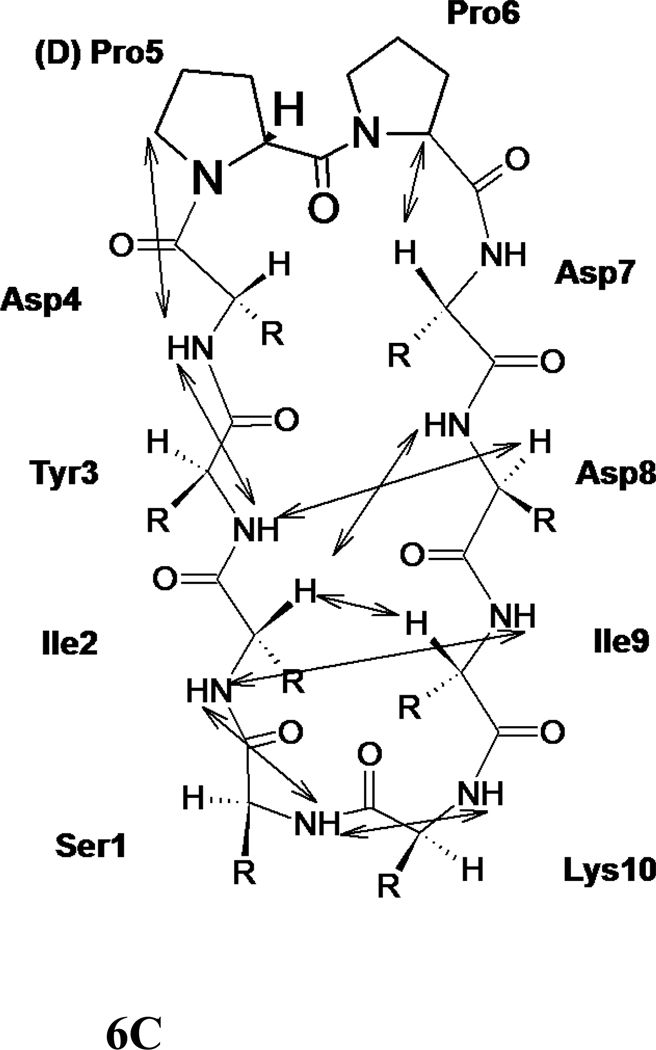
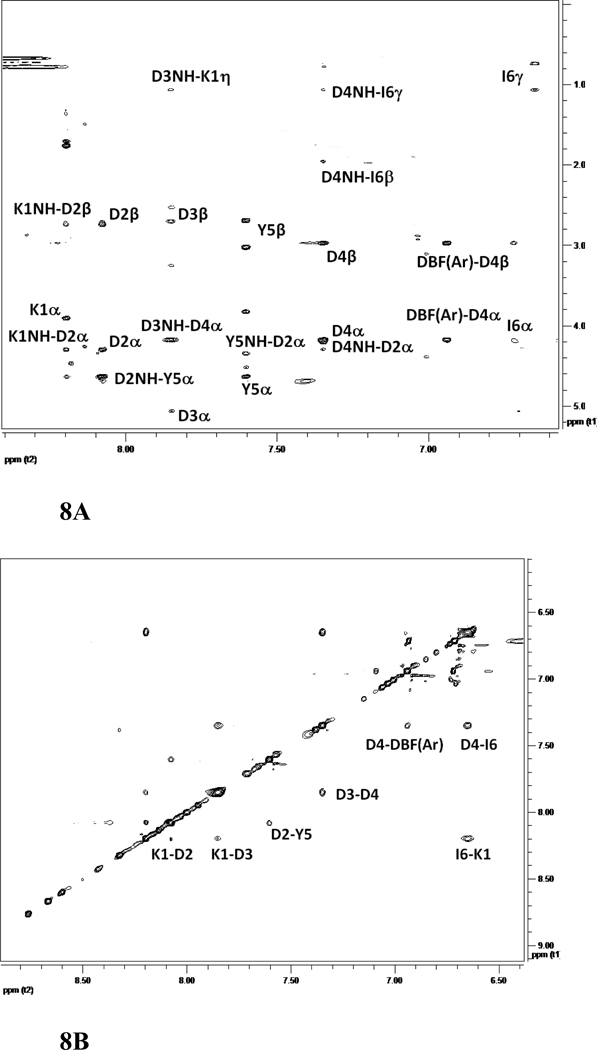
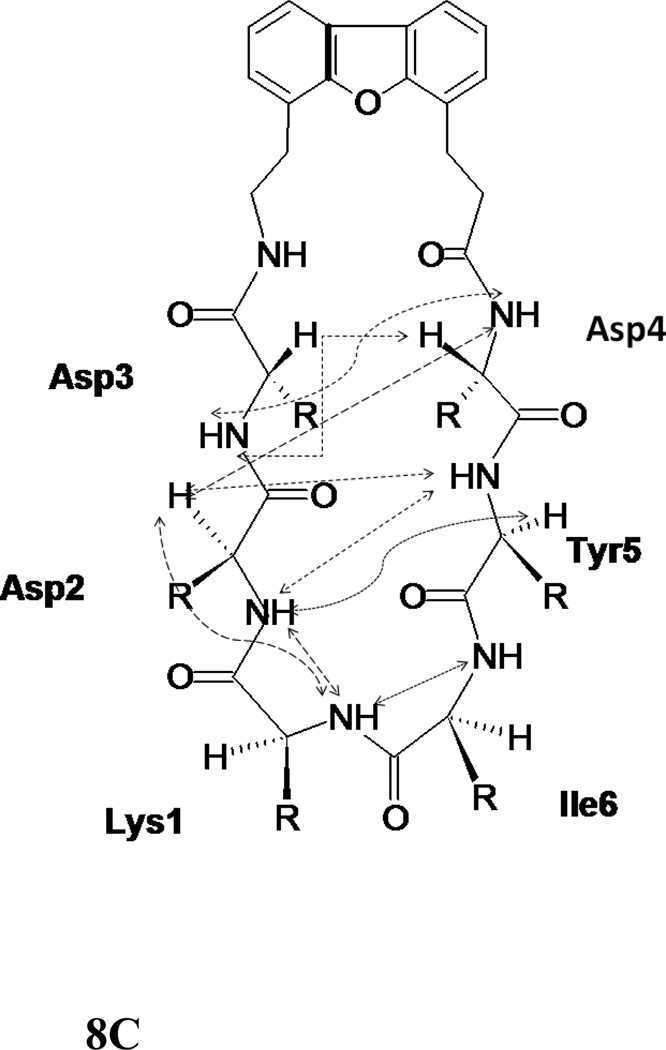
The NMR spectrum of peptide 6 was well resolved, and all the amino acids in the peptide could be identified with corresponding peaks in TOCSY 36, NOESY 37, ROESY 38, and DQF-COSY 39. The amide resonances of the peptide were spread over a range of 1.3 ppm, suggesting the well-defined structure of the peptide in solution 35. The NOESY and ROESY spectra of peptide 6 showed sequential connectivities between CαH (i)-NH (i+1) residues as well as long-range connectivities, which are indicative of a β-strand type of structure of the peptide 40. NOE connectivities in the NH-CαH and NH-NH region, a schematic diagram indicating the long-range NOE connectivities between NH-CαH and NH-NH are shown in Figure 6A–C. The characteristic NOEs of two strands of β-sheet, Ile2 NH-Ile9 NH and Tyr3 NH-Asp8 CαH, Ile2 CαH-Asp8 NH clearly indicate a β-sheet or β-hairpin structure of the peptide in solution. The NOE connectivity between Lys10 NH-Ser1 NH as well as Ser1 NH-Ile 2 NH is indicative of a possible β-turn around Lys 10 and Ser1. Chemical shift deviations (CSD) 41, 42 of CαH and NH resonances in peptides are used to measure the propensity of β-hairpin structure in solution. CSD of CαH of residues Ile2-Asp4, Pro6 and Asp7 were positive (Supporting information) suggesting the β-hairpin structure of the peptide 6. The coupling constant 3JHNα for Ile2-Asp4 and Asp7-Ile9 was in the range of 7.8–9.5 Hz. Such large coupling constants are indicative of β-strand structure of these residues in the peptide. Ser1 and Lys10 exhibit coupling constant of 6.06 and 5.90 respectively, suggested that these residues have conformational averaging or do not acquire stable β-stranded structure in solution. The temperature coefficients of the chemical shift of amide resonances of Ser1, Ile2, Asp4, Asp7, and Ile9 were > −4 ppb/K, suggesting that the amides of these residues may be intramolecularly hydrogen-bonded or solvent-shielded. Tyr3 showed a temperature coefficient of chemical shift value of −4.1 ppb/K suggesting a weak intramolecular hydrogen bonded amide or solvent shielded amide.
Figure 6.
500MHz 2D NMR data for peptide 6. A) NH-aliphatic region of NOESY spectrum of peptide in 90% H2O/10%D2O at 298 K showing NH-CαH connectivity. Sequential as well as long-range connectivities are shown. B) NH-NH region of NOESY spectrum of peptide 6. C) Schematic diagram showing the NOESY connectivity indicating the β-strand or β-hairpin structure of the peptide in solution. Observed NOEs are shown by double-ended arrows.
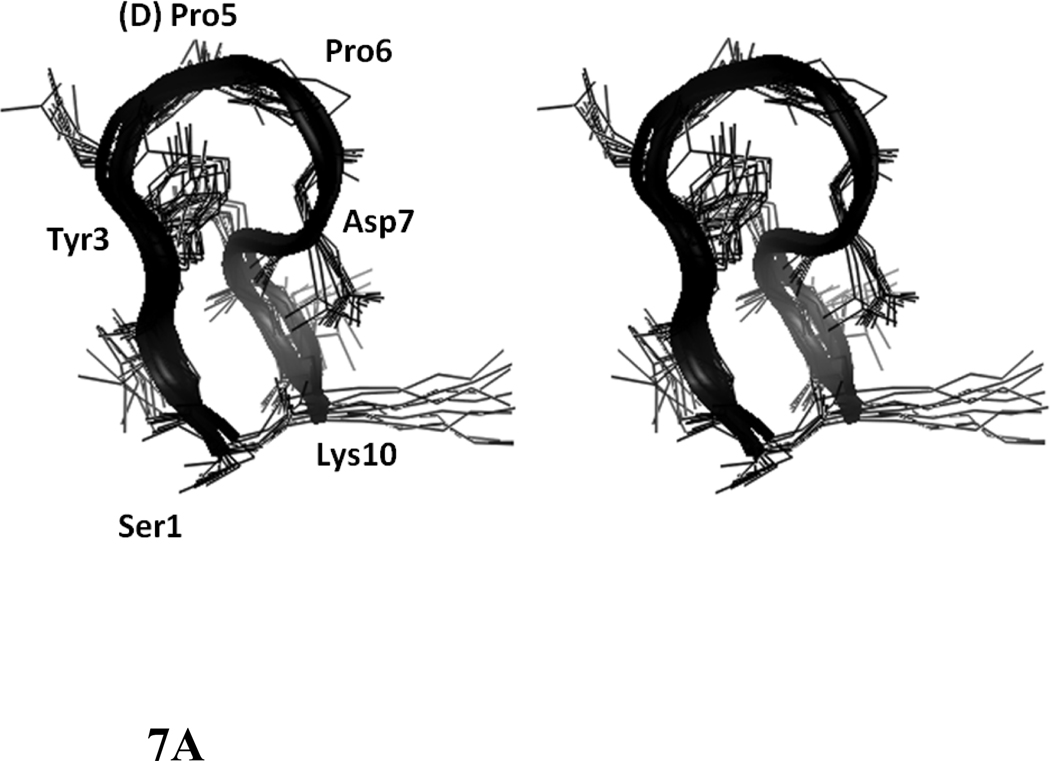
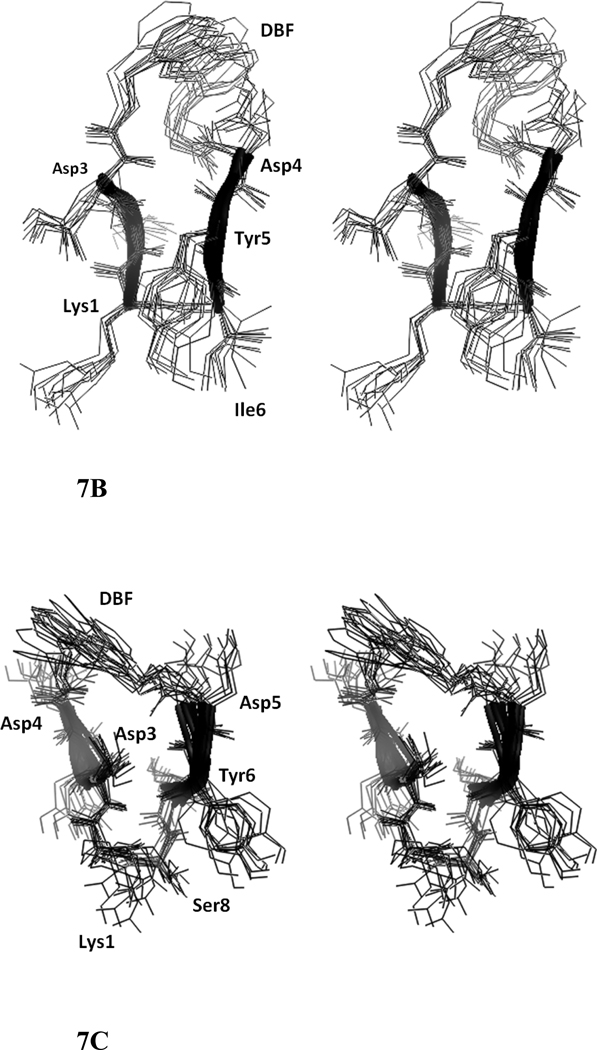
A total of 74 NOEs/ROEs were used to calculate the structure of peptide 6 using MD simulations and energy minimization. The ten low-energy structures obtained from NMR-restained MD simulations are shown in Figure 7A. The rms deviation of the backbone atoms of the structures compared to the average structure was 0.38 ± 0.1 Å. Analysis of Ramachandran plot suggested that 100 % of the residues were in the allowed region of the Φ, ψ plot. The structure exhibited a β-hairpin structure with a twist in the backbone around Asp8. Structural parameters for the low energy structures are shown in Table 2. (D)-Pro-Pro segment acquired a type II′ β-turn structure with (D)-Pro Φ= 88°, ψ= −109° and Pro Φ= −65°, ψ =−19°. The heterochiral (D)-Pro-Pro segment is known to exhibit type II′ structures. 24,40 The β-hairpin structure of the peptide was stabilized by two intramolecular hydrogen bonds between C=O of Tyr3 and NH of Asp7 and C=O of Asp7 and NH of Tyr3. A β-turn was observed around Ile9-Lys10-Ser1-Ile2. The Φ and ψ angles at Lys10 and Ser1 (Lys10 Φ = –69, ψ= 93; Ser1 Φ = 67, ψ = –19) indicated that there is a type I β-turn around these residues 43. The β-turn was stabilized by an intramolecular hydrogen bond between Ile9 C=O and Ile2 NH. Overall, the structure resembled a β-hairpin structure with D-Pro-Pro stabilizing and nucleating the hairpin structure 24.
Figure 7.
Stereo view of overlapped NMR-restrained energy-minimized structures of peptides 6, 7, and 8. Ten overlapped structures are shown. Residues are labeled with three letter amino acid codes. A) peptide 6, B) peptide 7, C) peptide 8.
Table 2.
Structural Parameters for the NMR restrained MD simulated, energy minimized final structure of peptide 6, 7 and 8.
| Parameter | Peptide 6 | Peptide 7 | Peptide 8 |
|---|---|---|---|
| Total number of NOE/ROEs and cyclization constraints used for calculations Number of intra residue NOE/ROEs Number of inter residue NOE/ROEs |
74 43 31 |
50 30 20 |
66 40 26 |
| Number of NOE Violations > 0.5 Å | 0 | 0 | 0 |
| Ramachandran plot | |||
| Number of residues in favored region | 88% | 100% | 100% |
| Number of residues in allowed region | 100% | 100% | 100% |
| Number of outliers | 0% | 0% | 0% |
| RMSD of backbone atoms (in Å) of 10 structures Compared to the average structure | 0.38 ± 0.10 | 0.63 ± 0.09 | 0.52 ± 0.10 |
Peptide 7
The amide region of NMR spectrum of peptide 7 was well resolved with amide resonances spanning a region of 1.5 ppm. Ile6 resonances were upfield shifted with amide resonance at 6.7 ppm. This may be due to the fact that the amide of Ile6 appears in the diamagnetic region of the aromatic ring of Tyr5 residue. There were few sequential connectivities in the NOESY spectrum of the peptide. However, the characteristic long-range connectivities indicated the β-hairpin structure of the peptide 7 in solution. Region of NOESY spectra with connectivities and a schematic diagram showing NOE connectivities are shown in Figures 8A–C. Some notable NOE connectivities are between Asp3 NH-Asp4 NH, Asp3 NH-Asp4 CαH, Asp2 CαH-Asp4 NH, Tyr5 CαH-Asp2 NH, and Tyr5 NH-Asp2 CαH, indicating the β-hairpin structure of the peptide. There were NOEs between Ile6 NH-Lys1 NH and Lys1 NH-Asp2 NH as well as Lys1 NH-Asp2 CαH, indicating a β-turn structure at Tyr5-Ile6-Lys1-Asp2. The DBF aromatic protons showed NOE connectivity with Asp4 NH and the DBF-aliphatic region, indicating the stable turn or hairpin nucleating structure of the DBF moiety. The coupling constant 3JHNα for Asp2, Asp4, Tyr5, and Ile6 was in the range of 7.2–8.3 Hz indicating the β-strand conformation of these residues. Asp3 had a coupling constant of 5.4 Hz indicating folded structure of the peptide near Asp3 residue. The temperature coefficient of chemical shift for Asp2, Asp3, Asp4, Tyr5, Ile6 were > −4 ppb/K, whereas for Lys1 it was < −4 ppb/K clearly suggesting the solvent shielding of Asp2, Asp3, Asp4, Tyr5 and Ile6. It should be noted that for peptide 7 NMR studies reported are in DMSO-d6 solvent. The data from chemical shift deviation had small positive value or near to zero for most of the amino acid residues in the peptide sequence. It is not clear whether DBF moiety in the peptide structure caused perturbation of chemical shift of residues.
Figure 8.
500MHz 2D NMR data for the peptide 7. A) NH-aliphatic region of NOESY spectrum of peptide in DMSO-d6 at 298 K showing NH-CαH connectivity. Sequential as well as long-range connectivities are shown. B) NH-NH region of NOESY spectrum of peptide 7. C) Schematic diagram showing the NOESY connectivity indicating the β-strand or β-hairpin structure of the peptide in solution. Observed NOEs are shown by dotted lines.
A total of 30 intra-residue and 20 inter-residue NOEs were used to calculate the structure of the peptide using NMR-restained MD simulation and energy minimization (Table 2). Ten low energy structures of peptide 7 are shown in Figure 7B. The rms deviation of the backbone of ten structures was 0.63 ± 0.09 Å. Analysis of Φ, ψ angles resulted in 100% of the amino acid residues in the peptide in allowed region of Ramachandran map. The structure of the peptide was stabilized by four intramolecular hydrogen bonds, Asp3 C=O to Tyr5 NH, Tyr5 C=O to Asp3 and Asp2 NHs, and the side chain of Asp4 to Asp 3 C=O. A β-turn was observed at Tyr5-Ile6-Lys1-Asp2, and the turn was stabilized by intramolecular hydrogen bonding between Tyr5 C=O and Asp2 NH. The Φ and ψ values (Ile6 Φ = −60, ψ = −40; Lys1 Φ = −70, ψ = 15) indicate that the turn is a type I β-turn. Overall, the structure exhibits a β-hairpin structure with a kink around Asp4.
Peptide 8
The amide resonances NMR spectra of peptide 8 were spread over a region of less than 1 ppm, suggesting the flexible conformation of the peptide. The NOESY/ROESY NMR spectra of the peptide showed sequential as well as amide-amide connectivity between adjacent residues. There were very few long-range connectivity NOEs in the NOESY spectrum. There was an NH-NH NOE between Ile2 and Tyr6, suggesting a possible β-strand type of structure (Supporting Information). There was an NOE between the β hydrogens of Ile2 and Ile 7. Apart from these two, there were no other NOEs showing strong evidence of a β-hairpin type of structure in the peptide. The coupling constant 3JHNα for Lys1, Asp3, Asp5, Ile7 Tyr5, was in the range of 6–7 Hz indicating the conformational averaging in the peptide structure in solution. Only Ile2, and Asp4 showed coupling constant around 7.7 Hz suggesting the extended structure of the peptide around these residues. Tyr6 and Ser8 coupling constants could not be measured accurately because of overlapping of resonances. Thus, results obtained from NMR studies of the peptide suggested that the peptide has a small population of β-hairpin type of structure in solution.
A total of 66 inter- and intra-residue NOEs were used to calculate the structure of the peptide (Table 2). Although the total number of NOEs are relatively large there were not many long range NOEs. The ten low-energy structures of the peptide are shown in Figure 7C. The peptide exhibited a possible β-turn at Ile7-Ser8-Lys1-Ile2 (Ser8 Φ = −120, ψ = −78; Lys1 Φ = −69, ψ = 3). However, the β-turn was not stabilized by an intramolecular hydrogen bond. The amino acid residues of the two strands (Lys1-Ile2-Asp3-Asp4 and Asp5-Tyr6-Ile7-Ser8) Asp4–Asp5, Asp3-Tyr6 were far apart in the structure, suggesting a flexible structure of the peptide. However, because of the rigid DBF moiety in the structure, the overall structure of the peptide appears to be a β-hairpin.
Peptides 5, 9, and 10
The 2D TOCSY NMR spectra of peptides 9 and 10 consisted of major and minor peaks. At least two sets of resonances could be identified for each amino acid in the peptide in the TOCSY spectrum. The NH-CαH region of NOESY and ROESY spectra consisted of several sets of cross-peaks, and assignments were ambiguous because of minor peaks in the spectra (Supporting Information). To confirm that the additional resonances in the NMR spectra were not from impurities, high-resolution mass spectrometry and HPLC were employed. All the peptides examined in the studies were ≥95% pure by HPLC, and high resolution mass spectrometry confirmed high intensity peaks corresponding to the molecular ion of the peptide for 9 and 10. Hence, it was concluded that peptides 9 and 10 exhibit multiple conformations in solution. Peptide 5 showed well-resolved TOCSY spectra in water as well as in DMSO-d6. While there were some minor resonances, the TOCSY spectrum could be assigned with major resonances. However, NOESY/ROESY spectra did not indicate sequential or long-range cross-peaks in the NH-NH region. The NH-CαH region showed few sequential cross-peaks and no long range cross-peaks. The total number of ROEs observed was less than 20. Hence, the structure of the peptide 5 was not elucidated. Moreover, peptides 5, 9, and 10 did not show any biological activity, suggesting that these peptides do not have a significant percentage of stable, biologically active secondary structure.
Discussion
Design of peptides
CD2–CD58 protein-protein interactions cover a surface area of 1,200 Å.2 Mutagenesis studies indicated that the residues in these F and C strands are important in binding of the CD2 to CD58 protein 25, 26. Point mutation studies suggested that, in human CD2 (hCD2), the Y86 to A86 mutation (F strand) resulted in a significant loss of binding of CD2 to CD58 (more than 1000-fold), while Y86 to F86 mutation in the same region did not affect the binding. In the crystal structure of the complex of CD2–CD58, there are 10 salt bridges and 5 hydrogen-bonding interactions between CD2 and CD58. Point mutation indicated that mutation of Y86 disrupted the interaction between the proteins significantly. Thus, the F and C strands of CD2 protein containing Y86 and aspartates are a good epitope for the design of peptides (Figure 1). We proposed that, in our peptide design from CD2 protein sequence, if we retain the C strand of CD2 with D31, D32, and K34 residues that are close to the hydrophobic region and the F strand with hot-spot Y86, the peptide will mimic the native structure of the protein. Based on the results mentioned above, and our previous studies 19–22, we designed a cyclized β-hairpin peptide assembling the two strands (residues 31–34 and 84–87) (Figure 1) for mimicking the CD2 interface with CD58. To mimic the β-strand structure of the F and C strands in the designed peptide, β-hairpin structure with β-turn was used. It is well known that β-turns nucleate β-hairpin structures and determine the structural stability of β-hairpin structures in model peptides 40. β-hairpins also form binding epitopes of protein-protein interactions 44–49. In our previous studies we have designed β-hairpin structures with a Pro-Gly sequence in the peptide. In the present study we wanted to introduce more rigid structures to nucleate the β-hairpin structure in the peptide designed from the F and C strands of CD2 protein. A (D)-Pro-Pro sequence or a dibenzofuran moiety 23 was inserted to connect the two strands between D31 and D87. The other end of the strand (K34-S84) was cyclized by main chain cyclization to acquire a stable peptide structure (Figure 1, Table 1). The use of (D)-Pro at the i+1 position of a β-turn facilitates the formation of type II' or I' turns. (D)-Pro-Pro sequences have been shown to be strong β-hairpin nucleators 24. The DBF moiety was also used to stabilize the β-hairpin structures. Thus, peptides with (D)-Pro-Pro and the DBF moiety were used to stabilize the peptide sequences designed from the F and C strands of CD2 protein. The designed peptides are shown in Table 1. Different peptides were designed based on the following criteria: a) Conformational constraints, b) orientation of F and C strand amino acids, c) Different lengths of F and C strands. Peptides 5 and 6 have (D)-Pro-Pro sequence. Peptide 5 is a truncated form of peptide 6. Peptides 7–10 are analogs of 5 and 6 and contain the DBF moiety. To design peptides 7–10, different orientations of the F and C strands were used. Peptides 7 and 8 contain anticlockwise orientation of F and C strands whereas 9 and 10 contain clockwise orientation of sequence. To design the control peptide, a 12-amino acid residue sequence was chosen from the hot-spot region of CD2 (containing Y86) 25, 26, and the sequence was reversed. Y86 and Y81 were replaced with Ala to generate the control peptide (Table 1).
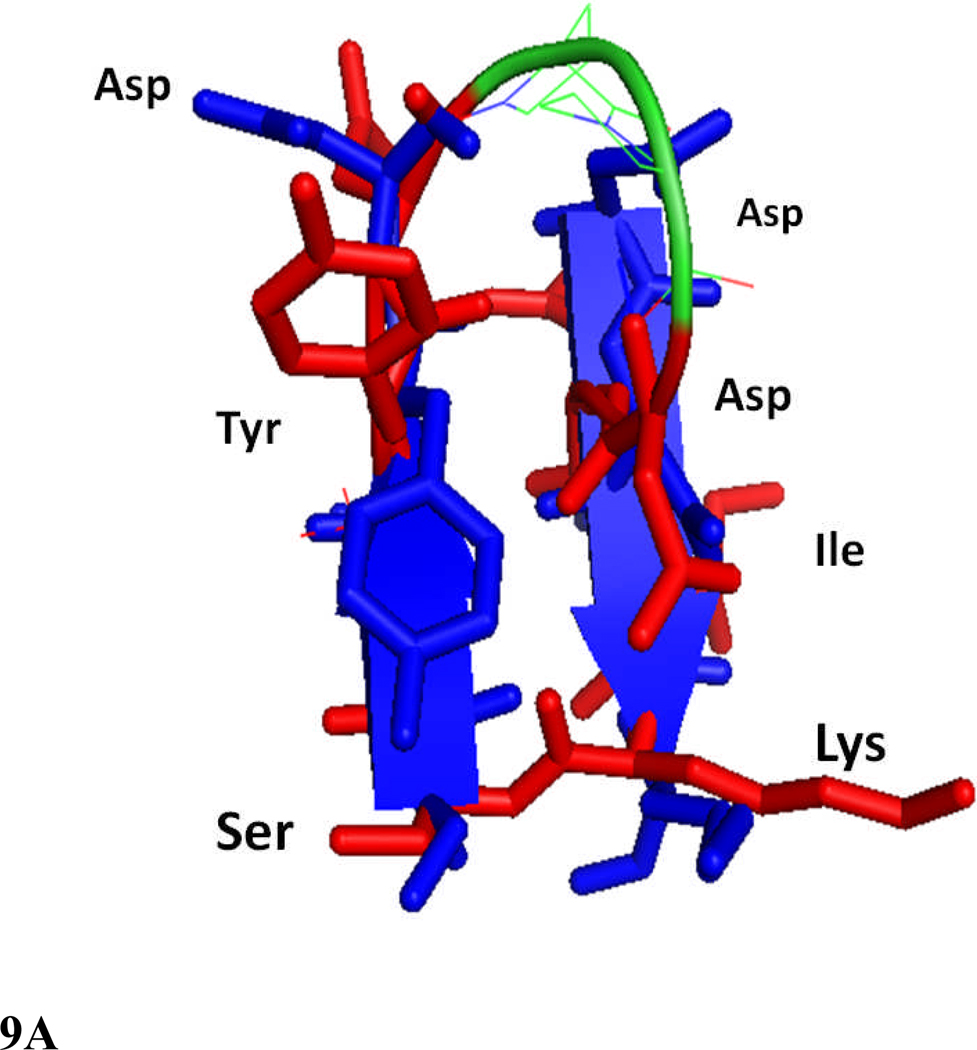
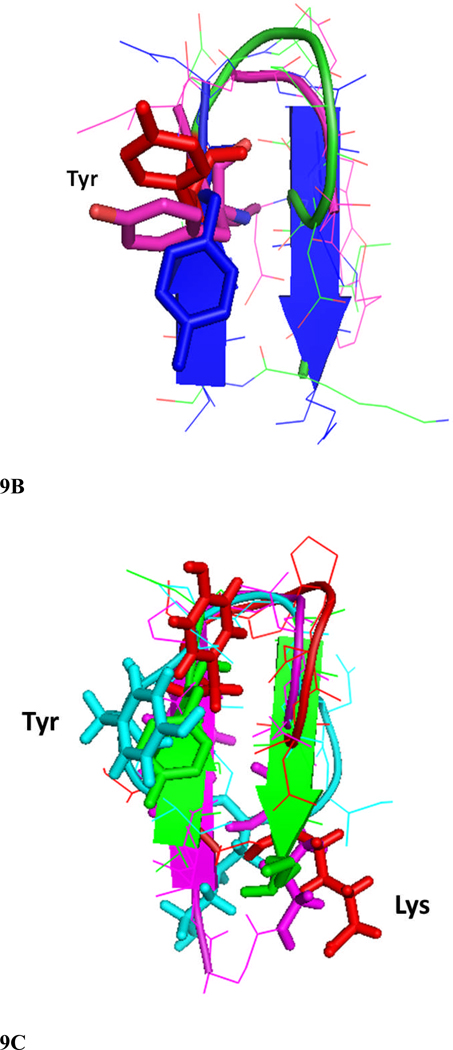
The peptides from the surface epitopes of CD2 protein (Tables 1) can block the adhesion interaction between T cells and epithelial cells, presumably by inhibiting CD2–CD58-mediated adhesion. We have used cellular assays to evaluate the activity of these peptides to block T-cell adhesion. Specifically, we have shown that these peptides could inhibit (a) adhesion between Jurkat cells and Caco-2 cells, and b) E-rosette formation between Jurkat cells and sheep red blood cells. 50 and c) suppress antigen-specific T cell response in LNC cells from transgenic mice susceptible to develop arthritis.28–30 All the peptides in the present studies are derived from the same region (F and C strands) of CD2 protein. However, the cell adhesion activities of the peptides varied over a wide range, depending on the number of amino acids in the peptide and the arrangement of β-strands used in the design. Peptides 6 and 7 showed inhibition of cell adhesion activity with IC50 values in the nanomolar range, whereas 5, 8, 9, and 10 showed less than 20% inhibition activity even at 150 µM concentration of the peptide. Furthermore, our observations with arthritis-susceptible humanized mice and peptide 6 suggested that peptide 6 could suppress antigen-specific T cell response in vitro in a recall response28, 29 (Figure 4). From these observations, one can speculate that peptide 6 derived from CD2 not only inhibits adhesion of T cells to CD58 expressing cells in a model system, but also has the ability to suppress CII-specific immune response. CD, NMR, and molecular dynamics simulations along with the biological activity of the peptides clearly indicated that stable secondary structure such as β-hairpin structure is necessary for the biological activity of this peptide among the peptides studied. CD studies clearly indicated a large negative band for peptide 6 with a negative maximum around 217 nm, which suggests a stable β-hairpin secondary structure of the peptide in solution (Figure 5). Peptide 7 (Figure 5 insert) exhibited a negative bands around 208 nm indicative of β-hairpin/β-turn structure of the peptide in solution34, 51, 52. NMR studies suggested that peptides 6 and 7 exhibited several NOE/ROE connectivities between the strands of the β-hairpin, indicating the stable secondary structure of the peptide in solution. The chemical shift index of CαH protons clearly indicated a positive chemical shift for more than 50% of residues in peptide 6(Supporting information), suggesting the β-hairpin structure of peptide 6. It is known that heterochiral (D)-Pro-Pro unit is an extremely efficient nucleator of β-hairpin structures and has high propensity to form type II′ β-turns in peptides and proteins. 24, 40 In peptide 6, (D)-Pro-Pro segment of the peptide exhibited a type II ′ β-turn. The peptides reported in this study have similar sequences. However, subtle changes in the sequence of amino acids in the peptide and clockwise or anti-clock wise direction of the conjugation of F and C strands or introduction of DBF moiety seem to have significant impact on the overall structure of the peptide and its biological activity. Peptides 5 and 9 have similar sequences except for (D) Pro-Pro and DBF moiety. Neither of these peptides showed stable major secondary structure in solution nor did they exhibit inhibition of adhesion activity. Peptides 6 and 10 were similar in sequence with (D) Pro-Pro and DBF moieties, respectively. Peptide 6 exhibited stable secondary structure in solution as seen in CD and NMR results. Peptide 10 exhibited flexible structure with the existence of major and minor conformers in solution. Peptides 5 and 7 had reversed sequences and were constrained by different strategies. Peptide 7 exhibited stable β-hairpin structure in solution and potent inhibition activity, whereas peptide 5 exhibited flexible structure with no inhibition activity. The CD spectrum of peptide 5 exhibited two negative bands around 220 and 208 nm, suggestive of a β-hairpin structure. However, NMR studies indicated that the number of NOE constraints was less than 20, and the NH-NH region of the NOESY spectrum of peptide 5 did not indicate any NOE cross-peaks. This suggests conformational interconversion or a less stable β-hairpin structure. Peptide 5 also showed resonances corresponding to the minor conformation in NMR spectra. It is possible that peptide 5 undergoes conformational interconversion via cis-trans isomerization of the X-Pro bond since the peptide contains two prolines. Peptides 8, 9, and 10 exhibited negative CD bands around 200 nm, which was suggestive of flexible structure. This was consistent with the biological activity of the peptides, where peptides 8, 9, and 10 did not show potent cell adhesion inhibition activity. Based on NMR and CD studies, molecular dynamics, and energy minimization, were carried out only on peptides 6, 7, 8 and the possible three-dimensional structures of these peptides were proposed. The proposed models were also consistent with observed NMR data such as low temperature coefficient of chemical shifts, relative large coupling constants (>7.5 Hz for peptide 6 and 7) and positive chemical shift deviations for CαH protons which is indicative of β-hairpin structure of the peptide. Peptides 6 and 7 exhibit β-hairpin structures stabilized by β-turns. However, the β-hairpin was twisted in one of the strands. A comparison of these β-hairpin structures of peptides 6 and 7 with the crystal structure of the F and C strands of CD2 protein is shown in Figures 9A and B. The overall structure of peptide 6 is similar to the F and C β-strand structure of CD2 in the crystal structure of the protein. Comparison of the backbone structure of the peptide with crystal structure of the protein by overlapping of backbone atoms indicated that there was a kink at Asp7 in the peptide structure and, hence, the backbone atoms of Asp6 and Asp7 did not overlap with the protein structure. However, amino acids Ser1-Asp4 and Asp8-Lys10 of peptide 6 overlapped with the F and C strands (Figure 9A). The rms deviation of overlap of Cα atoms between the F and C strands and peptide 6 was 0.5 Å for residues Ser1-Asp4 and Asp8-Lys10. The hot-spot residue Y86 of the CD2 protein overlapped with Tyr3 in the peptide 6 structure. Overlapped structures of peptides 6 and 7 with CD2 F and C strands are shown in Figure 9B. Peptide 7 has the DBF moiety and the sequence is anticlockwise compared to peptide 6. Hence, an rms deviation comparison of peptide 7 backbone atoms with F and C strands and peptide 6 was not calculated. Figure 9B indicates that the overall structure of peptide 7 is similar to the crystal structure of the CD2 F and C strands, and the important residue Tyr overlaps with Y86 of the CD2 crystal structure and Tyr 3 in peptide 6. The stable β-hairpin structures of peptides 6 and 7, along with the fact that their three-dimensional structures are comparable to the CD2 protein, suggests that these peptides mimic the CD58 binding structure of CD2 and presumably bind to CD58 protein on Caco-2 cells and sheep red blood cells. Furthermore, peptides 6 and 7 did not show any toxicity effects on Caco-2 cells and T cells.
Figure 9.
Comparison of structures of peptides 6 and 7 with the F and C strands of crystal structure of CD2 protein. A) Peptide 6 backbone overlapped with F and C strands of CD2 structure. CD2 is shown in blue, peptide 6 backbone is shown in green and residues are shown as red sticks. Notice the overlap of similar residues in the crystal structure and in peptide 6. B) Overlap of backbone atoms of CD2 F and C strands with peptides 6 and 7. F and C strands of CD2, blue; peptide 6, red; peptide 7, magenta. Notice that the tyrosine residue from the hot-spot region of CD2 protein overlaps with the Tyr residue in the β-hairpin peptides 6 and 7. C) Overlapping of peptide 6 (red) structure with F and C strands of CD2 protein (green), and peptides 2 (magenta) and 3 (cyan) from our previous report. Notice that Tyr of peptide 2 has a different orientation than peptide 6 and F and C strands in crystal structure of CD2, and the Lys side chain is cyclized in peptide 2 by side chain cyclization strategy. Peptide 3 has Lys and Tyr (cyan) in different orientation compared to peptide 6 and CD2 F and C strands. Sequence of peptide 2, Cyclo(1,11) H-E1S2I3Y4D5P6G7D8D9I10K11-OH (side chain cyclization); and 3, Cyclo(1,10) E1I2Y3D4P5G6D7D8I9K.10
In our previous studies we have reported the cell adhesion inhibition activity of peptides from CD2 F and C strands21. However, peptides 2, 3 and 4 had Pro-Gly sequence to induce the turn conformation instead of (D) Pro-Pro as seen in peptide 6. Peptides 2, 3, and 4 exhibited nearly 80% inhibition activity in the concentration range of 30–90 µM and their IC50 values were in the lower micromolar range (1–5 µM). Introduction of (D) Pro-Pro in the sequence (6) increased the potency of the peptide to inhibit cell adhesion by nearly 150 times. To investigate such a large change in IC50 value of peptide 6 compared to peptides 2 and 3, we overlaid the structures of 2 and 3 with 6 as well as C and F strands of CD2 protein (Figure 9C). The orientation of side chains of Tyr and Lys that are important in protein-protein interaction between CD2 and CD58 were compared in peptide structures. The orientation of side chain of Tyr defined in this study was based on number of NOE constraints and most probable conformation of χ1 of Tyr observed during 300K MD simulation. In peptide 3 (Figure 9C, cyan), the side chain orientation of Lys is different than the Lys orientation in peptide 6 (red) and the F and C strands of CD2 (green). The overall backbone is bulged in peptide 3 near the Tyr residue compared to peptide 6. In peptide 2 (Figure 9C, magenta), the Lys side chain is cyclized and thus orientation of the side chain is restricted and different compared to peptide 6 and F and C strands of CD2. Furthermore, the Tyr side chain in peptide 3 (magenta) is oriented on the opposite side of the peptide backbone compared to peptide 6. In peptide 6, Tyr and Lys side chain orientation is similar to that in the F and C strands of CD2 crystal structure. We believe these conformational features along with the stable, well-defined secondary structure of peptide 6 are the reason for the enhanced cell adhesion activity of peptide 6. Based on our cell adhesion inhibition studies and comparison of peptide structures, we can conclude that the orientation of side chains of important amino acids with respect to the backbone structures and the design of β-hairpin structure with proper conformational constraints such as (D) Pro-Pro sequence or DBF moiety to attain a stable secondary structure that can mimic the native protein structure (F and C strands of CD2 protein) are important for cell adhesion inhibition activity of the peptides.
Conclusions
Peptides were designed from the hot-spot β-strand region of CD2 protein that is important in binding to CD58. Conformational constraints were incorporated into the peptides by (D)-Pro-Pro or the dibenzofuran moiety to nucleate β-hairpin structure in the peptides. Cell adhesion inhibitory activities of the peptides were evaluated by two cell adhesion assays. Among the six peptides studied in this series, peptides 6 and 7 exhibited cell adhesion inhibition activity with IC50 values in the nanomolar range. The structures of the peptides studied by NMR and MD simulations suggested that peptides 6 and 7 have a stable β-hairpin structure. Peptide 6 also suppressed T cell response at nanomolar concentrations in cells derived from an arthritis-susceptible mouse model that is relevant to humans. Cell viability studies suggested that the designed peptides are not toxic to the cells studied and, hence, these peptides may form the lead compounds for designing potential cell adhesion inhibitors to inhibit immune response.
Experimental
Peptides
The control peptide (Table 1) was designed and synthesized in the laboratory using solid phase synthesis method 53,54; the cyclic peptides were purchased from Aroztech, LLC (Cincinnati, OH) or Polypeptide Laboratories (San Diego CA). The pure products were analyzed by HPLC, electrospray ionization mass spectrometry (ESI-MS), and high resolution mass spectrometry (HR-MS). HPLC chromatograms showed that the peptides were ≥ 95 % pure.
Cell Lines/cells
The human colon adenocarcinoma cell lines (Caco-2) and the T-leukemia Jurkat cell line were obtained from the American Type Culture Collection (Rockville, MD). Caco-2 cells were maintained in minimum essential medium-α containing 20% FBS, 1% nonessential amino acids, 1 mM Na-pyruvate, 1% L-glutamine, and 100 mg/L of penicillin/streptomycin. T cells were maintained in RPMI1640 (Gibco/BRL, Bethesda, MD) supplemented with 10% FBS, 2 mM L-glutamine, 100 mg/L of penicillin/streptomycin and 5 mg of bovine insulin in 500 mL medium. Sheep red blood cells (SRBC) were obtained from Colorado Serum Company (Denver, CO) and were prepared in Alsevers solution.
Lymphocyte-Epithelial Adhesion Assay
Caco-2 cells were plated onto 96-well plates at approximately 1 × 104 cells/well. When the cells reached confluency, the monolayers were washed once with MEM-α. Jurkat cells were labeled the same day as the adhesion assay by loading with 2 µM fluorescent dye BCECF-AM at 37 °C for 1 h. Stock solution of peptides were prepared by dissolving 2 mg of peptide in DMSO (in final solution DMSO was less than 1%). Dilution of the peptide solution was carried out by adding appropriate volume of the medium. Peptide diluted in MEM-α was added at various concentrations (0.0005 to 150 µM) to Caco-2 cell monolayers. After incubation at 37 °C for 30 min, the labeled Jurkat cells (1 × 106 cells/well) were added onto the monolayers. After incubation at 37°C for 45 min, non-adherent Jurkat cells were removed by washing three times with PBS, and the monolayer-associated Jurkat cells were lysed by incubating with 2% Triton X-100 in 0.2 M NaOH for about 10 minutes. Soluble lysates were read with a microplate fluorescence analyzer with emission wavelength 528 (±20) nm. Data are presented as relative fluorescence or percent inhibition as described previously 50. Relative fluorescence (FL) was found by reading the values of fluorescence intensity corrected for the reading of background (cell monolayers only). Cells with DMSO and only medium were used as controls.
Inhibition activity of the peptides was calculated by comparing it to cells without peptide. GraphPad software was used to plot a dose response curve and IC50 values were calculated using four parameter-fit equation of log of inhibitor concentration vs response assuming standard slope equal to Hill slope. IC50 value represented was an average of IC50 value calculated from three data sets. Statistical analysis of results from the cell adhesion assay was carried out using Microsoft Excel. Peptides were grouped into 6, 7, and 5, 8–10 and compared with control peptide and within each group. P values were compared for analysis.
Sheep red blood cells (SRBC) were pelleted by centrifuging about 15 mL of sheep blood in Alsevers solution at 200g for 7 min. The pellet was washed three times with phosphate buffered saline (PBS), removing supernatant and buffer coat on each wash. SRBC were then incubated with four volumes of 2-(2-aminoethyl)isothiourea dihydrobromide (AET) solution at 37°C for 15 min. The cells were washed three times with PBS and re-suspended in RPMI 1640 containing 10% FBS to give a 10% suspension. For use, this cell suspension was diluted twentyfold with RPMI 1640 (10% FBS). Peptide stock solutions were prepared by dissolving 2 mg of peptide in DMSO. Serial dilutions of peptides in PBS (0.0005 to 150 µM) were added to 0.2 mL of 0.5% (w/v) AET-treated SRBC and incubated at 37 C for 30 min. Then, 0.2 mL of Jurkat cell suspension (2 × 106 cells/mL) was added to the mixture and incubated for another 15 min. The cells were centrifuged (200g, 5 min, 4 C) and incubated at 4°C for 1 h, after which the cell pellet was gently re-suspended and the E-rosettes counted using a hemacytometer. Cells with five or more SRBC bound were counted as E-rosettes. At least 200 cells were counted to determine the percentage of E-rosette cells. The inhibitory activity was calculated by using the equation described previously 50.
In vitro T cell proliferation
Transgenic mice expressing functional HLA-DQ8 (DQA1*0301/DQB1*0302)) molecules have been characterized and described before 55. Mice used in this study were bred and maintained in the pathogen-free Immunogenetics Mouse Colony of Mayo Clinic. Pure native chick type II collagen (CII) was obtained by multiple step purification described previously 56. HLA-DQ8 Mice were immunized with 200 µg of CII emulsified 1:1 with CFA (Difco, Lawrence KS) by injecting intradermally at the base of the tail. Ten days postimmunization, draining popliteal, caudal, and lumbar lymph nodes were removed and prepared for in vitro culture. LNCs (1 × 106) were challenged in vitro with or without native collagen. Inhibition experiments were done by adding peptide 6 at four different concentrations (10–.01 µM) to the cells challenged in vitro with CII at 50 µg/mL. During the last 18 hours the cells were pulsed with 3H-thymidine (1 µCi/well). At the end of the assay, the cells were harvested using a plate harvester and incorporated radioactivity was determined using an automated counter (Microbeta, Perkin Elmer, Waltham, MA) for T-cell proliferation.
Cell Viability Assay
Peptides that exhibited potent cell adhesion inhibition activity on Jurkat-Caco-2 adherence were tested by CellTiter-Glo assay (Promega Corporation, Madison, WI) 31 to determine if their effects were due to toxicity. The cell viability of Caco-2 cells and Jurkat cells were determined in the presence of peptides. Stock solutions of peptides 6 and 7 were prepared in DMSO and diluted with serum-free medium to obtain the desired concentration. Different concentrations (200–0.1 µM) of peptide were added to Caco-2 cells and incubated for 2 and 3 hrs (which is more than the maximum time of exposure of Caco-2/Jurkat cells during the adherence assay). After the compounds were incubated with cells, the wells were washed with PBS, and 100 µL of CellTiter-Glo reagent was added to cells containing 100 µL medium. The plates were equilibrated for 10 minutes, and luminescence from the cells with and without the compounds was read on a Biotek microplate reader using blank, cells with 0.1% SDS, and 1% DMSO. The luminescence values obtained at each concentration (triplicates for each run and two independent runs were carried out) were averaged and adjusted by subtraction of blank values (wells without cells). Similar studies were carried out on Jurkat cell suspensions (Jurkat cells are non adherent cells and hence studies were performed in appendoff tubes and for washing steps, centrifugation was used. In the final step of the assay microplate was used). Cell viabilities were calculated.
Circular Dichroism Spectroscopy
Samples for circular dichroism (CD) were prepared by dissolving 0.5 mg of peptide in 1 mL of deionized water. CD spectra were collected in a JASCO J-815 spectrophotometer flushed with nitrogen and equipped with temperature control. All the CD spectra were collected at 298 K. CD spectra of the samples were collected from 320–190 nm using 1 mm path length of rectangular quartz cell. Each spectrum was the average of four scans (in the case of peptide 9, because of the nature of the spectrum, 10 scans were accumulated) taken at a scan rate of 50 nm/min with a spectral bandwidth of 1 nm. Spectra were obtained at different concentration of the peptide and spectrum of the peptide at 0.5 mM was used for representation. For the final representation, the baseline was subtracted from the spectrum.
NMR Spectroscopy
NMR studies of the peptides were carried out in two solvents, water and dimethylsulfoxide because of the limited solubility of peptides 7, 8, and 9 in water. The samples of peptides in water were prepared by dissolving 2 mg of the peptide in 0.7 mL of 90% H2O/10% D2O. Samples in DMSO were prepared by dissolving 2 mg of the peptide in deuterated DMSO (DMSO-d6). NMR spectra were collected using a 500 MHz Varian NMR spectrometer equipped with a variable temperature probe. Spectra in water were acquired from 288 K to 308 K unless otherwise specified. 2D NMR data of the peptides were collected at temperature at which 1D NMR spectra exhibited well-resolved amide resonances. TOCSY 36, NOESY 37, DQF-COSY,39 and rotating frame nuclear Overhauser spectroscopy (ROESY) 38 experiments were performed by WATERGATE pulse sequence. NMR spectra were processed using NMRPipe 57 and analyzed by SPARKY 58 software. Assignment of the spin system was carried out using the TOCSY and DQF-COSY spectra for each peptide. Cross-peaks from NOE/ROE were classified as strong, medium and weak based on intensities and were converted into distances (NH-CαH (i,i), 1.9–3.0 Å; NH-CαH (i, i+1), 2.0–3.6 Å; all other 2.0–5.0 Å). One-dimensional temperature-dependent NMR experiments were carried out from 293 to 308 K for DMSO-d6 samples.
Structure Calculations from NMR Data using Molecular Dynamics and Energy Minimization
Structure calculations of the peptides were performed on a Linux platform computer using InsightII software (Accelrys Inc. San Diego, CA). Linear structures of the peptides were built using InsightII, and NMR restraints were applied. For cyclizing the peptide, N-to-C terminal constraint was used (1.4 Å for peptide bond). A 300 K MD simulation 59, 60 with NMR constraints was performed to cyclize the peptide structure. The resulting structure was energy minimized using the steepest-descent method for 100 steps and was subjected to a simulated annealing procedure. MD simulations of the peptide were performed at 900 K for 10 ps, and several structures from the history file were randomly selected. These structures were subjected to MD simulations for 5 ps, and the temperature of the system was reduced by 100 K steps from 900 K. After the simulations reached 300 K, the peptide structure was soaked with a 10 Å layer of water molecules and subjected to 20 ps MD simulations at 300 K with NMR restraints. From the history file of 300 K MD simulations, a plot of energy vs. time was obtained and at least 10–12 low energy structures of the peptides were chosen for final minimization. An average structure was also chosen from the 300 K dynamics. For each peptide a total of 80–100 structures were chosen from simulated annealing procedure. These 80–100 structures were energy-minimized using the steepest descent and conjugate gradient methods until the rms derivative was 0.5 kcal/mol. The structures were verified for NMR-derived distances and 15–20 of the structures that were consistent with NOE/ROE data were chosen for the final representation. Structures were also validated with Molprobity for bond length and bond angle deviations as well as for Ramachandran plot analysis 61, 62. RMSD of the backbone atoms were calculated by superimposing 10 low energy structures with an average structure in that family.
Supplementary Material
Acknowledgements
This research was supported by Louisiana Board of Regents grant LEQSF(2009-12)-RD-A-23(SJ). The CD spectrometer used in the study was purchased from Louisiana Board of Regents grant LEQSF(2009-10)-ENH-TR-75 (SJ). The authors would like to thank the Mass Spectrometry Facility, Department of Chemistry, Louisiana State University, Baton Rouge for high resolution mass spectra of compounds. VT is supported by funds from NIH, AI60752 and AR60077.
Abbreviations
- AET
2-aminoethylisothiouronium hydrobromide
- APC
antigen presenting cell
- BCECF-AM
Bis-carboxyethyl-carboxyfluorescein acetoxymethyl
- BSA
bovine serum albumin
- Caco-2 cell line
colon adenocarcinoma cell line
- CFA
complete freund’s adjuvant
- CII
collagen type II
- CSD
chemical shift deviations
- DBF
dibenzofuran
- DSS
4,4-dimethyl-4-silapentane-1-sulfonic acid
- ESI-MS
electrospray ionization mass spectrometry
- LNC
Lymph node cells
- MD
Molecular Dynamics
- NK cells
natural killer cells
- pMHC
peptide-major histocompatibility complex
- Ps
pico-second
- PPI
protein-protein interactions
- rmsd
root mean square deviation
- SDS
sodium dodecyl sulfate
- SRBC
sheep red-blood cells
- TCR
T cell receptor
Three letter abbreviations are used for amino acids in peptides and single letter code for amino acid residues in protein in the text.
Footnotes
Supporting Information Available: 1H NMR, HPLC, MS, and purity data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Reichmann D, Rahat O, Cohen M, Neuvirth H, Schreiber G. The molecular architecture of protein-protein binding sites. Curr. Opin. Struct. Biol. 2007;17:67–76. doi: 10.1016/j.sbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Ryan DP, Mathews JM. Protein-protein interactions in human disease. Curr. Opin. Struct. Biol. 2005;15:441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 5.Moreira IS, Fernandes PA, Ramos MJ. Hot spots- a review of the protein-protein interface determinant amino acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 6.Chene P. Drugs targeting protein-protein interactions. ChemMedChem. 2006;1:400–411. doi: 10.1002/cmdc.200600004. [DOI] [PubMed] [Google Scholar]
- 7.Van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 8.Jois SD, Liu J, Nagarajarao LM. Targeting T-cell adhesion molecules for drug design. Curr. Pharm. Design. 2006;12:2797–2812. doi: 10.2174/138161206777947696. [DOI] [PubMed] [Google Scholar]
- 9.Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TL, van der Merwe PA. The nature of molecular recognition by T-cells. Nature Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- 10.Dustin ML, Springer TA. T cell receptor cross linking transiently stimulates adhesiveness through LFA- 1. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 11.Wingren AG, Parra E, Varga M, Kalland T, Sjogren HO, Hedlund G, Dohlsten M. T cell activation pathways: B7, LFA-3, and ICAM-1 shape unique T cell profiles. Crit. Rev. Immunol. 1995;15:235–253. doi: 10.1615/critrevimmunol.v15.i3-4.30. [DOI] [PubMed] [Google Scholar]
- 12.da Silva AJ, Brickelmaier M, Majeau GR, Li Z, Su L, Hsu YM, Hochman PS. Alefacept, an immunomodulatory recombinant LFA-3/IgG1 fusion protein, induces CD16 signaling and CD2/CD16-dependent apoptosis of CD2+ cells. J. Immunol. 2002;168:4462–4471. doi: 10.4049/jimmunol.168.9.4462. [DOI] [PubMed] [Google Scholar]
- 13.Mrowietz U. Treatment targeted to cell surface epitopes. Clin. Exp. Derm. 2002;27:591–596. doi: 10.1046/j.1365-2230.2002.01171.x. [DOI] [PubMed] [Google Scholar]
- 14.Braun J, Sieper J. Role of novel biological therapies in psoriatic arthritis: effects on joints and skin. Biodrugs. 2003;17:187–199. doi: 10.2165/00063030-200317030-00005. [DOI] [PubMed] [Google Scholar]
- 15.Spitzer TR, McAfee SL, Dey BR, Colby C, Hope J, Grossberg HTR, Preffer F, Shaffer J, Alexander SI, Sachs DH, Sykes M. Nonmyeloablative haploidentical stem-cell transplantation using anti-CD2 monoclonal antibody (MEDI-507)-based conditioning for refractory hematologic malignancies. Transplantation. 2003;75:1748–1751. doi: 10.1097/01.TP.0000064211.23536.AD. [DOI] [PubMed] [Google Scholar]
- 16.Weger W. Current status and new developments in the treatment of psoriasis and psoriatic arthritis with biological agents. Br. J. Pharmacol. 2010;160:810–820. doi: 10.1111/j.1476-5381.2010.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 18.Hruby VJ. Designing peptide receptor agonists and antagonists. Nat. Rev. Drug. Discov. 2002;1:847–856. doi: 10.1038/nrd939. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Ying J, Chow VT, Hruby VJ, Satyanarayanajois SD. Structure-activity studies of peptides from the "hot spot" region of human CD2 protein: development of peptides for immunomodulation. J. Med. Chem. 2005;48:6236–6249. doi: 10.1021/jm0503547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Li C, Ke S, Satyanarayanajois SD. Structure-based rational design of beta-hairpin peptides from discontinuous epitopes of cluster of differentiation 2 (CD2) protein to modulate cell adhesion interaction. J. Med. Chem. 2007;50:4038–4047. doi: 10.1021/jm0700868. [DOI] [PubMed] [Google Scholar]
- 21.Giddu S, Subramanian V, Yoon HS, Satyanarayanajois SD. Design of beta-hairpin peptides for modulation of cell adhesion by beta-turn constraint. J. Med. Chem. 2009;52:726–736. doi: 10.1021/jm8008212. [DOI] [PubMed] [Google Scholar]
- 22.Satyanarayanajois SD, Büyüktimkin B, Gokhale A, Ronald S, Siahaan TJ, Latendresse JR. A peptide from the beta-strand region of CD2 protein that inhibits cell adhesion and suppresses arthritis in a mouse model. Chem. Biol. Drug Des. 2010;76:234–244. doi: 10.1111/j.1747-0285.2010.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayo KH, Dings RP, Flader C, Nesmelova I, Hargittai B, van der Schaft DW, van Eijk LI, Walek D, Haseman J, Hoye TR, Griffioen AW. Design of a partial peptide mimetic of anginex with antiangiogenic and anticancer activity. J. Biol. Chem. 2003;278:45746–45752. doi: 10.1074/jbc.M308608200. [DOI] [PubMed] [Google Scholar]
- 24.Saha I, Chatterjee B, Shamala N, Balaram P. Crystal structures of peptide enantiomers and racemates: probing conformational diversity in heterochiral Pro-Pro sequences. Biopolymers. 2008;90:537–543. doi: 10.1002/bip.20982. [DOI] [PubMed] [Google Scholar]
- 25.Kim M, Sun ZY, Byron O, Campbell G, Wagner G, Wang J, Reinherz EL. Molecular dissection of the CD2-C58 counter-receptor interface identifies CD2 Tyr86 and CD58 Lys34 residues as the functional "hot spot.". J. Mol. Biol. 2001;312:711–720. doi: 10.1006/jmbi.2001.4980. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Smolyar A, Tan K, Liu J, Kim M, Sun ZJ, Wagner G, Reinherz EL. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell. 1999;97:791–803. doi: 10.1016/s0092-8674(00)80790-4. [DOI] [PubMed] [Google Scholar]
- 27.Albert-Wolf M, Meuer SC, Wallich R. Dual function of recombinant human CD58: inhibition of T-cell adhesion and activation via the CD2 pathway. Int. Immunol. 1991;3:1335–1347. doi: 10.1093/intimm/3.12.1335. [DOI] [PubMed] [Google Scholar]
- 28.Taneja V, Krco CJ, Behrens MD, Luthra HS, Griffiths MM, David CS. B -cells are important as antigen presenting cells for induction of MHC-restricted arthritis in transgenic mice. Mol. Immunol. 2007;44:2988–9296. doi: 10.1016/j.molimm.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taneja V, Taneja N, Behrens M, Griffiths MM, Luthra HS, David CS. Requirement for CD28 may not be absolute for collagen-induced arthritis: study with HLA-DQ8 transgenic mice. J. Immunol. 2005;174:1118–1125. doi: 10.4049/jimmunol.174.2.1118. [DOI] [PubMed] [Google Scholar]
- 30.Knutson KL, dela Rosa C, Disis ML. Laboratory analysis of T-cell immunity. Front Biosci. 2006;11:1932–1944. doi: 10.2741/1936. [DOI] [PubMed] [Google Scholar]
- 31.Riss T, Moravec R, Niles A. Selecting cell-based assays for drug discovery screening. Cell Notes. 2005;13:16–21. [Google Scholar]
- 32.Dyer RB, Maness SJ, Franzen S, Fesinmeyer RM, Olsen KA, Andersen NH. Hairpin folding dynamics: the cold–denatured state is predisposed for rapid refolding. Biochemistry. 2005;44:10406–10415. doi: 10.1021/bi050698z. [DOI] [PubMed] [Google Scholar]
- 33.Anderson NH, Olsen KA, Fesinmeyer M, Tan X, Hudson MF, Eidenschink LA, Farazi SR. Minimization and optimization of designed β-hairpin folds. J. Am. Chem. Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perczel A, Hollosi M. Turns. In: Fasman GD, editor. Circular dichroism and the conformational analysis of biomolecules. New York: Plenum Press; 1996. pp. 285–380. [Google Scholar]
- 35.Wuthrich K. NMR of proteins and nucleic acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 36.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 37.Kumar A, Wagner G, Ernst RR, Wuthrich K. Buildup rates of the nuclear overhauser effect measured by two-dimensional proton magnetic resonance spectroscopy: implications for studies of protein conformation. J. Am. Chem. Soc. 1981;103:3654–3658. [Google Scholar]
- 38.Bax A, Davis DG. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 1985;63:207–213. [Google Scholar]
- 39.Rance M, Sorensen OW, Bodenhausen G, Wagner G, Ernst RR, Wüthrich K. Improved spectral resolution in COSY 1H NMR spectra of protein via double quantum filtering. Biochem. Biophys. Res. Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 40.Rai R, Aravinda S, Kanagarajadurai K, Ragothama S, Shamala N, Balaram P. Diproline templates as folding nuclei in designed peptides. Conformational analysis of synthetic peptide helices containing amino terminal Pro-Pro segments. J. Am. Chem. Soc. 2006;128:7916–7928. doi: 10.1021/ja060674v. [DOI] [PubMed] [Google Scholar]
- 41.Fesinmeyer RM, Hudson FM, Olsen KA, White GW, Euser A, Andersen NH. Chemical shifts provide fold populations and register of beta hairpins and beta sheets. J. Biomol. NMR. 2005;33:213–231. doi: 10.1007/s10858-005-3731-7. [DOI] [PubMed] [Google Scholar]
- 42.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 43.Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv. Protein. Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- 44.Simpson ER, Meldrum JK, Bofill R, Crespo MD, Holmes E, Searle MS. Engineering enhanced protein stability through b-turn optimization: Insights for the design of stable peptide β-hairpin systems. Angew. Chem. Int. Ed. 2005;44:2–7. doi: 10.1002/anie.200500577. [DOI] [PubMed] [Google Scholar]
- 45.Schumachaer MA, Hurlburt BK, Brennan RG. Crystal structures of SarA, a pleiotropic regulator of virulence genes in S. aureus. Nature. 2001;409:215–219. doi: 10.1038/35051623. [DOI] [PubMed] [Google Scholar]
- 46.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 47.Stano NM, Patel SS. The intercalating beta-hairpin of T7 RNA polymerase plays a role in promoter DNA melting and in stabilizing the melted DNA for efficient RNA synthesis. J Mol Biol. 2002;315:1009–1025. doi: 10.1006/jmbi.2001.5313. [DOI] [PubMed] [Google Scholar]
- 48.Mahalakshmi R, Ragothama S, Balaram P. NMR analysis of aromatic interactions in designed peptide β-hairpins. J. Am. Chem. Soc. 2006;128:1125–1138. doi: 10.1021/ja054040k. [DOI] [PubMed] [Google Scholar]
- 49.Zavala-Ruiz Z, Strug I, Walker BD, Norris PJ, Stern LJ. A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition. Proc. Natl. Acad. Sci. U S A. 2004;101:13279–13284. doi: 10.1073/pnas.0403371101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Chow VT, Jois SD. A novel, rapid and sensitive heterotypic cell adhesion assay for CD2–CD58 interaction, and its application for testing inhibitory peptides. J. Immunol. Methods. 2004;291:39–49. doi: 10.1016/j.jim.2004.04.026. (2004) [DOI] [PubMed] [Google Scholar]
- 51.Anderson NH, Dyer RB, Fesinmeyer RM, Gai F, Liu Z, Neidigh JW, Tong H. Effect of hexafluoroisopropanol on the thermodynamics of peptide secondary structure formation. J. Am. Chem. Soc. 1999;121:9879–9880. [Google Scholar]
- 52.Greenfield NJ. Analysis of circular dichroism data. Meth. Enzymol. 2004;383:282–317. doi: 10.1016/S0076-6879(04)83012-X. (2004) [DOI] [PubMed] [Google Scholar]
- 53.Handbook of Combinatorial and Solid Phase Organic Chemistry: a Guide to Principles, Products and Protocols. Louisville, KY: Advanced Chemtech; 1998. pp. 329–372. [Google Scholar]
- 54.Lloyd-Williams P, Albericio F, Giralt E. Chemical Approaches to the Synthesis of Peptides and Proteins. Boca Raton, Florida: CRC; 1997. pp. 19–82. [Google Scholar]
- 55.Taneja V, Taneja N, Paisansinsup T, Behrens M, Griffiths M, Luthra H, David CS. CD4 and CD8 T cells in susceptibility/protection to collagen-induced arthritis in HLA-DQ8-transgenic mice: implications for rheumatoid arthritis. J. Immunol. 2002;168:5867–5875. doi: 10.4049/jimmunol.168.11.5867. [DOI] [PubMed] [Google Scholar]
- 56.Griffiths MM, Eichwald EJ, Martin JH, Smith CB, DeWitt CW. Immunogenetic control of experimental type II collagen-induced arthritis. I. Susceptibility and resistance among inbred strains of rats. Arthritis and Rheumatism. 1981;24:781–789. doi: 10.1002/art.1780240605. [DOI] [PubMed] [Google Scholar]
- 57.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Goddard TD, Kneller DG. SPARKY3. San Francisco: University of California; ( http://www.cgl.ucsf.edu/home/sparky/). [Google Scholar]
- 59.Sutcliffe MJ. Structure determination from NMR data II. Computational approaches. In: Roberts GCK, editor. NMR of Macromolecules: A Practical Approach. New York: Oxford University Press; 1993. pp. 359–390. [Google Scholar]
- 60.Dolenc J, Missimer JH, Steinmetz MO, van Gunsteren WF. Methods of NMR structure refinement: molecular dynamics simulations improve the agreement with measured NMR data of a C-terminal peptide of GCN4-p1. J. Biomol. NMR. 2010;47:221–235. doi: 10.1007/s10858-010-9425-9. [DOI] [PubMed] [Google Scholar]
- 61.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.