Summary
Since it was unexpectedly discovered that the anti-hypertensive agent, ifenprodil, has neuroprotective activity through effects to N-methyl-D-aspartate (NMDA) receptors1, enormous efforts have been made to understand the mechanism of action and to develop improved therapeutic compounds based on this knowledge2–4. Neurotransmission mediated by NMDA receptors is essential for basic brain development and function5. These receptors form heteromeric ion channels and become activated upon concurrent binding of glycine and glutamate to the GluN1 and GluN2 subunits, respectively. A functional hallmark of NMDA receptors is that their ion channel activity is allosterically regulated by binding of small compounds to the amino terminal domain (ATD) in a subtype specific manner. Ifenprodil and related phenylethanolamine compounds, which specifically inhibit GluN1/GluN2B NMDA receptors6,7, have been intensely studied for their potential use in treatment of various neurological disorders and diseases including depression, Alzheimer’s disease and Parkinson’s disease2,4. Despite great enthusiasm, mechanisms underlying recognition of phenylethanolamines and the ATD-mediated allosteric inhibition remain limited due to lack of structural information. Here we report that the GluN1 and GluN2B ATDs form heterodimer and that phenylethanolamine binds at the GluN1-GluN2B subunit interface rather than within the GluN2B cleft. The crystal structure of the GluN1b/GluN2B ATD heterodimer shows a highly distinct pattern of subunit arrangement that is different from those observed in homodimeric non-NMDA receptors and reveals the molecular determinants for phenylethanolamine binding. Restriction of domain movement in the bi-lobed structures of the GluN2B ATD by engineering an inter-subunit disulfide bond dramatically decreases ifenprodil-sensitivity indicating that conformational freedom in the GluN2B ATD is essential for ifenprodil-mediated allosteric inhibition in NMDA receptors.
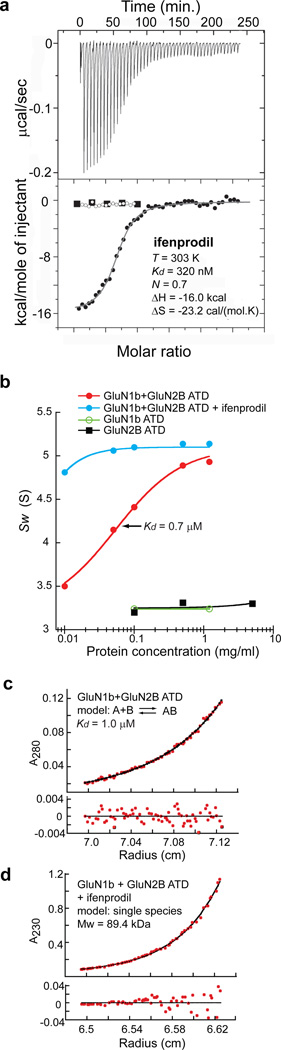
The consensus view emerged from functional studies using site directed mutagenesis and molecular modeling is that ifenprodil binds to the ATD of the GluN2B subunit. However, this has not been established directly and its mechanism of action is complicated by the obligate heteromeric assembly of NMDA receptors. To establish directly that phenylethanolamines bind to NMDA receptor ATDs, we measured binding of ifenprodil and Ro 25-6981 by isothermal titration calorimetry (ITC) using the purified recombinant Rattus norvegicus GluN2B and Xenopus laevis GluN1-4b ATDs (Supplementary Fig. 1). The GluN1-4b from Xenopus laevis8,9 was used in this study because of its superior biochemical stability compared to other orthologues. It is 93% identical in primary sequence to the Rattus norvegicus GluN1 ATD and is capable of forming functional NMDA receptor ion channels that undergo ifenprodil inhibition when combined with the Rattus norvegicus GluN2B9 (Supplementary Fig. 2). When the GluN1b ATD or GluN2B ATD proteins are individually injected with ifenprodil, there is no evidence of binding (Fig. 1a). However, when the mixture of the GluN1b and GluN2B ATD proteins are injected with ifenprodil or Ro 25-6981, a dose-dependent heat exchange is observed with Kd values of 320 nM and 60 nM, respectively (Fig. 1a and Supplementary Fig. 3). Thus, the experiment described above establishes that both GluN1b and GluN2B ATDs are required for binding of phenylethanolamines.
Figure 1. Binding of phenylethanolamine requires both GluN1b and GluN2B ATDs and stabilizes heterodimers.
a, Calorimetric titration of ifenprodil into a GluN1b-GluN2B ATD mixture (upper panel) and integrated heat as a function of ifenprodil/protein molar ratio for GluN1b ATD (open circles), GluN2B ATD (closed squares) and the GluN1b-GluN2B ATD mixture (closed circles; lower panel). b, Weighted-average sedimentation coefficient (Sw) for GluN1b ATD alone (green), GluN2B ATD alone (black), and the GluN1b-GluN2B ATD mixture in the presence (cyan) and absence (red) of 10 µM ifenprodil fitted with a monomer-dimer model (lines). c-d, Sedimentation equilibrium analysis of GluN1b and GluN2B ATDs in the presence (d) and absence (c) of 10 µM ifenprodil. Data points at a rotor speed of 18,000 rpm (red dots) are shown with a global fit (black line) of data. Residuals from the fit are shown in the lower panel.
The necessity of both GluN1b and GluN2B ATDs for recognition of phenylethanolamine implies that the binding takes place in the GluN1-GluN2B heteromer. To probe the association pattern of GluN1b and GluN2B ATDs, we determined the mass of the ATD proteins in solution by sedimentation experiments (Fig. 1b-d). While individual GluN1b ATD and GluN2B ATD are exclusively monomeric at 1.2 mg ml−1 (Fig. 1b), they form a heterodimer with a Kd of 0.7–1 µM when mixed together (Fig. 1b-c). Strikingly, when ifenprodil is included in the GluN1b/GluN2B ATD protein mixture, the heterodimerization is strengthened by at least 20-fold (Fig. 1b, d). These results establish that the GluN1b and GluN2B ATDs form heterodimers and that phenylethanolamines likely bind at the GluN1b-GluN2B subunit interface.
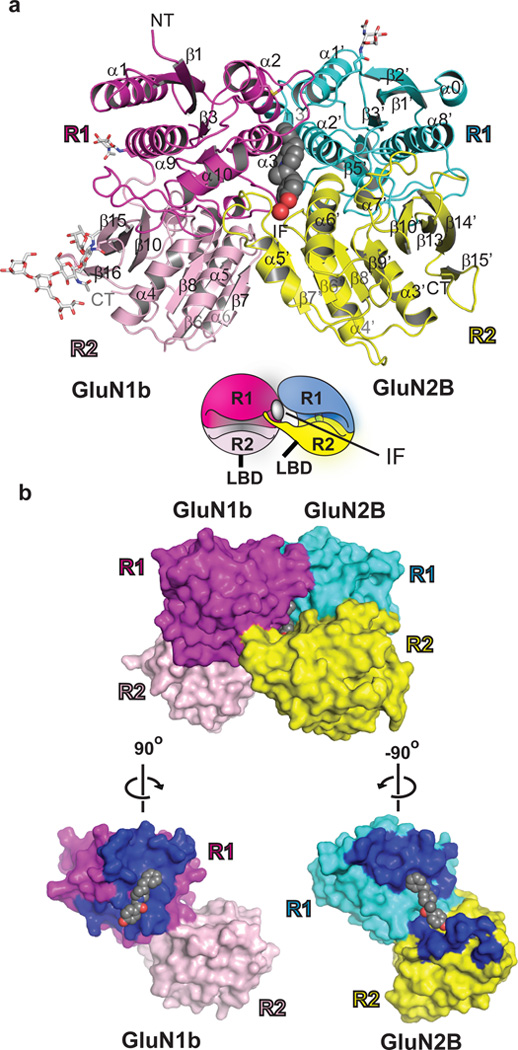
To understand the nature of the subunit interaction between GluN1b and GluN2B at ATD and to pinpoint the location of the phenylethanolamine binding site, we conducted crystallographic studies on the GluN1b and GluN2B ATD proteins (Supplementary Table 1). The crystallographic analysis showed that GluN1b and GluN2B ATDs exist as heterodimers in both ifenprodil-bound and Ro 25-6981-bound forms (Fig. 2). No significant structural difference was observed between the monomer of GluN1b ATD (Supplementary Fig. 4) or GluN2B ATD10 and the respective subunits in the GluN1b/GluN2B ATD complex indicating that dimerization did not cause changes in the overall conformation. Most importantly, the crystal structures clearly identified the phenylethanolamine binding site at the heterodimer interface (Fig. 2).
Figure 2. Structure of GluN1b/GluN2B ATD heterodimer in complex with ifenprodil at 2.6 Å resolution.
a, View from side of amino termini. GluN1b and GluN2B ATDs have bi-lobed architecture composed of R1 (magenta and cyan) and R2 (light pink and yellow) domains. Ifenprodil (gray sphere) sits at the heterodimer interface. N-glycosylation hains are shown in white. The cartoon shows an approximate orientation of the GluN1b and GluN2B ATDs with black sticks below R2 indicating the C-terminal ends where ligand-binding domains (LBDs) begin. b, Surface presentation of the GluN1b/GluN2B ATD heterodimer (upper panel) and each subunit (lower panel) showing residues at the subunit interface (dark blue). Note that ifenprodil (gray sphere) is occluded in the subunit interface. The heterodimer buries 1,191 Å2 of solvent accessible surface area per subunit with GluN1b R1-GluN2B R1 and GluN1b R1-GluN2B R2 interfaces contributing 62% and 38%, respectively.
Both GluN1b and GluN2B ATDs have bi-lobed clamshell-like architectures composed of R1 and R2 domains roughly similar in secondary structure distribution to non-NMDA receptor ATDs11–14. However, the structures of GluN1b and GluN2B ATD monomers are not superimposable onto non-NMDA receptor ATD monomers due to a major difference in the R1-R2 orientations as also observed previously in the study of the GluN2B ATD monomer10 (Supplementary Fig. 5). The unique R1-R2 orientations of the GluN1 and GluN2B ATDs result in a heterodimer assembly distinct from that observed in non-NMDA receptor ATD homodimers11–14 (Supplementary Fig. 6). While non-NMDA receptor ATD subunits form symmetrical homodimers through strong R1-R1 and R2-R2 interactions, GluN1b and GluN2B ATDs associate with each other asymmetrically through R1-R1 and R1 (GluN1b)-R2 (GluN2B) interactions11,12 (Fig. 2). No residue from GluN1b R2 is involved in the GluN1b-GluN2B interaction (Fig. 2b). The R1-R1 interface contains hydrophobic interactions mediated by residues from the cores of α2 and α3 in GluN1b and α1’ and α2’ in GluN2B, surrounded by polar interactions involving GluN1b α2, GluN2B α1’, and the hypervariable loops (HVL)10 (Supplementary Fig. 7). The R1-R2 interface involves mainly polar interactions involving residues on α10 and a loop extending from η2 in GluN1b and loops extending from β6’ and β7’ in GluN2B (Fig. 2d). The lack of R2-R2 interaction in GluN1b/GluN2B ATDs leaves a sufficient room for previously suggested conformational movement of the bi-lobed structure in GluN2B10,15, which is important in mediating allosteric regulation unique to NMDA receptors. In non-NMDA receptors such movement is prohibited due to strong R2-R2 interactions that lock the movement of R23,11–13.
The GluN1b/GluN2B heterodimeric arrangement creates a phenylethanolamine binding pocket composed of residues from GluN1b R1, GluN2B R1 and GluN2B R2 (Fig. 2). The phenylethanolamine binding site has no overlap with the zinc binding site that is located within the GluN2B ATD cleft10,16 (Supplementary Fig. 8). In the crystal structure, ifenprodil is buried in the dimer interface with no sufficient space for entering or exiting (Fig. 2b), which implies that the binding occurs through an induced fit mechanism and that unbinding perhaps involves opening of the GluN2B ATD bi-lobed structure. All of the residues at the binding sites are identical among Xenopus laevis, rat, and human orthologues indicating that phenylethanolamine-inhibition of NMDA receptors is a conserved feature observed among those species (Supplementary Fig. 9). Binding of both ifenprodil and Ro 25-6981 is mediated primarily through hydrophobic interactions between the benzylpiperidine group and a cluster of hydrophobic residues from GluN1b α2 and α3 and GluN2B α1’ and α2’, and between the hydroxylphenyl groups and GluN1b Leu135, GluN2B Phe176 and GluN2B Pro177 (Fig. 3a-b). Furthermore, the drugs make three direct polar interactions with GluN1b Ser132, GluN2B Gln110, and GluN2B Asp236. The superposition of the ifenprodil and Ro 25-6981 binding sites illustrates that the methyl and hydroxyl groups in the propanol moiety of both ligands face opposite directions while the benzylpiperidine groups sit in the binding pocket in the similar way (Fig. 3c). Consequently, Ro 25-6981 has a higher affinity than ifenprodil17 because the methyl group in Ro 25-6981 is in a favourable position to form a hydrophobic interaction involving Phe176 and Pro177 in the GluN2B subunit, while ifenprodil makes a weaker hydrophobic interaction with GluN1b involving Leu135. Extensive mutagenesis studies in the past have indicated GluN1b Tyr10918 and GluN2B Phe17619 and Asp23619 to be critical in mediating ifenprodil-inhibition but whether these were involved in binding or transducing the inhibitory effect was unknown. We performed additional mutagenesis studies on newly identified residues in both GluN1b and GluN2B at the ifenprodil binding site, measured macroscopic currents by two-electrode voltage clamp (TEVC), and revealed significant alterations in IC50 or the extent of inhibition (Fig. 3d-e, Supplementary Table 2) thereby confirming the physiological relevance of the binding site. Interestingly, disruption of the “empty” hydrophobic space formed by GluN1b Ala75, GluN2B Ile82 and GluN2B Phe114 (arrows in Fig. 3a-b) by site-directed mutations to hydrophilic residues has dramatic effects on ifenprodil-sensitivity (Fig. 3d-e). Thus, stabilization of this hydrophobic space by filling in with a hydrophobic moiety may be a valid strategy to improve the design of phenylethanolamine-based drugs.
Figure 3. Phenylethanolamine binding site.
a-b, Bindings of ifenprodil (a) and Ro 25-6981 (b) take place at the GluN1b-GluN2B subunit interface. Mesh represents Fo-Fc omit electron density map contoured at 3σ. Residues with asterisks in panel a are the ones previously shown to affect ifenprodil sensitivities. Adjacent to the binding pocket is an empty space surrounded by hydrophobic residues including GluN1b Ala75, GluN2B Ile82 and Phe114 (arrows). c, Comparison of binding patterns of ifenprodil (gray) and Ro 25-6981 (lime) in stereoview. The Ro 25-6981-bound structure is coloured as in panel b whereas the ifenprodil-bound structure is coloured white. d, New residues found to interact with phenylethanolamines in this study are mutated and analyzed for sensitivity to ifenprodil. Mutation of the residues surrounding the binding site caused changes in IC50 as well as extent of inhibition.
Why does phenylethanolamine bind specifically to the GluN1/GluN2B subunit combination? While inspection of the primary sequences shows non-conservation of the critical binding site residues between GluN2B and GluN2C or GluN2D (e.g. the residue equivalent to GluN2B Phe176), all of the residues in GluN2A except GluN2B Ile111 (Met112 in GluN2A) are conserved (Supplementary Fig. 10). Indeed, GluN2A Met112Ile or GluN2B Ile111Met does not confer or abolish ifenprodil sensitivity in GluN1/GluN2A or GluN1/GluN2B receptors, respectively (Supplementary Table 2). Thus, the insensitivity of the GluN1/GluN2A receptors to phenylethanolamine may stem from a fundamental difference in a mode of subunit association between GluN1/GluN2A and GluN1/GluN2B at ATD.
To further validate the physiological relevance of the hetero-dimeric assembly, we have engineered cysteine mutants at the subunit interface using the ifenprodil-bound GluN1b/GluN2B ATD structure as a guide in a context of the intact rat GluN1-4b/GluN2B receptor so that they would form spontaneous disulphide bonds if the mutated residues are proximal to each other. Towards this end, we have designed two pairs of cysteine mutants, GluN1-4b (Asn70Cys)/GluN2B (Thr324Cys) and GluN1-4b (Leu341Cys)/GluN2B (Asp210Cys), which “locks” the R1-R1 and R1-R2 interfaces, respectively (Fig. 4a). We then expressed the mutant receptors in mammalian cell cultures, and analyzed for formation of disulphide-linked oligomers in western blots. When mutant receptors of one subunit are co-expressed with wild-type receptors of the other, they give rise to monomeric bands (110 KDa and 170 KDa for GluN1-4b and GluN2B, respectively; Fig. 4b arrow 2 and 3) identical to wild-type GluN1-4b/GluN2B receptors in both reducing and non-reducing conditions. In contrast, co-expressing pairs of the GluN1-4b/GluN2B cysteine mutants gives rise to a heterodimeric ~280 KDa band recognized by both anti-GluN1 and anti-GluN2B antibodies in non-reducing conditions (Fig. 4b, arrow 1). This confirms that the R1-R1 and R1-R2 subunit interfaces observed in the GluN1-4b/GluN2B ATD crystal structures are physiological and that the heterodimer, but not homodimer, is the basic functional unit in the NMDA receptor ATD. Furthermore, the disulphide cross-linking is observed in the presence and absence of ifenprodil indicating that the ligand-free GluN1b/GluN2B ATDs may oscillate between the previously suggested open conformation15 and closed conformation represented by the current crystal structure.
Figure 4. Engineering of disulphide bonds at the subunit interface alters ifenprodil sensitivity.
a, Location of mutated residues at the R1-R1 and R1-R2 interfaces in GluN1b/GluN2B ATDs (sphere) and the ifenprodil binding pocket (asterisk). b, Observation of disulphide bonds by anti-GluN1 and anti-GluN2B blots in reducing (DTT) and non-reducing (−DTT) conditions. c, Macroscopic current recording of the wild-type and mutant receptors in the presence (red) and absence (black) of DTT (2 mM). d, Effect of disulphide bonds on ifenprodil sensitivity in the presence (red) and absence (black) of DTT. e, Possible model of ifenprodil binding and movement of ATD for allosteric inhibition. Ifenprodil binds to the open GluN2B clamshell and induces domain closure thereby favouring resulting in allosteric inhibition. In the GluN1-4b (N70C)/GluN2B (T324C) receptors, the GluN2B ATD is locked in the closed conformation, thus, ifenprodil cannot access the binding site.
To understand the functional effects of “locking” the R1-R1 and R1-R2 interactions in the GluN1b/GluN2B ATDs, we measured the macroscopic current responses from the ion channels of the cysteine mutant receptors by TEVC. First, we explored the effect of breaking the disulphide bonds on ion channel activity. Application of DTT has a minor inhibitory effect on the wild-type GluN1-4b/GluN2B receptors or the GluN1-4b (Asn70Cys)/GluN2B (Thr324Cys) receptors. In contrast, a 2.5-fold potentiation was observed upon breakage of the GluN1-4b (Leu341Cys)-GluN2B (Asp210Cys) disulphide bond at the R1-R2 interface (Fig. 4c and Supplementary Fig. 11). This implies that “locking” the closed conformation in the GluN2B ATD bi-lobed structure by the R1-R2 cross-link results in down-regulation of the ion channel activity. We next tested the effect of the disulphide bonds on ifenprodil sensitivity. While the R1-R1 cross-link has only a minor effect, the R1-R2 cross-link almost completely abolishes ifenprodil inhibition even at 3 µM (Fig. 4d). When this R1-R2 disulphide cross-link is broken by an application of DTT, the mutant receptors regain sensitivity to ifenprodil to a similar extent to that observed in GluN1-4b (WT)/GluN2B (Asp210Cys) in non-reducing conditions (Fig. 4d, Supplementary Fig. 12). This indicates that ifenprodil cannot bind to the GluN1b/GluN2B ATD when the R1-R2 interaction is “locked” and thus, when the GluN2B ATD clamshell is closed. Taken together, the experiments described above imply that binding of ifenprodil requires an opening of the GluN2B bi-lobed structure and that ifenprodil inhibition involves a closure of the clamshell through the GluN1b R1-GluN2B R2 interaction (Fig. 4e).
In conclusion, the current study shows that phenylethanolamine binds at the GluN1/GluN2B subunit interface through an induced-fit mechanism and that the allosteric inhibition involves stabilization of the GluN2B ATD clamshell structure in a closed conformation. The binding mechanism presented here provides a molecular blueprint for improving designs of therapeutic compounds targeting NMDA receptor ATD.
Methods Summary
GluN1b and GluN2B ATD proteins were expressed as secreted proteins using the insect cells/baculovirus system and purified using metal-chelate chromatography and size-exclusion chromatography. Crystallization was performed in hanging-drop vapor diffusion configuration in a buffer containing 20% PEG3350, 150 mM KNO3 and 50 mM HEPES-NaOH (pH 7.0) and 3.0–3.5 M NaFormate and 0.1M HEPES-NaOH (pH 7.5) for GluN1b ATD and GluN1b/GluN2B ATDs, respectively. Diffraction data sets obtained at 100K were indexed, integrated, and scaled using HKL2000. The GluN1b ATD structure was solved by the single anomalous diffraction (SAD) phasing method using Se-Met incorporated crystals and the GluN1b/GluN2B ATD structures were solved by molecular replacement using coordinates of GluN1b ATD and GluN2B ATD (PDB code: 3JPW)10. Model refinement was conducted using the program Phenix20. Experiments involving analytical ultracentrifugation and isothermal titration calorimetry were conducted using the purified protein samples in the glycosylated form. Ion channel activities of full-length NMDA receptors were measured by whole-cell recording using cRNA injected Xenopus laevis oocytes using a two electrode voltage-clamp configuration.
Methods
Expression, purification, and crystallization of GluN1b and GluN2B ATD
The Xenopus laevis GluN1b ATD (Met 1 to Glu 408) containing the C22S, N61Q and N371Q mutations was C-terminally fused to a thrombin cleavage site followed by an octa-histidine tag. The Xenopus laevis GluN1b ATD and rat GluN2B ATD10 constructs were individually expressed or co-expressed using the High Five (Trichoplusia ni)/baculovirus system (DH10multibac)21. Purification was performed using a similar method as previously described10 except that the proteins were deglycosylated by endoglycosidase F1 after purification by metal-chelate chromatography and 1 µM ifenprodil or 1 µM Ro 25-6981 was included in the running buffer of size-exclusion chromatography (Superdex200) for isolation of the GluN1b/GluN2B ATD complex. The proteins used for ITC and sedimentation experiments were purified without the endoF1 deglycosylation step and in the absence of ifenprodil or Ro 25-6981. SeMet-incorporated GluN1b ATD proteins were expressed using the methionine free media (ESF921) supplemented with DL-SeMet (Sigma) at 100 mg/L10. The GluN1b ATD and GluN1b/GluN2B ATDs were crystallized by hanging-drop vapor diffusion at 17°C by mixing the protein (8 mg/ml) with a reservoir solution containing 20% PEG3350, 150 mM KNO3 and 50 mM HEPES-NaOH (pH 7.0) at 1:1 ratio and 3.0–3.5 M NaFormate and 0.1M HEPES pH 7.5 at 2:1 ratio, respectively.
Data collection and structural analysis
Crystals of GluN1b ATD and the GluN1b/GluN2B ATDs were cryoprotected in buffers containing 20% PEG3350, 150 mM KNO3, 50 mM HEPES-NaOH (pH 7.0), and 20% glycerol and 5 M NaFormate and 0.1 M HEPES-NaOH (pH 7.5), respectively. X-ray diffraction data was collected at the X25 and X29 beamlines at National Synchrotron Light Source and processed using HKL200022. SAD data for the Se-Met incorporated GluN1b ATD crystals was collected at the peak wavelength (0.9788 Å) and used for phasing by the program SHARP23. Initial model was built using flex-wArp24. The crystal structure of GluN1b/GluN2B ATD was solved by molecular replacement using the coordinates of GluN1b ATD and GluN2B ATD10 (PDB code: 3JPW) with the program PHASER25. The models were built using COOT26 and structural refinement was performed using the program PHENIX20.
Isothermal titration calorimetry
Proteins were dialyzed against a buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.4) and 10% glycerol overnight prior to the experiment. ITC measurements were performed using VP-ITC (MicroCal) by successive injections of 5 µl of 0.15 mM ifenprodil to 0.01 mM GluN1b ATD, 10 µl of 0.25 mM ifenprodil to 0.007 mM GluN2B ATD, 5 µl of 0.15 mM ifenprodil to 0.01 mM GluN1b/GluN2B ATD complex and 5 µl of 0.05 mM Ro 25-6981 to 0.007 mM GluN1b/GluN2B ATD complex at 27°C. Data analysis was done using the software Origin 7.0 (Origin Labs).
Analytical ultracentrifugation
Both sedimentation velocity and equilibrium experiments were performed using a Beckman Coulter Optima XL-I analytical ultracentrifuge. Proteins were dialyzed against a buffer containing 150 mM NaCl, and 20 mM Tris pH 7.4 with or without 10 µM ifenprodil. Sedimentation velocity experiments were performed by centrifuging protein samples loaded on 2-sector centerpieces at 42,000 rpm at 20°C. Concentration gradients were measured using interference optics or absorbance optics at a wavelength of 280 nm or 230 nm depending on the loading protein concentrations (0.01, 0.05, 0.1, 0.5 and 1.2 mg/ml for GluN1b/GluN2B ATD in the presence and absence of ifenprodil, 0.1 and 1.2 mg/ml for GluN1b ATD and 0.1, 0.5 and 5 mg/ml for GluN2B ATD). Data was analyzed using the continuous c(s) and c(M) distribution models implemented in Sedfit27. Weighted-average sedimentation coefficient (Sw) was determined from the peak integration of c(s).
Sedimentation equilibrium experiments were performed using 6-channel centerpiece loaded with 100 µl of protein samples at protein concentrations 0.05, 0.1 and 0.3 mg/ml and in the presence and absence of 10 µM ifenprodil. The samples were centrifuged at 9,000, 13,000 and 18,000 rpm sequentially and allowed to reach equilibrium at each speed. Absorbance measurements were performed at wavelengths 230, 250 and 280 nm to obtain measurements at low and high protein concentrations. Global analysis of the data for multiple protein concentrations and rotor speeds was performed using single species and A+BA↔B models implemented in HETEROANALYSIS v1.1.44 (Univ. of Connecticut, Storrs, CT).
Electrophysiology
Recombinant GluN1/GluN2B NMDA receptors were expressed by co-injecting 0.1–0.5 ng of the wild-type or mutant rat GluN1 and GluN2B cRNAs into defolliculated Xenopus laevis oocytes. The two-electrode voltage-clamp recordings were performed using agarose-tipped microelectrodes (0.4–1.0 MΩ) filled with 3 M KCl at a holding potential of −40 mV for respectively. The bath solution contained 5 mM HEPES, 100 mM NaCl, 0.3 mM BaCl2 and 10 mM Tricine at pH 7.4 (adjusted with KOH). Currents were evoked by applications of 100 µM each of glycine and L-glutamate. Ifenprodil-inhibition was monitored in the presence of agonists and various concentrations of ifenprodil. For redox experiments, the oocytes were preincubated in the bath solution supplemented with 2 mM DTT for 3 min before recording in the continuous presence of 2 mM DTT. Data was acquired and analyzed by the program Pulse (HEKA).
Cysteine cross-linking and western blot
The single point mutations were incorporated to the full-length rat GluN1-4b and GluN2B in the pCI vector (Promega). Human embryonic kidney 293 cells were transfected with a mixture of 0.5 µg and 1 µg of the GluN1-4b and GluN2B DNA plasmids, respectively, by Fugene HD (Roche). Cells were harvested 24–48 h after transfection and resuspended in a buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% dodecyl-maltoside, and a protease-inhibitor cocktail (Roche) as previously described28. After centrifugation at 150,000g, the supernatant were subjected to SDS-PAGE (4–15%) in the presence and absence of 100 mM DTT. The proteins were transferred to the Hybond-ECL nitrocellulose membranes (GE Healthcare). The membranes were blocked by TBST (20 mM Tris-HCl pH 7.4, 150 mM NaCl, and 0.1% Tween20) containing 10% milk, then, incubated with GluN1 (MAB 1586; Millipore) or GluN2B (Invitrogen) mouse monoclonal antibodies followed by HRP-conjugated anti-mouse antibodies (GE Healthcare). Protein bands were detected by ECL detection kit (GE Healthcare).
Supplementary Material
Acknowledgement
We thank the staff at X25 and X29 at the National Synchrotron Light Source for the beamline support. M. Mayer is thanked for providing insightful comments on this work. We also thank D. Raleigh at Stony Brook University for the use of analytical ultracentrifugation. GluN1 clones from Xenopus laevis were kind gifts from M. Hollmann and H. Cline. This work was supported by NIH MH085926, Alzheimer’s association and a donation from the Fox family (to H.F.). H.F. was also funded by scientist development grant from American Heart Association. E.K. is supported by a NARSAD Lieber Young Investigator Award.
Footnotes
Author Contribution
The project was initiated by E.K. and H.F. All of the experiments were designed by E.K. and H.F. Crystallographic studies, isothermal calorimetry, and analytical ultracentrifugation were carried out by E.K. Electrophysiology and cross-linking experiments were conducted by H.F. Technical supports were given by N.S. The manuscript was written by E.K. and H.F.
Author Information
Structural coordinates are deposited to the Protein Data Bank with accession codes of 3QEK, 3QEL, and 3QEM, for GluN1b ATD, GluN1b/GluN2B ATD in complex with ifenprodil, and GluN1b/GluN2B ATD in complex with Ro, respectively. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Gotti B, et al. Ifenprodil and SL 82.0715 as cerebral anti-ischemic agents. I. Evidence for efficacy in models of focal cerebral ischemia. J Pharmacol Exp Ther. 1988;247:1211–1221. [PubMed] [Google Scholar]
- 2.Koller M, Urwyler S. Novel N-methyl-D-aspartate receptor antagonists: a review of compounds patented since 2006. Expert Opin Ther Pat. 2010;20:1683–1702. doi: 10.1517/13543776.2010.533656. [DOI] [PubMed] [Google Scholar]
- 3.Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010;78:535–549. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mony L, Kew JN, Gunthorpe MJ, Paoletti P. Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential. Br J Pharmacol. 2009;157:1301–1317. doi: 10.1111/j.1476-5381.2009.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher MJ, Huang H, Pritchett DB, Lynch DR. Interactions between ifenprodil and the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem. 1996;271:9603–9611. doi: 10.1074/jbc.271.16.9603. [DOI] [PubMed] [Google Scholar]
- 7.Williams K. Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: selectivity and mechanisms at recombinant heteromeric receptors. Mol Pharmacol. 1993;44:851–859. [PubMed] [Google Scholar]
- 8.Ewald RC, Cline HT. Cloning and Phylogenetic Analysis of NMDA Receptor Subunits NR1, NR2A and NR2B in Xenopus laevis Tadpoles. Front Mol Neurosci. 2009;2:4. doi: 10.3389/neuro.02.004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt C, Hollmann M. Molecular and functional characterization of Xenopus laevis N-methyl-d-aspartate receptors. Mol Cell Neurosci. 2009;42:116–127. doi: 10.1016/j.mcn.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin R, et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. Embo J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clayton A, et al. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- 14.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rachline J, Perin-Dureau F, Le Goff A, Neyton J, Paoletti P. The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J Neurosci. 2005;25:308–317. doi: 10.1523/JNEUROSCI.3967-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malherbe P, et al. Identification of critical residues in the amino terminal domain of the human NR2B subunit involved in the RO 25-6981 binding pocket. J Pharmacol Exp Ther. 2003;307:897–905. doi: 10.1124/jpet.103.056291. [DOI] [PubMed] [Google Scholar]
- 18.Masuko T, et al. A regulatory domain (R1-R2) in the amino terminus of the N-methyl-D-aspartate receptor: effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol Pharmacol. 1999;55:957–969. doi: 10.1124/mol.55.6.957. [DOI] [PubMed] [Google Scholar]
- 19.Perin-Dureau F, Rachline J, Neyton J, Paoletti P. Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors. J Neurosci. 2002;22:5955–5965. doi: 10.1523/JNEUROSCI.22-14-05955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.de La Fortelle E, Bricogne G. Maximum-Likelihood Heavy-Atom Parameter Refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods. Methods in Enzymology. 1997;276:472–492. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 24.Cohen SX, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D Biol Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 25.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.