Abstract
To understand the genetic heterogeneity underlying developmental delay, we compare copy-number variants (CNVs) in 15,767 children with intellectual disability and various congenital defects to 8,329 adult controls. We estimate that ~14.2% of disease in these individuals is due to large CNVs > 400 kbp. We find greater CNV enrichment in patients with craniofacial anomalies and cardiovascular defects than epilepsy or autism. We identify 59 pathogenic CNVs including 14 novel or previously weakly supported candidates. We refine the critical interval for several genomic disorders such as the 17q21.31 microdeletion syndrome and identify 940 candidate dosage-sensitive genes. We also develop methods to opportunistically discover small, disruptive CNVs within the large and growing diagnostic array datasets. This evolving CNV morbidity map combined with exome/genome sequencing will be critical for deciphering the genetic basis of developmental delay, intellectual disability, and autism spectrum disorders.
INTRODUCTION
Large copy number variants (CNVs) are enriched in the aggregate among severe cases of pediatric disease including neurological and congenital birth defects1,2 as well as neuropsychiatric diseases3–5. Clinical interpretation of individual loci has been problematic for several reasons. First, except for CNV “hotspots” flanked by duplications prone to unequal crossing over and elevated de novo mutation rates6,7, disease associations for many individual CNVs remain unclear due to their rarity and the need to screen extraordinarily large sample sizes. Second, even for CNVs with clear pathogenicity, the dosage-sensitive genes that underlie the phenotypes observed have generally not been identified because the CNVs are large and encompass many genes. Finally, considerable variation in expressivity is often observed, with the same lesion contributing to different disease outcomes8–12. Thus, while their disease risk in general is well established, the phenotypic consequences for most large CNVs are not well characterized nor have these effects been fine mapped. Here, we leverage a collection of data from 15,767 children with various developmental and intellectual disabilities and compare them to a CNV map we generated from 8,329 adult controls. We present the first detailed genome-wide morbidity map of developmental delay and congenital birth defects. Striking differences in the CNV landscape are revealed including potentially pathogenic genes, refinement of known disease-causing mutations, and the discovery of potentially novel genes, including the development of methods to opportunistically discover smaller disruptive CNVs from clinical datasets.
RESULTS
We analyzed 15,767 DNA samples from children referred to Signature Genomic Laboratories, LLC, with a general diagnosis of intellectual disability (ID) and/or developmental delay (DD), although we note that this ID/DD cohort also includes a constellation of phenotypes including, but not restricted to, congenital malformation, hypotonia and feeding difficulties, speech and motor deficits, growth retardation, cardiovascular and renal defects, epilepsy, hearing impairment, craniofacial and skeletal features, and behavioral issues. Overall 73% of cases suffer from ID/DD and/or autism spectrum disorder, while 12% of cases were not annotated. The remainder were classified with various congenital abnormalities. Detailed phenotypic information is limited to 48.4% of the cases where specific subclassifications could be made, including 575 cases with cardiovascular defects, 1,776 cases with epilepsy/seizure disorder, 1,379 with autism spectrum disorder, and 3,898 with craniofacial defects (Supplementary Tables 1 & 2).
DNA samples obtained from whole blood were analyzed by customized array comparative genomic hybridization (CGH) at an average probe density of ~97,000 oligonucleotides, sufficient for reliable genome-wide detection of CNVs >300 kbp and for targeted detection of events >40 kbp for approximately one-fourth of the genome13. After filtering, a total of 16,526 rare (< 1% population frequency) autosomal CNV calls were made with an average of 1.05 CNV events per individual (median size 213 kbp). Using a customized higher density microarray and fluorescent in situ hybridization, we validated 402/425 CNVs (precision of 0.945) greater than 150 kbp (Supplementary Note, and Supplementary Table 3). Similarly, manual inspection of calls with low log ratios or z-scores (absolute values of <0.25 and <1.5, respectively) suggests a false discovery rate of 0.0138. For comparison, we identified CNVs from a control set of 8,329 adult samples assayed using multiple Illumina genome-wide single-nucleotide polymorphism (SNP) microarrays. These samples were studied as part of genome-wide association studies (dbGaP) for phenotypes unrelated to neurological disease (e.g. lipid concentration levels, blood pressure, asthma, etc.) (Supplementary Table 4). CNVs were called using a Hidden Markov Model (HMM)–based discovery method 14 with an overall precision of 0.892 in identifying large CNVs (>100 kbp) (validation rates of 6/615 and 19/2216). From this dataset, we identified 446,736 CNVs with an average of 53.6 events (rare and common) per individual (median size 1.9 kbp). Due to the increased probe density (most >550,000 probes), our control dataset provides increased CNV detection power and resolution when compared to the disease dataset, reducing the potential for spurious CNV enrichments within cases (see Methods).
CNV burden
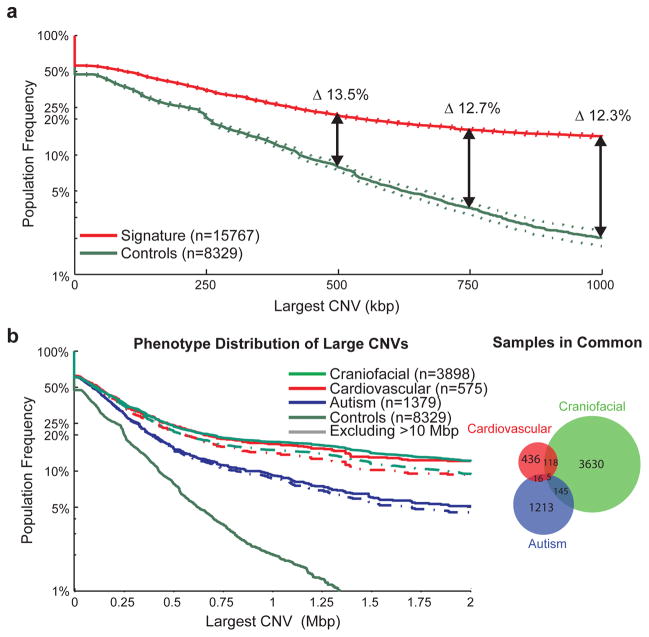
We compared CNV content between the cases and controls excluding common CNVs (>1% population frequency). Consistent with previous studies of pediatric neurological disease3–5,17,18, we find a significant excess of large CNVs among cases relative to controls. This excess is evident at 250 kbp and becomes more pronounced with increasing CNV size (Figure 1A). For example, at a threshold of 400 kbp, ~25.7% (4,047 cases) of ID/DD children harbor an event of at least this size compared to 11.5% of controls, suggesting that an estimated 14.2% of ID and DD is due to the presence of CNVs >400 kbp in length (OR = 2.7, p = 5.86×10−158). At a threshold of 1.5 Mbp, we identify 1,782 (11.3%) affected individuals versus only 52 (0.6%) controls (OR = 20.3, p = 6.87×10−266) and at a threshold of 3.0 Mbp the odds ratio jumps to 47.7 (p = 1.68×10−197). There is a remarkably strong correlation (R2 = 0.97) with the de novo rate as a function of increasing CNV size, with 50% of events at 1 Mbp reported as inherited (Supplementary Figure 1).
Figure 1. CNV size distributions in affected and unaffected individuals.
The population frequency of the largest CNV in a sample is displayed as a survivor function with the proportion of samples carrying a CNV of a given size displayed as a curve, with 95% confidence intervals indicated by dotted lines. (A) The distribution of large CNVs in the Signature set (filtered to only contain events detectable by the Illumina 550K array) versus our control population (downsampled to only events detectable by the Signature 97K array) is indicated for the overall population. After corrections for different array densities, we observed a >13.5% increase in CNV burden beyond 500 kbp in cases with a proportion of the burden representing potentially novel loci. (B) We also performed a similar analysis on subphenotypes; in this analysis, we included all Signature CNVs in conjunction with downsampled control CNVs as we are highlighting interphenotype differences rather than case versus control frequencies. This is demonstrated here for the autism, cardiovascular and craniofacial phenotypes, which represent fairly distinct sample sets and show an increased burden for the cardiovascular and craniofacial phenotypes, even after exclusion of karyotypically visible (>10 Mbp) events.
We find 1,492 CNVs in 1,400 individuals within 45 known genomic disorder regions (Table 1, Supplementary Table 5). Among these, deletions are twice as common (n = 954 deletions vs. 538 duplications) and show greater average penetrance (96.3%) when compared to duplications (94.3%). We note that “classic,” phenotypically well-defined syndromes known to result from CNVs (e.g. Smith-Magenis, Williams syndrome, etc.) are underrepresented here relative to other cohorts of individuals with similar phenotypes (Supplementary Table 6), suggesting that our estimate of CNV burden in ID/DD is not upwardly biased by ascertainment for known CNV carriers.
Table 1.
Frequency of known genomic disorders in cases and controls.
| Deletions (<10 Mbp) | Duplications (<10 Mbp) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| chr | start | end | Deletion | Cases (n=15767) |
Control (n=8329) |
p-value | Penetrance | Duplication | Cases (n=15767) |
Control (n=8329) |
p -value | Penetrance |
| chr1 | 0.00 | 10.00 | 1p36 deletion syndrome (GABRD)a | 79 | 0 | 2.62E-15 | 1.00 | 1p36 duplication (GABRD)a | 16 | 1 | 0.0074 | 0.94 |
| chr1 | 144.00 | 144.34 | TAR deletion (HFE2) | 13 | 2 | 0.0659 | 0.87 | 1q21.1 duplication (HFE2) | 25 | 6 | 0.0511 | 0.81 |
| chr1 | 145.04 | 145.86 | 1q21.1 deletion (GJA5) | 47 | 2 | 3.28E-07 | 0.96 | 1q21.1 duplication (GJA5) | 26 | 1 | 0.0002 | 0.96 |
| chr2 | 96.09 | 97.04 | 2q11.2 deletion (LMAN2L, ARID5A) | 2 | 0 | 0.4282 | 1.00 | 2q11.2 duplication (LMAN2L, ARID5A) | 1 | 0 | 0.6543 | 1.00 |
| chr2 | 100.06 | 107.81 | 2q11.2q13 deletion (NCK2, FHL2) | 0 | 0 | 1.0000 | NA | 2q11.2q13 duplication (NCK2, FHL2) | 2 | 0 | 0.4282 | 1.00 |
| chr2 | 110.18 | 110.34 | 2q13 deletion (NPHP1) | 78 | 30 | 0.0813 | 0.72 | 2q13 duplication (NPHP1) | 118 | 32 | 0.0003 | 0.79 |
| chr2 | 239.37 | 242.12 | 2q37 deletion (HDAC4)a | 22 | 0 | 0.0001 | 1.00 | 2q37 duplication (HDAC4)a | 0 | 0 | 1.0000 | NA |
| chr3 | 197.23 | 198.84 | 3q29 deletion (DLG1) | 6 | 0 | 0.0785 | 1.00 | 3q29 duplication (DLG1) | 4 | 0 | 0.1833 | 1.00 |
| chr4 | 1.84 | 1.98 | Wolf-Hirschhorn deletion (WHSC1, WHSC2)a | 21 | 0 | 0.0001 | 1.00 | Wolf-Hirschhorn region duplication | 7 | 0 | 0.0513 | 1.00 |
| chr5 | 175.65 | 176.99 | Sotos syndrome deletion (NSD1) | 8 | 0 | 0.0336 | 1.00 | 5q35 duplication (NSD1) | 0 | 0 | 1.0000 | NA |
| chr6 | 100.92 | 101.05 | 6q16 deletion (SIM1)a | 1 | 0 | 0.6543 | 1.00 | 6q16 duplication (SIM1)a | 1 | 0 | 0.6543 | 1.00 |
| chr7 | 72.38 | 73.78 | Williams syndrome deletion (ELN, GTF2I) | 42 | 0 | 1.80E-08 | 1.00 | Williams syndrome duplication (ELN, GTF2I) | 16 | 0 | 0.0011 | 1.00 |
| chr7 | 74.80 | 76.50 | WBS-distal deletion (RHBDD2, HIP1) | 2 | 0 | 0.4282 | 1.00 | WBS-distal duplication (RHBDD2, HIP1) | 0 | 0 | 1.0000 | NA |
| chr8 | 8.13 | 11.93 | 8p23.1 deletion (SOX7, CLDN23) | 7 | 0 | 0.0513 | 1.00 | 8p23.1 duplication (SOX7, CLDN23) | 7 | 0 | 0.0513 | 1.00 |
| chr9 | 136.95 | 140.20 | 9q34 deletion (EHMT1)a | 60 | 0 | 8.54E-12 | 1.00 | 9q34 duplication (EHMT1)a | 4 | 0 | 0.1833 | 1.00 |
| chr10 | 81.95 | 88.79 | 10q23 deletion (NRG3, GRID1) | 8 | 0 | 0.0336 | 1.00 | 10q23 duplication (NRG3, GRID1) | 1 | 0 | 0.6543 | 1.00 |
| chr11 | 43.94 | 46.02 | Potocki-Shaffer syndrome (EXT2)a | 5 | 0 | 0.1199 | 1.00 | 11p11.2 duplication (EXT2)a | 0 | 0 | 1.0000 | NA |
| chr11 | 67.51 | 70.96 | SHANK2 FGFs deletion | 1 | 0 | 0.6543 | 1.00 | SHANK2 FGFs duplication | 0 | 0 | 1.0000 | NA |
| chr12 | 63.36 | 66.93 | 12q14 deletion syndrome (GRIP1, HMGA2)a | 2 | 0 | 0.4282 | 1.00 | 12q14 duplication (GRIP1, HMGA2)a | 0 | 0 | 1.0000 | NA |
| chr13 | 19.71 | 19.91 | 13q12 deletion (CRYL1)a | 14 | 12 | 0.9240 | 0.54 | 13q12 duplication (CRYL1)a | 4 | 0 | 0.1833 | 1.00 |
| chr15 | 20.35 | 20.64 | 15q11.2 deletion (NIPA1) | 94 | 19 | 2.13E-05 | 0.83 | 15q11.2 duplication (NIPA1) | 64 | 36 | 0.6614 | 0.64 |
| chr15 | 22.37 | 26.10 | Prader-Willi/Angelman | 16 | 0 | 0.0011 | 1.00 | Prader-Willi/Angelman region duplication | 27 | 0 | 1.06E-05 | 1.00 |
| chr15 | 28.92 | 30.27 | 15q13.3 deletion (CHRNA7) | 42 | 0 | 1.8E-08 | 1.00 | 15q13.3 duplication (CHRNA7) | 20 | 3 | 0.0200 | 0.87 |
| chr15 | 70.70 | 72.20 | 15q24 BP0-BP1 deletion (BBS4, NPTN, NEO1) | 4 | 0 | 0.1833 | 1.00 | 15q24 BP0-BP1 duplication (BBS4, NPTN, NEO1) | 1 | 0 | 0.6543 | 1.00 |
| chr15 | 70.70 | 73.58 | 15q24 BP0-BP1 (PML) | 4 | 0 | 0.1833 | 1.00 | 15q24 BP0-BP1 (PML) | 4 | 0 | 0.1833 | 1.00 |
| chr15 | 73.76 | 75.99 | 15q24 BP2-BP3 deletion (FBXO22, TPSAN3) | 1 | 0 | 0.6543 | 1.00 | 15q24 BP2-BP3 duplication (FBXO22, TPSAN3) | 0 | 0 | 1.0000 | NA |
| chr15 | 80.98 | 82.53 | 15q25.2 deletion (HOMER2, BNC1) | 1 | 0 | 0.6543 | 1.00 | 15q25.2 duplication (HOMER2, BNC1) | 0 | 0 | 1.0000 | NA |
| chr15 | 97.18 | 100.34 | None | 10 | 1 | 0.0641 | 0.91 | None | 1 | 0 | 0.6543 | 1.00 |
| chr16 | 3.72 | 3.80 | Rubinstein-Taybi Syndromea | 7 | 0 | 0.0513 | 1.00 | Rubinstein-Taybi region duplication | 6 | 0 | 0.0785 | 1.00 |
| chr16 | 15.41 | 16.20 | 16p13.11 deletion (MYH11) | 18 | 3 | 0.0361 | 0.86 | 16p13.11 duplication (MYH11) | 24 | 10 | 0.3315 | 0.71 |
| chr16 | 21.26 | 29.35 | 16p11.2p12.1 deletion | 2 | 0 | 0.4282 | 1.00 | 16p11.2p12.1 duplication | 2 | 0 | 0.4282 | 1.00 |
| chr16 | 21.85 | 22.37 | 16p12.1 deletion (EEF2K, CDR2) | 37 | 3 | 0.0001 | 0.93 | 16p12.1 duplication (EEF2K, CDR2) | 4 | 1 | 0.4368 | 0.80 |
| chr16 | 28.68 | 29.02 | 16p11.2 distal deletion (SH2B1) | 15 | 1 | 0.0107 | 0.94 | 16p11.2 distal duplication (SH2B1) | 14 | 2 | 0.0484 | 0.88 |
| chr16 | 29.56 | 30.11 | 16p11.2 deletion (TBX6) | 64 | 3 | 3.39E-09 | 0.96 | 16p11.2 duplication (TBX6) | 28 | 2 | 0.0004 | 0.93 |
| chr17 | 0.05 | 2.54 | 17p13.3 deletion (both YWHAE and PAFAH1B1)a | 7 | 0 | 0.0513 | 1.00 | 17p13.3 duplication (both YWHAE and PAFAH1B1)a | 2 | 0 | 0.4282 | 1.00 |
| chr17 | 0.50 | 1.30 | 17p13.3 deletion (including PAFAH1B1)a | 8 | 0 | 0.0336 | 1.00 | 17p13.3 duplication (including PAFAH1B1)a | 6 | 0 | 0.0785 | 1.00 |
| chr17 | 2.31 | 2.87 | 17p13.3 deletion (including YWHAE)a | 7 | 0 | 0.0513 | 1.00 | 17p13.3 duplication (including YWHAE)a | 4 | 0 | 0.1833 | 1.00 |
| chr17 | 14.01 | 15.44 | HNPP (PMP22) | 3 | 0 | 0.2801 | 1.00 | CMT1A (PMP22) | 9 | 2 | 0.2086 | 0.82 |
| chr17 | 16.65 | 20.42 | Smith-Magenis syndrome deletion | 16 | 0 | 0.0011 | 1.00 | Potocki-Lupski syndrome | 9 | 0 | 0.0220 | 1.00 |
| chr17 | 26.19 | 27.24 | NF1 deletion syndrome | 5 | 0 | 0.1199 | 1.00 | NF1 duplication | 2 | 0 | 0.4282 | 1.00 |
| chr17 | 31.89 | 33.28 | RCAD (renal cysts and diabetes) (TCF2) | 14 | 2 | 0.0484 | 0.88 | 17q12 duplication | 18 | 3 | 0.0361 | 0.86 |
| chr17 | 41.06 | 41.54 | 17q21.31 deletion (MAPT) | 23 | 0 | 0.0001 | 1.00 | 17q21.31 duplication (MAPT) | 2 | 0 | 0.4282 | 1.00 |
| chr22 | 17.40 | 18.67 | DiGeorge/VCFS deletion | 96 | 0 | 0.0000 | 1.00 | 22q11.2 duplication | 50 | 5 | 1.26E-05 | 0.91 |
| chr22 | 20.24 | 21.98 | 22q11.2 distal deletion (BCR, MAPK1) | 13 | 0 | 0.0040 | 1.00 | 22q11.2 distal duplication (BCR, MAPK1) | 7 | 0 | 0.0513 | 1.00 |
| chr22 | 49.46 | 49.52 | Phelan-McDermid syndrome deletion (SHANK3)a | 45 | 0 | 0.0000 | 1.00 | 22q13 duplication (SHANK3)a | 7 | 0 | 0.0513 | 1.00 |
All coordinates are according to build36. The genes in parentheses are potential candidate genes and identifiers of the genomic locations.
Rearrangements not mediated by segmental duplications; VCFS – velocardiofacial syndrome, WBS – Williams-Beuren syndrome, HNPP – hereditary neuropathy with liability to pressure palsies, CMT1A – Charcot-Marie-Tooth disease type 1A. No CNVs were identified in 2p15p16.1 (VRK2), 15q24 (BP1-BP2) (CLK3), 15q24 (SIN3A), 17q23 (TUBD1), and 17q23.1q23.2 (TBX2, TBX4). Note that a single CNV may encompass more than one genomic disorder.
Examining the size distribution of CNVs in the context of major subphenotypes shows that the large CNV burden is increased in more severe developmental phenotypes associated with multiple congenital abnormalities. We find, for example, that children also diagnosed with craniofacial and cardiovascular defects show a significantly increased burden of large CNVs when compared to children with autism spectrum disorder (p = 4.99×10−10 and 6.45×10−5, respectively, at >400 kbp) (Figure 1B). Children with an additional diagnosis of epilepsy/severe seizure disorder tend to have a more intermediate CNV burden when compared to individuals with autism or more severe ID (Supplementary Figure 2). These distinctions remain significant even after excluding CNVs larger than 10 Mbp (which would have been detectable by karyotype analysis) and when the CNV burden among the subset of controls screened for psychiatric disease is used as the baseline, demonstrating a role for large CNVs in more severe phenotypic variation.
Locus-specific enrichments
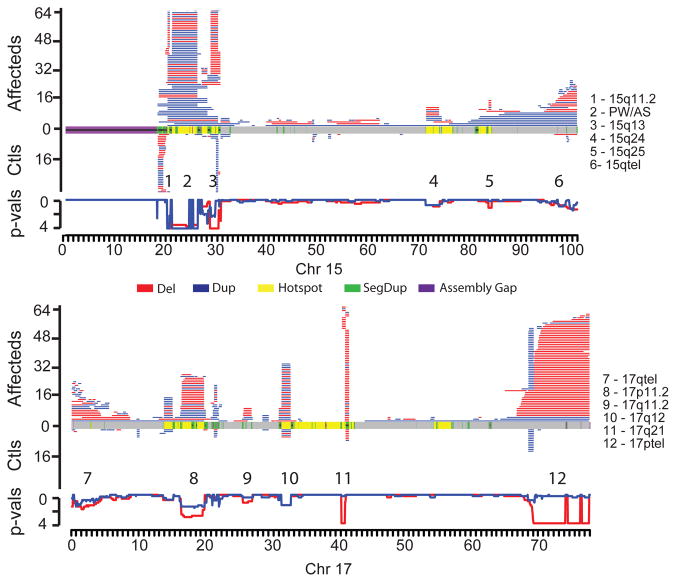
A comparison of the CNV landscape between cases and controls reveals striking differences and some general genomic architectural features (Figure 2). To ameliorate the effects of breakpoint imprecision and multi-platform comparisons, we contrasted the number of deletions (or duplications) present in cases versus controls in 200 kbp windows along the human genome using a Fisher’s exact test (Supplementary Table 7, Supplementary Figure 3). This analysis identified 80 genomic regions that were at least weakly enriched for CNVs (counting deletions and duplications separately) among cases (at least five windows with p < 0.1), 27 of which exhibit strong evidence for enrichment (p < 0.001). Notably, 27.5% (22/80) of the enriched CNV-loci reside at genomic hotspots flanked by large (>10 kbp) blocks of highly similar (>90%) segmental duplication (SD) and include most known genomic disorders (Supplementary Table 7). An additional 46 enrichments represent large CNVs near telomeres (Supplementary Figure 4). While we observed enrichments at one or both ends of all chromosomes, 12 chromosome ends showed particularly strong (p < 0.001) enrichment. Of the 80-CNV loci, 15 are novel or are supported by isolated case reports (Table 2). Additional phenotypic details for CNV carriers, including ethnicity and inheritance status, at each of these 15 CNV-loci is available in Supplementary Table 8, in some cases with comparison to similar CNVs observed in case reports from the DECIPHER database19. We note that one of these 15 (duplications at 10p15.3) appears to be enriched among cases as a consequence of allelic stratification between African and European populations and was eliminated from further consideration (see Methods and Supplementary Note).
Figure 2. Maps of CNV locations for chromosomes 15 (top) and 17 (bottom).
CNVs (>400 kbp) in affected individuals are shown in the upper portion for each chromosome with control CNVs shown in the lower portion. Disease enrichment p-values are plotted just below the control CNV maps, computed in 200 kbp windows along each chromosome (step size of 50 kbp). Deletions and duplications are red and blue, respectively, with the p-value wiggle plots colored accordingly and plotted on a negative log scale. In the middle of each plot, chromosomal features are colored as depicted. Significantly enriched regions are numbered and named on the right-hand side.
Table 2.
Novel, potentially pathogenic loci identified by sliding window analysis.
| Chr | Start (Mb) | End (Mb) | Size (Mb) | CNV | p value (adjusted) | Cases (adjusted)a | Controls (adjusted)a | Description | Ethnicityb |
|---|---|---|---|---|---|---|---|---|---|
| chr2c,d | 111.05 | 112.95 | 1.9 | del | 0.006 (0.032) | 12 (12) | 0 (1) | 2q13 | 10C,1A |
| chr10c | 81.6 | 88.9 | 7.3 | del | 0.014 (0.064) | 10 (10) | 0 (1) | 10q23.1 | 6C,1O |
| chr2 | 45.2 | 45.9 | 0.7 | dup | 0.022 (0.022) | 9 (9) | 0 (0) | 2p21 | 8C |
| chr2b,c | 111.05 | 112.85 | 1.8 | dup | 0.034 (0.022) | 8 (9) | 0 (0) | 2q13 | 5C,2O |
| chr4 | 9.45 | 10.45 | 1 | dup | 0.034 (0.051) | 8 (7) | 0 (0) | 4p16.1 | 6C,1A,1O |
| chr4 | 81.95 | 83.35 | 1.4 | del | 0.034 (0.034) | 8 (8) | 0 (0) | 4q21.21 - q21.22 | 6C,1A |
| chr2 | 3.25 | 3.45 | 0.2 | dup | 0.051 (0.051) | 7 (7) | 0 (0) | 2p25.3 | 3C,1O |
| chr2 | 165.4 | 166.1 | 0.7 | del | 0.051 (0.051) | 7 (7) | 0 (0) | 2q24.3 | 5C,1O |
| chr21 | 19.95 | 20.25 | 0.3 | del | 0.051 (0.079) | 7 (6) | 0 (0) | 21q21.1 | 1C,1A,2O |
| chr8 | 53.45 | 54.05 | 0.6 | dup | 0.051 (0.051) | 7 (7) | 0 (0) | 8q11.23 | 6C,1O |
| chr1 | 170 | 170.6 | 0.6 | del | 0.079 (0.079) | 6 (6) | 0 (0) | 1q24.3 | 5C |
| chr12 | 8.05 | 8.25 | 0.2 | dup | 0.079 (0.051) | 6 (7) | 0 (0) | 12p13.31 | 6C |
| chr15c,d | 82.9 | 83.6 | 0.7 | del | 0.079 (0.12) | 6 (5) | 0 (0) | 15q25 | 1C,2A,2O |
| chr6 | 20.85 | 21.25 | 0.4 | del | 0.079 (0.079) | 6 (6) | 0 (0) | 6p22.3 | 1E,1A,1O |
The counts and p-values are based on the single most significant 200 kb window, while the 'adjusted' counts include all samples with a CNV overlapping the region but exclude all related samples (see Supplementary Table 7).
C – Caucasian (primarily European descent), A - African-American, O – other.
Hotspot regions.
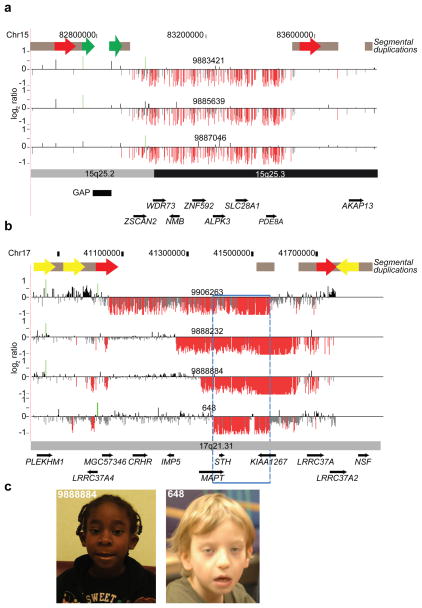
Among the 14 novel ID/DD CNV-loci, we identified a 660 kbp deletion mapping to chromosome 15q25.2 flanked by SDs (69.8 kbp, 98.6% identity) (Figure 3A). The deletion is absent in the controls analyzed here and the Database of Genomic Variants (http://projects.tcag.ca/variation/), but present in five affected individuals (including two siblings) among the ID/DD sample set. Clinical aspects of the probands were variable consisting of neurologic features and DD (Supplementary Table 9); one female had only mild motor delay associated with a congenital myopathy but was otherwise cognitively normal. The two brothers with the deletion both had autism spectrum disorders but additional family members were not tested (Supplementary Note). A previous meta-analysis of patients found this deletion in 4 of 6,860 cases16 with schizophrenia and autism compared to 0 of 5,674 controls (combined with this study, p = 0.037 after excluding one sibling). Thus, while statistical significance remains modest and population stratification cannot be definitively ruled out (see Supplementary Note), these data suggest a potentially new genomic disorder that will be observed at a frequency of 1/3,000 referred cases.
Figure 3. Discovery of novel microdeletions associated with genomic disorders.
(A) A novel microdeletion on chromosome 15q25.2q25.3. Array CGH analysis for three individuals with a 660 kbp (chr15:82,889,423–83,552,890) deletion is shown. This microdeletion maps within a genomic hotspot flanked by high-identity SD blocks. Intrachromosomal SDs of high similarity relevant to this hotspot region are depicted as red (69.8 kbp, 98.6% identity) and green (17.6 kbp, 98.6% identity) block arrows. Note that the directly orientated SDs (red block arrows) likely mediate the underlying 15q25 rearrangements by non-allelic homologous recombination (NAHR). This region also contains a 60 kbp (chr15:82,775,465–82,835,495) gap in the current builds (build36 and build37) of the reference genome assembly. (B) Atypical 17q21.31 microdeletions refine critical interval genes. High-density array CGH for the 17q21.31 microdeletion region is shown for three individuals. Probes with log2 ratios below a threshold of 1.5 standard deviations from the normalized mean log2 ratio denote deletions (red). The typical deletions (top panel) were identified in 23 individuals while atypical deletions were identified in three individuals. Note that the smallest deletion (blue box) refines the phenotype-associated critical region (chr17:41,356,798–41,631,306) to encompass only five RefSeq genes. (C) Photographs of two individuals (9888884 and 648) with atypical deletions are shown. Patient #9888884 is a 5-year-old female child with clinical features typical of 17q21.31 microdeletion syndrome and includes distinctive dysmorphic features with a bulbous nasal tip, upslanting and almond-shaped palpebral fissures, long face, strabismus, epicanthal folds, and prominent ears; DD with limited speech; hypotonia in infancy; and a friendly disposition. Additional features are low birth weight, short stature, microcephaly, long fingers, and heart defects. She also presented with postaxial polysyndactyly, neonatal cholestasis, resolved leucopenia, dry skin with some hyperpigmented lesions, and an anteriorly split tongue. Patient #648 is 9-year-old male child and has a clinical history of generalized hypotonia, seizures, autism, mental retardation, motor DD, and dysmorphic features consistent with the 17q21.31 microdeletion syndrome (epicanthal folds; ptosis; long, pear-shaped nose; long, tapering fingers). Informed consent was obtained to publish the photographs.
One of the most common genomic hotspots in this study is 15q11.2 (NIPA1), a 292 kbp deletion whose pathogenicity has been considered uncertain4,20. In terms of frequency, the 15q11.2 deletion is second only to VCF/DGS deletion, and our data indicate it is significantly enriched (OR = 2.36, p = 2.5×10−5) albeit at lower penetrance (0.83) than most other genomic disorders. In addition, we find support for the pathogenicity of duplications of obesity-associated 16p11.2 (SH2B1)21,22 and epilepsy-associated 15q13.3 (CHRNA7)23. We also analyzed 111 regions of the human genome predicted to be prone to recurrent microdeletions and microduplications based on the presence of homologous SDs at their flanks in the reference assembly6. Of these potential hotspots, 62 harbored CNVs likely mediated by NAHR between the flanking SDs (“active hotspots”), while the remaining 49 did not. The presence of SDs in direct, as opposed to inverted, orientation is a key distinction between active and inactive hotspots (46/54 direct vs. 16/57 inverted; OR = 3.04). We also found that SDs flanking active hotspots are larger and show higher sequence identity compared to inactive hotspots (Kolmogorov-Smirnov test, p = 0.0022) (Supplementary Figure 5). Interestingly, eight regions were identified that showed no evidence of copy number variation in cases or controls despite the presence of large, highly similar, and directly oriented SDs at their flanks (Supplementary Table 10). These may be regions that are mutationally active but in which dosage imbalance is lethal (e.g. 7p14.3 flanked by 19.9 kbp duplications and containing BBS9 and BMPER).
In addition to identifying new potentially pathogenic loci, the large number of cases provides the opportunity to identify atypical deletions (i.e. characterized by noncanonical breakpoints and likely generated by a non-NAHR mutational mechanism) and refine the critical region of known genomic disorders. For example, we identified three individuals with smaller, atypical deletions within the 17q21.31 microdeletion syndrome region18,24,25 (Figure 3B). These patients’ breakpoints contrast with those of 23 patients carrying the canonical 480 kbp deletion mediated by unequal crossover between directly orientated SDs—a genomic architecture largely restricted to individuals of European descent26. Detailed clinical information on two individuals with the atypical deletion (Figure 3C), showed strong phenotypic similarity with the known syndrome including a pronounced philtrum, epicanthic folds, cupped ears and skeletal defects of the hand (Supplementary Note, Supplementary Table 11). The strong phenotypic similarity refines the dosage-sensitive region to only three genes (Figure 3B), including MAPT, which is disrupted by one of these atypical deletions.
Gene content analysis
Encouraged by the additional refinement provided by atypical deletion events, we performed a gene-based analysis on the complete ID/DD dataset, as well as on patient subsets partitioned by additional phenotypic data. We identified 615 genes as significantly deleted in any phenotype (Benjamini-Hochberg corrected p < 0.05; Supplementary Table 12), the vast majority of which associated with known pathogenic loci or subtelomeric alterations. An Ingenuity Pathways Analysis (IPA) (www.ingenuity.com) showed significant enrichment in expected functional categories (e.g. cardiovascular disease, developmental, endocrine system and developmental disorders).
We then expanded our analysis to include candidate associations with nominal significance, as the above analysis is likely to be overly conservative due to the high level of dependence between neighboring genes. An IPA of genes with a nominal p < 0.02 identified the same functional categories as above suggesting that a large proportion of the nominally significant genes are likely relevant to morbidity. In addition to identifying genes within known genomic disorders, this analysis identified genes outside of these intervals. For example, we observed an excess of smaller deletions of SCN1A specifically in patients with epilepsy (p = 0.019), consistent with the literature27. CD44 deletions on 11p13 are significant in craniofacial cases (p = 0.010) and have previously been linked to cleft lip and palate in SNP and expression microarray studies28,29. A region on 9p24 containing five genes is significant in craniofacial cases, with the peak significance focused at SLC1A1 (peak p = 0.00172), a high affinity glutamate transporter previously implicated in multiple neurological conditions30. This peak, specific to SLC1A1, is also significant in neurological, craniofacial and epilepsy cases. A 2q37 deletion immediately proximal to the 2q37 deletion region (Table 1) containing 15 genes is enriched primarily in neurological (modal p = 0.00479) and epilepsy (modal p = 0.00542) phenotypes and contains genes associated with neurodevelopmental and sleep phase disturbances (GBX2 and PER2)31,32. Finally, the deletion of PARD3 is significant in autism (p = 0.01023). PARD3 has been previously associated with bipolar disease33 and is involved in both tight junctions formation and axonal fate determination34.
We also identified 325 duplicated genes (Supplementary Table 12) significantly enriched among the patients (Benjamini-Hochberg corrected p < 0.05). As for deletions, nearly all genes enriched among duplications at this stringent threshold were within known pathogenic duplications and were overrepresented (IPA) in categories that fit well with the expected phenotypic abnormalities (e.g. cardiovascular disease, developmental, endocrine system and developmental disorders). Expanding our analysis to enrichments with nominal significance identified IPA functions identical to the conservative approach as well as several promising candidate gene regions. We observed duplications containing three genes (SH3YL1, ACP1 and FAM150B) on chromosome 2p in cases with craniofacial disorders (p = 0.01032). Notably, large 2p distal duplications have been associated with facial dysmorphism in multiple case reports35,36. Similarly, we observed duplication of two genes (RSPO4 and PSMF1) on distal chromosome 20p in cases with cardiac defects (p = 0.01195), and larger duplications of 20p have been associated with cardiac defects37. The results suggest a potential role for these small subtelomeric regions in disease. Finally, we observed duplication of proximal 8p extending to include two genes in cases with neurological disorders (p = 0.00479), one of which (FNTA) has been shown to be more highly expressed in schizophrenia38.
Discovery of smaller gene-disrupting CNVs
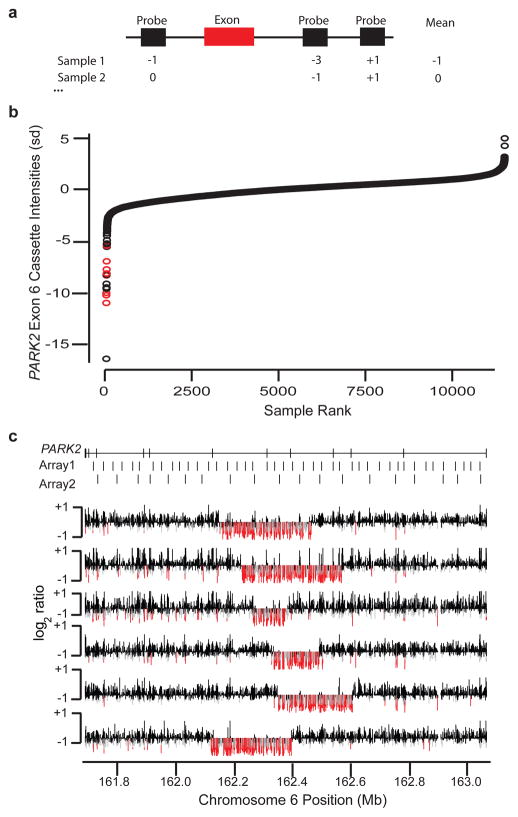
While the data suggest that as much as 14.2% of DD may be explained by large CNVs, many causal mutations remain to be identified. We sought to determine if novel, smaller CNVs could be identified among these patients assuming that breakpoints would not necessarily be recurrent and individually relevant events would be rare (<0.1%); such variants may, in principle, identify novel candidate genes, refine the molecular basis for the phenotypic consequences of larger CNVs, and broaden the predictive power of a given microarray experiment. Therefore, we conducted a directed search for small, exon-affecting CNVs, reasoning that such variants are more likely to have disease relevance and be amenable to follow-up. For each consensus coding sequence (CCDS) exon39, we determined the average intensity for the three closest probes (termed a “cassette”) in each sample and, in turn, identified cassettes exhibiting outlier intensities that may be indicative of deletions (see Methods, Supplementary Figure 6). Note that because this strategy is exon-centric, it is partially platform and breakpoint independent. We analyzed 186,014 autosomal coding exons using 65,704 cassettes (multiple exons are often targeted by the same cassette), excluding exons within known common CNVs16,40,41. After a series of data normalization and quality-control steps, we identified 829 cassettes in which a small (10–100) set of samples exhibited probe intensities that clustered well below the population-wide mean. Each of these was manually reviewed to eliminate artifacts and select for genes with greater potential for disease involvement; 19 were selected for follow-up and organized into two subjectively defined tiers of quality (Table 3).
Table 3.
Validation of smaller deletions.
| Chrom | Start | Stop | Gene | Confirmation | Identical BP |
|---|---|---|---|---|---|
| Tier 1 | |||||
| chr12 | 113316929 | 113317081 | TBX5 | 3 of 4 | Ambiguous |
| chr1 | 40001351 | 40013297 | BMP8 | 6 of 6 | Ambiguous |
| chr1 | 233932670 | 233932900 | LYST | 6 of 6 | Yes |
| chr12 | 12868741 | 12873755 | DDX47 | 6 of 6 | Yes |
| chr11a | 43729037 | 43732247 | HSD17B12 | 6 of 6 | Yes |
| chr20 | 45205105 | 45205194 | EAB1 | 6 of 6 | Yes |
| chr13 | 21173329 | 21173574 | FGF9 | 4 of 6 | Yes |
| chr6 | 162314324 | 162314439 | PARK2 | 6 of 6 | No |
| chr9a,b | 93525765 | 93527210 | NTRKR2 | 6 of 6 | No |
| chr1 | 166548570 | 166548864 | TBX19 | 6 of 6 55 of 58 |
Yes |
| Tier 2 | |||||
| chr18 | 148699 | 148714 | USP14 | 3 of 4 | Yes |
| chr2 | 166518441 | 166518461 | TTC21B | 0 of 5 | NA |
| chr10 | 26889040 | 26896423 | APBB1IP | 2 of 3 | No |
| chr4 | 110114972 | 110115164 | COL25A1 | 4 of 5 | Yes |
| chr4a,c | 77301890 | 77308653 | SCARB2 | 2 of 4 | Yes |
| chr9 | 883912 | 884195 | DMRT1 | 5 of 5 | Yes |
| chr12 | 31835960 | 31836367 | H3F3C | 4 of 4 | Yes |
| chr13 | 97907423 | 97907559 | MST3 | 0 of 4 | NA |
| chr9 | 86546627 | 86546662 | NTRK2 | 5 of 5 25 of 40 |
Yes |
Exon-altering variants.
Five samples harbor a non-exonic copy number polymorphism; one sample has a unique, exon-altering deletion.
Overlaps neighboring gene: FAM47D. Note that annotations are based on the UCSC gene model and not RefSeq genes. BP: breakpoints.
Among the “first tier” of predicted deletions, we found that 55 of 58 individual (i.e. sample-level) predictions validated, with at least one validated event for all 10 examined genes, and for the “second tier,” we found that 25 of 40 predictions validated across seven of the nine examined genes. A total of 44 of the validated deletions spanned only a single probe on the originally used array (Supplementary Figure 7). Deletion events at three genes were determined to be polymorphisms42–44. Interestingly, we found PARK2 to contain at least six distinct exon-affecting deletions ranging in size from 118 to 315 kbp (Figure 4, Supplementary Note, Supplementary Figure 8). However, there is no evidence for CNV enrichment at this locus among cases as this phenomenon also holds true for control samples (Supplementary Figure 9), suggesting that PARK2 is a fragile gene prone to recurrent deletion events. We also identified small deletions in TBX5, a gene known to cause Holt-Oram syndrome45 (a disorder characterized by upper limb abnormalities and congenital heart defects; OMIM #142900). We found that 7 of 15 samples predicted to harbor a TBX5 event were fetal samples, a rate significantly greater than the background proportion of fetal samples (13.4%, p = 0.0019), consistent with the observations that TBX5 mutations can result in prenatal abnormalities detectable by ultrasound46.
Figure 4. Discovery of novel, exon-altering CNVs using the Signature CGH data.
(A) For each coding exon (red bar), the three probes (black rectangles) nearest the exon for any given individual are used to define a cassette score. (B) Distribution of cassette intensities for exon 6 of PARK2 are sorted from lowest to highest (measured in standard deviations, Y-axis) across all samples (X-axis). Red points correspond to known, large deletion events that span the exon. (C) Validation results for the most strongly negative samples from (B) not previously known to carry deletions. Log2 ratio values (Y-axis for each individual row) for PARK2 (coordinates on the X-axis) in each of six tested samples are shown. Probes with very low intensities (< −0.5) are colored red, while those with moderately low values (< −0.3) are gray. Locations of PARK2 exons and probes on two of the most commonly used original oligonucleotide arrays are shown at the top.
DISCUSSION
We present one of the largest studies investigating the role of rare CNVs in ID and DD, analyzing data from 15,767 affected individuals and 8,329 controls. These data quantify the massive contribution of large CNVs to pediatric disease, with 25.7% of affected individuals harboring CNVs >400 kbp in contrast with only 11.5% of controls. Disease risk increases steadily in relation to CNV size, with an odds ratio >20 for carriers of CNVs larger than 1.5 Mbp and nearly 50 at a threshold of 3 Mbp. We find that the CNV burden differs significantly depending on the nature of the primary clinical referral, with craniofacial abnormalities and structural defects of the heart being especially enriched for large CNVs relative to epilepsy and autism spectrum disorder (Figure 1, Supplementary Figure 2). As has been observed in model organisms and predicted based on theory47,48, haploinsufficiency appears more common and penetrant than triplosensitivity for severe developmental phenotypes. While this cohort does not represent a random sampling of individuals with ID/DD and includes some individuals without ID or DD, our estimates are likely applicable to ID/DD in general. For example, the average CNV burden across 15 genome-wide studies of ID/DD (combined sample size of 1,021) was estimated to be ~13.7%, similar to our estimate of 14.2%, in a literature survey by Miller et al.49 (note that this estimate was derived by averaging the diagnostic yields for all studies with a genome-wide resolution of 1 Mbp or better as indicated in Table 2 of Miller et al.). Furthermore, the observed enrichment for many loci known to contribute to ID/DD risk (Table 1) and individual genes previously identified to be disrupted among affected individuals (Supplementary Table 12) clearly supports the applicability of the inferences generated here for both ID/DD specifically and neurological disease (e.g. schizophrenia, autism, etc.) in general.
Practically, these data serve as a clinical resource useful in diagnostics (Tables 1 and 2). The large number of controls and cases provides estimates of penetrance for 60 pathogenic CNVs (accounting for ~10% of cases) and sheds light on either ambiguous or previously unknown pathogenic variants, including 14 novel or previously marginally supported CNV loci that collectively represent ~0.7% (112 of 15,767, Table 2 and Supplementary Note) of the individuals studied here. We note that while one CNV-locus (10p15.3 duplications) appeared to be enriched among cases as a result of ancestry differences between cases and controls, the aggregate ethnic composition of the 14 loci in Table 2 matched closely our control dataset (see Supplementary Note, Supplementary Figures 10 and 11), suggesting that population stratification for rare variants is unlikely to explain the enrichment at these loci. The size distribution (median of 940 kb), inheritance rate (15 of 34 tested CNVs are de novo, with at least 1 de novo variant observed in 6 of the 14 loci), and overlap with DECIPHER entries further support the disease risk for these CNV-loci.
Among these potentially novel CNVs, we provide additional support for a genomic disorder mapping to 15q25.2, which we find in five affected individuals (including two affected siblings) and zero controls (Supplementary Figure 12). Our results combined with earlier studies of schizophrenia and autism (four cases vs. zero controls)16 implicate this CNV as a high-risk allele for pediatric neurological disease with variable outcomes (Supplementary Note, Supplementary Table 9) as well as neuropsychiatric disease (p = 0.037). In addition, our data support the pathogenicity of CNVs at 2q13 whose significance was uncertain because they were observed in a small number of control samples50. In our study, we observed 12 deletions (p = 0.032) and 9 duplications (p = 0.022) on chromosome 2q13 in patients but only one deletion in controls. We furthermore find an enrichment of the deletion in cardiovascular cases (peak p = 0.012) and the duplication in cases with craniofacial features (peak p = 0.010). These results are consistent with two previously reported deletion cases with multiple heart defects and two duplication patients with various facial and skeletal features50. Additionally, our data support the pathogenicity of duplications at 16p11.2 (SH2B1), duplications at 15q13.3 (BP3-BP5; CHRNA7), and deletions at 15q11.2 (BP1-BP2; NIPA1). The latter are present in ~1 in 167 affected individuals studied here and, although incompletely penetrant (0.83), are likely strong risk factors for DD in addition to schizophrenia4,51.
Finally, the discovery of atypical and smaller deletions among patients with virtually identical phenotypes helps to refine the smallest region of overlap for known syndromes. The atypical deletions of 17q21.31 exclude deletions of CRHR1 as playing a role in this syndrome (although deletions of long-range regulatory elements that change CRHR1 expression cannot be ruled out) and narrow the likely candidates to three genes, including MAPT, which is disrupted by proximal breakpoints in two cases (Figure 3B). Overall, we identified 615 deleted genes and 325 duplicated genes significantly enriched in cases when compared to controls. The dosage imbalance of these genes should not be considered as proven but rather as candidates with higher prior probability of dosage sensitivity for future studies. It is encouraging that this set includes a number of previously hypothesized and novel associations between genes and particular traits (Supplementary Table 12). In addition, our data show that even older, low-resolution microarray data afford discovery opportunities for CNVs that have not previously been detectable. Indeed, we successfully identified and confirmed dozens of small deletion events, several of which have plausible disease roles (e.g. TBX5 deletions and Holt-Oram syndrome), including many detected by only a single probe in the original microarray experiment. As the underlying raw data from diagnostic laboratories becomes released, prospectively, there will be great potential for finding additional exon-altering deletions. Further validation of these and other novel candidates will yield new insights into the specific phenotypes affected by the loss or gain of individual genes. While most arrays cannot robustly capture the small deletions we identified, such as those adjacent to exons of FGF9 and LYST (associated with Chediak-Higashi Syndrome), control screening using PCR or other targeted high-throughput assays may be used to follow-up individually interesting candidates (Supplementary Note).
We predict that this map of CNVs and potentially dosage-sensitive genes will be invaluable for both clinical and research purposes in the future. For example, Boone et al. used an exon-targeted microarray to identify a number of individual gene disruptions in individuals with ID/DD of plausible but uncertain pathogenicity given their rarity. We find support for a number of these genes, including two—CCREBBP and SLC1A1—that are significantly enriched among individuals here with similar phenotypes to those previously described (Supplementary Note). As genomic discovery efforts—especially exome sequencing—expand, the results described here should prove increasingly important to clinicians and researchers faced with the challenges of linking rare disruptive mutations to pediatric diseases.
METHODS
Cases
Samples from individuals with ID/DD and related phenotypes were submitted to Signature Genomic Laboratories, LLC, mostly from the U.S. and Canada, for clinical microarray-based CGH; a total of 15,767 samples were analyzed and 16,526 rare autosomal CNV calls were detected (Supplementary Table 1) and deposited into dbVar (dbVar study accession nstd54)52. Informed consent was obtained to publish clinical information and photographs and to further characterize the CNVs present in the individuals with detailed information presented in this paper, using a protocol approved by the Institutional Review Board. Although not a random set of children with ID/DD, the presentations are representative of those observed in a clinical diagnostic setting. The majority of the individuals have an ID/DD phenotype; however, clinical features such as craniofacial and skeletal features, growth retardation, cardiovascular and renal defects, hypotonia, speech and motor deficits, hearing impairment, epilepsy, and behavioral problems were also documented. We identified 575 cases with cardiovascular defects, 1,776 cases with epilepsy/seizure disorder, 1,379 cases with autism spectrum disorder, 3,898 cases with craniofacial defects, and 8,772 cases with general neurological defects; many individuals had multiple subclassifications (Supplementary Table 2). Self-reported ethnicity was available for 144 individuals, with 75% (95% CI 67.3–81.4%, 108/144) reporting Caucasian (primarily European descent), 6.9% (95% CI 3.8–12.3% 10/144) African American, and 18.1% (95% CI 12.6–25.1% 26/144) as other. These samples were analyzed across nine custom array-CGH platforms, with most tested on an Agilent array with ~97,000 probes (Supplementary Figure 13).
Controls
Controls were not ascertained specifically for neurological disorders, but all were obtained from adult samples providing informed consent so developmental disorders should be exceedingly rare. Of individuals with known ethnicity, 81.2% are Caucasian (primarily European descent), 2% are African/African American, and 16.5% are other/mixed ancestry. Due to the slight enrichment of African-American cases compared to our control samples, we modeled the potential impact of large CNV stratification and found no evidence for an overall enrichment of unique large CNVs in the African cohort (Supplemental Figure 10). DNA was obtained from cell lines and blood-derived samples generated for association studies of various phenotypes. Datasets are detailed in Supplemental Table 4. Data were obtained from the following sources: HGDP16,53; NINDS (dbGaP accession no. phs00008916,54; PARC/PARC2)55,56; London (parents of asthmatic children)15; FHCRC (pre-release data provided courtesy of Aaron Aragaki, Charles Kooperberg, and Rebecca Jackson as part of an ongoing genome-wide association study to identify genetic components of hip fracture in the Women's Health Initiative); InCHIANTI (data provided by InCHIANTI study of aging; http://www.inchiantistudy.net15,57); and WTCCC2 (NBS)58. Control CNV arrays were analyzed as described previously16. Briefly, a Hidden Markov Model (HMM) based on both allele frequencies and total intensity values was used to identify putative alterations, followed by manual inspection of large CNVs (>100 probes and >1 Mbp) in conjunction with user guided merging of nearby (<1 Mbp between for arrays with <1 million probes and <200 kbp for arrays with >1 million probes) calls, which represent a single region broken up by the HMM, or gaps. All samples on arrays with densities <1M probes were filtered by a maximal genome-wide LogR standard deviation of 0.25, while the high density 1.2 million probe WTCCC2 data was filtered using an increased standard deviation cut-off of 0.37. Large alterations with noncanonical allele frequencies indicative of mosaics were excluded due to the high likelihood of these resulting from cell culture immortalization. For the two datasets where the Illumina array mapping corresponded to build35 (NHGRI), we utilized the autosomal calls generated previously16 and mapped the coordinates to build36 using the UCSC LiftOver tool (http://genome.ucsc.edu).
Multi-platform CNV comparison
Microarray platform heterogeneity may yield false CNV enrichments signals as a function of differential detection power related to probe density, data quality, analysis methods, etc. We made a number of efforts to control for such potential effects and believe our study design is robust to this source of error for a number of reasons. First, the control data for this study were generated on higher resolution platforms (317,000 to 1,200,000 probe Illumina SNP arrays, with 88% of controls being profiled on 550,000 probe or higher density platforms) compared to the case data (median array is ~97,000 probes, highest density is ~130,000). As a result, our CNV detection power is substantially higher for cases than controls; notably, such differences will tend to manifest as false positive enrichments for CNVs in controls while we are focused exclusively on enrichments within cases. Second, we rigorously eliminated potential sources of errors in the case CNV data with a combination of both manual and automated filters, including calls with low probe counts, high degrees of overlap with segmental duplications in the reference assembly, and likely reference-sample CNVs. Third, for the sliding window enrichment tests we eliminated all CNVs in cases that spanned fewer than 10 probes on the lowest resolution (HH317K) control SNP array. Fourth, we have validated 402/425 CNVs and determined the precision in cases to be high in general (0.945) and higher in cases relative to controls (0.892). Fifth, we specifically analyzed the 14 potentially pathogenic CNVs (Table 2) for control SNP microarray performance. 11/14 loci harbored small CNV calls within the region of interest from multiple control studies; as CNV calling algorithms tend to demonstrate increased sensitivity to larger alterations, we consider this to indicate sufficient control sensitivity within these loci to detect large CNVs. The remaining three loci are split between the minimal common region on 1q24.3, which demonstrates a single 72 kbp CNV in controls (again suggesting detectability of larger events), and two loci that harbor very small CNVs detectable only on the highest resolution 1.2M probe arrays. These two regions have high probe coverage on the 550K control array (46 probes within the smallest 6p22.3 Signature call and 40 probes in the MCR of 2q24.3). Further, all of these regions demonstrate de novo CNVs in our samples, supporting the hypothesis that these are pathogenic loci and not simply common copy number variants that we failed to detect with SNP platforms.
Control CNV burden
Control CNVs were merged into CNVRs by comparing each CNV to all of its overlapping partners and merging those with 50% reciprocal overlap. These CNVRs were then analyzed in the context of sliding 300 kbp genomic windows to identify regions of high variability (Supplementary Figure 9, Supplementary Table 13). Regions of high SNP diversity were obtained from Kidd et al.44 and used to identify regions where the breakpoint variability is likely to result from general sequence variation (such as the HLA locus on 6p). To perform a gene-based search for highly variable loci, we first generated a merged RefSeq list that combined overlapping splice variants into a single, large gene definition. We then analyzed these loci in the context of overlapping gain and loss CNVs that either contained the entire gene, overlapped the transcript (gene breaking or exon hits), or were contained within an intron. Finally, we analyzed each gene in the context of the number of unique CNVRs that overlapped the gene space (exonic or intronic).
Novel, exon-altering CNV discovery
For a subset of 11,529 samples, we identified for each coding exon39 the three closest probes, requiring at least one probe on both sides within 100 kbp of the exon. We required that all probes map within 200 kbp, yielding 65,704 unique cassettes targeting 186,014 autosomal coding exons. We then determined the average cassette intensity for each sample and normalized this by array type. Subsequently, we considered filtered cassettes by the following criteria: 10–100 samples with scores at least 5 standard deviations below average; the subset of samples at less than 5 standard deviations below average compose at least 10% of samples with scores less than 3 standard deviations below average (a measure of cluster separation); and no overlap of the target exon (note, individual probes were not filtered given the heterogeneity of platforms and potential for atypical CNVs) with common copy number polymorphisms or deletions seen in multiple control individuals16,42,43,59. This yielded 829 candidates for follow-up, each of which were manually reviewed to eliminate cassettes in which all candidate deletions clustered within a single array type suggestive of a batch artifact and noisy cassettes resulting from probes embedded within SDs (for examples, see Supplementary Figure 6). Subsequently, 19 cassettes were chosen for validation, manually divided into two qualitative tiers based on the totality of the evidence (follow-up potential of the affected gene, visual analysis of probe intensity distributions, etc.). We designed a custom NimbleGen oligonucleotide array, spanning each of the 19 genes and their flanks at very high density (Supplementary Note), and performed CGH on 98 samples, chosen by cassette score and availability and predicted to carry a deletion at one of the 19 genes.
Supplementary Material
Acknowledgments
We thank Niklas Krumm, Maika Malig, Laura Vives, and Jason Luu for assistance in validation experiments. We also thank Megan Dennis, Can Alkan, Emre Karakoc and Tonia Brown for useful discussions and editing the manuscript. B.P.C. is supported by a fellowship from the Canadian Institutes of Health Research. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under awards 076113 and 085475. We would also like to thank Aaron Aragaki, Charles Kooperberg, and Rebecca Jackson for access to SNP data (FHCRC control dataset) generated as part of an ongoing Genome-wide Association Study to Identify Genetic Components of Hip Fracture in the Women's Health Initiative. This work was supported by NIH HD065285 to E.E.E. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION CODES
All CNV calls have been submitted to dbVar under accession nstd54.
AUTHOR CONTRIBUTIONS
This study was designed by G.M.C., B.P.C., S.G., E.E.E., J.A.R., B.C.B., and L.G.S. L.G.S. supervised array-CGH experiments at Signature Genomics. J.A.R. and B.C.B. coordinated clinical data collection. G.M.C. and B.P.C. performed data analysis and curated control CNV data. SG curated genomic disorders data. S.G., T.V., and C.B. performed array CGH and PCR validations. C.W., H.S., R.H., V.H., H.A.H., P.B., E.M., D.N., K.L., H.T., M.H., N.A., J.G., J.K., V.S., K.J., and C.R. provided clinical information. G.M.C., B.P.C., S.G. and E.E.E. wrote the manuscript. All authors have read and approved the final version of the manuscript.
COMPETING FINANCIAL INTERESTS
E.E.E. is a member of the Scientific Advisory Board of Pacific Biosciences. J.A.R., B.C.B., and L.G.S are employees of PerkinElmer.
References
- 1.Greenway SC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–5. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mefford HC, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–69. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebat J, et al. Strong Association of De Novo Copy Number Mutations with Autism. Science. 2007 doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 6.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girirajan S, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mefford HC, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Bon BW, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–23. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shprintzen RJ. Velocardiofacial syndrome and DiGeorge sequence. J Med Genet. 1994;31:423–4. doi: 10.1136/jmg.31.5.423-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karayiorgou M, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–6. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coe BP, et al. Resolving the resolution of array CGH. Genomics. 2007;89:647–53. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008 doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itsara A, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–81. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itsara A, et al. Population analysis of large copy number variants and their relationship to hotspots of human genetic disease. American Journal of Human Genetics. 2009 doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries BB, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–16. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006;7:407–42. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 19.Firth HV, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mefford HC, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009 doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–5. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochukova EG, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helbig I, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–2. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 25.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–7. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 26.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–83. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suls A, et al. Microdeletions involving the SCN1A gene may be common in SCN1A-mutation-negative SMEI patients. Hum Mutat. 2006;27:914–20. doi: 10.1002/humu.20350. [DOI] [PubMed] [Google Scholar]
- 28.Baroni T, et al. Human cleft lip and palate fibroblasts and normal nicotine-treated fibroblasts show altered in vitro expressions of genes related to molecular signaling pathways and extracellular matrix metabolism. J Cell Physiol. 2010;222:748–56. doi: 10.1002/jcp.22006. [DOI] [PubMed] [Google Scholar]
- 29.Park JW, et al. High throughput SNP and expression analyses of candidate genes for non-syndromic oral clefts. J Med Genet. 2006;43:598–608. doi: 10.1136/jmg.2005.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCullumsmith RE, Meador-Woodruff JH. Striatal excitatory amino acid transporter transcript expression in schizophrenia, bipolar disorder, and major depressive disorder. Neuropsychopharmacology. 2002;26:368–75. doi: 10.1016/S0893-133X(01)00370-0. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Chatterjee M, Li JY. The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J Neurosci. 2010;30:14824–34. doi: 10.1523/JNEUROSCI.3742-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 33.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem. 2004;279:12804–11. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- 35.Stalker DJ, Vigneswaren S, Sharples PM, Lunt PW. Distal trisomy 2p and arachnodactyly. J Med Genet. 2000;37:974–6. doi: 10.1136/jmg.37.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, Batista DA, Maumenee I, Wang T. An unbalanced translocation between chromosomes 2p and 6p associated with Axenfeld-Rieger anomaly type 3, hearing loss, developmental delay, and distinct facial dysmorphism. Am J Med Genet A. 2010;152A:1318–21. doi: 10.1002/ajmg.a.33355. [DOI] [PubMed] [Google Scholar]
- 37.Chaabouni M, et al. De novo trisomy 20p of paternal origin. Am J Med Genet A. 2007;143A:1100–3. doi: 10.1002/ajmg.a.31704. [DOI] [PubMed] [Google Scholar]
- 38.Bowden NA, Scott RJ, Tooney PA. Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genomics. 2008;9:199. doi: 10.1186/1471-2164-9-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruitt KD, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–23. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 41.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 42.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 43.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basson CT, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–5. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 46.Brons JT, et al. Prenatal ultrasound diagnosis of the Holt-Oram syndrome. Prenat Diagn. 1988;8:175–81. doi: 10.1002/pd.1970080303. [DOI] [PubMed] [Google Scholar]
- 47.Turner DJ, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–5. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher E, Scambler P. Human haploinsufficiency--one for sorrow, two for joy. Nat Genet. 1994;7:5–7. doi: 10.1038/ng0594-5. [DOI] [PubMed] [Google Scholar]
- 49.Miller DT, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudd MK, et al. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum Mol Genet. 2009;18:2957–62. doi: 10.1093/hmg/ddp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 54.Simon-Sanchez J, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 55.Albert MA, Danielson E, Rifai N, Ridker PM. Effect of statin therapy on C-reactive protein levels: the pravastatin inflammation/CRP evaluation (PRINCE): a randomized trial and cohort study. JAMA. 2001;286:64–70. doi: 10.1001/jama.286.1.64. [DOI] [PubMed] [Google Scholar]
- 56.Simon JA, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–50. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 57.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Consortium WTCC. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–20. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.