Abstract
Antigenic variation is an immune evasion strategy used by African trypanosomes, in which the parasites periodically switch the expression of VSG genes that encode their protective variant surface glycoprotein coat. Two main routes exist for VSG switching: changing the transcriptional status between an active and an inactive copy of the site of VSG expression, called the bloodstream VSG expression site, or recombination reactions that move silent VSGs or VSG copies into the actively transcribed expression site. Nothing is known about the proteins that control and catalyze these switching reactions. This study describes the cloning of a trypanosome gene encoding RAD51, an enzyme involved in DNA break repair and genetic exchange, and analysis of the role of the enzyme in antigenic variation. Trypanosomes genetically inactivated in the RAD51 gene were shown to be viable, and had phenotypes consistent with lacking functional expression of an enzyme of homologous recombination. The mutants had an impaired ability to undergo VSG switching, and it appeared that both recombinational and transcriptional switching reactions were down-regulated, indicating that RAD51 either catalyzes or regulates antigenic variation. Switching events were still detectable, however, so it appears that trypanosome factors other than RAD51 can also provide for antigenic variation.
Keywords: Antigenic variation, trypanosome, recombination, RAD51
African trypanosomes, such as Trypanosoma brucei, are single-celled eukaryotic parasites of mammals that reside in the bloodstream and tissue fluids and cause the disease trypanosomiasis, or sleeping sickness in humans. To avoid being eliminated by the host's immune response, trypanosomes have evolved an evasion strategy termed antigenic variation. It is now recognized that antigenic variation is a general survival strategy used by a diverse array of pathogenic microorganisms, and a wide spectrum of molecular mechanisms are used to this end (Borst and Greaves 1987; Deitsch et al. 1997). In African trypanosomes, this consists of spontaneous, periodic changes in the Variant Surface Glycoprotein (VSG) species that acts as a protective coat on the surface of the parasite (Cross 1975). A change of VSG coat by a subpopulation of infecting trypanosomes allows a portion of the trypanosome population to escape the host's immune response directed against the previous VSG species, thereby prolonging the infection. For recent reviews of trypanosome antigenic variation see Cross (1996), Barry (1997), Borst et al. (1998), Cross et al. (1998), Pays and Nolan (1998).
Each trypanosome normally expresses a VSG coat composed of only one VSG species at a given time. It can select that single VSG species from ∼1000 VSG genes present within its genome (van der Ploeg et al. 1982). To achieve exclusive expression, a VSG can only be transcribed if it resides within a specialized telomeric transcription unit called the expression site (Johnson et al. 1987; Kooter et al. 1987; Pays et al. 1989). The expression sites used in the bloodstream are very large (40–60 kb), and contain a number of expression site-associated genes (ESAGs) in addition to the VSG itself (Cully et al. 1985; Xong et al. 1998). Each cell contains ∼20 bloodstream expression sites, but only one is transcribed at a time (Navarro and Cross 1996; Chaves et al. 1998). The remainder of the 1000 VSGs act as a reservoir of silent genes, and must be moved into the expression sites before they can be expressed.
Because of the complex arrangement of VSG expression described above, a switch from one VSG coat to another can be accomplished in a number of ways. One way is by activating transcription from one of the silent VSG expression sites and silencing transcription from the active site. This reaction is termed an in situ switch, and recent work has suggested that it may involve epigenetic mechanisms of transcriptional control (Horn and Cross 1995; Rudenko et al. 1995). The other way to execute a switch is by DNA recombination reactions that move a silent VSG into the active expression site and remove the resident VSG. A number of such reactions have been described. The most commonly observed reaction is a gene conversion process, termed duplicative transposition, in which the VSG in the expression site is deleted and replaced by a copy made from a silent VSG (Hoeijmakers et al. 1980; Bernards et al. 1981; Pays et al. 1981, 1983a). Silent VSGs in a number of genomic locations can act as gene donors in this reaction: transposition of genes from silent expression sites, from silent chromosome-internal arrays of VSGs, and from the telomeres of minichromosomes (of which ∼100 are present) have been described (for review, see Borst et al. 1996; Barry 1997). The amount of VSG sequence copied during duplicative transposition is variable and normally extends beyond the boundaries of the VSG ORF. A number of sequence motifs around the VSG might provide the sequence homology that drives the recombination reaction. 3′ of the gene, and within the carboxyl terminus of the ORF, short stretches of sequence homology have been found in most VSGs (Liu et al. 1983; Michels et al. 1984); further downstream, telomere (and other) repeats could provide sequence homology if the VSG donor is also telomeric (de Lange et al. 1983; Aline et al. 1989; Scholler et al. 1989). 5′ of almost all VSGs are runs of imperfect 70-bp repeats, and this normally provides the only upstream homology for transposition of chromosome-internal genes (Liu et al. 1983; Campbell et al. 1984); if the recombination occurs between two expression sites, however, the ESAGs and surrounding expression site sequence can provide many kilobases of upstream homology (Laurent et al. 1983; Pays et al. 1983b; Myler et al. 1984). Another form of recombination is also possible for VSG switching, but has been less commonly observed. This is a reciprocal genetic exchange that occurs upstream of the VSG and moves the active gene into a silent telomere, and moves a silent telomeric VSG into the active expression site (Pays et al. 1985; Rudenko et al. 1996).
Nothing is known about the trypanosome genes that regulate antigenic variation, or encode the enzymes that catalyze the switching reactions. Moreover, it is not known what pathways of recombination are involved. For example, we do not know whether a classical double-strand break repair mechanism (Shinohara and Ogawa 1995; Eggleston and West 1996) is involved, or whether more specialized means of genetic exchange are utilized (perhaps analogous to intron mobility or yeast mating-type switching; Grivell 1996; Haber 1998). There has been considerable debate over the event(s) that initiates recombinogenic VSG switching: Is it due to random DNA breaks around the VSG (Borst et al. 1996), or is it a result of specific breaks catalyzed by a discrete enzyme (Barry 1997)? To address these questions, this study describes the identification of a T. brucei gene putatively encoding a homolog of eukaryotic RAD51 and analysis of the role that this enzyme plays in antigenic variation.
RAD51 is a eukaryotic homolog of bacterial RecA, and is highly conserved in a diverse array of eukaryotes (Basile et al. 1992; Shinohara et al. 1992, 1993). Like RecA, RAD51 has a pivotal role in the repair of double-strand DNA breaks and in homologous recombination. Comparison of the catalytic properties of purified Escherichia coli RecA with RAD51 from Saccharomyces cerevisiae and Homo sapiens has shown that the enzymes are functionally related. All of the enzymes bind single- or double-stranded DNA in the presence of ATP and form nucleoprotein filaments in which the DNA is extended and underwound relative to its normal structure (Ogawa et al. 1993b; Benson et al. 1994; Sung and Robberson 1995). On relaxed double-stranded DNA, the three filaments have a highly similar structure. All of the enzymes display ATPase activity in the presence of single-stranded DNA and, on binding, can catalyze the transfer of a single-stranded DNA end into duplex DNA. By use of ATP-hydrolysis, the enzymes then generate homology-dependent strand exchange products in which the single-stranded DNA is basepaired with the complementary strand of the duplex and displaces the noncomplementary strand (Sung 1994; Baumann et al. 1996; Baumann and West 1997). In performing these strand transfer and exchange reactions, RAD51 and RecA initiate the process of genetic exchange and ensure that it occurs between homologous DNA sequences. By examining the role of RAD51 in trypanosome antigenic variation, we assess whether VSG switching is effected by homologous recombination or some other pathway of genetic exchange, and begin to examine the pathways of recombination that are available to this ancient eukaryotic parasite.
Results
Cloning the trypanosome RAD51 gene
The amino acid sequences of RAD51 proteins from a diverse range of eukaryotes have been shown to be highly conserved, particularly in a central domain of ∼200 residues that has sequence similarities with bacterial RecA (Ogawa et al. 1993a; Shinohara et al. 1993; Dunderdale and West 1994). This high level of conservation allowed us to isolate a gene from T. brucei putatively encoding a homolog of RAD51. A comparison of the predicted T. brucei polypeptide with RAD51 sequences from a number of eukaryotes is shown in Figure 1. Southern mapping (Fig. 2a) suggested that the putative RAD51 gene is present in single copy within the T. brucei genome, and probing of poly(A)+ RNA from bloodstream and procyclic (tsetse fly midgut) T. brucei suggested that RAD51 is expressed at a low level in both stages as a 2.4-kb mRNA (data not shown).
Figure 1.

Alignment of the amino acid sequences of some eukaryotic RAD51 proteins. Amino acid residues are shown in single letter code, and the predicted T. brucei polypeptide (bottom) is compared with RAD51 polypeptide sequences from H. sapiens (EMBL accession no. D13804), X. laevis (D38488), Schizosaccharomyces pombe (D13805), S. cerevisiae (M88470), and Drosophila melanogaster (L41342). Numbers correspond to the largest polypeptide (from S. cerevisiae), and residues that are identical in all the sequences are in a black background. Black dots identify amino acid residues shown to be invariant or highly conserved in the active site of E. coli RecA, and the peptides identified as A and B are putative conserved nucleotide-binding motifs. The region identified under the T. brucei sequence corresponds to the region of the RAD51 gene that was PCR amplified for cloning the complete gene; the solid lines show the conserved regions used for generating primers, and the dotted line corresponds to the region that was amplified.
Figure 2.
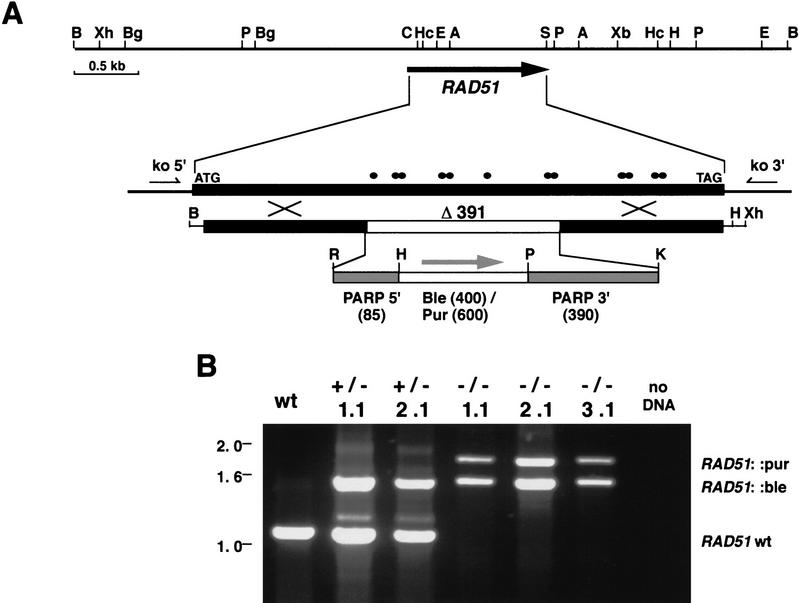
Generation of trypanosome RAD51 knockout mutants. (A) A restriction enzyme map of the genomic environment of the RAD51 ORF and, below, the constructs generated to disrupt RAD51 expression. Highly conserved residues in the RAD51 amino acid sequence are shown as black dots, and the 300-bp RAD51 5′ and 3′ sequences that allow for integration of the constructs are indicated with crosses. An amount of 391 bp of the RAD51 ORF is replaced by expression cassettes that give phleomycin (BLE) or puromycin (PUR) resistance; these contain 5′ and 3′ sequences from the PARP locus (gray shading) that allow transcripts of the resistance genes to be processed into mature mRNAs. Restriction enzyme recognition sites, (A) AccI; (B) BamHI; (Bg) BglII; (C) ClaI; (E) EcoRV; (H) HindIII; (Hc) HincII; (K) KpnI; (P) PstI; (S) SalI; (R) EcoRI; (Xb) XbaI; (Xh) XhoI. (B) PCR analysis of bloodstream T. brucei transformed with RAD51 knockout constructs. PCR used the primers ko5′ and ko3′. Size markers (in kilobases) are indicated at left of the agarose gel. (wt) Untransformed cells; (+/−) cells transformed with the construct containing the BLE cassette; (−/−) cells transformed with both the BLE and PUR cassette constructs. The origins of the PCR products are indicated at right of the gel photograph.
The overall level of sequence identity between the putative trypanosome RAD51 polypeptide and other RAD51s is extremely high: 55% when compared with the S. cerevisiae enzyme, ranging to 67% for the Xenopus laevis protein. In database searches with the trypanosome polypeptide sequence, RAD51 proteins were identified ahead of other RecA homologs that have been discovered in eukaryotes. Therefore, it seems likely that the gene cloned encodes a bonafide RAD51 homolog, rather than one of a number of related eukaryotic proteins, which appear to have no role in general homologous exchange (e.g., DMC1; Bishop et al. 1992; Pittman et al. 1998; Yoshida et al. 1998), or have accessory functions in RAD51 recombination (e.g., RAD55 and 57; Sung 1997). Two nucleotide-binding consensus motifs have been identified within the published RAD51-like sequences (Bishop et al. 1992; Shinohara et al. 1992), and both of these are conserved within the T. brucei polypeptide. An exhaustive comparison by Story et al. (1993) of 23 bacterial RecA sequences, the DMC1 protein of S. cerevisiae and the bacteriophage T4 UVSX protein identified 12 invariant or highly conserved residues. From the three-dimensional structure of E. coli RecA, these residues are believed to be involved in the enzyme's active site, and all are conserved in the putative T. brucei enzyme. This suggests that the trypanosome will encode a polypeptide with RecA/RAD51 enzymatic properties.
Generation of T. brucei RAD51 knockout mutants
The strategy that we used to examine the function of RAD51 was to create transgenic trypanosomes that no longer express active RAD51. To make inactive derivatives of the RAD51 gene, we generated constructs that deleted the central 391 bp of the ORF and replaced them with cassettes for bleomycin phosphotransferase (providing phleomycin resistance; BLE) and puromycin N-acetyltransferase (puromycin resistance; PUR) expression (Fig. 2a). After transforming these constructs into T. brucei, the RAD51 sequences flanking the expression cassettes provide sequence homology, enabling them to integrate into, and replace, the endogenous RAD51 locus. The RAD51 deletion encompasses 8 of the 12 conserved RecA catalytic residues, and both of the putative ATP-binding motifs, so we predicted that any residual RAD51 peptide expressed should be inactive. A similar strategy for disruption of the mouse RAD51 gene deleted a smaller region of the ORF than described here, and created a lethal phenotype (Lim and Hasty 1996; Tsuzuki et al. 1996).
Trypanosomes are predominantly diploid, and so we disrupted both RAD51 alleles by performing two rounds of transformation; first, the RAD51::BLE construct was electroporated and then, once these transformants were established, the RAD51::PUR construct was transformed. Figure 2b shows PCR analysis of bloodstream transformants with primers that flank the RAD51 ORF. In the first round transformants, a 1.10-kb PCR product from the RAD51 wild-type allele and a 1.57-kb product corresponding to the integrated BLE cassette were detected, suggesting that the construct had integrated into one allele to generate putative heterozygous mutants (RAD51+/−). A 1.80-kb product corresponding to the integrated PUR cassette was seen in the second round transformants, and there was no longer any sign of the PCR product from the wild-type allele, suggesting that integration of both cassettes had removed RAD51 wild type from this locus. Note that the two second round transformants shown here (and a third discussed below) were generated completely independently; each was created in a separate experiment, from different starting populations of untransformed cells, involving two rounds of transformation and cloning. Southern blots of genomic DNA from the transformants, when probed with RAD51 wild type sequence (data not shown), confirmed that no intact RAD51 alleles were present in the putative homozygous mutants (RAD51−/−). Identical results were obtained for procyclic transformants (data not shown), suggesting that the RAD51 gene can be disrupted in both life-cycle stages and the cells remain viable.
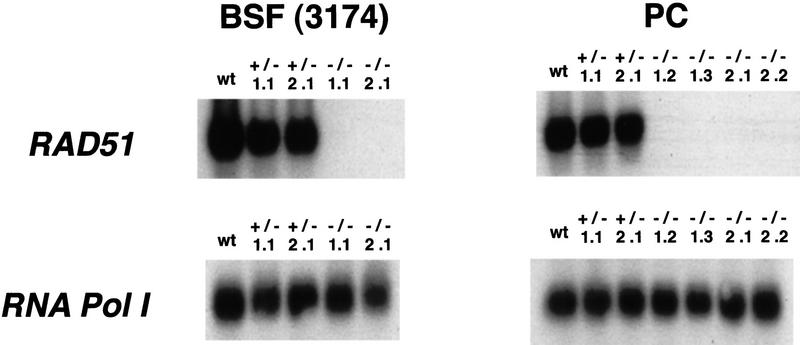
To check that there was no wild-type RAD51 mRNA being made in the putative homozygous mutants, we isolated total RNA from the various transformants, generated cDNA by reverse transcription, and performed PCR (Fig. 3). The PCR used primers complementary to RAD51 ORF sequences outside of the deleted region, and so even a small amount of residual wild-type mRNA should be detected. For both life-cycle stages a PCR product of the expected size was readily amplified from both the RAD51 wild-type and RAD51+/− cells, but none could be detected in the RAD51−/− cells. We therefore conclude that it is possible to remove all transcripts that can express more than the first third of the RAD51 protein and trypanosomes remain viable.
Figure 3.
Reverse transcription PCR of trypanosome RAD51 transformants. cDNA representing the RNA from bloodstream form (BSF; these are the transgenic line 3174) and procyclic (PC) transformants was tested by PCR for the presence of either RAD51 or, as a control for the integrity of the substrate, RNA polymerase I (RNA pol I) mRNA. The PCR reactions were separated by agarose gel electrophoresis, Southern blotted, and probed with either the RAD51 or RNA Pol I ORF. (wt) Untransformed cells; heterozygous or homozygous RAD51 transformants are indicated by +/− and −/−, respectively.
Trypanosome RAD51−/− cells are more sensitive to DNA damage than RAD51 wild-type or RAD51+/− cells
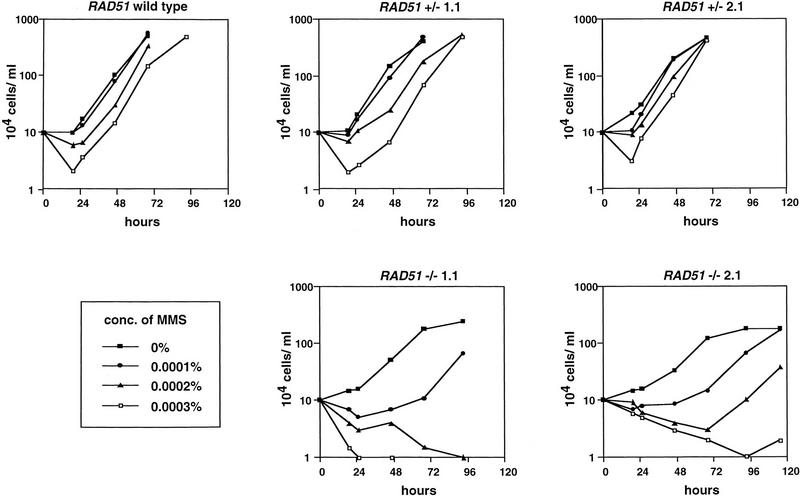
Homologous recombination is one of a number of DNA repair pathways that are available to cells to reverse the potentially lethal effects of damage to their genomes. Therefore, it has been observed that RAD51 mutants in yeast, and recA mutants in bacteria, display increased sensitivity to DNA damaging agents such as ultraviolet (UV) light or the radiomimetic alkylating agent methyl methanesulphonate (MMS; Ogawa et al. 1993a). To test whether or not our putative trypanosome RAD51 mutants display a phenotype consistent with the loss of active RAD51, we assayed the transformants for their sensitivity to such agents. Figure 4 shows growth curves of RAD51 wild-type cells and two independently generated RAD51+/− and RAD51−/− bloodstream cell lines in the presence of increasing concentrations of MMS.
Figure 4.
Growth of RAD51 knockout trypanosomes in the presence of MMS. Bloodstream trypanosomes that had been unaltered in their RAD51 gene (RAD51 wild type) had one allele disrupted (RAD51+/−) or had both alleles disrupted (RAD51−/−) were grown in medium containing increasing concentrations of MMS (as shown). The concentration of trypanosomes in the medium was counted at the given times.
It is apparent that, in the absence of mutagen (in this case MMS), the RAD51−/− cells have a reduced growth rate relative to the RAD51 wild-type or RAD51+/− cells, and reach slightly lower maximal densities in culture medium. The average population doubling time during exponential growth in vitro was 9.6 hr for the RAD51 wild-type cells and 8.1 and 8.7 hr for the RAD51+/− cell lines, compared with 10.6 and 11.1 hr for the RAD51−/− cell lines. In other words, the RAD51−/− mutation increased the generation time of the cells in vitro by 12% to 30% (relative to wild-type or +/−). Next, we checked whether the RAD51−/− cells are capable of establishing an infection in mice, where they are exposed to host defenses (Table 1). The reduction in growth rate was not as severe during in vivo growth as was seen in vitro, but was still apparent: The RAD51−/− cells had an increased doubling time of 10% to 14% relative to the wild-type or +/− cells in either immunosuppressed or immunocompetent hosts. Although significant, this is a modest effect on growth when compared with either yeast RAD51 or E. coli recA mutants (which show 60% and 40% increased generation times, respectively; Capaldo et al. 1974; Fingerhut et al. 1984). We assume that this is a population phenomenon, whereby a larger portion of the RAD51−/− than the RAD51 wild-type or RAD51+/− cells fails to complete cell division, although we have not excluded that individual RAD51−/− cells simply take longer to undergo their cell cycle. Clearly, however, a RAD51 mutation in trypanosomes is not lethal, as it is mice (Lim and Hasty 1996; Tsuzuki et al. 1996), because the cells are capable of replicating efficiently in vivo.
Table 1.
Population doubling times (hr) of bloodstream trypanosome RAD51 knockouts in mice
| Strain
|
Immunocompromised
|
Immunocompetent
|
|---|---|---|
| RAD51 wild type | 7.8 ± 0.3 | 6.6 ± 0.2 |
| RAD51+/− 1.1 | 7.7 ± 0.2 | 6.6 ± 0.2 |
| RAD51−/− 2.1 | 8.8 ± 0.7 | 7.45 ± 0.25 |
| RAD51−/− 3.1 | 8.6 ± 0.5 | 7.3 ± 0.2 |
Comparing the growth curves in increasing MMS concentrations (Fig. 4) shows that the RAD51−/− cells are more sensitive to the drug than either RAD51 wild-type or RAD51+/− cells. Growth of the wild-type and +/− cells was barely affected by 0.0001%MMS, but 0.0002%and 0.0003% caused some growth retardation. In contrast, growth of the RAD51−/− cells was substantially retarded even at 0.0001%, and severely affected at higher concentrations. These results are consistent with the RAD51−/− cells being less able to repair DNA damage caused by MMS. To test this hypothesis further, we compared the cells' sensitivity with a second compound, 3-aminobenzamide. This is a competitive inhibitor of the nuclear enzyme poly (ADP-ribose) polymerase (ADPRT), which is responsible for recognizing single-strand breaks in DNA and coordinating DNA repair (Jeggo 1998). Chemical inhibition of ADPRT causes increased levels of sister chromatid exchange. This has been interpreted as meaning that inhibition prevents normal repair of single-strand breaks, and allows them to be processed to double-strand breaks, which are then the target for homologous recombination (Lindahl et al. 1995). As with MMS, we found that the RAD51−/− cells are more sensitive to 3-aminobenzamide (5–15 mm) than the RAD51 wild-type or RAD51+/− cells during growth (R. McCulloch, unpubl.). Together, these data suggest that the RAD51−/− mutation we have made impairs the ability of the trypanosomes to repair damage to their DNA, which is most simply explained by an absence of functional RAD51 in the cells.
If RAD51 is the trypanosome enzyme responsible for DNA strand transfer and exchange, we would expect that the RAD51−/− cells would be deficient in general homologous recombination. Unlike in bacteria and yeast, in which a plethora of genetic assays have revealed a number of different pathways of genetic exchange, very little is known about the recombination mechanisms used by trypanosomes. However, it appears that integration of DNA into the trypanosome genome occurs only into sites with homologous sequences, as no examples of nonhomologous insertions have been described (Lee and van der Ploeg 1990; Eid and Sollner-Webb 1991; Blundell et al. 1996). Therefore, we sought to determine whether the RAD51−/− mutation results in a deficiency in the integration of transformed DNA. This was first assayed by measuring the transformation efficiencies of procyclic cell lines electroporated with the construct pUCtubneo3 (Ten Asbroek et al. 1990), which integrates a neomycin phosphotransferase cassette into the tubulin gene array. For both RAD51 wild-type and RAD51+/− cells the measured transformation efficiencies were between 1 × 10−5 and 0.3 × 10−5. In contrast, the maximum transformation efficiency obtained for the RAD51−/− cells was 1 × 10−6, indicating a reduction of at least 10-fold. The bloodstream cells used here already contain an integrated neomycin resistance cassette (see below), making pUCtubneo3 transformation impossible. Instead, we used clonal growth on semisolid agarose plates to measure the transformation efficiency of a construct (tub::RAD51–BSR; see below) designed to integrate the complete RAD51 gene into the tubulin locus, allowing its re-expression in the knockout mutants. The RAD51 wild-type cells had an average transformation frequency of 8 × 10−7. In three independent experiments with the RAD51−/−2.1 transformant, each time selecting 1 × 107 cells, we recovered no transformants, indicating a reduction in transformation frequency of at least 24-fold. Bloodstream transformants could, however, be recovered by selecting in liquid medium rather than on agarose plates (see below). Together, these data show that in both life-cycle stages the absence of RAD51 impairs, but does not eliminate, the trypanosome's ability to integrate transformed DNA.
Mutating RAD51 reduces the frequency of trypanosome antigenic variation
To assess the effect that the RAD51−/− mutation has on trypanosome antigenic variation, we made the gene knockouts in a transgenic strain of trypanosomes that allows us to assay for the frequency of VSG switching as well as score the relative contributions of in situ and gene conversion switching reactions. This strain has been described previously (McCulloch et al. 1997); it contains antibiotic resistance markers for hygromycin and G418 within its active expression site (Fig. 5). By immunizing mice against the VSG coat that this strain expresses (called VSG221) and then injecting measured numbers of trypanosomes into the immunized mice, we can estimate the number of trypanosomes that survive the immune selection, and thereby calculate the frequency with which the cells switch to another VSG coat. Furthermore, having isolated and cloned the surviving switched trypanosomes, we can use the resistance markers within the expression site to determine the switching reaction they have undergone (see Fig. 5).
Figure 5.
Use of the transgenic trypanosome line 3174 to characterize VSG switching events. The telomeric region of the actively transcribed expression site in 3174 bloodstream cells is indicated. VSG221 and the most proximal ESAG (ESAG1) are separated by resistance markers for hygromycin (Hyg) and G418 (Neo), which are in turn separated by a stretch of 70-bp repeats; transcription of the site is depicted by a dashed arrow, and the position of primers used to assay for the presence of the genes within the expression site are indicated by small arrows. Three different switching events are observed in this trypanosome strain. In an in situ switch, another expression site (containing an unknown VSG; X) is transcriptionally activated and the VSG221 site is inactivated. These cells are sensitive to Hygromycin (Hygs) and G418 (G418s), and each marker gene can be amplified by PCR (Hyg+, Neo+, 221+). A long range gene conversion event from another expression site deletes all the marker genes from the VSG221 site, and replaces them with VSGX, these are Hygs and G418s, and the genes cannot be amplified by PCR (Hyg−, Neo−, 221−). In a gene conversion event extending from the 70-bp repeats to the 3′ end of VSG221, VSGX replaces only the 221 and Neo markers; these switchers are hygromycin resistant (HygR) and G418s, and are Hyg+, Neo− , 221− in PCR.
A comparison of the switching frequencies measured for the RAD51 transformants is shown in Table 2. The RAD51 wild-type cells underwent antigenic variation at a frequency of ∼0.9 × 10−6 (±0.2) switches/cell/generation. Disruption of one RAD51 allele appeared to have no effect on this switching frequency. In contrast, disruption of both alleles appeared to impair the ability of the cells to undergo switching, because a reduction in the switching frequency of the RAD51−/− cells was apparent in all the measurements made. The extent of this reduction was highly variable, however, and could be as little as 2-fold or as great as 130-fold. The reason for this variability is not clear, but is not due to genetic changes in one particular RAD51−/− transformant: Three independently generated RAD51−/− cell lines were tested, and all displayed the same variable phenotype. A likely explanation is experimental fluctuation in the assay, because switched cells can arise at any cell division during expansion of the population prior to injection into the immunized mice. If a switching event occurred at an early division, this would result in a greater number of that switched variant than if it had occurred at a late division. For the RAD51 wild-type and RAD51+/− cells such fluctuations are likely to be less obvious, because active RAD51 will cause switchers to arise with greater frequency (and hence consistency) at each cell division.
Table 2.
Effect of RAD51 mutation on the frequency of VSG switching in the 3174 trypanosome strain
| Strain
|
No. of injected tryps
|
No. of wells growinga
|
Estimated VSG switching frequency (×10−6)b
|
|---|---|---|---|
| RAD51 | 5 × 107 | 75/96 | 0.95 |
| wild type | 5 × 107 | 56/96 | 0.70 |
| 4.5 × 107 | 76.5/96 | 1.1 | |
| 4.5 × 107 | 72/96 | 1.0 | |
| 5 × 107 | 67/96 | 0.83 | |
| RAD51+/− 1.1 | 5 × 107 | 76/96 | 0.95 |
| 5 × 107 | 53.5/96 | 0.66 | |
| RAD51−/− 1.1 | 5 × 107 | 1.5/96 | 0.02 |
| 5 × 107 | 1.5/96 | 0.02 | |
| 1.6 × 108 | 41/96 | 0.18 | |
| RAD51−/− 2.1 | 5 × 107 | 0.5/96 | 0.007 |
| 5 × 107 | 6.5/96 | 0.09 | |
| 1 × 108 | 2/96 | 0.01 | |
| 1 × 108 | 19/96 | 0.13 | |
| RAD51−/− 3.1 | 7.5 × 107 | 8.5/96 | 0.08 |
| 7.5 × 107 | 43.5/96 | 0.41 | |
| RAD51−/− 2.1 | 5 × 107 | 88/96 | 1.1 |
| tub::RAD51-BSR | 5 × 107 | 82/96 | 1.0 |
| 5 × 107 | 24/96 | 0.3 | |
| 5 × 107 | 25/96 | 0.31 |
Counted 7 days after cloning for wild-type and +/− cells, and up to 10 days after cloning for −/− cells.
Events/cell/generation: determined as described in McCulloch et al. (1997), and as discussed in Materials and Methods.
To test that the reduction in VSG switching frequency is due to the RAD51 mutation and not incidental changes the arose during selection and cloning of the transformants, we examined the effect of re-expressing RAD51 in the homozygous mutants. To do this, we transformed RAD51−/−2.1 cells with the construct tub::RAD51–BSR; this should integrate the complete RAD51 gene into the tubulin locus, with a coexpressed blasticidin resistance cassette allowing selection. Transformation occurred at the expected frequencies in the RAD51 wild-type cells, but was more inefficient in the RAD51−/− cells; no blasticidin-resistant clones were recovered on plate selection (see above), and in nine independent experiments, each selecting 107 cells in liquid medium, only five drug-resistant cell lines arose. Southern analysis showed that only one of the five had integrated the construct into the expected genomic location, and RT–PCR showed that it was expressing intact RAD51 RNA (data not shown). The VSG switching frequency for this clone was ∼0.7 × 10−6 (±0.4) switches/cell/generation (Table 2), indicating that it was the absence of functional RAD51, and no other genetic changes, that caused an impairment in VSG switching in the RAD51−/− cells.
Next, we took a selection of switched variants derived from the RAD51 wild-type cells and from one RAD51−/− cell line and compared the switching mechanisms that they used (Table 3; variants arising from three independent switching experiments were examined). No significant differences in the ratios of the different reactions were observed. For both cell lines, transcriptional switches accounted for around one-half of the total events analyzed, and the remainder were apparent gene conversions that removed the marker genes from the active expression site. These gene conversions can remove both markers, and hence involve extensive changes to the expression site, or can remove only the neomycin marker and VSG221 gene downstream of the 70-bp repeats: the relative amounts of these were also unaltered by the RAD51−/− mutation. These data suggest that both in situ transcriptional switching and gene conversion switching were down-regulated by the RAD51 mutation to cause the observed reduction in the frequency of antigenic variation.
Table 3.
VSG switching mechanisms used by RAD51 wild-type and RAD51 homozygous mutant trypanosomes
| Strain (no. of switched variants analyzed)
|
In situ switch
|
Expression site gene conversion
|
VSG gene conversion
|
|---|---|---|---|
| RAD51 wild type (34) | 20/34 | 9/34 | 5/34 |
| 59% | 26% | 15% | |
| RAD51−/− 1.1 (45) | 23/45 | 14/45 | 8/45 |
| 51% | 31% | 18% |
Types of switching event were assayed as described in Fig. 5 and McCulloch et al. (1997).
Discussion
To begin to determine the genetic requirements of antigenic variation in African trypanosomes, we have cloned a T. brucei gene encoding a homolog of eukaryotic RAD51 and have analyzed its role in this immune evasion process. Our contention that the cloned gene encodes a RAD51-like enzyme is based on phenotypes that we observe when we make trypanosomes mutant in this gene, and also on the very high level of conservation in primary amino acid sequence between the putative T. brucei polypeptide and those characterized from other eukaryotes.
Analysis of the phenotypes of the RAD51−/− mutants indicated that we were able to generate transgenic trypanosomes that lack functional RAD51 enzyme. The RAD51−/− mutation described rendered bloodstream-stage cells more sensitive to two DNA damaging agents tested: methylmethane sulfonate and 3-aminobenzamide. This is consistent with an impairment in the mutant cells' ability to repair DNA damage through homologous recombination. We could not, however, detect the same increased sensitivity to UV light (data not shown). This may be because repair of UV damage can occur efficiently by nucleotide excision repair (Sancer 1994) in trypanosomes, thereby masking any defects in homologous recombination. Alternatively, the simple growth curves we have used may not be a sensitive enough assay, as some yeast RAD51 mutants display only slight UV sensitivity (Game 1983). Genetic transformation of RAD51−/− cells by DNA electroporation was less efficient than either RAD51 wild-type or RAD51+/− cells. It is likely that this is because the absence of functional RAD51 reduces the cells' ability to recombine transformed DNA into their genomes. It is interesting to note, however, that transformants could still be generated. Many different assays of genetic exchange in bacteria, yeast, and higher eukaryotes have suggested that organisms contain multiple pathways of homologous and nonhomologous recombination (Haber 1995; Shinohara and Ogawa 1995; Baumann and West 1998), some of which involve RAD51 or RecA and some of which do not (see below). Most of the trypanosome RAD51−/− transformants analyzed here had integrated the electroporated constructs into unexpected genomic locations (data not shown), indicating that the normal pathway of homologous integration had been altered by the absence of RAD51. This suggests that RAD51-independent pathways of genetic exchange are also present in trypanosomes. The viability of trypanosome RAD51 homozygous mutants is comparable with the same mutants in yeast (and recA in bacteria), and contrasts with the lethality of RAD51 mutations in mice (Lim and Hasty 1996; Tsuzuki et al. 1996). Whether this reflects RAD51 functions beyond DNA strand transfer and exchange that are present (or critical) in mice but not in single-cell organisms, or is a reflection of the greater necessity for homologous recombination during multicellular development as opposed to clonal outgrowth of a single-cell population, remains to be determined (Edelmann and Kucherlapati 1996; Baumann and West 1998).
Homozygous knockout of RAD51 reduced the efficiency with which trypanosomes switch from one VSG coat to another. This suggests either that VSG switching is, at least in part, catalyzed by RAD51 and involves homologous recombination, or that RAD51 has a regulatory role in VSG switching. The absence of functional RAD51 did not render the trypanosomes completely unable to undergo antigenic variation. In all the experiments, some RAD51−/− trypanosomes that had succeeded in switching their VSG coat were present; in some experiments, VSG switchers arose nearly as efficiently as in RAD51 wild-type cells. Nevertheless, this is the first time that disruption of a specific gene in trypanosomes has been shown to have an effect on antigenic variation. Previous attempts to assess the mechanisms involved in VSG switching have relied on indirect experimental approaches. Cornelissen et al. (1985) used 3-aminobenzamide to inhibit trypanosome poly (ADP-ribose) polymerase in vivo, and showed that this caused a reduction in VSG switching frequency. These experiments were later repeated in vitro, with conflicting results (Mhlanga 1994; Lamont and Cross 1987). van Leeuwen et al. (1998) showed that growth of trypanosomes in the presence of the nucleoside analog hydroxymethyldeoxyuridine caused a 15-fold increase in the amount of the modified base J in the genome, and a reduction in the incidence of chromosome breakage events that have been associated with expression site transcriptional switching in vitro (M. Cross et al. 1998; Rudenko et al. 1998). Whether these treatments affect the RAD51 pathway of switching, or whether they disrupt independent facets of the process, remains to be determined.
Antigenic variation occurs by two main routes, switching of transcription between the single active bloodstream expression site and an inactive site, or movement of silent VSGs into the active expression site. Because only the latter of these routes has been shown to involve DNA recombination, we expected that disruption of RAD51 would affect only this form of VSG switching. What we saw, however, was that the reduction in the frequency of antigenic variation was due to a uniform decrease in both routes of switching. This inference is complicated somewhat by the variability in the measured level of this reduction but, even in those experiments in which only very small numbers of switched variants were recovered from the RAD51−/− cells, we found examples of both types of switching event (data not shown). Switched variants in the 3174 RAD51 wild-type trypanosomes are generated by transcriptional switching events ∼50%of the time. If disruption of RAD51 affected only the recombination-based pathways, the maximum reduction we should have seen in the frequency of antigenic variation would be twofold. Because we saw reductions of at least fivefold in all but one experiment, it appears that in situ transcriptional switching is also affected by the RAD51 mutation. It should be noted, however, that our assay would not distinguish between transcriptional switching and reciprocal recombination events in which the active and inactive expression site exchange ends upstream of the markers. It seems unlikely, though, that this recombination reaction would be unaffected by RAD51 inactivation, whereas all other reactions are.
An alternative explanation for these results is that no VSG switching pathway is affected by the RAD51−/− mutation, and the apparent reduction in switching frequency is an indirect consequence of the absence of functional RAD51 (e.g., due to a generalized growth defect). This hypothesis is not supported by any phenotypes we observed, however. The RAD51−/− cells retain infectivity in mice, and the observed reduction in their growth rate was accounted for in calculating the switch frequencies. Furthermore, we find no evidence that the RAD51−/− cells are significantly impaired in their ability to grow in vitro after recovery from mice. It is also unlikely that the RAD51 mutation affects a VSG-switching mechanism that is undetectable in our assay, as all switching events thus far characterized fit with the scheme predicted in Figure 4 (see also McCulloch et al. 1997). Therefore, we interpret these results as meaning that the RAD51 mutation affects both pathways of switching equally, and the low level of residual events is due to RAD51-independent mechanisms. We suggest that the variability in the level of reduction in switching is due to a founder effect in the assay we have used. RAD51-independent switchers arise at random during the growth of the RAD51−/− cells prior to selection of switchers, and in some instances, these can accumulate to such sufficiently large numbers, that it appears antigenic variation is little affected by the RAD51 mutation.
The fact that we can detect switched variants that have deleted the VSG and associated marker genes from the active expression site suggests that gene conversion-like recombination events can occur in the absence of RAD51. How these reactions have occurred has not been examined in detail, but a number of recombination pathways that could potentially generate these types of events have been described in yeast and other organisms. RAD51 is normally considered to catalyze homologous recombination reactions that proceed from a double-strand DNA break and use a Holliday junction intermediate (so called double-strand break repair recombination). Other recombination reactions exist that do not conform to this pathway, and some have been shown not to involve RAD51 (Shinohara and Ogawa 1995; Baumann and West 1998). Single-strand annealing is a recombination reaction that has been shown to occur between directly repeated DNA sequences, and may be catalyzed by a conserved eukaryotic recombination enzyme called RAD52 (Mortensen et al. 1996). Work in Drosophila has detailed reactions associated with P-element transposition that also appear not to involve Holliday intermediates and can occur within, and between, chromosomes (Nassif et al. 1996). The genetic requirements of this so-called synthesis-dependent strand-annealing model have not yet been determined, but it has been noted that a number of the features of this reaction are salient for the requirements of trypanosome VSG switching (Borst et al. 1996). In yeast, DNA breaks have been shown to be repaired by DNA replication in the absence of RAD51, resulting in nonreciprocal transfer of sequence information from homologous sites (Malkova et al. 1996) up to and including the telomere; such events could readily provide new VSGs being expressed with concomitant deletion of sequence within the expression site as we see here. To establish whether or not any of the above types of recombination reaction are occurring in the RAD51−/− switchers, further experiments will be required. Interestingly, in one switch that we have analyzed by pulsed-field gel electrophoresis, we can see that the newly expressed VSG has made a new copy, but the VSG221 expression-site chromosome appears to have changed size (R. McCulloch, unpubl.), suggesting that these can involve nonstandard means of gene conversion.
How do we account for our observation that the RAD51 mutation affects in situ transcriptional switching, which has not been considered previously to require genetic exchange? The simplest explanation is that RAD51 is directly involved, and switching occurs by undetected recombination reactions between expression sites. This seems unlikely. Although DNA rearrangements can be detected around the expression site promoter during in situ switching (for review, see Pays et al. 1994), and alteration of expression-site sequences by recombination can apparently precipitate an in situ switch (Davies et al. 1997), a number of studies have detailed transcriptional switches that show no structural rearrangements either at the promoter or in upstream sequence elements (Horn and Cross 1997; Rudenko et al. 1998). A second explanation might be that the transcriptional switching reaction does not require RAD51 for recombination, but instead for transient association between the expression sites. Although there is no direct evidence that this is the case, most work has focused on the proposed mechanisms that control the activity of the expression sites, and has not considered whether association between the active site and an inactive site is needed to effect a switch (Horn and Cross 1995; Rudenko et al. 1995). Whether this would require some form of RAD51-dependent homology sensing is conjecture, however, and is argued against by the fact that the expression-site promoter can be changed to a nonhomologous ribosomal promoter without affecting switching (Rudenko et al. 1995). On the other hand, a number of reports have shown that pairing of chromosomes in homologous regions, or pairing of a chromosome with an injected plasmid (Ashe et al. 1997), can cause up- or down-regulation of transcription in a number of organisms (for review, see Hensikoff 1997; Grant 1999). A final explanation would be that it is not the absence of RAD51 itself that causes an impairment in in situ switching, but the effect this has on a protein(s) that RAD51 interacts with. In higher eukaryotes, RAD51 interacts with a number of cellular factors that are not directly involved in recombination, for example, the tumor suppressers p53 and BRCA1 and 2 (Sturzbecher et al. 1996; Scully et al. 1997; Sharan et al. 1997). It is possible that RAD51 provides a means by which the cell can integrate DNA repair and recombination with a number of other processes, such as cell cycle progression and transcriptional control (Baumann and West 1998). In other words, disruption of RAD51 may have an indirect effect that disrupts the normal way that trypanosomes co-ordinate in situ transcriptional switching, and VSG recombination, with other aspects of cell metabolism. Further studies of the genetics of trypanosome antigenic variation will be required to address these complex issues.
Materials and methods
Trypanosome strains and transformation
T. brucei 221a trypanosomes (MITat1.2a) of strain 427 (Cross 1975) were used. Bloodstream cells were grown in vitro at 37°C in HMI-9 medium (Hirumi and Hirumi 1989), whereas procyclic cells were grown at 27°C in SDM-79 (Brun and Schonenberger 1979). Trypanosomes were transformed by electroporating 5 × 107 cells with ∼5 μg of plasmid DNA that had been restriction digested, phenol/chloroform treated, and ethanol precipitated. Electroporation was performed in 0.5 ml of Zimmerman Post-Fusion medium (132 mm NaCl, 8 mm Na2HPO4, 1.5 mm KH2PO4, 0.5 mm Mg acetate, 0.09 mm Ca acetate at pH7.0), supplemented with 1%glucose for the bloodstream cells, with a Biorad Gene Pulser II set at 1.4 kV and 25 μF capacitance. Cells were allowed to recover for 18 hr before transformants were selected. Bloodstream RAD51 transformants were selected on semisolid agarose plates, whereas procyclic transformants were selected in 5–10 ml of liquid medium supplemented with ∼106 untransformed cells.
Cloning and sequencing the T. brucei RAD51 gene
To obtain a probe to clone the T. brucei RAD51 gene, PCR was performed on T. brucei genomic DNA with oligonucleotides directed against two conserved regions of the RAD51 protein family identified by Shinohara et al. (1993): 5′RAD51, 5′-CCGAATTCTT(C/T)GGIGA(A/G)TT(T/C)(A/C)GIACIGG(G/A/T/C)AA-3′ (EcoRI underlined); 3′RAD51, 5′-CCGGATCCIGTICC(T/C)TCIGT(A/G)TC(G/A/T/C)AT-3′ (BamHI underlined). PCR was carried out with ∼30 pmoles of each primer with an initial 5 min denaturation at 94°C followed by 30 cycles of 94°C for 1 min, 42°C for 1 min, and 72°C for 1 min A 120-bp product was generated and cloned into pBluescript (Stratagene) which, on sequencing, was predicted to encode the expected region of T. brucei RAD51 (Fig. 1). The probe was then used to screen two T. brucei mini-libraries generated in pBluescript with 2–3 kb of PstI-digested, and 5–6 kb of BamHI-digested, genomic DNA. This allowed us to isolate a 2.5-kb PstI fragment (Fig. 2) that contained the complete RAD51 ORF, as well as a larger 5.5-kb BamHI fragment. The sequence of the PstI fragment was determined on both strands by ABI-PRISM automated sequencing with custom-designed primers. The sequence predicts a polypeptide with 60 amino acids more at its carboxyl terminus than suggested by a database entry from Haag (accession no. Y13144). We believe ours to be correct, as the start codon we have used is the first that is present ∼180-bp downstream of a large polypyrimidine tract (Matthews et al. 1990; Schurch et al. 1990) that is likely to dictate the trans-splicing that creates the 5′ end of RAD51 mRNA (corroborated by RT–PCR; data not shown).
Generation and analysis of T. brucei RAD51 knockouts
For mapping of the transformants, genomic DNA was restriction digested and separated on 1.0% agarose gels in 1× TBE (90 mm Tris-borate, 2 mm EDTA). Intact chromosomes were separated by pulsed-field electrophoresis in 90 mm Tris-borate, 0.2 mm EDTA on 1.2% agarose gels with a Bio-Rad CHEF-DRIII system. Conditions for electrophoresis were from S. Melville (University of Cambridge, UK), 144 hr at 15°C and 2.5 V/cm with 1400–700 sec switch times. Each gel lane contained ∼2.5 × 107 cells embedded in 0.5% low melting point agarose. All DNA was blotted onto Hybond N (Amersham) following the manufacturer's instructions. Hybridization of probes (labeled with [α-32P]dATP by random priming) was as described in Sambrook et al. (1989), and all blots were washed to a stringency of 0.2× SSC, 0.1% SDS at 65°C. All PCR for generating and analyzing transformants (and VSG switchers) used an initial 5 min denaturation at 95°C followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1–2 min.
The constructs used to generate T. brucei RAD51 knockout mutants contain 300 bp of 5′ and 3′ RAD51 sequences (that act as targeting flanks) separated by one of two antibiotic resistance cassettes. The RAD51 sequences were amplified by PCR from genomic DNA with Pfu DNA polymerase, and cloned into pBluescript. 5′ flank primers: 5′-CCGGATCCATGCAGCAGCAAGTTGGTGA-3′ (BamHI), and 5′-CCGAATTCCTCACGGCTGCCTGTCGTGA-3′ (EcoRI). 3′ flank primers, 5′-CCGGTACCGCACGGCAGATGCATCTTGG-3′ (KpnI), and 5′-CCAAGCTTGTCCCTAACGTCTCCCACA-3′ (HindIII). The antibiotic resistance cassettes contained either the 400-bp bleomycin phosphotransferase ORF or the 600-bp puromycin N-acetyltransferase ORF (both gifts of J. Mottram, University of Glascow) flanked by 100 bp of 5′ and 300 bp of 3′ processing signals from the PARP locus (gifts from S. Graham, University of Glascow). The cassettes were cloned between the RAD51 targeting sequences to generate ΔRAD51::BLE and ΔRAD51::PUR constructs, respectively. The plasmids were digested with BamHI and XhoI prior to being transformed into T. brucei, and transformants were selected with 2.0 μg/ml bleomycin or 1.0 mg/ml puromycin. Note that the constructs contain no promoter elements to drive the antibiotic resistance cassette expression (and therefore rely on endogenous RAD51 transcription), and the RAD51 trans-splicing and polyadenylation signals are not altered by the knockout strategy because only internal RAD51 coding sequence is removed.
Correct integration of the constructs into RAD51 was determined in two ways. First, PCR was performed on the transformants' genomic DNA with primers outside of the RAD51 sequence included in the constructs (Fig. 2), ko5′, 5′-GATGATGCCGCAGTGGATGCGG-3′, and ko3′, 5′-GCACTTCTCCGCTTTCGGAGTG-3′. Because of the high GC content of the resistance cassettes, this PCR was performed with 5%DMSO. Second, to determine that RAD51 wild-type was removed from the genome in the puromycin and bleomycin double-resistant clones, genomic DNA from the transformants was digested with either PstI or EcoRV and probed with RAD51 wild-type ORF amplified by PCR with the BamHI and HindIII construct primers (above). In both digests a 2.5-kb fragment corresponding to intact RAD51 was detected in both the wild-type and heterozygous cells, but was absent in the homozygous mutants. The only other hybridising bands corresponded to the ΔRAD51::BLE and ΔRAD51::PUR alleles (data not shown).
Total RNA from the RAD51 transformants was prepared with TRIzol, and cDNA was synthesized with random hexamers and Superscript reverse transcriptase (all reagents from Life Technologies). To assess whether or not intact RAD51 mRNA was present in the knockouts, PCR was performed on the cDNAs with primers rad51d6 (5′-TGGTGACGCTGCCGGTGGGC- 3′) and rad51u3 (5′-TTTCCAAGATGCATCTGCCG-3′). The primers used to perform a control PCR on cDNA from RNA polymerase I mRNA have been described before (Rudenko et al. 1996).
Re-expressing RAD51 in the homozygous knockouts
The construct tub::RAD51–BSR contains 240 bp of β-α tubulin and 330 bp of α-β tubulin intergenic sequence (derived from the construct hygrotub; Blundell et al. 1996) at its 5′ and 3′ flanks, respectively; these allow integration into the tubulin locus, replacing an α tubulin gene. Between the flanks, a 400-bp Blasticidin S deaminase ORF (BSR; a gift from M. Cross, Netherlands Cancer Institute) was cloned and, downstream, a 2.27-kb BglII–PstI T. brucei genomic DNA fragment containing the complete RAD51 ORF and ∼1.0-kb of 5′ sequence. On correct integration, the endogenous RAD51 signals should dictate the 5′ splicing of RAD51 RNA and 3′ polyadenylation of BSR RNA, whereas the β tubulin gene signals 3′ of RAD51 will dictate its polyadenylation. We did not test whether this ectopic expression strategy resulted in the same steady-state levels of RAD51 mRNA as endogenous RAD51. tub::RAD51–BSR was digested with XbaI and XhoI prior to electroporation, and transformants were selected on semisolid agarose plates containing 2 μg/ml Blasticidin (Invitrogen), or in 10 ml of HMI-9 medium containing 5 or 10 μg/ml. Integration of the constructs was examined by Southern analysis of EcoRV or BamHI-digested genomic DNA, and RT–PCR to check for intact RAD51 mRNA was performed as described in the previous section (data not shown).
Measuring the growth of trypanosome RAD51 mutants
To measure the growth rate of the RAD51 transformants in vivo, 2 × 105 trypanosomes growing at mid-log in vitro were injected into an ICR mouse (which, in some experiments, had been immunocompromised 1–24 hr previously with 300 mg/kg of cyclophosphamide). At intervals thereafter, small blood samples were removed from the mouse tail into heparinized capillary tubes, and 1 μl of the blood was diluted into 99 μl of 0.85%ammonium chloride. Erythrocytes were allowed to lyse for 5 min at room temperature before the concentration of trypanosomes in the blood was measured with a bright-line hemacytometer (Sigma). For each growth rate measurement, three mice were independently infected.
In vitro growth rates were measured by diluting mid-log bloodstream trypanosomes to a concentration of 1 × 105/ml in 2.5 ml of HMI-9 medium in 6-well culture dishes. Cell concentrations were measured at time intervals thereafter with a hemacytometer. Methyl methanesulphonate (Sigma) was diluted in HMI-9 from a 99%stock solution, and 3-aminobenzamide (Sigma) was dissolved in 100%DMSO and then diluted in HMI-9.
Generation and analysis of trypanosome variants that have switched their VSG coat
The approach adopted to isolate VSG-switched variants is essentially that described in McCulloch et al. (1997), but with some modifications. To generate mice with immunity to the VSG221 coat, a number of ICR mice were intraperitoneally injected with 2 × 105 to 4 × 105 3174 RAD51 wild-type trypanosomes (McCulloch et al. 1997; Fig. 5) growing on hygromycin (5.0 μg/ml) and G418 (2.5 μg/ml); 3–4 days later, the infections were cured by injection of cymelarsan (Rhone Merieux; 5 mg/kg). The immunized mice were used between 7 and 35 days post-curing. To select for switch variants, the 3174 RAD51 transformants were passaged from medium containing hygromycin and G418 into nonselective medium at a density of 0.5 × 104 to 1.0 × 104 cells/ml and grown until they reached densities of 1 × 106 to 3 × 106/ml. Measured numbers were then intraperintoneally injected into immunized mice in 300 ml of HMI-9. Twenty-four hours later, the mice were bled by cardiac puncture and trypanosomes isolated and cloned as described (McCulloch et al. 1997). In calculating the VSG switching frequencies from the numbers of clones that grew out, we have assumed (as before, McCulloch et al. 1997) that the 3174 RAD51 wild-type and 3174 RAD51+/− cell, populations double every 8 hr; for the 3174 RAD51-/− cells, we have assumed that the doubling time is increased by 15%, which is slightly greater than was measured in vivo. The drug sensitivities of the switched variant clones, as well as the presence or absence of the marker genes within the marked VSG221 expression site, were assayed as described previously (McCulloch et al. 1997).
Acknowledgments
We thank Piet Borst for helpful discussions, and Jo Bell, Pat Blundell, and Marshall Stark for critical reading of the manuscript. This work was supported by fellowships from the Royal Society and Wellcome Trust to R.M., and grants from The Wellcome Trust to J.D.B.; part of the work that was conducted in the Netherlands Cancer Institute was supported by the Netherlands Organization for Scientific Research (NWO/SON).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The EMBL/GenBank accession no. for the sequence reported is AF174136.
Footnotes
E-MAIL rmc9z@udcf.gla.ac.uk; FAX 0044-141-330-5422.
References
- Aline RF, Stuart KD. Trypanosoma brucei-conserved sequence organization 3′ to telomeric variant surface glycoprotein genes. Exp Parasitol. 1989;68:57–66. doi: 10.1016/0014-4894(89)90008-8. [DOI] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human B-globin locus. Genes & Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JD. The relative significance of mechanisms of antigenic variation in African trypanosomes. Parasitol Today. 1997;13:212–218. doi: 10.1016/s0169-4758(97)01039-9. [DOI] [PubMed] [Google Scholar]
- Basile G, Aker M, Mortimer RK. Nucleotide sequence and transcriptional regulation of the yeast recombination repair gene RAD51. Mol Cell Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, West SC. The human RAD51 protein: Polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. Human RAD51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E.coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A, van der Ploeg LHT, Frasch ACC, Borst P, Boothroyd JC, Coleman S, Cross GAM. Activation of trypanosome surface antigen genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. doi: 10.1016/0092-8674(81)90391-3. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E.coli RecA required for recombination, synaptonemal complex formation and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Blundell PA, Rudenko G, Borst P. Targeting of exogenous DNA into Trypanosoma brucei requires a high degree of homology between donor and target DNA. Mol Biochem Parasitol. 1996;76:215–229. doi: 10.1016/0166-6851(95)02560-x. [DOI] [PubMed] [Google Scholar]
- Borst P, Greaves DR. Programmed gene rearrangements altering gene expression. Science. 1987;235:658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- Borst P, Rudenko G, Taylor MC, Blundell PA, van Leeuwen F, Bitter W, Cross MA, McCulloch R. Antigenic variation in trypanosomes. Arch Med Res. 1996;27:379–388. [PubMed] [Google Scholar]
- Borst P, Bitter W, Blundell PA, Chaves I, Cross MA, Gerrits H, van Leeuwen F, McCulloch R, Taylor MC, Rudenko G. Control of VSG gene expression sites in Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:67–76. doi: 10.1016/s0166-6851(97)00184-9. [DOI] [PubMed] [Google Scholar]
- Brun R, Schoenenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Campbell DA, van Bree MP, Boothroyd JC. The 5′ limit of transposition and upstream barren region of a trypanosome VSG gene: Tandem 76 bp repeats flanking (TAA)90. Nucleic Acids Res. 1984;12:2759–2774. doi: 10.1093/nar/12.6.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldo FN, Ramsey G, Barbour SD. Analysis of the growth of recombination-deficient strains of Escherichia coli K12. J Bacteriol. 1974;118:242–249. doi: 10.1128/jb.118.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Zomerdijk J, Dirks-Mulder A, Dirks RW, Raap AK, Borst P. Subnuclear localization of the active variant surface glycoprotein expression site in Trypanosoma brucei. Proc Natl Acad Sci. 1998;95:12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen AWCA, Michels PAM, Borst P, Spanjer W, Versluis-Broers JAM, van der Meer C, Farzaneh F, Shall S. Effect of 3-aminobenzanide on antigenic variation of Trypanosoma brucei. Biochem Pharmacol. 1985;34:4151–4156. doi: 10.1016/0006-2952(85)90208-4. [DOI] [PubMed] [Google Scholar]
- Cross GAM. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- ————— Antigenic variation in trypanosomes: Secrets surface slowly. BioEssays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- Cross GAM, Wirtz LE, Navarro M. Regulation of VSG gene expression site transcription and switching in Trypanosoma brucei. Mol Biochem Parasitol. 1998;91:77–91. doi: 10.1016/s0166-6851(97)00186-2. [DOI] [PubMed] [Google Scholar]
- Cross M, Taylor MC, Borst P. Frequent loss of the active site during variant surface glycoprotein expression site switching in vitro in Trypanosoma brucei. Mol Cell Biol. 1998;18:198–205. doi: 10.1128/mcb.18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully DF, Ip HS, Cross GAM. Coordinate transcription of variant surface glycoprotein genes and an expression site associated gene family in Trypanosoma brucei. Cell. 1985;42:173–182. doi: 10.1016/s0092-8674(85)80113-6. [DOI] [PubMed] [Google Scholar]
- Davies KP, Carruthers VB, Cross GAM. Manipulation of the VSG co-transposed region increases expression site switching in Trypanosoma brucei. Mol Biochem Parasitol. 1997;86:163–177. doi: 10.1016/s0166-6851(97)02853-3. [DOI] [PubMed] [Google Scholar]
- Deitsch KW, Moxon ER, Wellems TE. Shared themes of antigenic variation and virulence in bacterial, protozoal and fungal infections. Microbiol & Mol Biol Rev. 1997;61:281–293. doi: 10.1128/mmbr.61.3.281-293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, Kooter JM, Michels PA, Borst P. Telomere conversion in trypanosomes. Nucleic Acids Res. 1983;11:8149–8165. doi: 10.1093/nar/11.23.8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunderdale HJ, West SC. Recombination genes and proteins. Curr Biol. 1994;4:221–228. doi: 10.1016/s0959-437x(05)80048-6. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Kucherlapati SC. Role of recombination proteins in mammalian cell survival. Proc Natl Acad Sci. 1996;93:6225–6227. doi: 10.1073/pnas.93.13.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston AK, West SC. Exchanging partners: Recombination in E.coli. Trends Genet. 1996;12:20–26. doi: 10.1016/0168-9525(96)81384-9. [DOI] [PubMed] [Google Scholar]
- Eid J, Sollner-Webb B. Stable integrative transformation of Trypanosoma brucei that occurs exclusively by homologous recombination. Proc Natl Acad Sci. 1991;88:2118–2121. doi: 10.1073/pnas.88.6.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerhut R, Kiefer J, Otto F. Cell cycle parameters in radiation sensitive strains of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:192–194. doi: 10.1007/BF00327437. [DOI] [PubMed] [Google Scholar]
- Game JC. Radiation-sensitive mutants and repair in yeast. In: In: Spencer J, Spencer D, Smith A, editors. Yeast genetics: Fundamental and applied aspects. New York, NY: Springer-Verlag; 1983. pp. 109–137. [Google Scholar]
- Grant SR. Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell. 1999;96:303–306. doi: 10.1016/s0092-8674(00)80541-3. [DOI] [PubMed] [Google Scholar]
- Grivell LA. Transposition: Mobile introns get into line. Curr Biol. 1996;6:48–51. doi: 10.1016/s0960-9822(02)00420-7. [DOI] [PubMed] [Google Scholar]
- Haber JE. In vivo biochemistry: Physical monitoring of recombination by site-specific endonucleases. BioEssays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- ————— Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- Hensikoff S. Nuclear organization and gene expression: Homologous pairing and long-range interactions. Curr Opin Cell Biol. 1997;9:388–395. doi: 10.1016/s0955-0674(97)80012-9. [DOI] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Hoeijmakers JHJ, Frasch ACC, Bernards A, Borst P, Cross GAM. Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature. 1980;284:78–80. doi: 10.1038/284078a0. [DOI] [PubMed] [Google Scholar]
- Horn D, Cross GAM. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- ————— Analysis of Trypanosoma brucei. VSG expression site switching in vitro. Mol Biochem Parasitol. 1997;84:189–201. doi: 10.1016/s0166-6851(96)02794-6. [DOI] [PubMed] [Google Scholar]
- Jeggo PA. PARP - another guardian angel? Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- Johnson PJ, Kooter JM, Borst P. Inactivation of transcription by UV irradiation of Trypanosoma brucei provides evidence of a muligene transcription unit, including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- Kooter JM, van der Spek HJ, Wagter R, d'Oliveira CE, van der Hoeven F, Johnson PJ, Borst P. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell. 1987;51:261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- Lamont GS, Cross GAM. Effect of 3-aminobenzanide on the frequency of antigenic switching of Trypanosoma brucei. in vitro. Biochem Pharmacol. 1987;36:1750–1751. doi: 10.1016/0006-2952(87)90065-7. [DOI] [PubMed] [Google Scholar]
- Laurent M, Pays E, Magnus E, van Meirvenne N, Matthyssens G, Williams RO, Steinert M. DNA rearrangemements linked to the expression of a predominant sufrace antigen gene of trypanosomes. Nature. 1983;302:263–266. doi: 10.1038/302263a0. [DOI] [PubMed] [Google Scholar]
- Lee MG-S, Van der Ploeg LHT. Homologous recombination and stable transfection in parasitic protozoan Trypanosoma brucei. Science. 1990;250:1583–1587. doi: 10.1126/science.2177225. [DOI] [PubMed] [Google Scholar]
- Lim DS, Hasty P. A mutation in mouse RAD51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhal T, Satoh MS, Poirier GG, Klugland A. Post-translational modification of poly (ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- Liu AYC, Van der Ploeg LHT, Rijsewijk FAM, Borst P. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences. J Mol Biol. 1983;167:57–75. doi: 10.1016/s0022-2836(83)80034-5. [DOI] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. Double strand break repair in the absence of Rad51 in yeast: A possible role for break-induced DNA replication. Proc Natl Acad Sci. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre- mRNA in trypanosomes. Genes & Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- McCulloch R, Rudenko G, Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in Variant Surface Glycoprotein expression sites lacking 70 bp repeat sequences. Mol Cell Biol. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhlanga J. “Antigenic variation in Trypanosome brucei, a relationship with poly ADP-ribose polymerase.” PhD thesis. UK: University of Sussex; 1994. [Google Scholar]
- Michels PAM, van der Ploeg LHT, Liu AYC, Borst P. The activation and reactivation of an expression-linked copy for a variant surface glycoprotein in Trypanosoma brucei. EMBO J. 1984;3:1345–1351. doi: 10.1002/j.1460-2075.1984.tb01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjeravic I, Rothstein R. DNA strand annealing is promoted by the yeast protein Rad52. Proc Natl Acad Sci. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler PJ, Nelson RG, Agabian N, Stuart KD. Two mechanisms of expression of a predominant variant antigen gene of Trypanosoma brucei. Nature. 1984;309:282–284. doi: 10.1038/309282a0. [DOI] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels W, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Cross GAM. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol Cell Biol. 1996;16:3615–3625. doi: 10.1128/mcb.16.7.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu K, Egelman EH, Ogawa H. RecA-like recombination proteins in eukaryotes: Functions and structures of RAD51 genes. Cold Spring Harb Symp Quant Biol. 1993a;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu K, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993b;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Pays E, Nolan DP. Expression and functions of surface proteins in Typanosoma brucei. Mol Biochem Parasitol. 1998;91:3–36. doi: 10.1016/s0166-6851(97)00183-7. [DOI] [PubMed] [Google Scholar]
- Pays E, Meirvenne van N, Le Ray D, Steinert M. Gene duplication and transposition linked to antigenic variation in T. brucei. Proc Natl Acad Sci. 1981;78:2673–2677. doi: 10.1073/pnas.78.5.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, van Assel S, Laurent M, Darville M, Vervoort T, van Meirvenne N, Steinert M. Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell. 1983a;34:371–381. doi: 10.1016/0092-8674(83)90371-9. [DOI] [PubMed] [Google Scholar]
- Pays E, Delauw MF, van Assel S, Laurent M, Vervoort T, van Meirvenne N, Steinert M. Modifications of a Trypanosoma b. brucei antigen gene repertoire by different DNA recombinational mechanisms. Cell. 1983b;35:721–731. doi: 10.1016/0092-8674(83)90105-8. [DOI] [PubMed] [Google Scholar]
- Pays E, Guyaux M, Aerts D, van Meirvenne N, Steinert M. Telomere reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature. 1985;316:562–564. doi: 10.1038/316562a0. [DOI] [PubMed] [Google Scholar]
- Pays E, Tebabi P, Pays A, Coquelet H, Revelard P, Salmon D, Steinert M. The genes and transcripts of an antigen gene expression site from T. brucei. Cell. 1989;57:835–845. doi: 10.1016/0092-8674(89)90798-8. [DOI] [PubMed] [Google Scholar]
- Pays E, Vanhamme L, Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. Annu Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric variant surface glycoprotein gene expression site can be efficiently switched on and off in Trypanosoma brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- Rudenko G, McCulloch R, Dirks-Mulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Chaves I, Dirks-Mulder A, Borst P. Selection for activation of a new variant surface glycoprotein expression site in Trypanosoma brucei can result in deletion of the old one. Mol Biochem Parasitol. 1998;95:97–109. doi: 10.1016/s0166-6851(98)00099-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sancer A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- Scholler JK, Myler PJ, Stuart KD. A novel telomeric gene conversion in T. brucei. Mol Biochem Parasitol. 1989;35:11–19. doi: 10.1016/0166-6851(89)90137-0. [DOI] [PubMed] [Google Scholar]
- Schurch N, Hehl A, Vasella E, Braun R, Roditi I. Accurate poyadenylation of procyclic mRNAs in T. brucei. is determined by pyrimidine-rich elements in the intergenic regions. Mol Cell Biol. 1994;14:3668–3675. doi: 10.1128/mcb.14.6.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen JJ, Plug A, Xiao YH, YH, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with RAD51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht A, Lim D-S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking BRCA2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Homologous recombination and the role of double strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikep K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- Sturzbecher H-M, Donzelmann B, Henning W, Knippschild U, Buchop S. p53 is linked directly to homologous recombination processes via Rad51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- Story RM, Bishop DK, Kleckner N, Steitz TA. Structural relationship of bacterial recA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- ————— Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Ten Asbroek ALMA, Ouellette M, Borst P. Targeted insertion of the neomycin phosphotransferase gene into the tubulin gene cluster of Trypanosoma brucei. Nature. 1990;348:174–175. doi: 10.1038/348174a0. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekigughi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ploeg LHT, Valerio D, de Lange T, Bernards A, Borst P, Grosveld F. An analysis of cosmid clones from Trypanosoma brucei shows that the genes for variant surface gclycoproteins are clustered in the genome. Nucleic Acids Res. 1982;10:5905–5923. doi: 10.1093/nar/10.19.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen F, Kieft R, Cross M, Borst P. Biosynthesis and function of the modified DNA base B-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol Cell Biol. 1998;18:5643–5651. doi: 10.1128/mcb.18.10.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, van den Abbeele J, Pays A, van Meirvenne N, Hamers R, de Baetselier P, Pays E. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]