Abstract
The UNC-4 homeoprotein and the Groucho-like corepressor UNC-37 specify synaptic choice in the Caenorhabditis elegans motor neuron circuit. In unc-4 mutants, VA motor neurons are miswired with inputs from interneurons normally reserved for their lineal sisters, the VB motor neurons. Here we show that UNC-4 and UNC-37 function together in VA motor neurons to repress VB-specific genes and that this activity depends on physical contact between UNC-37 and a conserved Engrailed-like repressor domain (eh1) in UNC-4. Missense mutations in the UNC-4 eh1 domain disrupt interactions between UNC-4 and UNC-37 and result in the loss of UNC-4-dependent repressor activity in vivo. A compensatory amino acid substitution in UNC-37 suppresses specific unc-4 alleles by restoring physical interactions with UNC-4 as well as UNC-4-dependent repression of VB-specific genes. We propose that repression of VB-specific genes by UNC-4 and UNC-37 is necessary for the creation of wild-type inputs to VA motor neurons. The existence of mammalian homologs of UNC-4 and UNC-37 indicates that a similar mechanism could regulate synaptic choice in the vertebrate spinal cord.
Keywords: Groucho, unc-4, repression, C. elegans, synaptic specificity
Animal movement is controlled by axial arrays of motor neurons that transmit signals from the brain to muscles. The function of these circuits is defined by the creation of precise sets of connections between motor neurons and their synaptic partners. These include interneurons that extend processes from the brain into the nerve cord to synapse with particular motor neurons (Brodal 1998) and specific muscles that are targeted by motor axons emanating from the nerve cord (Landmesser 1978). The synaptic choices that lead to the creation of these stereotypical networks are likely to depend on cell surface markers or secreted signals that facilitate target recognition. Although the molecular identities of these factors are largely unknown, it is reasonable to propose that the cell specificity of their expression is defined by coordinated actions of transcription factors. In Drosophila, Caenorhabditis elegans, and vertebrate organisms, motor neurons that synapse with particular muscles express unique combinations of LIM-type homeodomain proteins (Ericson et al. 1992; Tsuchida et al. 1994; Appel et al. 1995; Hobert et al. 1998). Furthermore, subsets of these motor neurons can be redirected to alternative muscle targets by altering the pattern of LIM-HD gene expression (Sharma et al. 1998; Thor et al. 1999). The specificity of synaptic inputs to motor neurons is also correlated with expression of particular transcription factors. Sensory neurons in the dorsal root ganglion and their postsynaptic motor neuron partners in the ventral horn express common sets of ETS-type transcription factors (Ghosh and Kolodkin 1998; Lin et al. 1998). Direct evidence for transcriptional regulation of presynaptic input to motor neurons is provided by genetic experiments in C. elegans, demonstrating that the UNC-4 homeoprotein controls the pattern of interneuron synapses with motor neurons (White et al. 1992; Miller et al. 1992; Miller and Niemeyer 1995).
In C. elegans, adjacent pairs of VA and VB motor neurons arise from common precursor cells at periodic intervals along the length of the ventral nerve cord (Sulston and Horvitz 1977) (Fig. 1). Both VA and VB motor neurons are excitatory and cholinergic (Rand and Nonet, 1997) but adopt different axonal trajectories and receive inputs from separate sets of command interneurons (White et al. 1986). The cell bodies of these interneurons reside in the head or tail and extend processes into the nerve cord to synapse with appropriate classes of motor neurons. Laser ablation experiments have demonstrated that the VA motor neuron circuit is required for backward movement whereas the VBs and their specific presynaptic partners regulate forward locomotion (Chalfie et al. 1985). Mutations in the unc-4 gene differentially affect backward movement and have been shown to specifically perturb the pattern of interneuron inputs to VA motor neurons (Fig. 1). Electron microscopy of the presumptive null allele, unc-4(e120), has revealed that VAs adopt input from interneurons that are normally reserved for their VB sister cells. VA axonal trajectory and morphology are not altered, however. Thus, unc-4 regulates the specificity of synaptic input to the VAs but not other traits that distinguish VAs from their VB sister cells (White et al. 1992).
Figure 1.
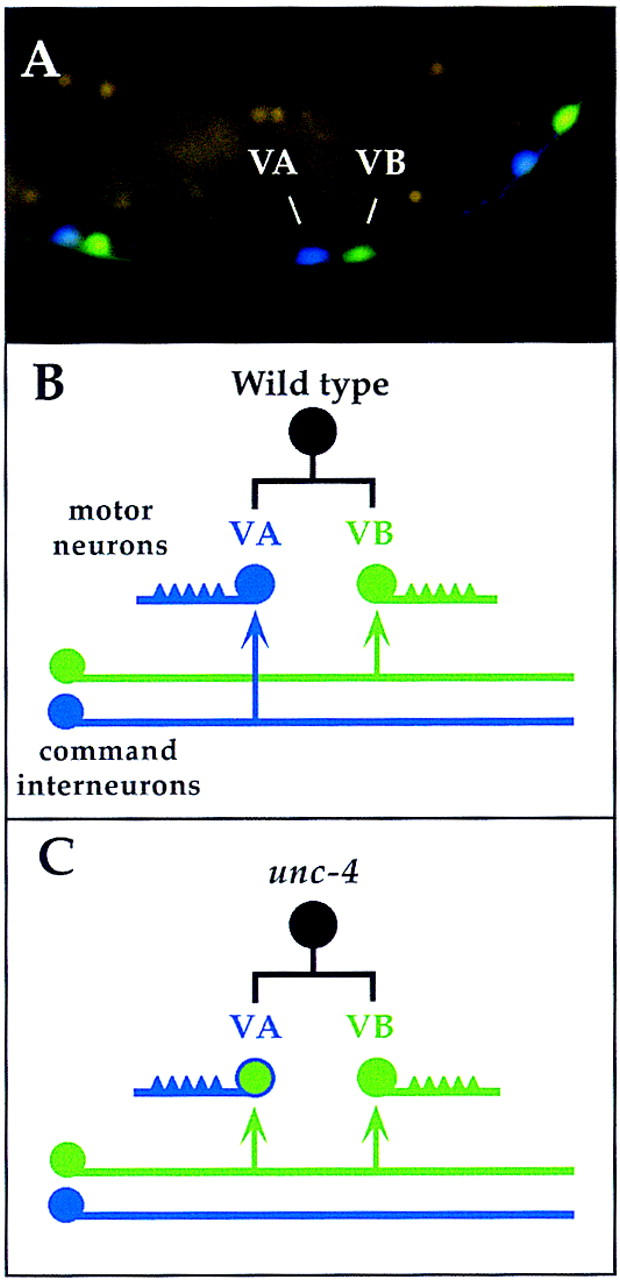
unc-4 specifies synaptic input to VA motor neurons. (A) Adjacent VA and VB motor neurons are visualized in this transgenic animal with neuron-specific promoters driving expression of the cyan GFP variant in VA motor neurons (unc-4::CFP) and the yellow GFP mutant protein in VB motor neurons (del-1::YFP) (Miller et al. 1999). (B) Most VA and VB motor neurons arise from a common precursor cell (black circle). In wild-type animals, VA neurons, which extend axons anteriorly, receive inputs from VA-type command interneurons (blue): AVA (chemical synapse and gap junction); AVD and AVE (chemical synapse). VB axons project posteriorly and are innervated by the VB-type interneurons (green): AVB (gap junction); PVC (chemical synapse). (C) In unc-4 mutants, VA neurons are miswired with VB-type inputs (i.e., form gap junctions with AVB). The arrows denote en passant synapses between side-by-side processes in the tightly packed ventral nerve cord and therefore are not indicative of axonal branches.
The UNC-4 homeoprotein is expressed in VA motor neurons but is not detected in the VBs (Miller et al. 1992; Miller and Niemeyer 1995). We have therefore hypothesized that UNC-4 functions in the VAs to block expression of target genes that lead to VB-type synaptic inputs. The proposal that UNC-4 mediates transcriptional repression is supported by our recent finding that unc-4 function in VA motor neurons depends on the simultaneous expression of a Groucho-like protein encoded by the unc-37 gene (Pflugrad et al. 1997).
A screen for suppressors of an unc-4 homeodomain missense allele revealed a rare, dominant mutation, unc-37(d), which restores normal movement (Miller et al. 1993). Conversely, the loss-of-function mutant, unc-37(e262), displays an Unc-4-like VA neuron wiring defect (D. Hall and E. German, pers. comm.) and is also defective in backward locomotion (Miller et al. 1993). Both of these unc-37 alleles alter specific amino acids within a WD40 repeat region and are therefore predicted to affect protein–protein interactions (Pflugrad et al. 1997).
Drosophila Groucho is the founding member of a highly conserved family of transcriptional corepressor proteins (Fisher and Caudy 1998). Groucho proteins are defined by an amino-terminal dimerization domain and a carboxy-terminal region of WD40 repeats. The transcriptional repressor activity of Groucho depends on physical association with DNA-specific binding proteins. Groucho-interacting transcription factors are surprisingly diverse and include TCF/LEF-1 as well as proteins containing bHLH, Rel, Runt and homeobox domains (Paroush et al. 1994; Aronson et al. 1997; Dubnicoff et al. 1997; Jimenez et al. 1997; Cavallo et al. 1998; Roose et al. 1998). The Engrailed homeoprotein, for example, interacts physically with Groucho to repress target genes in Drosophila. A conserved repressor domain, known as eh1, is required for Engrailed-mediated repression and for binding to Groucho (Smith and Jaynes 1996; Tolkunova et al. 1998; Jimenez et al. 1999).
In this paper we report the identification of an eh1-like repressor domain in nematode UNC-4 proteins and in UNC-4-like homologs from other species. Missense mutations in the UNC-4 eh1 domain disable UNC-4-dependent repression of VB-specific genes and perturb physical interactions with UNC-37. Conversely, the unc-37(d) suppressor allele simultaneously restores repressor activity and UNC-37 binding to these UNC-4 mutant proteins. We also show that the hypomorphic WD40 missense mutation unc-37(e262) results in derepression of VB-specific genes in VA motor neurons. These findings support a model in which UNC-4 and UNC-37 function together in the VA motor neurons to block expression of genes that lead to the creation of VB-type synaptic inputs. Furthermore, the evident conservation of both UNC-4 (Rovescalli et al. 1996; Saito et al. 1996; Mansouri et al. 1997; Tabuchi et al. 1998) and UNC-37 (Stifani et al. 1992; Dehni et al. 1995) indicates that a similar molecular mechanism may define synaptic inputs to motor neurons in the vertebrate spinal cord.
Results
The C. elegans motor neuron circuit
The motor neuron circuit is generated during two distinct developmental periods (White et al. 1986). The basic architecture of the ventral nerve cord is established in the embryo when interneurons located in the head and tail send out axons along the ventral midline to synapse with nascent motor neurons. Three classes of motor neurons are generated in the embryo: DA, DB, and DD. Combinations of chemical synapses and gap junctions are created en passant between interneuron and motor neuron processes in the tightly packed ventral cord fascicle (Durbin 1987). After hatching and in the first larval stage (L1), five new classes of motor neurons (VA, VB, VC, AS, VD) arise by stereotypical divisions of an axial array of ectodermal blast cells (P cells) (Sulston and Horvitz 1977). unc-4 is selectively expressed in the A-class motor neurons, DA and VA, so named because they show similar axonal polarities (anterior), receive inputs from a common set of interneurons, and are both required for backward movement (Chalfie et al. 1985; White et al. 1986; Miller et al. 1992; Miller and Niemeyer 1995). DB and VB motor neurons comprise the B-class, which project posteriorly directed axons and synapse with a separate set of command interneurons that drive forward locomotion (White et al. 1986).
The majority of VA motor neurons (9 of 12) arise as lineal sisters of VB motor neurons (Sulston and Horvitz 1977) and are selectively miswired with B-type motor neuron inputs in unc-4 mutants (White et al. 1992). Synaptic inputs to the embryonically derived DA motor neurons and to VAs that do not arise from a cell division that produces a VB sister (3 of 12) are not altered, however.
A carboxy-terminal eh1-like repressor domain is conserved in UNC-4-related proteins.
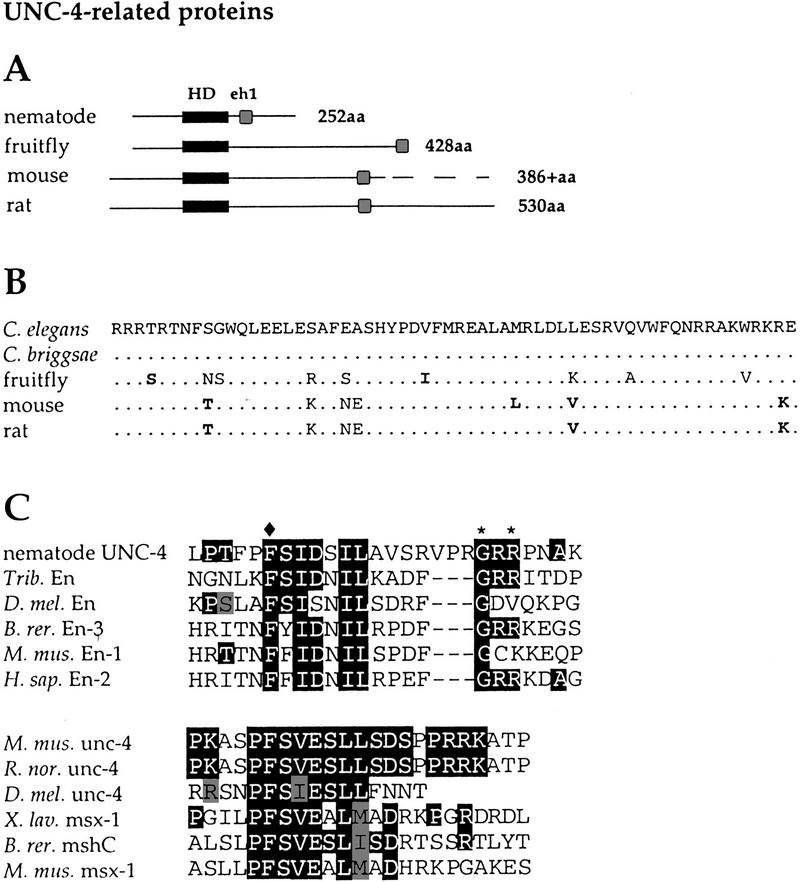
We isolated an unc-4 gene from a related nematode species by screening a Caenorhabditis briggsae fosmid grid with an unc-4 cDNA probe. The Genome Sequencing Consortium at Washington University (St. Louis, MO) then determined the complete sequence of the C. briggsae unc-4 gene. C. briggsae and C. elegans unc-4 are highly conserved in the homeodomain (100%) and in a carboxy-terminal region (97%) spanning 60 residues (Fig. 2). Close inspection of the conserved carboxy-terminal sequence revealed a region of high similarity to the eh1 repressor domain in the Engrailed homeoprotein (Smith and Jaynes 1996). This 23-residue sequence is required for Engrailed-mediated repression of target genes (Smith and Jaynes 1996; Tolkunova et al. 1998; Jimenez et al. 1999). Variants of the eh1 domain have been identified in several additional classes of homeodomain-containing proteins. All of these eh1 domains contain a conserved phenylalanine residue (position 4) that is required for Engrailed eh1 repressor function (Smith and Jaynes 1996).
Figure 2.
Comparison of UNC-4-related proteins. (A) Schematic of UNC-4-related proteins showing relative positions of the homeodomains (solid rectangles) and eh1 domains (shaded boxes). A complete sequence of the mouse UNC-4 protein has not been reported. (B) Homeodomain sequences of UNC-4-related proteins. Dots indicate identity to C. elegans UNC-4 sequence. Bold residues show similarity to UNC-4. (C) eh1 domains of nematode UNC-4 proteins (C. elegans and C. briggsae) are most similar to eh1 domains of Engrailed family members. Vertebrate and Drosophila UNC-4 eh1 domains most closely resemble the msh homeoprotein class of eh1 sequences. For nematode UNC-4 eh1 comparison, solid boxes indicate identity to nematode UNC-4 and shaded boxes show similarity. (♦) The position of the invariant phenylalanine; (*) residues mutated in the unc-4 alleles e26, e2307, and e2323. For the vertebrate and Drosophila unc-4 eh1 alignments, the solid boxes indicate identity between the mouse and rat UNC-4 proteins and at least one other protein in the comparison; the shaded boxes show similarity.
Vertebrate proteins from mouse (uncx-4.1), rat (PHD1), and zebrafish (Zunc-4) with homeodomains most similar (90%) to UNC-4 have eh1-like repressor domain sequence in their carboxy-terminal regions (Fig. 2; Rovescalli et al. 1996; Saito et al. 1996; Mansouri et al. 1997; P. Sordino and N. Holder, pers. comm.). The recently identified Drosophila UNC-4-like protein contains a truncated but still recognizable eh1 domain at its carboxy-terminal end (Fig. 2; Tabuchi et al. 1998). All of these UNC-4-like proteins exhibit the conserved phenylalanine at position 4. These findings are consistent with the proposal that UNC-4 functions as a transcriptional repressor protein in C. elegans and support the idea that other UNC-4 family members may also function as negative regulators of gene expression in more complex nervous systems.
Amino acid substitutions in the UNC-4 homeodomain and in the UNC-4 eh1 repressor domain perturb unc-4 function
The temperature-sensitive mutation unc-4(e2322ts) changes a conserved leucine to a phenylalanine (L121F) in helix 2 of the UNC-4 homeodomain (Fig. 3A; Table 1) (Miller et al. 1992). In an effort to identify other functionally important UNC-4 residues, we sequenced 12 additional unc-4 alleles. These mutations include premature stop codons (wd44, e2308), splice junction defects (e120, jd16), deletions (e887, e2320, wd1), and amino acid substitutions (e26, e2307, e2323, e521, e2321) (Fig. 3; Table 1).
Figure 3.
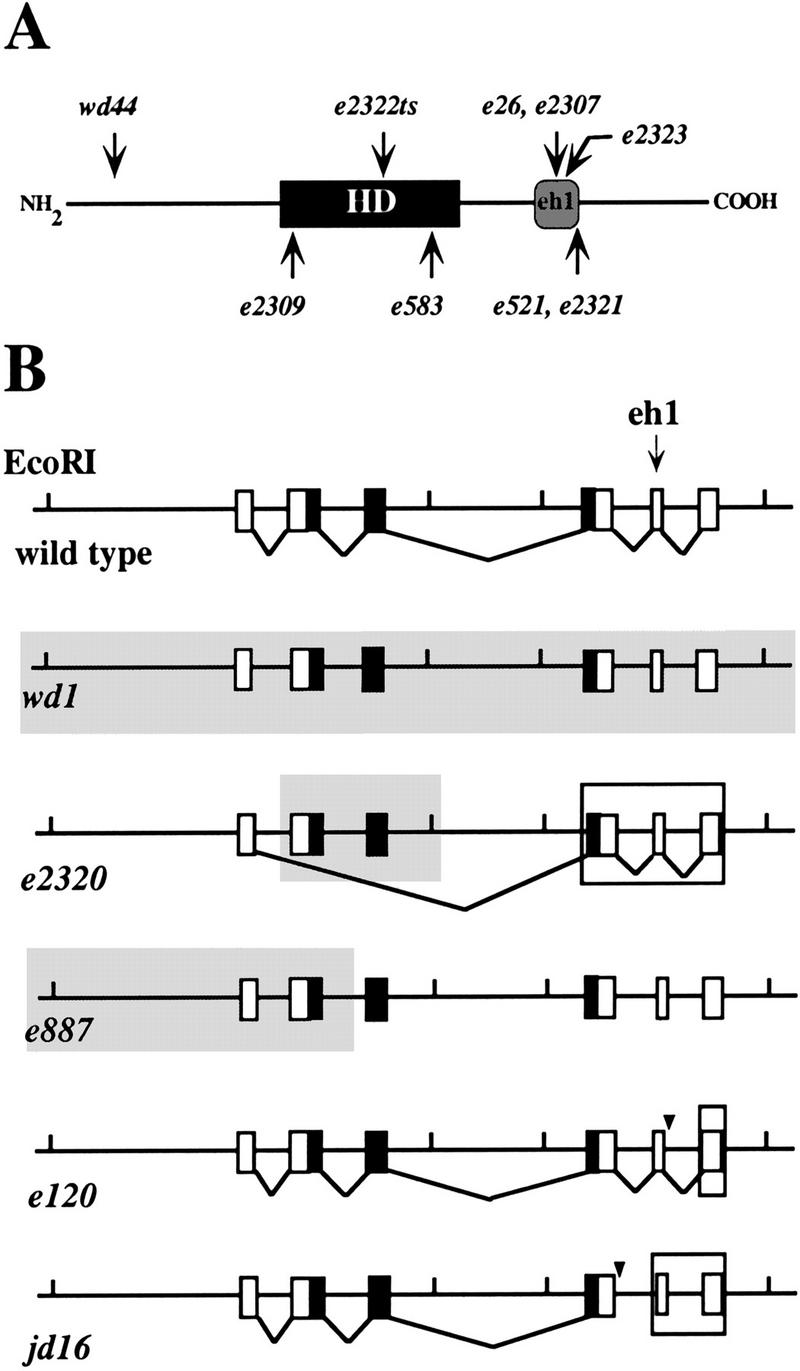
unc-4 mutations. (A) Approximate locations of UNC-4 point mutations. (HD) Homeodomain; (eh1) Engrailed-like repressor domain. (B) unc-4 frameshifts and deletions. Vertical bars denote EcoRI sites; boxes represent unc-4 exons; solid boxes show exons encoding the homeodomain (exon five contains the eh1 domain); shaded overlays denote deleted regions. Arrowheads indicate splice donor site mutations in e120 and jd16. Large open rectangles around e2320, e120, and jd16 exons denote frameshifted coding sequence.
Table 1.
UNC-37(E580K) suppresses specific unc-4 missense mutations
| Allele
|
Mutation
|
Suppresseda
|
|---|---|---|
| e2322ts | L121F | + |
| e26 | G188D | − |
| e2307 | G188S | − |
| e2323 | R190Q | + |
| e521 | R197K | + |
| e2321 | R197K | + |
| wdEx65 | F174E | − |
| wd44 | W20-stop | − |
| e2309 | W98-stop | − |
| e583 | Q137-stop | − |
| jd16 | splice donor site mutation | − |
| e120 | splice donor site mutation | − |
| e887 | deletion | − |
| e2320 | deletion | − |
| wd1 | deletion | − |
(+) unc-4 alleles that are suppressed by UNC-37 (E580K); (−) nonsuppressed unc-4 mutations.
Five amino acid substitutions in UNC-4 (e26, e2307, e2323, e521, e2321) alter residues in the vicinity of the UNC-4 eh1 domain (Fig. 3A; Table 1). It is noteworthy that these five independently derived mutations affect only three residues. Two of these amino acids, G188 and R190, are conserved in the Engrailed subclass of eh1 repressor domains (Fig. 2C; Smith and Jaynes 1996). Site-directed mutagenesis of the conserved UNC-4 eh1 phenylalanine (F174) to glutamic acid also results in a loss of unc-4 function (data not shown). Furthermore, the two unc-4 splice junction mutants jd16 and e120 are predicted to disrupt translation of the UNC-4 eh1 domain and/or sequences immediately adjacent to this repressor domain (Fig. 3B). Thus, the elimination of unc-4 activity is correlated with mutations that either truncate or alter sequence within the eh1-containing region of the UNC-4 protein.
A dominant mutation in UNC-37 (Groucho) suppresses specific missense mutations in the UNC-4 homeodomain and UNC-4 eh1 repressor domain
The C. elegans Groucho-related protein UNC-37 is required for UNC-4 function in VA motor neurons (Pflugrad et al. 1997). Four unc-37(d) alleles were originally isolated in a screen for dominant suppressors of the L121F homeodomain mutation in the unc-4(e2322ts) mutant (Miller et al. 1993). Sequence analysis revealed that all four of these unc-37(d) mutations correspond to an E580K missense mutation in the sixth WD40 repeat of UNC-37 (Pflugrad et al. 1997). The UNC-37(E580K) mutation also suppresses specific unc-4 missense mutations in the eh1 region (Table 1); wild-type movement is restored to mutants bearing the amino acid substitutions R190Q or R197K. Interestingly, the two GLY188 point mutations in the UNC-4 eh1 domain, G188D and G188S, are not suppressed by UNC-37(E580K). The movement defect of animals expressing an UNC-4 eh1 F174E mutant transgene is also not suppressed by UNC-37(E580K) (Table 1; data not shown). In addition, UNC-37(E580K) does not suppress mutants with unc-4 deletions (e887, e2320, wd1) or truncations (e120, wd44, e2308, e583) (Table 1). The allele specificity of UNC-37(E580K)-dependent suppression is consistent with the idea that UNC-4 function depends on interactions between UNC-37 and the UNC-4 homeodomain and eh1 repressor domain.
UNC-4 and UNC-37 repress B-type neuronal genes in the A-type motor neurons
The loss-of-function alleles unc-4(e120) and unc-37(e262) exhibit similar backward movement defects (Miller et al. 1993), and VA motor neurons are miswired with VB-type inputs in both mutants (White et al. 1992) (D. Hall and E. German, pers. comm.). A simple hypothesis explains this effect: UNC-4 and UNC-37 normally repress VB-specific genes; in unc-4 and unc-37 mutants, these VB genes are inappropriately activated in the VA motor neurons, thereby leading to VB-type synaptic inputs (Pflugrad et al. 1997). The identification of an eh1-like repressor domain in UNC-4 and its genetic and physical interactions (see below) with the transcriptional corepressor protein UNC-37 are consistent with this model.
In this scenario, an authentic unc-4 target gene should be expressed in VB motor neurons in wild-type animals but not in VA motor neurons. Furthermore, in either unc-4 or unc-37 mutants, these unc-4 target genes should be ectopically expressed in VA motor neurons. Using these criteria, we evaluated existing green fluorescent protein (gfp) reporters to identify two VB-specific genes, del-1 and acr-5, that are negatively regulated by UNC-4 and UNC-37 in VA motor neurons.
The DEL-1 protein is most closely related to the degenerin class of sodium channel subunits (Tavernarakis et al. 1997). Degenerins are membrane proteins that are hypothesized to form mechanically gated ion channels (Driscoll and Chalfie 1991). Using a gfp reporter gene under the control of 1.8 kb of del-1 upstream sequence, we determined that del-1 is expressed in the VB motor neurons beginning in the L2 larval stage (Fig. 4A–C; Table 2). By the end of L2, del-1::GFP is also visible in a few VA motor neurons at the anterior end of the nerve cord. Expression of del-1::GFP in the VAs progresses in a wave from anterior to posterior, with all VAs expressing del-1::GFP by the adult stage. Thus, del-1::GFP is not expressed in the VAs during the L2 period in which unc-4 functions in these cells to establish synaptic inputs (Miller et al. 1992) but is expressed in the VAs after they have been wired into the ventral cord circuit (Fig. 4A–C; Table 2). del-1::GFP is also expressed in five neurons (VB1, VB2, SABVR, SABVL, VA1) in the retrovesicular ganglion at the anterior end of the ventral nerve cord (data not shown).
Figure 4.
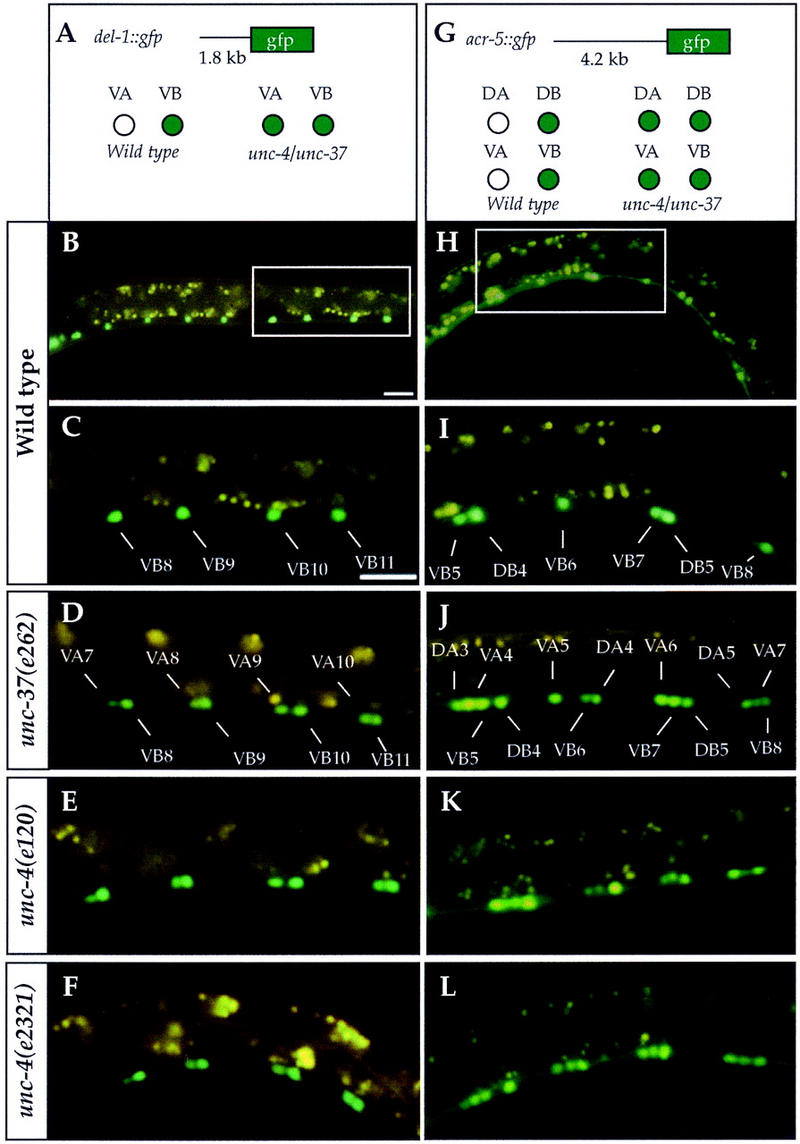
del-1::GFP and acr-5::GFP are expressed ectopically in A-type motor neurons of unc-4 and unc-37 mutants. GFP expression (green) was visualized in mid- to late L2 larval animals. Autofluorescent gut granules are yellow. (A–F) The del-1::gfp reporter gene is regulated by 1.8 kb of del-1 upstream sequence. In the ventral nerve cord of wild-type L2 animals, del-1::GFP is expressed in the VB motor neurons but is prematurely expressed in the VAs during this period in unc-37(e262) and in unc-4 mutants. (G–L) The acr-5::gfp reporter gene is regulated by 4.2 kb of acr-5::gfp upstream sequence. acr-5::GFP is expressed in B-type motor neurons (DB, VB) in ventral nerve cords of wild-type animals. In unc-4 and unc-37 mutants, acr-5::GFP is ectopically expressed in A-type motor neurons (DA, VA). Genotypes for each row are indicated at left. unc-37(e262) corresponds to UNC-37(H539Y); unc-4(e2321) corresponds to UNC-4(R197K). (C,I) Enlargements of areas indicated by insets in B and H, respectively. Unlabeled motor neurons in E, F, K, and L are identified at top. Bars, 20 μm. Bar in B applies to B and H; bar in C applies to C–L.
Table 2.
UNC-4/UNC-37-dependent repression of del-1::GFP and acr-5::GFP
| Genotype
|
del-1::GFP
|
acr-5::GFP
|
||||
|---|---|---|---|---|---|---|
| %VAs
|
%VBs
|
%DAs
|
%DBs
|
%VAs
|
%VBs
|
|
| Wild type | 17 | 100 | 5 | 100 | 1 | 95 |
| unc-37(e262) (H539Y) | 98 | 100 | 94 | 97 | 76 | 84 |
| unc-4(wd1) | 96 | 100 | 94 | 100 | 79 | 94 |
| unc-4(e521) (R197K) | 100 | 100 | 98 | 99 | 85 | 89 |
| unc-4(e2307) (G188S) | 98 | 100 | 96 | 99 | 83 | 89 |
| unc-4(e2323) (R190Q) | 96 | 100 | 93 | 100 | 49 | 94 |
| unc-4(e2322ts) (L121F) | 94 | 100 | 63 | 94 | 54 | 81 |
| unc-4(e120) | 97 | 100 | 98 | 97 | 82 | 87 |
| unc-37(d);aunc-4(e120) | 96 | 100 | 98 | 99 | 70 | 84 |
| unc-4(e26) (G188D) | 95 | 99 | 98 | 96 | 77 | 92 |
| unc-37(d);unc-4(e26) | 98 | 100 | 99 | 100 | 84 | 95 |
| unc-4(e2321) (R197K) | 95 | 100 | 95 | 99 | 82 | 89 |
| unc-37(d);unc-4(e2321) | 50 | 100 | 88 | 97 | 35 | 85 |
| unc-37(d) | 3 | 100 | 0 | 97 | 0 | 87 |
Larval (L2) animals (n ≥ 15) were scored for each genotype. Neurons scored per animal: 9–10 VAs; 9 VBs, 6 DAs; 5 DBs. unc-4 and unc-37 genotypes are shown at left; allele names and amino acid substitutions are indicated in parentheses. Percentage of GFP-positive neurons for each genotype are listed in the columns for each motor neuron class.
unc-37(d) corresponds to the UNC-37(E580K) mutation.
During the mid-L2 larval stage, del-1::GFP expression in the ventral nerve cord is largely restricted to the VB class of motor neurons (Fig. 4A–C; Table 2). In the unc-4 null alleles wd1 and e120, del-1::GFP is also expressed ectopically in the VA motor neurons in mid-L2 larvae (Table 2; Fig. 4E). An identical pattern of premature del-1::GFP expression in the VAs was observed in the hypomorphic unc-37(e262) mutant (Fig. 4D; Table 2). Thus, in both unc-4 and unc-37 mutants, del-1::GFP expression is seen in most VA and VB motor neurons in L2 larvae, whereas in wild-type animals, del-1::GFP expression is limited to VB motor neurons during this period.
acr-5 encodes an α-like subunit of a nicotinic acetylcholine receptor. An acr-5::gfp reporter gene containing a 4.2-kb acr-5 promoter fragment is expressed exclusively in the nervous system (Fig. 4G). Expression is initiated in the embryo and persists throughout adult life. In the ventral nerve cord of L1 larvae, acr-5::GFP expression is restricted to the embryonically derived DB motor neurons. VB motor neurons, which arise during the late L1 stage, begin to express acr-5::GFP in L2 larvae (Fig. 4G–I; Table 2). acr-5::GFP is also expressed in other unidentified neurons in the head and tail. We reasoned that the differential expression of acr-5::GFP in B-type neurons (DB, VB), but not in A-type motor neurons (DA, VA), could result from negative regulation by UNC-4 and UNC-37. This prediction was confirmed by examining the acr-5::gfp reporter gene in unc-4 and unc-37 mutant backgrounds. In unc-4(wd1), unc-4(e120), and unc-37(e262) mutants, acr-5::GFP is expressed in the A-type motor neurons (DA, VA), as well as in the B-type motor neurons (DB, VB) (Table 2; Fig. 4J,K). We conclude that both UNC-4 and UNC-37 are required to prevent expression of del-1::GFP and acr-5::GFP in A-type motor neurons. The simplest explanation for this effect is that UNC-4 and UNC-37 act directly on the promoter regions of these genes. Alternatively, the UNC-4/UNC-37 complex may repress an activator of del-1 and acr-5. Additional experiments will be required to distinguish between these possibilities.
The eh1 domain is required for UNC-4 repressor activity
In the experiments above, del-1::GFP and acr-5::GFP are expressed ectopically in the A-type motor neurons of unc-4 null mutants. Missense mutations in the UNC-4 eh1-like repressor domain and in the UNC-4 homeodomain also disrupt UNC-4 regulation of del-1 and acr-5. The unc-4 homeodomain mutation L121F results in derepression of del-1::gfp and acr-5::gfp in the A-type motor neurons (Table 2). In addition, missense mutations within the eh1 domain (G188D, G188S, R190Q) or in a region adjacent to the UNC-4 ehl repressor domain (R197K) disrupt repression of these B-type motor neuron-specific genes (Table 2; Fig. 4F,L). These data support the hypothesis that the UNC-4 carboxy-terminal missense mutations perturb a repressor domain and that UNC-4 normally functions in A-type motor neurons to prevent the expression of genes that are specific to B-type motor neurons. The UNC-37-dependence of UNC-4-mediated repression in C. elegans parallels the finding that Groucho is required for Engrailed repressor activity in Drosophila (Jimenez et al. 1997; Tolkunova et al. 1998).
The UNC-37(E580K) suppressor mutation restores UNC-4 repression of B-type motor neuron genes
Dominant mutations in unc-37 result in an E580K substitution that can suppress specific unc-4 missense mutations in the UNC-4 homeodomain and eh1 repressor domain (Table 1). Suppressed animals exhibit wild-type backward movement, suggesting that wild-type synaptic inputs have been restored to the VA neurons (Miller et al. 1993). If this effect is due to the restoration of UNC-4 repressor activity, then UNC-37(E580K)-mediated suppression should result in a concomitant extinction of ectopic acr-5::gfp and del-1::gfp expression in VA motor neurons. In UNC-37(E580K); UNC-4(R197K) animals, del-1::gfp is repressed in 50% of the VA neurons, as opposed to 5% of the VA neurons in UNC-37+; UNC-4(R197K) (Table 2; Fig. 5B). acr-5::gfp repression is also observed in a higher percentage (65%) of VA motor neurons in UNC-37(E580K); UNC-4(R197K) in comparison to VA neurons of UNC-37+; UNC-4(R197K) (19%) (Table 2; Fig. 5D). Thus, suppression of the Unc-4 movement phenotype is correlated with repression of B-type neuron-specific transgenes in the VA motor neurons. We note, however, that UNC-37(E580K) does not restore acr-5::gfp repression in the DA motor neurons of UNC-4(R197K) animals (Table 2; Fig. 5D). These data may indicate that the regulation of acr-5::gfp by UNC-4 differs between the DA and VA motor neurons. It is also consistent with the conclusion that the Unc-4 backward movement defect is not a consequence of unc-4 dysfunction in the adult DA motor neurons (White et al. 1992).
Figure 5.
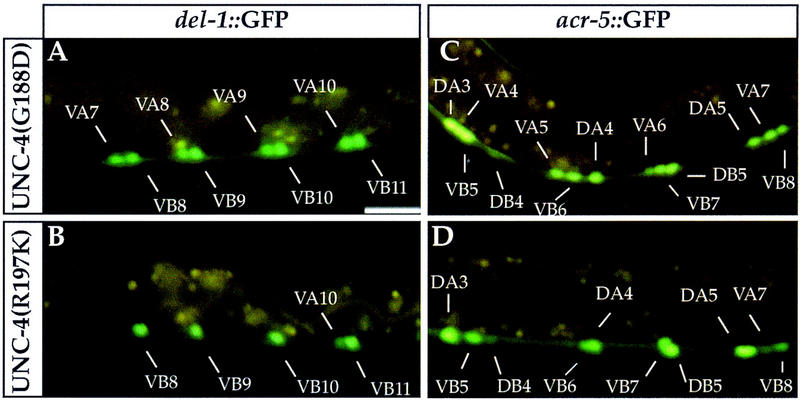
UNC-37(E580K) restores UNC-4-dependent repressor activity to specific unc-4 mutants. (A,C) UNC-37(E580K); UNC-4(G188D). UNC-4 fails to repress the B-type-specific genes in A-type motor neurons in this genetic background. (B,D) UNC-37(E580K); UNC-4(R197K). UNC-37(E580K) suppresses the Unc-4 phenotype by restoring UNC-4 repression of del-1::gfp and acr-5::gfp in A-type motor neurons (cf. Figs. 5, B,D and 4, F,L). Bar, 20 μm.
acr-5::GFP and del-1::GFP expression in two nonsuppressible mutants, UNC-4(G188D) or unc-4(e120), is not significantly altered by UNC-37(E580K) (Table 2; Fig. 5A,C). As a control, we showed that the UNC-37(E580K) mutation also has no effect on acr-5::GFP and del-1::GFP expression in B-type neurons of unc-4(+) animals (Table 2). Thus, we conclude that the UNC-37(E580K) mutation effectively restores UNC-4 function to specific unc-4 missense mutations and that this unc-4 activity represses VB-specific genes.
The UNC-4 eh1 domain is required for physical interactions with UNC-37
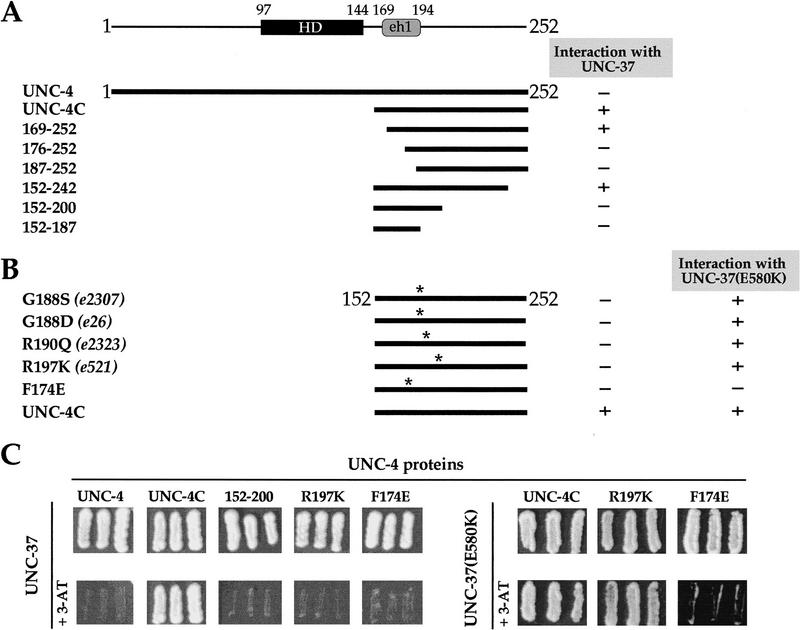
The genetic interactions between the UNC-4 eh1 domain and UNC-37 suggest specific physical contacts between UNC-4 and UNC-37 residues. Full-length UNC-4 and UNC-37 proteins do not interact in yeast two-hybrid assays or in in vitro experiments with recombinant proteins (Fig. 6A–C; Pflugrad et al. 1997). However, the UNC-4 carboxyl terminus, including the eh1 domain, strongly interacts with full-length UNC-37 in the yeast two-hybrid assay (Fig. 6A–C). Deletions that remove the UNC-4 eh1 domain dramatically reduce this interaction (compare UNC-4C with the truncated fragments 176–252 and 187–252). In addition, missense mutations in the UNC-4 eh1 region (Fig. 2C), including a glutamate substitution for the invariant eh1 phenylalanine, significantly weaken interactions with UNC-37 (Fig. 6B,C). From these results, we conclude that UNC-4/UNC-37 interactions require an intact eh1 domain.
Figure 6.
UNC-4 and UNC-37 interact in yeast two-hybrid assays. UNC-4 proteins are fused to the GAL4 DNA-binding domain; UNC-37 proteins are fused to the GAL4 activation domain. Assays for growth on His− plates were performed in the presence or absence of 3-AT: (+) growth on 100 mm 3-AT; (−) weak or no growth on 100 mm 3-AT. (A) Interactions between full-length UNC-37 and UNC-4 deletion mutants. UNC-4 refers to the full-length protein; UNC-4C represents the carboxy-terminal truncated protein (152–252). Other UNC-4 truncations are denoted by amino acid boundaries. (B) Interactions of UNC-37 and UNC-37(E580K) with UNC-4 eh1 region missense mutations. The allele numbers of previously identified unc-4 mutations are indicated. Asterisks indicate relative positions of mutations. (C) Selected growth assays ± 100 mm 3-AT from A and B.
UNC-4/UNC-37 interactions may also depend on UNC-4 carboxy-terminal sequences flanking the eh1 domain (Fig. 6A,C). Of particular note is the observation that UNC-37 interactions are perturbed by a deletion of the carboxy-terminal region (corresponding to truncated fragment 152–200) that is also eliminated by the unc-4(e120) mutation. This result is consistent with our in vivo data showing that the unc-4(e120) mutation disrupts unc-4 repressor activity, although the predicted e120 mutant protein contains an intact eh1 domain (Figs. 3B and 4E,K; Table 2).
As described previously, the UNC-37(E580K) mutation suppresses the Unc phenotype of specific unc-4 missense mutations (Table 1). One model to account for this suppression is that UNC-37(E580K) restores disrupted physical contacts between UNC-4 and UNC-37. Our yeast two-hybrid experiments confirm this prediction. UNC-37(E580K) restores interaction with the suppressible UNC-4 mutant proteins R190Q and R197K (Fig. 6B, C). Curiously, the UNC-37(E580K) mutation also restores interaction with UNC-4(G188S), although this unc-4 allele is not suppressible in vivo (Table 1). We attribute this discrepancy to differences in the sensitivity of these two assays to weak interactions. The UNC-4 F174E eh1 mutant fails to interact with UNC-37(E580K), however, which may imply a critical role for this residue in governing UNC-4 binding to UNC-37.
The simplest interpretation of the yeast two-hybrid results is that UNC-4 and UNC-37 interact directly. This interpretation is supported by recent experiments showing that UNC-4 and UNC-37 recombinant proteins bind to each other in vitro (J.Y.-J. Meir and D.M. Miller, unpubl.).
Discussion
UNC-4 and UNC-37 function together to mediate transcriptional repression
unc-4 mutants are unable to execute backward locomotion because the usual inputs to VA motor neurons are replaced with connections from interneurons normally reserved for their VB sister cells (White et al. 1992). Thus, unc-4 specifies the wild-type pattern of synaptic choice by preventing VA motor neurons from adopting VB-type presynaptic inputs (see Fig. 1). We have shown previously that unc-4 encodes a homeodomain protein (Miller et al. 1992) that is expressed in the VA motor neurons (Miller and Niemeyer 1995) and that UNC-4 activity in these cells depends on the Groucho-like UNC-37 protein (Miller et al. 1993; Pflugrad et al. 1997). Because Groucho is known to function as a transcriptional corepressor protein in Drosophila (Fisher and Caudy 1998), we have proposed that UNC-4 and UNC-37 act together as negative regulators of genes that lead to the creation of VB-type synaptic inputs (Pflugrad et al. 1997). Here we provide evidence that UNC-4 function depends on physical interaction with UNC-37 and that the resultant repressor complex prevents expression of VB-specific genes in VA motor neurons (Fig. 7A). We have identified two VB-specific genes, acr-5 and del-1, that are repressed by UNC-4 and UNC-37 in the VA motor neurons. In unc-4 mutants, ectopic expression of these genes in the VAs is associated with a backward movement defect, as well as with the miswiring of the VA neurons with VB-type inputs (Fig. 7B). UNC-4-mediated repression requires the function of a conserved Engrailed-like repressor domain, eh1, in the UNC-4 carboxyl terminus. An E580K substitution in the sixth WD40 repeat of UNC-37 restores VB-specific gene repressor activity to UNC-4 proteins with eh1 missense mutations. This dominant unc-37 allele concomitantly suppresses the backward movement defect that these unc-4 eh1 point mutants display. Furthermore, these genetic interactions are consistent with yeast two-hybrid assays which suggest that UNC-4 eh1 mutations and the UNC-37(E580K) substitution alter physical interactions between these two proteins. Our findings provide a molecular model that can explain the phenotypic similarity between unc-4 and unc-37 loss-of-function alleles. UNC-4 eh1 mutant proteins may fail to repress VB-type genes because the corepressor UNC-37 cannot be recruited to the transcription complex. Analogously, mutations in UNC-37 could prevent interactions with UNC-4 and/or perturb UNC-37 repressor activity. Thus, we propose that UNC-4/UNC-37-dependent repression of VB-specific genes in the VA neurons is necessary for the differentiation of these two sister cells during synaptic partner selection.
Figure 7.
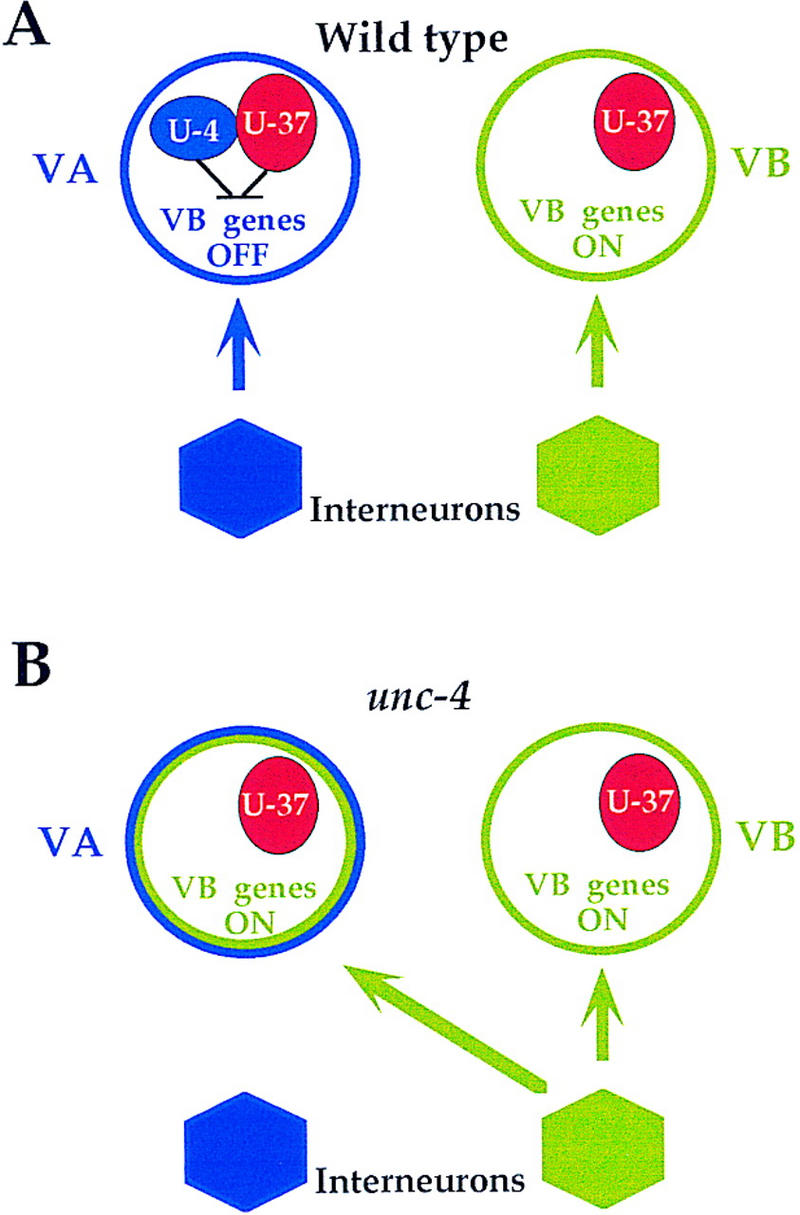
Model for synaptic choice. (A) In wild-type animals, UNC-4 and UNC-37 prevent the adoption of VB-type presynaptic inputs by repressing VB-specifying genes. (B) In the absence of unc-4 function, VB-specific genes are derepressed in the VAs thereby imposing inputs from VB-type command interneurons.
UNC-4 physically interacts with UNC-37 (Groucho)
The observation that the eh1 domain of UNC-4 mediates interactions with UNC-37 parallels the conclusions of earlier studies showing that the Drosophila Engrailed eh1 domain is necessary for interactions with Groucho (Jimenez et al. 1997; Tolkunova et al. 1998). However, our findings also differ in one important aspect. We have shown that the UNC-4 eh1 domain is not adequate for UNC-37-dependent binding or for mediating UNC-37-dependent repression. Mutant UNC-4 proteins in which the eh1 domain is intact, but are missing the flanking carboxy-terminal amino acid sequences, fail to interact with UNC-37 in the yeast two-hybrid assay and do not exhibit in vivo repressor function. In contrast, insertion of a 7 amino-acid-long eh1 core sequence within the carboxy-terminal domain of Hairy is sufficient to confer Groucho-dependent repression (Jimenez et al. 1999). At least two hypotheses may explain these differences. The UNC-4 sequence flanking the eh1 domain may have a repressor function, or alternatively, these residues may be critical for UNC-4 protein stability or conformation.
UNC-4 repressor function is effectively eliminated by a single amino acid substitution (H539Y) of UNC-37 in the hypomorphic allele e262 (Pflugrad et al. 1997). This mutation, in the fifth WD repeat of UNC-37, is predicted to disrupt a hydrogen-bonding network that stabilizes the propeller-like modular structure that individual WD domains have been shown to adopt (Wall et al. 1995; Sondek et al. 1996). The limited disruption of other unc-37-dependent functions by the e262 allele indicates that the principal effect of this mutation is to perturb unc-4 function. This conclusion is consistent with the observation that the strong backward movement defect that e262 mutants display can be rescued by unc-4 promoter-driven expression of UNC-37 in VA motor neurons (Pflugrad et al. 1997). A simple explanation for these effects is that the e262 mutation perturbs physical interactions of UNC-37 with UNC-4 but not with other classes of transcription factors. However, this prediction has not been substantiated by yeast two-hybrid assays, which detect robust binding of UNC-4 to the UNC-37(H539Y) mutant protein (J.Y.-J. Meir and D.M. Miller, unpubl.). Alternatively, the e262 mutation could disrupt an UNC-37-dependent transcriptional repressor mechanism that is uniquely employed by UNC-4. Given that this critical histidine residue is conserved in all known Groucho family members (Pflugrad et al. 1997), it will be important to distinguish between these possibilities.
A different UNC-37 point mutation, E580K, restores in vivo repressor function to specific UNC-4 missense mutations, as well as physical interaction with UNC-4. This invariant Groucho residue is predicted to reside on the surface of the sixth WD propeller domain and is therefore potentially involved in protein–protein interactions (Wall et al. 1995; Sondek et al. 1996). However, UNC-37 proteins bearing the E580K mutation do not display a mutant phenotype on their own, which indicates that interaction with the wild-type UNC-4 protein is not perturbed by this mutation (Miller et al. 1993; Pflugrad et al. 1997).
UNC-4 amino-terminal sequences regulate UNC-4/UNC-37 interactions
Full-length UNC-4 and UNC-37 proteins do not interact in yeast two-hybrid assays or in vitro (Fig. 7A; data not shown). However, we have shown here that an UNC-4 carboxy-terminal fragment, lacking homeodomain and amino-terminal residues, does associate with UNC-37. These data suggest that UNC-4 has adopted an amino-terminal-dependent intramolecular mechanism to regulate its interactions with UNC-37. The Drosophila Runt DNA-binding protein appears to use a similar mechanism to regulate its associations with the Groucho corepressor (Aronson et al. 1997). Runt exhibits both Groucho-dependent as well as Groucho-independent repressor activities. Furthermore, Runt proteins appear to function as activators in some cellular contexts, requiring strict regulation of associations with widely expressed Groucho family members. The Rel protein Dorsal, which has been shown to interact with Groucho, also exhibits activator as well as repressor functions (Dubnicoff et al. 1997). Thus, specific DNA-binding proteins may regulate their interactions with Groucho-like corepressor proteins to perform activator versus repressor functions. The proposed UNC-4 amino-terminal inhibition of UNC-37 interaction may indicate that UNC-4 is capable of UNC-37-independent gene regulation. Consistent with this idea, we have observed that UNC-4, but not UNC-37, is required for expression of del-1::GFP in specific neurons of the retrovesicular ganglion (data not shown).
UNC-4/UNC-37-dependent gene repression may be conserved
The predicted DNA-binding motif of UNC-4, the homeodomain, is 100%conserved in a nematode species (C. briggsae) that diverged from C. elegans 50 million years ago (Butler et al. 1981) and 85%–90%conserved in UNC-4-related proteins from Drosophila, zebrafish, mouse, and rat (Rovescalli et al. 1996; Saito et al. 1996; Mansouri et al. 1997; Tabuchi et al. 1998; P. Sordino and N. Holder, pers. comm.). In addition, a carboxy-terminal region that is similar to the eh1 repressor domain of Drosophila Engrailed (Smith and Jaynes 1996) is present in all of these UNC-4-like proteins. The evolutionary conservation of both of these domains within the family of UNC-4-related proteins suggests that UNC-4-dependent repressor activity may also be conserved.
The expression patterns of Drosophila and vertebrate unc-4-related genes are consistent with these proteins having nervous system functions (Saito et al. 1996; Mansouri et al. 1997; P. Sordino and N. Holder, pers. comm.). The Drosophila UNC-4-like protein DPHD-1 is selectively expressed in specific postmitotic neurons (Tabuchi et al. 1998). Similarly, the murine unc-x-4.1 transcript is expressed along the length of the neural tube in bilaterally positioned groups of cells immediately adjacent to the floor plate (C.-M. Fan, pers. comm.). The expression domains of unc-4-related genes in Drosophila and vertebrate species are not restricted to the nervous system, however. Murine unc-4, for example, is highly expressed in the kidney (Mansouri et al. 1997). This finding indicates that UNC-4 proteins may have adopted additional functions during evolution.
The Groucho-like UNC-37 corepressor has also been conserved. We have shown that the WD repeat regions of a human Groucho protein and UNC-37 are functionally interchangeable in C. elegans (Pflugrad et al. 1997). Thus, UNC-4/Groucho-dependent gene repression could represent a common mechanism for regulating synaptic target selection during neuronal development.
UNC-4 represses VB-specific genes that encode ion channel components and cell surface proteins
According to our model, UNC-4 and UNC-37 specify VA-type inputs by repressing target genes that normally mediate recognition events between VB motor neurons and appropriate interneuron partners. del-1 and acr-5 encode predicted cell surface proteins and are therefore candidates for molecules that interact with command interneurons to specify VB-type synaptic inputs. Furthermore, del-1::gfp and acr-5::gfp are expressed during the time in which VA and VB motor neurons receive synaptic inputs, indicating that the endogenous del-1 and acr-5 genes could also be regulated by UNC-4 and UNC-37 to specify synaptic choice in this motor neuron circuit.
acr-5 encodes a nicotinic acetycholine receptor subunit most similar to mammalian neuronal α-type acetylcholine receptors (nAChR) (Le Novere and Changeux 1995, Mongan et al. 1998). nAChRs are known to be expressed in terminally differentiated neurons, where they can modulate synaptic transmission (McGehee and Role 1995). There is also evidence that AChRs may function in nervous system development as well (Young and Poo 1983; Corriveau and Berg 1993; Role and Berg 1996).
del-1 encodes a member of the degenerin class of sodium channel proteins (Tavernarakis et al. 1997). Genetic studies of the degenerin family members, UNC-8, MEC-4, and MEC-10, have led to proposals that these proteins are components of mechano-transducing sodium channels (for reviews, see Chalfie 1995; Corey and Garcia-Anoveros 1996; Garcia-Anoveros and Corey 1996). In particular, UNC-8 and DEL-1 have been proposed to contribute to a stretch receptor that modulates motor neuron excitability (Tavernarakis et al. 1997). Analysis of del-1::GFP expression indicates that del-1 is expressed in both VA and VB motor neurons in the adult but is selectively repressed in VAs during the larval period (L2) in which UNC-4 functions to prevent VAs from adopting VB-type inputs (Miller et al. 1992). Thus, it is reasonable to imagine that DEL-1-dependent effects on motor neuron activity could also define presynaptic inputs during this critical period.
Alternatively, del-1 and acr-5 do not have a role in the creation of motor neuron-specific inputs but, rather, contribute to other traits that distinguish VA and VB motor neuron physiology. In that case, studies of UNC-4 and UNC-37 regulation of acr-5 and del-1 may identify new requirements for these transcription factors in motor neuron differentiation.
Repression of motor neuron-specific genes defines VA synaptic inputs
The data that we have presented in this paper provide strong support for a model in which UNC-4 and UNC-37 function together in the VA motor neurons to repress genes that promote the creation of VB-type synaptic inputs. Our findings are consistent with the hypothesis that UNC-4/UNC-37-dependent repression of these VB-specific genes is sufficient to ensure the adoption of normal inputs to the VA neurons. Alternatively, VA-type inputs may also require the activation of genes that regulate the recognition of the VA neurons by their appropriate presynaptic partners. We have no evidence that either UNC-4 or UNC-37 is required for the positive regulation of VA-specific genes, however. Thus, to delineate the mechanisms of synaptic choice in this motor neuron circuit, it may be necessary to identify VA-specific traits and their regulators as well as to define the roles of the VB-specifying genes that are inhibited by UNC-4 and UNC-37.
Materials and methods
Nematode strains
Nematodes were grown as described (Brenner 1974). All genetic experiments were performed at 25°C. The wild-type strain is N2 (Bristol). The unc-4 alleles used in this work are wd1, e521, e887, e2307, e2309, e2320, e2321, e2322ts, e2323 (Miller et al. 1992), e26, e120, e583 (Brenner 1974), wd44, and jd16. The unc-37 alleles used in this work are wd17 (unc-37(d)) (Miller et al. 1993) and e262 (Brenner 1974). The following morphological markers were used: linkage group I, dpy-5(e61), bli-4(e937); linkage group II, rol-6(e187); linkage group IV, dpy-20(e1282) (Brenner 1974).
Identification of C. briggsae unc-4
A 750-bp unc-4 cDNA probe spanning the unc-4 coding region was random primer-labeled with [α-32P]CTP (Stratagene) and hybridized to a C. briggsae high-density fosmid nylon filter (Genome Systems, Inc). Hybridization (6× SSC, 0.5%SDS) was performed at 62°C overnight. Separate autoradiographs were obtained after low stringency (2× SSC, 0.5%SDS) and high-stringency washes (0.1× SSC, 0.5%SDS at 68°C). A comparison of the X-ray films revealed five positive clones that were present after the low-stringency wash but not after the high-stringency wash. This approach was necessitated by a large number of nonspecific and strongly hybridizing clones on this filter (data not shown). Sequence analysis of one of these clones by The Genome Sequencing Consortium confirmed the presence of the C. briggsae unc-4 gene.
Sequencing of unc-4 alleles
Total RNA was isolated from mixed age populations of animals as described in Miller et al. (1992). unc-4 cDNA fragments were generated by RT–PCR with unc-4-specific primers and purified using Qiaquick spin columns (Qiagen) followed by isolation from low-melt GTG agarose (Seaplaque). Purified PCR products were subjected to automated sequencing (Applied Biosystems) using unc-4 primers. To analyze unc-4 splice junction mutants, genomic DNA from single mutant worms was PCR-amplified using unc-4 primers (Williams et al. 1992). Resultant PCR products were purified and sequenced as described above. unc-4 deletions were analyzed using a combination of the above strategies, as well as by comparing PCR product sizes to Southern blots of EcoRI-digested DNA to determine the boundaries of the deleted sequences (Miller et al. 1992). Descriptions of unc-4 primer sequences are available upon request.
Testing for unc-37(d)-dependent suppression of unc-4 mutants
The following unc-4 alleles were tested for suppression by unc-37(d) (represented by the allele unc-37(wd17)): e26, e2307, e2323, e2321, e521, wd44, e2308, e583, e887, e2320, and e120. unc-37(d) suppresses the missense allele unc- 4(e2322ts) but does not suppress the null mutation unc-4(wd1) such that the following genotype—unc-37(d) dpy-5(e61) (I); unc-4(wd1) rol-6(e187) (II)—is Unc. The morphological markers dpy-5 (dumpy) and rol-6 (roller) are closely linked to unc-37(I) and unc-4(II), respectively. Using standard genetic crosses, chromosome II of the above strain [unc-4(wd1)rol-6(e187)] was replaced by other unc-4 mutant alleles to test for suppression by unc-37(d). After confirming the homozygosity of the unc-4 mutations (i.e. no Roller progeny) as well as the presence of the unc-37(d) suppressor mutation (Dpy animals), backward mobility of the resulting animals was assessed as described previously (Miller et al. 1993).
Generation of del-1::gfp and acr-5::gfp transgenic strains
The acr-5::gfp plasmid pJR7 was constructed by subcloning a 4.2-kb SphI fragment of the acr-5 promoter into the gfp expression plasmid pPD95.70. del-1::gfp and acr-5::gfp transgenic lines (Mello and Fire 1995) were generated by injecting 50 ng/μl of each plasmid with 25 ng/μl dpy-20(+) cotransformation marker pMH86 (Clark et al. 1995) into the gonads of adult dpy-20(e1282) hermaphrodites. The del-1::gfp chromosomal integrant wdIs3 (X) was obtained by γ irradiation (4000 Rad). Standard genetic crosses were used to transfer the reporter genes wdEx60(acr-5::gfp) and wdIs3(del-1::gfp) into unc-4 and unc-37 mutant backgrounds. gfp reporter gene expression patterns in mid-L2 larval animals of unc-4 and unc-37 mutant transgenic strains were determined. Larvae were classified as mid- to late L2 by determining the number of seam cells posterior to P6.p and by scoring for the division of the postdereid cells (Sulston and Horvitz 1977). Microscopy was performed as described in Pflugrad et al. and Miller et al. (1999).
Yeast two-hybrid assays
Manipulation of the yeast strain Y190 was performed as described in Durfee et al. (1993). Yeast strains were transformed with plasmids containing full-length UNC-37 fused to the pACT2 GAL4 activation domain, as well as plasmids encoding various UNC-4 mutant proteins fused to the GAL4 DNA-binding domain from pAS2. Two-hybrid interactions were assessed by growth on selection plates (Trp−, Leu−, His−) in the presence of increasing concentrations of 3-AT (0, 10, 50, 100, 150 mm) (Durfee et al. 1993). Three independent transformants were examined for each combination. Mutant UNC-4 fusion protein stability was analyzed by Western blots of yeast protein extracts using an anti-GAL4 DNA-binding domain polyclonal antibody (Santa Cruz, Inc.) All UNC-4 fusion proteins were stably expressed (data not shown). Details of plasmid constructions are available upon request.
Acknowledgments
We thank D. Greenstein and K. Lickteig for helpful discussions and comments on the manuscript; S. Elledge for yeast strains; D. Hall, E. German, P. Sordino, N. Holder, and C.-M. Fan for sharing unpublished data; A. Fire for providing gfp vectors; G. Garriga and B. Walthall for gifts of unc-4 alleles wd44 and id16, respectively; and the Vanderbilt–Ingram Cancer Center (V.-I. Cancer Ctr) DNA Core supported by (Cancer Center Support Grant 1P30 CA68485) and the Vanderbilt Center for Molecular Neuroscience for DNA sequencing. Some of the strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, St. Paul) which is funded by National Institutes of Health (NIH) National Center for Research Resources. This work was supported by research grants from the NIH [NS 26115 (D.M.M.) and MH-11831 (A.R.W.)], from the Howard Hughes Medical Institute (J.M.R), and from the National Cancer Institute (CA 68485) (V.-I. Cancer Ctr).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL david.miller@mcmail.vanderbilt.edu; FAX (615) 343-4539.
References
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodal P. The central nervous system. Structure and Function. 2nd ed. New York, NY: Oxford University Press; 1998. [Google Scholar]
- Butler MH, Wall SM, Luehrsen KR, Fox GE, Hecht RM. Molecular relationships between closely related strains and species of nematodes. J Mol Evol. 1981;18:18–23. doi: 10.1007/BF01733207. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signaling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chalfie M. The differentiation and function of the touch receptor neurons of Caenorhabditis elegans. Prog Brain Res. 1995;105:179–182. doi: 10.1016/s0079-6123(08)63293-8. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thompson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabitis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DV, Suleman DS, Beckenbach KA, Gilchrist EJ, Baillie DL. Molecular cloning and characterization of the dpy-20 gene of Caenorhabditis elegans. Mol Gen Genet. 1995;247:367–378. doi: 10.1007/BF00293205. [DOI] [PubMed] [Google Scholar]
- Corey DP, Garcia-Anoveros J. Mechanosensation and the DEG/ENaC ion channels. Science. 1996;273:323–324. doi: 10.1126/science.273.5273.323. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Berg DK. Coexpression of multiple acetylcholine receptor genes in neurons: Quantification of transcripts during development. J Neurosci. 1993;13:2662–2671. doi: 10.1523/JNEUROSCI.13-06-02662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehni G, Liu T, Husain J, Stifani S. TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination. Mech Dev. 1995;53:369–381. doi: 10.1016/0925-4773(95)00452-1. [DOI] [PubMed] [Google Scholar]
- Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of dorsal from an activator to a repressor by the global corepressor groucho. Genes & Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. “Studies on the development and organization of the nervous system of Caenorhabditis elegans.” Ph.D. dissertation. Cambridge, UK: University of Cambridge; 1987. [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene islet-1. Science. 1992;256:1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Garcia-Anoveros J, Corey DP. Touch at the molecular level. Mechanosensation Curr Biol. 1996;6:541–543. doi: 10.1016/s0960-9822(02)00537-7. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Kolodkin AL. Specification of neuronal connectivity: ETS marks the spot. Cell. 1998;95:303–306. doi: 10.1016/s0092-8674(00)81762-6. [DOI] [PubMed] [Google Scholar]
- Hobert O, D'Alberti T, Liu Y, Ruvkun G. Control of neural development and function in a thermoregulatory network by the LIM homeobox gene lin-II. J Neurosci. 1998;18:2084–2096. doi: 10.1523/JNEUROSCI.18-06-02084.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including hairy and engrailed. Genes & Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Verrijzer CP, Ish-Horowicz D. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol Cell Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L. The development of motor projection patterns in AA chick hindlimb. J Physiol. 1978;284:391–414. doi: 10.1113/jphysiol.1978.sp012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Changeux JP. Molecular evolution of the nicotinic acetylcholine receptor: an example of multigene family in excitable cells. J Mol Evol. 1995;40:155–172. doi: 10.1007/BF00167110. [DOI] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TA, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Yokota Y, Wehr R, Copeland NG, Jenkins NA, Gruss P. Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev Dyn. 1997;210:53–65. doi: 10.1002/(SICI)1097-0177(199709)210:1<53::AID-AJA6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- McGehee DS, Role LW. Physiological diversity of nicotinic acetylcholine receptors expressed by vertebrate neurons. Annu Rev Physiol. 1995;57:521–546. doi: 10.1146/annurev.ph.57.030195.002513. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. In: Epstein H F, Shakes D C, editors. Caenorhabditis elegans: Modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. pp. 451–482. [Google Scholar]
- Miller DM, III, Niemeyer CJ. Expression of the unc-4 homeoprotein in Caenorhabditis elegans motor neurons specifies presynaptic input. Development. 1995;121:2877–2886. doi: 10.1242/dev.121.9.2877. [DOI] [PubMed] [Google Scholar]
- Miller DM, III, Shen MM, Shamu CE, Bürglin TR, Ruvkun G, Dubois ML, Ghee M, Wilson L. C. elegans unc-4 gene encodes a homeodomain protein that determines the pattern of synaptic input to specific motor neurons. Nature. 1992;355:841–845. doi: 10.1038/355841a0. [DOI] [PubMed] [Google Scholar]
- Miller DM, III, Niemeyer CJ, Chitkara P. Dominant unc-37 mutations suppress the movement defect of a homeodomain mutation in unc-4, a neural specificity gene in Caenorhabditis elegans. Genetics. 1993;135:741–753. doi: 10.1093/genetics/135.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, III, Desai NS, Hardin DC, Piston DW, Patterson GH, Fleenor J, Xu S, Fire A. Two-color GFP expression system for C. elegans. BioTechniques. 1999;26:914–921. doi: 10.2144/99265rr01. [DOI] [PubMed] [Google Scholar]
- Mongan NP, Baylis HA, Adcock C, Smith GR, Sansom MS, Satelle DB. An extensive and diverse gene family of nicotinic acetylcholine receptor alpha subunits in Caenorhabditis elegans. Receptors Channels. 1998;6:213–218. [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pflugrad A, Meir JY-J, Barnes TM, Miller DM., III The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development. 1997;124:1699–1709. doi: 10.1242/dev.124.9.1699. [DOI] [PubMed] [Google Scholar]
- Rand J, Nonet M. Synaptic transmission. In: Riddle DL, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1997. pp. 611–643. [PubMed] [Google Scholar]
- Role LW, Berg DK. Nicotinic receptors in the development and modulation of CNS synapses. Neuron. 1996;16:1077–1085. doi: 10.1016/s0896-6273(00)80134-8. [DOI] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Rovescalli AC, Asoh S, Nirenberg M. Cloning and characterization of four murine homeobox genes. Proc Natl Acad Sci. 1996;93:10691–10696. doi: 10.1073/pnas.93.20.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Lo L, Anderson DJ, Mikoshiba K. Identification of a novel paired homeodomain protein related to C. elegans UNC-4 as a potential downstream target of MASH1. Dev Biol. 1996;180:143–155. doi: 10.1006/dbio.1996.0291. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Smith ST, Jaynes JB. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein dimer at 2.1A resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Yoshikawa S, Yuasa Y, Sawamoto K, Okano H. A novel Drosophila paired-like homeobox gene related to Caenorhabditis elegans unc-4 is expressed in subsets of postmitotic neurons and epidermal cells. Neurosci Lett. 1998;257:49–52. doi: 10.1016/s0304-3940(98)00799-x. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Shreffler W, Wang S, Driscoll M. unc-8, a DEG/ENaC family member, encodes a subunit of a candidate mechanically gated channel that modulates C. elegans locomotion. Neuron. 1997;18:107–119. doi: 10.1016/s0896-6273(01)80050-7. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Tolkunova EN, Fujioka M, Kobayashi M, Deka D, Jaynes JB. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol Cell Biol. 1998;18:2804–2814. doi: 10.1128/mcb.18.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baidassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Young SH, Poo MM. Spontaneous release of transmitter from growth cones of embryonic neurones. Nature. 1983;305:634–637. doi: 10.1038/305634a0. [DOI] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc Lond B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN. Mutations in the Caenorhabditis elegans unc-4 gene alter the synaptic input to ventral cord motor neurons. Nature. 1992;355:838–841. doi: 10.1038/355838a0. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic-mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]