Abstract
Questions of understanding and quantifying the representation and amount of information in organisms have become a central part of biological research, as they potentially hold the key to fundamental advances. In this paper, we demonstrate the use of information-theoretic tools for the task of identifying segments of biomolecules (DNA or RNA) that are statistically correlated. We develop a precise and reliable methodology, based on the notion of mutual information, for finding and extracting statistical as well as structural dependencies. A simple threshold function is defined, and its use in quantifying the level of significance of dependencies between biological segments is explored. These tools are used in two specific applications. First, they are used for the identification of correlations between different parts of the maize zmSRp32 gene. There, we find significant dependencies between the 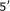 untranslated region in zmSRp32 and its alternatively spliced exons. This observation may indicate the presence of as-yet unknown alternative splicing mechanisms or structural scaffolds. Second, using data from the FBI's combined DNA index system (CODIS), we demonstrate that our approach is particularly well suited for the problem of discovering short tandem repeats—an application of importance in genetic profiling.
untranslated region in zmSRp32 and its alternatively spliced exons. This observation may indicate the presence of as-yet unknown alternative splicing mechanisms or structural scaffolds. Second, using data from the FBI's combined DNA index system (CODIS), we demonstrate that our approach is particularly well suited for the problem of discovering short tandem repeats—an application of importance in genetic profiling.
Contributor Information
Hasan Metin Aktulga, Email: haktulga@cs.purdue.edu.
Ioannis Kontoyiannis, Email: yiannis@aueb.gr.
L Alex Lyznik, Email: alex.lyznik@pioneer.com.
Lukasz Szpankowski, Email: lszpanko@bioinf.ucsd.edu.
Ananth Y Grama, Email: ayg@cs.purdue.edu.
Wojciech Szpankowski, Email: spa@cs.purdue.edu.
References
- Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18(supplement 2):S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- Dawy Z, Goebel B, Hagenauer J, Andreoli C, Meitinger T, Mueller JC. Gene mapping and marker clustering using Shannon's mutual information. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2006;3(1):47–56. doi: 10.1109/TCBB.2006.9. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L. et al. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Osada Y, Saito R, Tomita M. Comparative analysis of base correlations in
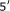 untranslated regions of various species. Gene. 2006;375(1-2):80–86. doi: 10.1016/j.gene.2006.02.018. [DOI] [PubMed] [Google Scholar]
untranslated regions of various species. Gene. 2006;375(1-2):80–86. doi: 10.1016/j.gene.2006.02.018. [DOI] [PubMed] [Google Scholar] - Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. doi: 10.1016/S0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Reddy DA, Mitra CK. Comparative analysis of transcription start sites using mutual information. Genomics, Proteomics and Bioinformatics. 2006;4(3):189–195. doi: 10.1016/S1672-0229(06)60032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy DA, Prasad BVLS, Mitra CK. Comparative analysis of core promoter region: information content from mono and dinucleotide substitution matrices. Computational Biology and Chemistry. 2006;30(1):58–62. doi: 10.1016/j.compbiolchem.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Research. 2004;32(5):1774–1782. doi: 10.1093/nar/gkh313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Brunak S, Frasconi P, Soda G, Pollastri G. Exploiting the past and the future in protein secondary structure prediction. Bioinformatics. 1999;15(11):937–946. doi: 10.1093/bioinformatics/15.11.937. [DOI] [PubMed] [Google Scholar]
- Battail G. Should genetics get an information-theoretic education? Genomes as error-correcting codes. IEEE Engineering in Medicine and Biology Magazine. 2006;25(1):34–45. doi: 10.1109/memb.2006.1578662. [DOI] [PubMed] [Google Scholar]
- Gao H, Gordon-Kamm WJ, Lyznik LA. ASF/SF2-like maize pre-mRNA splicing factors affect splice site utilization and their transcripts are alternatively spliced. Gene. 2004;339(1-2):25–37. doi: 10.1016/j.gene.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Cover TM, Thomas JA. Elements of Information Theory. John Wiley & Sons, New York, NY, USA; 1991. [Google Scholar]
- Good PI. Resampling Methods. Birkhäuser, Boston, Mass, USA; 2005. [Google Scholar]
- Manly B. Randomization, Bootstrap and Monte Carlo Methods in Biology. Chapman & Hall/CRC, Boca Raton, Fla, USA; 1977. [Google Scholar]
- Lehmann EL, Romano JP. Testing Statistical Hypotheses. 3. Springer, New York, NY, USA; 2005. [Google Scholar]
- Schervish MJ. Theory of Statistics. Springer, New York, NY, USA; 1995. [Google Scholar]
- Hagenauer J, Dawy Z, Göbel B, Hanus P, Mueller J. Genomic analysis using methods from information theory. Proceedings of IEEE Information Theory Workshop (ITW '04), San Antonio, Tex, USA, October 2004. pp. 55–59.
- Goebel B, Dawy Z, Hagenauer J, Mueller JC. An approximation to the distribution of finite sample size mutual information estimates. Proceedings of IEEE International Conference on Communications (ICC '05), Seoul, Korea, May 2005. pp. 1102–1106.
- Hutter M. Advances in Neural Information Processing Systems 14. MIT Press, Cambridge, Mass, USA; 2002. Distribution of mutual information; pp. 399–406. [Google Scholar]
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends in Genetics. 2006;22(3):119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Åberg J, Shtarkov YuM, Smeets BJM. Multialphabet coding with separate alphabet description. Proceedings of the International Conference on Compression and Complexity of Sequences, Positano, Italy, June 1997. pp. 56–65.
- Orlitsky A, Santhanam NP, Viswanathan K, Zhang J. Limit results on pattern entropy. IEEE Transactions on Information Theory. 2006;52(7):2954–2964. [Google Scholar]


