Abstract
The identification and analysis of repetitive patterns are active areas of biological and computational research. Tandem repeats in telomeres play a role in cancer and hypervariable trinucleotide tandem repeats are linked to over a dozen major neurodegenerative genetic disorders. In this paper, we present an algorithm to identify the exact and inexact repeat patterns in DNA sequences based on orthogonal exactly periodic subspace decomposition technique. Using the new measure our algorithm resolves the problems like whether the repeat pattern is of period  or its multiple (i.e., 2
or its multiple (i.e., 2 , 3
, 3 , etc.), and several other problems that were present in previous signal-processing-based algorithms. We present an efficient algorithm of
, etc.), and several other problems that were present in previous signal-processing-based algorithms. We present an efficient algorithm of 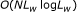 , where
, where  is the length of DNA sequence and
is the length of DNA sequence and 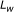 is the window length, for identifying repeats. The algorithm operates in two stages. In the first stage, each nucleotide is analyzed separately for periodicity, and in the second stage, the periodic information of each nucleotide is combined together to identify the tandem repeats. Datasets having exact and inexact repeats were taken up for the experimental purpose. The experimental result shows the effectiveness of the approach.
is the window length, for identifying repeats. The algorithm operates in two stages. In the first stage, each nucleotide is analyzed separately for periodicity, and in the second stage, the periodic information of each nucleotide is combined together to identify the tandem repeats. Datasets having exact and inexact repeats were taken up for the experimental purpose. The experimental result shows the effectiveness of the approach.
Contributor Information
Ravi Gupta, Email: rgcsedec@iitr.ernet.in.
Divya Sarthi, Email: samaypec@iitr.ernet.in.
Ankush Mittal, Email: ankumfec@iitr.ernet.in.
Kuldip Singh, Email: ksd56fec@iitr.ernet.in.
References
- Hahn WC. Telomerase and cancer: where and when? Clinical Cancer Research. 2001;7(10):2953–2954. [PubMed] [Google Scholar]
- Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL, Shlyakhtenko LS. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. Journal of Biosciences. 2002;27(1, supplement 1):53–65. doi: 10.1007/BF02703683. [DOI] [PubMed] [Google Scholar]
- Siyanova EY, Mirkin SM. Expansion of trinucleotide repeats. Molecular Biology. 2001;35(2):168–182. doi: 10.1023/A:1010431232481. [DOI] [PubMed] [Google Scholar]
- Tamaki K, Jeffreys AJ. Human tandem repeat sequences in forensic DNA typing. Legal Medicine. 2005;7(4):244–250. doi: 10.1016/j.legalmed.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpakov R, Bana G, Kucherov G. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Research. 2003;31(13):3672–3678. doi: 10.1093/nar/gkg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau GM, Schmidt JP, Sokol D. An algorithm for approximate tandem repeats. Journal of Computational Biology. 2001;8(1):1–18. doi: 10.1089/106652701300099038. [DOI] [PubMed] [Google Scholar]
- Adebiyi EF, Jiang T, Kaufmann M. An efficient algorithm for finding short approximate non-tandem repeats. Bioinformatics. 2001;17(1):S5–S12. doi: 10.1093/bioinformatics/17.suppl_1.S5. [DOI] [PubMed] [Google Scholar]
- Hauth AM, Joseph DA. Beyond tandem repeats: complex pattern structures and distant regions of similarity. Bioinformatics. 2002;18(1):S31–S37. doi: 10.1093/bioinformatics/18.suppl_1.S31. [DOI] [PubMed] [Google Scholar]
- Sharma D, Issac B, Raghava GPS, Ramaswamy R. Spectral repeat finders (SRF): identification of repetitive sequences using Fourier transformation. Bioinformatics. 2004;20(9):1405–1412. doi: 10.1093/bioinformatics/bth103. [DOI] [PubMed] [Google Scholar]
- Tran TT, Emanuele VA, II, Zhou GT. Techniques for detecting approximate tandem repeats in DNA. Proceedings of IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP '04), Montreal, Quebec, Canada, May 2004. pp. 449–452.
- Buchner M, Janjarasjitt S. Detection and visualization of tandem repeats in DNA sequences. IEEE Transactions on Signal Processing. 2003;51(9):2280–2287. doi: 10.1109/TSP.2003.815396. [DOI] [Google Scholar]
- Muresan DD, Parks TW. Orthogonal, exactly periodic subspace decomposition. IEEE Transactions on Signal Processing. 2003;51(9):2270–2279. doi: 10.1109/TSP.2003.815381. [DOI] [Google Scholar]
- Anastassiou D. Genomic signal processing. IEEE Signal Processing Magazine. 2001;18(4):8–20. doi: 10.1109/79.939833. [DOI] [Google Scholar]
- Tiwari S, Ramachandran S, Bhattacharya A, Bhattacharya S, Ramaswamy R. Prediction of probable genes by Fourier analysis of genomic sequences. Computer Applications in the Biosciences. 1997;13(3):263–270. doi: 10.1093/bioinformatics/13.3.263. [DOI] [PubMed] [Google Scholar]
- Otten AD, Tapscott SJ. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(12):5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem Repeat Finder. http://tandem.bu.edu/trf/trf.html.
- Hauth AM. Identification of tandem repeats simple and complex pattern structures in DNA, Ph.D. dissertation.
- Bussey H, Kaback DB, Zhong W. et al. The nucleotide sequence of chromosome I from Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(9):3809–3813. doi: 10.1073/pnas.92.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]


