Abstract
The modeling of genetic regulatory networks is becoming increasingly widespread in the study of biological systems. In the abstract, one would prefer quantitatively comprehensive models, such as a differential-equation model, to coarse models; however, in practice, detailed models require more accurate measurements for inference and more computational power to analyze than coarse-scale models. It is crucial to address the issue of model complexity in the framework of a basic scientific paradigm: the model should be of minimal complexity to provide the necessary predictive power. Addressing this issue requires a metric by which to compare networks. This paper proposes the use of a classical measure of difference between amplitude distributions for periodic signals to compare two networks according to the differences of their trajectories in the steady state. The metric is applicable to networks with both continuous and discrete values for both time and state, and it possesses the critical property that it allows the comparison of networks of different natures. We demonstrate application of the metric by comparing a continuous-valued reference network against simplified versions obtained via quantization.
[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23]
Contributor Information
Marcel Brun, Email: mbrun@tgen.org.
Seungchan Kim, Email: skim@tgen.org.
Woonjung Choi, Email: Woonjung.Choi@asu.edu.
Edward R Dougherty, Email: e-dougherty@tamu.edu.
References
- De Jong H. Modeling and simulation of genetic regulatory systems: a literature review. Journal of Computational Biology. 2002;9(1):67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- Srivastava R, You L, Summers J, Yin J. Stochastic vs. deterministic modeling of intracellular viral kinetics. Journal of Theoretical Biology. 2002;218(3):309–321. doi: 10.1006/jtbi.2002.3078. [DOI] [PubMed] [Google Scholar]
- Albert R, Barabási A-L. Statistical mechanics of complex networks. Reviews of Modern Physics. 2002;74(1):47–97. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- Kim S, Li H, Dougherty ER. et al. Can Markov chain models mimic biological regulation? Journal of Biological Systems. 2002;10(4):337–357. doi: 10.1142/S0218339002000676. [DOI] [Google Scholar]
- Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. Journal of Theoretical Biology. 2003;223(1):1–18. doi: 10.1016/S0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aburatani S, Tashiro K, Savoie CJ. et al. Discovery of novel transcription control relationships with gene regulatory networks generated from multiple-disruption full genome expression libraries. DNA Research. 2003;10(1):1–8. doi: 10.1093/dnares/10.1.1. [DOI] [PubMed] [Google Scholar]
- Goutsias J, Kim S. A nonlinear discrete dynamical model for transcriptional regulation: construction and properties. Biophysical Journal. 2004;86(4):1922–1945. doi: 10.1016/S0006-3495(04)74257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhan M. Systematic intervention of transcription for identifying network response to disease and cellular phenotypes. Bioinformatics. 2006;22(1):96–102. doi: 10.1093/bioinformatics/bti752. [DOI] [PubMed] [Google Scholar]
- Datta A, Choudhary A, Bittner ML, Dougherty ER. External control in Markovian genetic regulatory networks. Machine Learning. 2003;52(1-2):169–191. doi: 10.1093/bioinformatics/bth008. [DOI] [PubMed] [Google Scholar]
- Choudhary A, Datta A, Bittner ML, Dougherty ER. Control in a family of boolean networks. IEEE International Workshop on Genomic Signal Processing and Statistics (GENSIPS '06), College Station, Tex, USA, May 2006.
- Devroye L, Györffi L, Lugosi G. A Probabilistic Theory of Pattern Recognition. Springer, New York, NY, USA; 1996. [Google Scholar]
- Ivanov I, Dougherty ER. Modeling genetic regulatory networks: continuous or discrete? Journal of Biological Systems. 2006;14(2):219–229. doi: 10.1142/S0218339006001763. [DOI] [Google Scholar]
- Ivanov I, Dougherty ER. Reduction mappings between probabilistic boolean networks. EURASIP Journal on Applied Signal Processing. 2004;2004(1):125–131. doi: 10.1155/S1110865704309182. [DOI] [Google Scholar]
- Ott S, Imoto S, Miyano S. Finding optimal models for small gene networks. Proceedings of the Pacific Symposium on Biocomputing (PSB '04), Big Island, Hawaii, USA, January 2004. pp. 557–567. [DOI] [PubMed]
- Wessels LF, van Someren EP, Reinders MJ. A comparison of genetic network models. Proceedings of the Pacific Symposium on Biocomputing (PSB '01), Lihue, Hawaii, USA, January 2001. pp. 508–519. [PubMed]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. Oxford University Press, New York, NY, USA; 1993. [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science, New York, NY, USA; 2002. [Google Scholar]
- Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of Theoretical Biology. 1969;22(3):437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Lynn PA. An Introduction to the Analysis and Processing of Signals. John Wiley & Sons, New York, NY, USA; 1973. [Google Scholar]
-
Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage
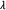 -infected Escherichia coli cells. Genetics. 1998;149(4):1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
-infected Escherichia coli cells. Genetics. 1998;149(4):1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar] - Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(11):5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO Journal. 1999;18(23):6705–6717. doi: 10.1093/emboj/18.23.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]


