Abstract
The cyclin-dependent kinases 4 and 6 (Cdk4/6) that drive progression through the G1 phase of the cell cycle play a central role in the control of cell proliferation, and CDK deregulation is a frequent event in cancer. Cdk4/6 are regulated by the D-type cyclins, which bind to CDKs and activate the kinase, and by the INK4 family of inhibitors. INK4 proteins can bind both monomeric CDK, preventing its association with a cyclin, and also the CDK-cyclin complex, forming an inactive ternary complex. In vivo, binary INK4–Cdk4/6 complexes are more abundant than ternary INK4–Cdk4/6–cyclinD complexes, and it has been suggested that INK4 binding may lead to the eventual dissociation of the cyclin. Here we present the 2.9-Å crystal structure of the inactive ternary complex between Cdk6, the INK4 inhibitor p18INK4c, and a D-type viral cyclin. The structure reveals that p18INK4c inhibits the CDK–cyclin complex by distorting the ATP binding site and misaligning catalytic residues. p18INK4c also distorts the cyclin-binding site, with the cyclin remaining bound at an interface that is substantially reduced in size. These observations support the model that INK4 binding weakens the cyclin's affinity for the CDK. This structure also provides insights into the specificity of the D-type cyclins for Cdk4/6.
Keywords: Cdk6, INK4, Viral cyclin, CDK inhibition, cell cycle
Cyclin-dependent kinases (CDKs) are a family of closely related Ser/Thr protein kinases that coordinate the cell's progression through the cell cycle by switching between active and inactive states. CDKs, inactive in the monomeric state, are activated by the binding of cyclins, which impart basal activity to the kinase, and by phosphorylation, which fully activates the kinase. CDKs are inactivated by the binding of CDK inhibitors (CKIs) or by additional phosphorylation (for review, see Morgan 1995; Pavletich 1999). The closely related cyclinD-dependent kinases Cdk4 and Cdk6 drive the cell's progression through the G1 phase of the cell cycle. G1 progression is dependent on extracellular mitogenic signals that activate Cdk4/6 by up-regulating levels of their cyclinD activators (Sherr 1994). Cdk4/6 in turn can be inactivated by the INK4 family of CKIs, which are induced by a variety of antiproliferative signals such as TGF-β (p15INK4b), senescence (p16INK4a), and terminal differentiation (p18INK4c and p19INK4d) (for review, see Sherr and Roberts 1999). Cdk4/6 and their regulators have a central role in the control of cell proliferation (Sherr 1996), as underscored by their frequent alteration in cancer (for review, see Hall and Peters 1996): An INK4 inhibitor, p16INK4a (p16), is a tumor suppressor that is altered in >50% of certain tumor types (Kamb et al. 1994; Nobori et al. 1994; Hall and Peters 1996); cyclinD is often amplified in breast and prostate cancer (Hall and Peters 1996); mutations in Cdk4 that render it refractory to inhibition by INK4 have been identified in several cancer cases (Wolfel et al. 1995; Zuo et al. 1996). In addition, certain γ herpes viruses including Kaposi's sarcoma-associated herpes virus (KSHV/HHV8; Chang et al. 1994) express an oncogenic D-type cyclin that can bind to and activate Cdk4/6 and contribute to the deregulation of the cell cycle (Godden-Kent et al. 1997; Li et al. 1997).
INK4 inhibitors can bind both monomeric Cdk4/6 (Serrano et al. 1993; Hall et al. 1995; Parry et al. 1995) and cyclinD-bound Cdk4/6 (Hirai et al. 1995; Adachi et al. 1997; Reynisdottir and Massague 1997) and appear to use multiple mechanisms to inhibit the CDK and halt the progression of the cell cycle. INK4 binding to the monomeric Cdk4/6 subunit interferes with the subsequent binding of cyclinD and renders the CDK nonactivatable (Parry et al. 1995; Guan et al. 1996; McConnell et al. 1999). This is, at least in part, because of the requirement of an Hsp90–Cdc37 heat shock protein complex for the assembly of the Cdk4/6–cyclinD complex (Stepanova et al. 1996) and the ability of INK4s to compete with Cdc37 binding to Cdk4/6 (Lamphere et al. 1997; Russo et al. 1998). However, if the Cdk4/6–cyclinD complex is assembled first, INK4s can bind the Cdk4/6–cyclin complex and inhibit it without dissociating the cyclin (Hirai et al. 1995; Adachi et al. 1997). The relative contribution of these two INK4 activities to cell cycle arrest has been a point of debate, as several studies have shown that the majority of p16 is found in binary complexes with Cdk4/6 in vivo (Parry et al. 1995; McConnell et al. 1999). However, it has also been shown that induction of p15INK4b or p19INK4d (p19) to levels that arrest cell growth results in the formation of INK4–cyclinD–Cdk4/6 complexes without causing immediate dissociation of cyclinD from Cdk4/6 (Adachi et al. 1997; Reynisdottir and Massague 1997).
Previous crystallographic studies of the p16–Cdk6 (Russo et al. 1998) and p19–Cdk6 (Brotherton et al. 1998; Russo et al. 1998) binary complexes showed how these inhibitors bind the monomeric CDK and, in comparison with structures of monomeric Cdk2 (De Bondt et al. 1993) and Cdk2–cyclinA complexes (Jeffrey et al. 1995; Russo et al. 1996), showed that INK4 binding to Cdk6 caused conformational changes that distorted the catalytic cleft and allosterically altered the cyclin binding site. However, the way in which INK4s bind the preformed CDK–cyclin complex, how they inhibit its activity, and how the cyclin remains bound to the CDK has not been clear.
To address these questions, we have determined the structure of a p18INK4c–Cdk6–D-type cyclin ternary complex. We used the D-type cyclin encoded by the Kaposi's sarcoma–associated herpesvirus (K-cyclin; Godden-Kent et al. 1997; Li et al. 1997), as we were not able to produce a cellular D-type cyclin in a form suitable for crystallization. The Cdk6–K-cyclin complex has been shown to be resistant to inhibition by INK4s (Swanton et al. 1997), but in this study, we show that when Cdk6 is unphosphorylated, the Cdk6–K-cyclin complex is susceptible to inhibition by INK4s.
The structure of the ternary p18–Cdk6–K-cyclin complex shows that Cdk6 adopts a conformation where residues involved in ATP binding and catalysis are misaligned. This active site distortion is similar to that seen in the binary INK4–Cdk6 complexes. The cyclin-binding site is also distorted, with the cyclin remaining bound in a nonfunctional way, via an interface that is 30% smaller in size. This suggests that INK4 binding reduces the stability of the CDK–cyclin interface. The structure also provides insights into how phosphorylated Cdk6 bound to the viral cyclin may evade inhibition by INK4s.
Results and Discussion
Biochemical characterization of the p18–Cdk6–K-cyclin complex
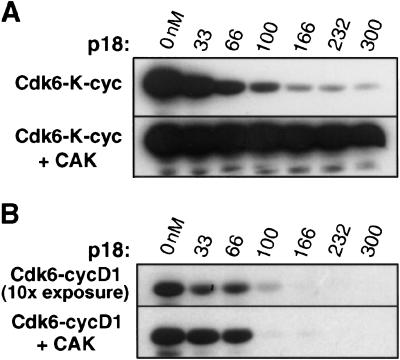
Unphosphorylated Cdk6 bound to K-cyclin has basal kinase activity toward the retinoblastoma protein (Rb) that is only slightly lower than that of the fully active phosphorylated Cdk6–K-cyclin and Cdk6–cyclinD complexes (Fig. 1A,B), suggesting that K-cyclin is more effective than cyclinD as an activator of Cdk6. Previous studies had shown that the Cdk6–K-cyclin complex is resistant to inhibition by the INK4 inhibitors (Swanton et al. 1997), but we find that this is the case only when Cdk6 is phosphorylated. As seen in Figure 1A, the unphosphorylated complex is inhibited by approximately one molar equivalent p18, whereas the phosphorylated complex is uninhibited under the same conditions. We thus used the structure of the unphosphorylated Cdk6 bound to p18 and K-cyclin as a model system to understand inhibition of Cdk4/6–cyclinD complexes.
Figure 1.
The Rb kinase activity of the Cdk6–K-cyclin complex is inhibited by p18 when Cdk6 is unphosphorylated but not when Cdk6 is phosphorylated by the Cdk7–cyclinH CDK-activating kinase. (A) Phosphorylation of the Rb C-terminal fragment by the unphosphorylated Cdk6–K-cyclin complex (top panel) and the phosphorylated Cdk6–K-cyclin complex (lower panel). Both Cdk6 complexes are at 100 nM concentration. The lanes in the two panels contain p18 at 0, 33, 66, 100, 166, 232, and 300 nM concentrations. (B) Comparison with the Rb kinase activity of the Cdk6–cyclinD complex. Reactions contain the same concentration (100 nM) of Cdk6 and p18 as in the corresponding lanes in A. The gel in the bottom panel was exposed for the same length of time as the panels in A; the top panel showing the activity of unphosphorylated Cdk6–cyclinD complex was exposed 10-fold longer because of the low kinase activity of the unphosphorylated complex.
Overall structure of the p18INK4c–Cdk6–K-cyclin complex
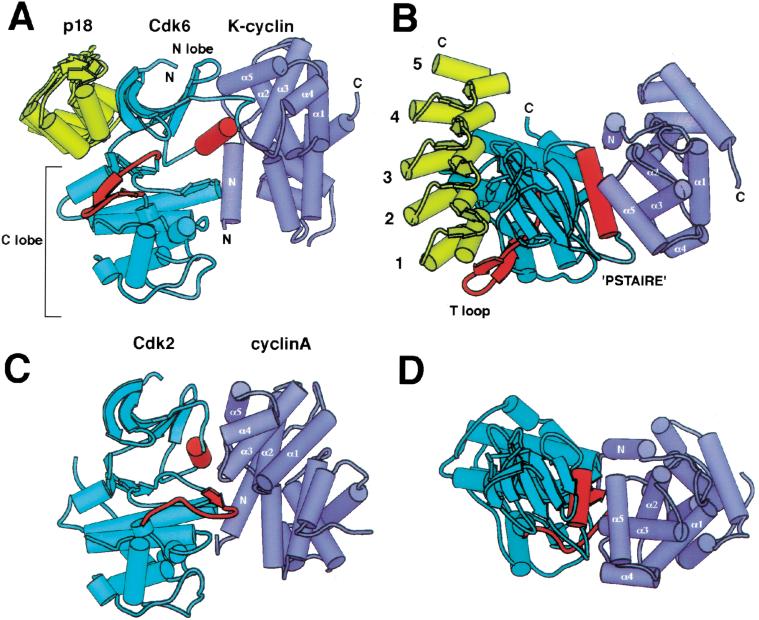
In the crystal structure of the p18–Cdk6–K-cyclin ternary complex, p18 and K-cyclin bind to opposite sides of the Cdk6 catalytic cleft and do not interact with each other (Fig. 2A,B). The p18–Cdk6 interface is similar in size (1850 Å2 surface area buried) to the interfaces in the binary p16–Cdk6 (2570 Å2) and p19–Cdk6 (2070 Å2) complexes (Brotherton et al. 1998; Russo et al. 1998). The Cdk6–K-cyclin interface, however, is much less extensive than that of the active unphosphorylated Cdk2–cyclinA complex: 2504 Å2 compared with 3540 Å2 (Jeffrey et al. 1995) surface area buried.
Figure 2.
Overall structure of the p18–Cdk6–K-cyclin complex and comparison with Cdk2–cyclinA. (A) Schematic view of p18–Cdk6–K-cyclin. p18 is shown in yellow, Cdk6 in cyan, K-cyclin in purple. The T loop and PSTAIRE elements of Cdk6 are highlighted in red, and the helices of the first cyclin repeat are labeled. N and C termini are labeled where visible. The p18–Cdk6 and K-cyclin–Cdk6 interfaces do not overlap and lie on opposite sides of the kinase, burying a total of 4350 Å2 of surface area. (B) Top view of the p18–Cdk6–K-cyclin complex, approximately orthogonal to view in A. The ankyrin repeats of p18 are numbered. The PSTAIRE helix is central to the Cdk6–K-cyclin interface, but the T loop packs on the other side of the kinase. (C) View of Cdk2–cyclinA complex superimposed on the C lobe of Cdk6 in the same orientation as in A. Both the PSTAIRE helix and T loop, in red, pack against cyclinA. (D) View of superimposed Cdk2–cyclinA complex from same viewpoint as B.
The structure of Cdk6, in common with Cdk2 and other eukaryotic kinase structures (Taylor and Radzio-Andzelm 1994), consists of an N-terminal lobe rich in β sheet (N lobe, residues 9–104) and a larger C-terminal lobe that is predominantly α-helical (C lobe, residues 105–301; Fig. 2A). The catalytic cleft lies at the junction of these two lobes, with active site residues being contributed by both lobes.
p18, which consists of five ankyrin repeats (Hirai et al. 1995), binds next to the ATP binding site of the Cdk6 catalytic cleft and interacts with both the N and C lobes of the kinase (Fig. 2A,B). The relative arrangement of p18 and Cdk6 and the intermolecular contacts are very similar to those seen in the p16–Cdk6 and p19–Cdk6 binary complexes (Brotherton et al. 1998; Russo et al. 1998), indicating that the presence of the cyclin does not affect the INK4–Cdk6 interface significantly.
K-cyclin binds to the side of the Cdk6 catalytic cleft opposite from where p18 binds and interacts predominantly with the N lobe of the kinase (Fig. 2A,B). The interactions with the N lobe are centered on the PSTAIRE helix (residues 55–66), which contains the CDK-specific PSTAIRE sequence motif (sequence is PLSTIRE in Cdk6; Pines 1994) and that forms the side wall of the catalytic cleft. The interactions are very similar to those seen in the active Cdk2–cyclinA structure (Jeffrey et al. 1995; Fig. 2C,D), and this underscores the prominent role the PSTAIRE helix has in cyclin binding. However, unlike the active Cdk2–cyclinA complex, there are no significant contacts between the cyclin and the CDK C lobe in the p18–Cdk6–K-cyclin complex.
The Cdk6 structure in the p18–Cdk6–K-cyclin complex has a large number of conformational changes compared with the active conformation of Cdk2 (Jeffrey et al. 1995; Fig. 2C,D) or of other protein kinases. In this inactive Cdk6 structure, the N and C lobes are rotated 13° away from each other, resulting in the misalignment of ATP-binding residues. The N-lobe PSTAIRE helix, which contains an invariant active site residue (Glu 61), is displaced by 4.5 Å away from the active site and is rotated by 16°. A C-lobe loop (T loop, residues 162–182), which contains the threonine that is phosphorylated (Thr 177) on the full activation of the kinase (Morgan 1995; Russo et al. 1996) and that forms part of the polypeptide substrate-binding site (Brown et al. 1999), is displaced by >30 Å. Finally, an additional loop at the back of the catalytic cleft (residues 99–102), which would hydrogen bond to ATP, is displaced by several Ångstroms.
The Cdk2–cyclinA structure (Jeffrey et al. 1995) showed that cyclinA binding to Cdk2 caused conformational and positional changes in the PSTAIRE helix and T loop and that these changes activated the kinase by correctly aligning certain active site residues and reorganizing the polypeptide substrate binding site. In the p18–Cdk6–K-cyclin complex, not only does the K-cyclin fail to carry out most of these conformational changes but p18 causes the misalignment of additional residues involved in ATP binding and catalysis.
Structure of the p18INK4c–Cdk6 interface
p18 binds Cdk6 through an interface consisting primarily of its second and third ankyrin repeats. Residues from the β hairpin and the first helix of each repeat contact the β sheet of the Cdk6 N lobe, and residues from the turn between the two helices of each repeat contact the Cdk6 C lobe. A few additional contacts are contributed by the first ankyrin repeat interacting with the N lobe β sheet (Fig. 2A,B), but there are no interactions between the fourth and fifth ankyrin repeats and Cdk6.
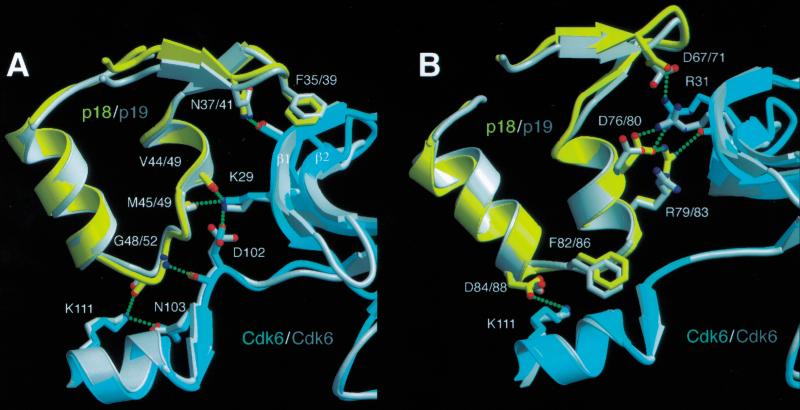
Contacts between the second and third ankyrin repeats of p18 and Cdk6 involve both hydrogen-bond networks and hydrophobic interactions, and the majority of these contacts are the same as those found in the binary INK4–Cdk6 complexes (Brotherton et al. 1998; Russo et al. 1998; Fig. 3A,B). There are three major intermolecular hydrogen bond networks linking Asp 67, Asp 76, and Arg 79 of p18 with Arg 31 and Gly 36 of Cdk6; the backbone carbonyl groups of Val 44 and Met 45 of p18 with Lys 29 of Cdk6; and Asp 84 and Gly 48 of p18 with Lys 111 of Cdk6. Additional hydrogen bonds are formed at the periphery of the interface. Hydrophobic interactions between p18 and Cdk6 are less extensive than the hydrogen bond networks. The most significant interaction involves Phe 82, which wedges in between two polypeptide segments of the C lobe at the back of the catalytic cleft, packing with backbone and side chain groups of Asp 102 and Ser 155 (Fig. 3B). This forces the peptide backbone in the vicinity of Asp 102 to move into the ATP binding site by 3 Å. Phe 82 is conserved in all INK4 family members, and it also causes the same conformational changes in Asp 102 of Cdk6 in the binary INK4–Cdk6 complexes (Brotherton et al. 1998; Russo et al. 1998).
Figure 3.
Interactions between p18 and Cdk6 mirror those made between p19 and Cdk6. (A) Hydrogen bond networks between the second ankyrin repeat of p18 and Cdk6 involve equivalent residues to those in p19. p19–Cdk6 is superimposed on p18–Cdk6 using residues from the second ankyrin repeat and adjacent residues in the Cdk6 N lobe. The p18–Cdk6 residues are colored in yellow and cyan, that of p19–Cdk6 in gray. Hydrogen bonds in the p18–Cdk6 complex are indicated by green dashed lines. The same hydrogen bonds are made in the p19–Cdk6 complex but omitted here for clarity. Residue numbering for the INK4 inhibitors is given as p18/p19. (B) Interface between the third ankyrin repeat of p18 and Cdk6 showing the extensive hydrogen bonding network centered around Arg 31 of Cdk6. Residues from the second and third ankyrin repeats of p18 contribute most of the interactions with Cdk6.
Structure of the Cdk6–K-cyclin interface
The K-cyclin structure consists of two tandem repeats of the five-helix cyclin fold plus an additional N-terminal α helix. The arrangement is very similar to that in cyclinA (the relative positions of the two cyclin repeats in K-cyclin and cyclinA differ by only 11° and 2.5 Å; Jeffrey et al. 1995) and to the structures of the squirrel monkey herpes virus cyclin (Schulze-Gahmen et al. 1999) and murine γ-herpes virus 68 (MHV68; Card et al. 2000). Although both K-cyclin and the squirrel monkey herpes virus cyclin share the ability to bind and activate Cdk6, MHV68 is unable to do so (Card et al. 2000). The first cyclin repeat of K-cyclin is more similar to that of cyclinA (RMS deviation of 0.71 Å for all 96 Cα atoms) than would be expected based on the 28% sequence identity (Chothia and Lesk 1986), underlining the critical role of this domain in CDK binding.
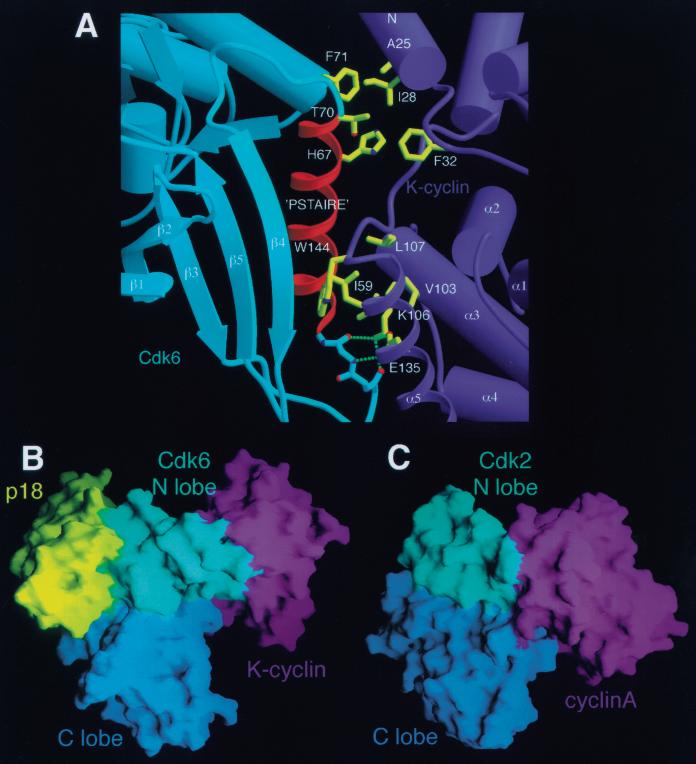
In the inhibited complex, K-cyclin binds the N lobe of Cdk6 using the first cyclin repeat and the N-terminal helix (Fig. 2A,B) and interacts predominantly with the PSTAIRE helix of Cdk6. The interactions between K-cyclin and the Cdk6 N lobe are similar to those observed in the Cdk2–cyclin complexes (Jeffrey et al. 1995; Card et al. 2000), except near the C terminus of the PSTAIRE helix, where Cdk6 has a three-residue insertion (Fig. 4A). Contacts equivalent to those in Cdk2–cyclinA include a hydrogen bond network between the Lys 106–Glu 135 pair in K-cyclin and backbone groups of residues 52–54 of Cdk6, immediately before the PSTAIRE helix. This Lys–Glu pair is found in all human cyclins and likely is a conserved structural feature in CDK–cyclin complexes. Interactions with the PSTAIRE helix are also similar. Ile 59, at the center of the helix, inserts into a hydrophobic pocket formed by Val 103, Lys 106, Leu 107, Leu 139, Trp 144, and Thr 146 of K-cyclin (Fig. 4A). Overall, the CDK-cyclin interactions in this portion of the interface involve residues that are either conserved or conservatively substituted in the CDK and cyclin families.
Figure 4.
The K-cyclin–Cdk6 interface. (A) The PSTAIRE helix of Cdk6 is a central feature of the Cdk6–K-cyclin interface. The viewpoint shown corresponds approximately to that in B. Three sets of interactions are shown: hydrogen bonds between the Cdk6 main-chain preceding the PSTAIRE helix and the conserved Lys–Glu pair of K-cyclin (K106, E135); the conserved Ile 59 of Cdk6 inserts into a hydrophobic pocket in K-cyclin; residues at the end of the PSTAIRE helix, one turn longer in Cdk4 and Cdk6 than in Cdk2, interact with residues on the N-terminal helix of K-cyclin and may play a role in cyclin–CDK specificity. (B) Surface representation of p18–Cdk6–K-cyclin complex illustrating the minimal interactions between K-cyclin and the Cdk6 C lobe. p18 is colored yellow, the Cdk6 N lobe is cyan, the Cdk6 C lobe is blue, and the K-cyclin is purple. The viewpoint is approximately the same as Fig. 2A. The only contacts between K-cyclin and the C lobe of Cdk6 arise from interactions with the N-terminal helix of K-cyclin. (C) Surface representation of Cdk2–cyclinA in the equivalent orientation as that in A, showing significantly greater interactions between the C lobe of the Cdk2 and the cyclinA, giving rise to a much more extensive cyclin–CDK interface.
The differences with respect to Cdk2–cyclinA arise from contacts at the C terminus of the PSTAIRE helix caused by a three residue insertion in Cdk6 (residues 70–72) resulting in one additional helical turn of 3.10 type. The longer PSTAIRE helix of Cdk6 would collide with the N-terminal helix of cyclinA (Thr 70 and Phe 71 of Cdk6 would clash with Met 189 and Tyr 185 of cyclinA). The longer Cdk6 PSTAIRE helix is accommodated in K-cyclin by a small shift of the N-terminal helix relative to cyclinA and by the substitution of smaller amino acids (Asn 24 of K-cyclin instead of Tyr 185 of cyclinA). This results in contacts between Thr 70 and Phe 71 in the Cdk6 insertion and Asn 24, Ile 28, and Phe 32 of K-cyclin. This extension of the PSTAIRE helix would also be present in Cdk4, which contains the three-residue insertion, and could contribute to the specificity of Cdk4/6 for the D-type cyclins.
Despite the overall similarities in the N lobe-cyclin interactions between the inhibited p18–Cdk6–K-cyclin complex and the active Cdk2–cyclinA complex, there is a large difference in the position and orientation of the cyclin relative to the kinase C lobe. When the two complexes are compared by superimposing their CDK C lobes, K-cyclin is rotated by ≈40°, and its center of gravity is shifted by 15 Å relative to cyclinA. This is caused in part by the rotation between the kinase N and C lobes in p18–Cdk6–K-cyclin and in part by the rotation of the PSTAIRE helix relative to the N lobe. The shift in K-cyclin leads to a lack of significant contacts between K-cyclin and the C lobe and T loop of Cdk6 (Fig. 4B). In the Cdk2–cyclinA complex, there are extensive contacts between the first cyclin repeat and the T loop and between the N-terminal helix and other parts of the Cdk2 C lobe (Fig. 4C; Jeffrey et al. 1995). In the inhibited Cdk6–K-cyclin complex, there are no contacts with the T loop and only a few minor contacts with the C lobe.
Conformation of Cdk6
The structure of Cdk6 in the p18–Cdk6–K-cyclin complex differs from the structure of cyclinA-activated Cdk2 in the orientation of the N and C lobes of the kinase and in the positions of the PSTAIRE helix and T loop. Compared to the Cdk2–cyclinA complex, the kinase N and C lobes of the p18–Cdk6–K-cyclin complex are rotated by 13° about an axis that passes through the back of the catalytic cleft and is approximately perpendicular to the plane of the ATP that would bind there (vertical axis in Fig. 5). This lateral rotation is very similar to that seen in the binary INK4–Cdk6 complexes, suggesting that it is caused by the contacts the INK4s make to the two lobes rather than reflecting a flexibility in the complex.
Figure 5.
Schematic representation of the different conformations of the CDK. CDKs undergo extensive conformational changes on binding of activating or inhibiting subunits. The major determinants of activity are the positions and conformation of the PSTAIRE helix and T loop, as well as the relative disposition of the kinase N and C lobes. The PSTAIRE helix adopts a position further away from the catalytic cleft in inactive CDKs (labeled as ‘out’) than in active CDKs (‘in’). The PSTAIRE helix conformation correlates with the location of a conserved active site residue (Cdk2, Glu 51; Cdk6, Glu 61) either inside or outside the catalytic cleft.
The rotation between the N and C lobes causes the PSTAIRE helix, which is attached to the N lobe, to move away from the catalytic cleft (Fig. 5). The PSTAIRE helix remains packed against the N lobe but loses about half of its packing interactions with the C lobe. The PSTAIRE helix contains a glutamic acid (Glu 61 in Cdk6), invariant among eukaryotic kinases, that contributes to ATP binding in the active kinase structures (Taylor and Radzio-Andzelm 1994; Jeffrey et al. 1995), and its position is thus critical for kinase activity. When p18–Cdk6–K-cyclin and Cdk2–cyclinA are compared by superimposing their C lobes, the N terminus of the PSTAIRE helix in the inactive complex is 4.5 Å away from its position in the active complex, placing the glutamic acid side chain too far from the other catalytic cleft residues on the C lobe to carry out its function.
In the inactive INK4–Cdk6 and monomeric Cdk2 structures (de Bondt et al. 1993), the PSTAIRE helix is also positioned away from the catalytic cleft (Fig. 5), similar to the p18–Cdk6–K-cyclin complex. However, in INK4–Cdk6 and monomeric Cdk2, the PSTAIRE helix is also rotated 90° about its axis relative to the active conformation, resulting in the glutamic acid residue pointing away from the catalytic cleft. In the p18–Cdk6–K-cyclin complex, K-cyclin orients the PSTAIRE helix such that it is rotated only 16° from the active conformation, resulting in the glutamic acid pointing toward the catalytic cleft.
The rotation of the N lobe and the PSTAIRE helix away from the C lobe is also associated with the T loop not adopting the conformation needed for substrate binding and kinase activity (Fig. 5). In the Cdk2–cyclinA complex, the T loop makes multiple contacts with the PSTAIRE helix, the cyclin, and other parts of the C lobe. As these contacts would not be possible in p18–Cdk6–K-cyclin because of the misalignment of the lobes and PSTAIRE helix, the T loop instead adopts a β hairpin structure and packs on the C lobe in a position near the first ankyrin repeat of p18 (Fig. 2A,B), which is opposite from its position in the active CDK. The T-loop phosphorylation site (Thr 177) is displaced by 23 Å from the equivalent residue in the unphosphorylated Cdk2–cyclinA complex. This inactive conformation of the T loop and the packing interactions it makes are essentially identical to those in the p16–Cdk6 and p19–Cdk6 complexes (Brotherton et al. 1998; Russo et al. 1998).
Mechanism of Cdk4/6–cyclin inhibition by INK4s
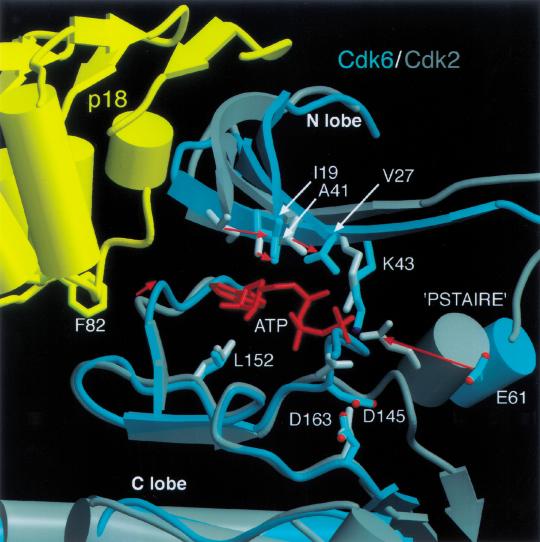
The INK4-induced conformational changes in Cdk6 would interfere with the binding of ATP and polypeptide substrate and would also misalign any weakly bound substrates with respect to phosphotransfer. On the basis of studies of Cdk2 (de Bondt et al. 1993; Jeffrey et al. 1995), the binding of the ATP purine group involves hydrophobic residues from both the N lobe (Ile 19, Val 27, and Ala 41 in Cdk6) and C lobe (Leu 154), which sandwich the ATP purine ring between them, and also backbone amide and carbonyl groups (Glu 99, Val 101) at the back of the catalytic cleft that hydrogen bond to the N1 and N6 groups of the adenine ring. In the p18–Cdk6–K-cyclin complex, the twist in the N and C lobes results in a displacement of as much as 4.5 Å for the hydrophobic residues, and the dislocation of the catalytic cleft loop (residues 99–102) by p18 displaces the backbone groups by up to 3 Å. The phosphate groups of the ATP are bound by a network of electrostatic interactions involving a lysine from the N lobe (Lys 43 in Cdk6), a glutamic acid from the PSTAIRE helix (Glu 61), and a magnesium ion coordinated by an aspartic acid (Asp 163) from the C lobe (de Bondt et al. 1993; Jeffrey et al. 1995). These interactions also help orient the γ phosphate group with respect to the putative catalytic base (Taylor and Radzio-Andzelm 1994) on the C lobe (Asp 145) and the polypeptide substrate on the T loop for phosphotransfer. In the p18–Cdk6–K-cyclin complex, the misalignment of the N lobe, C lobe, and PSTAIRE helix results in large displacements of these residues with respect to their configuration in the active CDK structures (e.g., the Cγ atom of Glu 61 moves by 7 Å relative to that of Glu 51 in Cdk2–cyclinA; Fig. 6).
Figure 6.
The ATP-binding site of p18–Cdkl6–K-cyclin and Cdk2–cyclinA. Active site residues implicated in ATP binding and catalysis are displaced in the p18–Cdk6–K-cyclin complex relative to the active Cdk2–cyclinA conformation. Cdk2 and Cdk6 were superimposed on their C lobes. Cdk6 is shown in cyan, p18 in yellow, Cdk2 in gray. Movement of active site residues is indicated by red arrows. p18 displaces the N lobe relative to the C lobe, causing the hydrophobic residues (Ile 19, Val 27, Ala 41, Leu 152) that sandwich the adenine ring of ATP to move by up to 4.5 Å. The p18 inhibitor also distorts the edge of the active site via Phe 82, affecting hydrogen bonding interactions with the edge of the ATP ring. The related shift of the PSTAIRE helix on the other side of the active site displaces an active site residue (Glu 61). The T loop of Cdk6 diverges from that of Cdk2 between Phe 164 and Val 181, and these residues are omitted for clarity.
This distortion of the catalytic cleft would not exclude ATP binding, as there is space remaining to accommodate it, but would reduce the affinity for ATP and would misalign any bound ATP for phosphotransfer. This has been supported by cross-linking studies using the ATP analog FSBA, which showed that INK4 binding to either free Cdk6 or cyclinD-bound Cdk6 reduced the cross-linking efficiency by a factor of ∼20 (Russo et al. 1998).
Resistance to INK4 inhibition by the K-cyclin-bound, phosphorylated Cdk6
The Cdk6–K-cyclin complex becomes resistant to inhibition by p18 when Cdk6 is phosphorylated at Thr 177, although the p18–Cdk6–K-cyclin ternary complex still forms and can be purified by gel filtration chromatography without any apparent dissociation (data not shown). In the Cdk2–cyclinA complex, phosphorylation of the equivalent residue (Thr 160) causes the T loop to undergo a local conformational change (Russo et al. 1996) that completes the organization of the substrate-binding site (Brown et al. 1999) and increases the catalytic activity 300-fold (Russo et al. 1996). In addition, phosphorylation stabilizes the active conformation of the kinase through two mechanisms (Russo et al. 1996): The phosphothreonine further links the PSTAIRE helix, C lobe, and T loop together by coordinating an arginine side chain from each of these regions (Arg 60, Arg 144, and Arg 168 in Cdk6); and the reorganized T loop makes more extensive contacts with the cyclin and the C lobe.
We suggest that the ability of phosphorylated Cdk6–K-cyclin to evade inhibition by INK4s (Swanton et al. 1997) is the result of increased interactions between the N and C lobes of the kinase via the phosphothreonine group and is also because of increased interactions between K-cyclin and the Cdk6 C lobe, thus stabilizing the active relative arrangement of the two lobes and the PSTAIRE helix. These additional interactions should give rise to an active complex that is more resistant to the inhibitory conformational changes that would otherwise result on INK4 binding. Although the p18–Cdk6–K-cyclin complex still forms (data not shown), it is unlikely that p18 is bound to Cdk6 in the same manner as in the inactive complex. We presume that it may remain bound through interactions with the N lobe only, forgoing its interactions with the C lobe. In this respect, it may be analogous to the way the cyclin remains bound to the N lobe in a nonfunctional way in the inhibited ternary complex.
Conclusion
Our results demonstrate that binding of INK4 inhibitors to the preformed active Cdk6–K-cyclin complex results in a competition between the cyclin and inhibitor to stabilize structures corresponding to an activated or inhibited kinase. The ability of p18 to inhibit Cdk6–K-cyclin and its inability to inhibit the same complex once Cdk6 is phosphorylated is a graphic illustration of the interplay between these competing components. Our structure also shows that the CDK–cyclin interface in the inhibited ternary complex is smaller than in the active binary complex, and this suggests that INK4s may increase the rate of cyclin dissociation. This, together with the short half-life of D-type cyclins relative to Cdk4/6 and INK4s (Sherr and Roberts 1999) and the ability of INK4s to prevent the assembly of new Cdk4/6–cyclinD complexes (Sherr and Roberts 1999), offers an explanation of how INK4–Cdk4/6 complexes can be significantly more abundant than the ternary INK4–Cdk4/6–cyclinD complexes in the cell.
Materials and methods
Protein expression and purification
K-cyclin and human p18 were expressed in Escherichia coli as glutathione S-transferase (GST) fusion proteins at room temperature and were isolated by glutathione affinity chromatography. Human Cdk6 (Meyerson and Harlow 1994) was overexpressed using a baculovirus vector in Hi5 (Invitrogen) insect cells in suspension in serum-free media (Sf900, GIBCO). Cdk6 was isolated by affinity chromatography using a column of the GST–K-cyclin fusion protein. The complex of Cdk6 bound to GST–K-cyclin was purified using anion exchange chromatography, treated with thrombin at 4°C to cleave the GST fusion protein, and further purified using anion exchange chromatography. To form the ternary complex, purified Cdk6–K-cyclin was incubated with a threefold molar excess of purified p18 at a total protein concentration of 10 mg/mL, in 200 mM NaCl, 5 mM dithiothreitol (DTT), 25 mM Tris-Cl (pH 7.5), for 1 h at 4°C. The ternary complex was purified by gel filtration and was concentrated by ultrafiltration (Amicon) to 15 mg/mL and further purified by gel-filtration chromatography before concentration to 20 mg/mL. To prepare the Thr 177–phosphorylated p18–Cdk6–K-cyclin complex, the p18–Cdk6–K-cyclin complex (124 mg) was incubated with the CDK-activating kinase Cdk7-cyclinH (12.5 mg) and expressed and purified as described (Russo et al. 1996) in 40 mM NaCl, 5 mM DTT, 5 mM MgCl2, 0.5 mM ATP, 25 mM Tris-Cl (pH 7.5) at a total protein concentration of 1.0 mg/mL. The reaction, monitored by the increased SDS gel mobility of phosphorylated Cdk6, was complete after 12 h at 6°C and 1 h at 16°C.
Crystallization and structure determination
Crystals were grown using the hanging drop vapor diffusion method from 38% saturated ammonium sulfate, 6% (v/v) dioxane, 5mM DTT, 100 mM Tris-Cl (pH 7.6). They form in space group P212121 with a = 77.8, b = 147.0, c = 164.8 Å and contain two complexes per asymmetric unit. Data were collected at −160°C from a crystal flash-frozen in 40% (w/v) ammonium sulfate, 6% (v/v) dioxane, 100 mM Tris-Cl (pH 7.6), 30% (v/v) glycerol using a Rigaku RAXIS IV detector mounted on a Rigaku Ru200 generator equipped with mirror optics. Data were processed using the HKL package (Otwinowski and Minor 1997). The structure of the p18–Cdk6–K-cyclin complex was determined by molecular replacement using AMORE (CCP4 1994). The models used were the Cdk6 subunit from the p19–Cdk6 complex structure (Russo et al. 1998), the monomeric p18 structure (Venkataramani et al. 1998), and the first cyclin repeat of cyclinA (Jeffrey et al. 1995). The location of K-cyclin was confirmed using twofold averaged 2Fo-Fc model-phased maps calculated using the p18 and Cdk6 coordinates alone. The structure was refined with CNS (Brünger et al. 1998) using tight restraints between molecules related by noncrystallographic symmetry. Initially, the amino acid side chains for the K-cyclin model were deleted and built back in to twofold averaged 2Fo-Fc electron density maps calculated using CNS. Omit maps of the PSTAIRE helix and T loop confirmed the positioning of these segments. The K-cyclin N-terminal helix was clearly defined in model-phased difference maps and added to the model during early stages of refinement. Several N- and C-terminal amino acids of each subunit are disordered and not observed in the electron density maps. A disordered internal loop (residues 214–218) in K-cyclin was omitted from the structure. The current model consists of residues 8–301 of Cdk6, 16–253 of K-cyclin, and 6–160 of p18. The crystallographic data were very anisotropic, and an overall correction (B11 = −15.6, B22 = 0.9, B33 = 14.7 Å2) was applied to the observed data during refinement and map calculation. The coordinates have been deposited at the Protein Data Bank with accession code 1G3N.
CDK activity assays
p18 (18, 36, 54, 90, 124, 162 ng) was preincubated with either 200 ng of the Cdk6–K-cyclin complex or 200 ng of the Cdk6–cyclinD1 complex and purified as described (Russo et al. 1998) in the presence or absence of 40 ng purified CAK (Russo et al. 1996) in 30 μL of 25 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.5 mM ATP and [γ-32P]ATP for 1 h at room temperature. As the substrate, 1 μg of a C terminal fragment of Rb (residues 773–874) was then added. After incubation at room temperature for 1 h, the reaction was stopped by the addition of SDS-PAGE sample buffer.
Table 1.
Data processing and refinement statistics
| Data Processing | ||
| Resolution (Å) | 100.–2.9 | (3.0–2.9) |
| Observations | 131920 | |
| Unique reflections | 40272 | |
| R-sym | 0.057 | (0.299) |
| Completeness (%) | 0.950 | (0.919) |
| Refinement | ||
| Resolution (Å) | 15.–2.9 | |
| Reflections, ‖F‖ > 1ςF | 37749 | |
| Data completeness (%) | 93.0 | |
| No. of atoms | 10652 | |
| R-free | 0.268 | |
| Residual | 0.224 | |
| RMSD bond (Å) | 0.013 | |
| RMSD angle (°) | 1.76 | |
| RMSD B-factor (Å2) | 2.2 | |
R-sym = Σh Σi ‖Ih,i − <Ih>‖/Σh Σi Ih,i for the intensity (I) of i observations of a reflection h. R-factor = Σ‖Fobs-Fcalc‖/Σ‖Fobs‖, where Fobs and Fcalc are the observed and calculated structure factors, respectively. R-free = R-factor calculated using 5% of the reflection data chosen randomly and omitted from refinement. RMSD: root mean square deviations from ideal geometry and root mean square variation in B-factor of covalently bonded atoms.
Acknowledgments
We thank Ethel Cesarman for the gift of KSHV cDNA and Christine Murray for administrative assistance. Supported by the NIH, the Howard Hughes Medical Institute, the Dewitt Wallace Foundation and the Samuel and May Rudin Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nikola@xray2.mskcc.org; FAX (212) 717-3135.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.851100.
References
- Adachi M, Roussel MF, Havenith K, Sherr CJ. Features of macrophage differentiation induced by p19INK4d, a specific inhibitor of cyclin D-dependent kinases. Blood. 1997;90:126–137. [PubMed] [Google Scholar]
- Brotherton DH, Dhanaraj V, Wick S, Brizuela L, Domaille PJ, Volyanik E, Xu X, Parisini E, Smith BO, Archer SJ, et al. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstieve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Card GL, Knowles P, Laman H, Jones N, McDonald NQ. Crystal structure of a γ-herpesvirus cyclin–cdk complex. EMBO J. 2000;19:2877–2888. doi: 10.1093/emboj/19.12.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- Godden-Kent D, Talbot SJ, Boshoff C, Chang Y, Moore P, Weiss RA, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Jenkins CW, Li Y, O'Keefe CL, Noh S, Wu X, Zariwala M, Matera AG, Xiong Y. Isolation and characterization of p19INK4d, a p16-related inhibitor specific to CDK6 and CDK4. Mol Biol Cell. 1996;7:57–70. doi: 10.1091/mbc.7.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Hall M, Bates S, Peters G. Evidence for different modes of action of cyclin-dependent kinase inhibitors: p15 and p16 bind to kinases, p21 and p27 bind to cyclins. Oncogene. 1995;11:1581–1588. [PubMed] [Google Scholar]
- Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Lamphere L, Fiore F, Xu X, Brizuela L, Keezer S, Sardet C, Draetta GF, Gyuris J. Interaction between Cdc37 and Cdk4 in human cells. Oncogene. 1997;14:1999–2004. doi: 10.1038/sj.onc.1201036. [DOI] [PubMed] [Google Scholar]
- Li M, Lee H, Yoon DW, Albrecht JC, Fleckenstein B, Neipel F, Jung JU. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Gregory FJ, Stott FJ, Hara E, Peters G. Induced expression of p16(INK4a) inhibits both CDK4- and CDK2-associated kinase activity by reassortment of cyclin–CDK-inhibitor complexes. Mol Cell Biol. 1999;19:1981–1989. doi: 10.1128/mcb.19.3.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parry D, Bates S, Mann DJ, Peters G. Lack of cyclin D–Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumour suppressor gene product. EMBO J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: Structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- Pines J. The cell cycle kinases. Semin Cancer Biol. 1994;5:305–313. [PubMed] [Google Scholar]
- Reynisdottir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes & Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- Schulze-Gahmen U, Jung JU, Kim SH. Crystal structure of a viral cyclin, a positive regulator of cyclin-dependent kinase 6. Struct Fold Des. 1999;7:245–254. doi: 10.1016/s0969-2126(99)80035-5. [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. G1 phase progression: Cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- ————— Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes & Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- Swanton C, Mann DJ, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Radzio-Andzelm E. Three protein kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Venkataramani R, Swaminathan K, Marmorstein R. Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations. Nat Struct Biol. 1998;5:74–81. doi: 10.1038/nsb0198-74. [DOI] [PubMed] [Google Scholar]
- Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Buschenfelde KH, Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]