Abstract
The Wingless signaling pathway directs many developmental processes in Drosophila by regulating the expression of specific downstream target genes. We report here that the product of the trithorax group gene osa is required to repress such genes in the absence of the Wingless signal. The Wingless-regulated genes nubbin, Distal-less, and decapentaplegic and a minimal enhancer from the Ultrabithorax gene are misexpressed in osa mutants and repressed by ectopic Osa. Osa-mediated repression occurs downstream of the up-regulation of Armadillo but is sensitive both to the relative levels of activating Armadillo/Pangolin and repressing Groucho/Pangolin complexes present and to the responsiveness of the promoter to Wingless. Osa functions as a component of the Brahma chromatin-remodeling complex; other components of this complex are likewise required to repress Wingless target genes. These results suggest that altering the conformation of chromatin is an important mechanism by which Wingless signaling activates gene expression.
Keywords: Osa, Brahma, SWI/SNF, Wingless, transcription, repression, chromatin, Eyelid
The Drosophila segment polarity gene wingless (wg) encodes a secreted signaling molecule that provides positional information for the growth and patterning of numerous structures during both embryonic and imaginal development. Wg and its vertebrate homologs (Wnt proteins) activate a conserved signal transduction pathway and influence cell fate decisions by regulating the expression of specific downstream target genes (Cadigan and Nusse 1997).
Binding of Wg/Wnt proteins to cell surface receptors of the Frizzled (Fz) family (Bhanot et al. 1996; Yang-Snyder et al. 1996; Bhat 1998) triggers the stabilization and accumulation of the effector proteins Armadillo (Arm) in Drosophila (Riggleman et al. 1990; Peifer et al. 1994) and β-catenin (β-cat; Larabell et al. 1997) in vertebrates. In the absence of a Wg/Wnt signal, Arm and β-cat are maintained at low levels through the activity of a complex consisting of the serine/threonine kinase Shaggy/Zeste-white 3 (Sgg) or glycogen synthase kinase 3β (GSK-3β) in vertebrates (Siegfried et al. 1992; Peifer et al. 1994; Yost et al. 1996), the scaffold protein Axin (Zeng et al. 1997; Behrens et al. 1998; Hamada et al. 1999; Willert et al. 1999), and the product of the adenomatous polyposis coli tumor suppressor gene (APC; Rubinfeld et al. 1996; McCartney et al. 1999). Reception of the Wg/Wnt signal inhibits the kinase activity of Sgg/GSK-3β, preventing the complex from phosphorylating Arm/β-cat and targeting it for degradation (Ruel et al. 1999).
The posttranslational up-regulation of Arm and β-cat permits these proteins to translocate to the nucleus (Orsulic and Peifer 1996; Yost et al. 1996) and bind the HMG box proteins Pangolin/dTCF (Pan; Brunner et al. 1997; van de Wetering et al. 1997) in Drosophila and Lef-1/TCF (Behrens et al. 1996; Molenaar et al. 1996) in vertebrates. These Pan/Arm and TCF/β-cat complexes are thought to comprise bipartite transcription factors that regulate the expression of Wg- and Wnt-responsive genes, respectively. However, the mechanism by which these complexes activate gene expression is poorly understood.
It has recently been reported that β-cat can interact in vitro with the TATA binding protein (TBP; Hecht et al. 1999), as well as with Pontin52 (Bauer et al. 1998), a protein that interacts with TBP. This suggests that Pan/Arm and TCF/β-cat complexes may regulate gene expression by targeting components of the basal transcriptional machinery to promoters of Wg/Wnt-responsive genes. However, these complexes do not appear to be sufficient to activate the expression of target genes in vivo (Riese et al. 1997; Prieve and Waterman 1999). Thus, regulation of gene expression by Pan/Arm and TCF/β-cat complexes is likely to be far more complex than the simple recruitment of TBP.
An interesting common theme emerging from recent research is the connection between the regulation of chromatin architecture and the expression of Wg/Wnt target genes. Pan, TCF, and Lef-1 are members of the HMG family of transcription factors. The HMG box is a DNA-binding domain that induces a sharp bend in DNA; this DNA-bending activity has been shown to be important for the regulation of gene expression by many HMG box proteins (for review, see Bustin 1999), including Lef-1 (Giese et al. 1997). Interestingly, Lef-1 bound to β-cat induces a sharper bend in DNA than does Lef-1 alone (Behrens et al. 1996). β-cat can also bind to CBP/p300 histone acetyltransferases, and this interaction stimulates β-cat dependent gene expression in vitro (Hecht et al. 2000). Furthermore, it has been shown that Pan and TCF are required for the repression of target gene expression in the absence of the Wg/Wnt signals (Brannon et al. 1997; Riese et al. 1997; Yang et al. 2000). Pan and TCF mediate this repression though an interaction with the Drosophila Groucho (Gro) and vertebrate TLE transcriptional corepressors (Cavallo et al. 1998; Roose et al. 1998). Gro has been shown to repress gene expression by directly interacting with the amino tail of histone H3 (Palaparti et al. 1997) and by recruiting the histone deacetylase Rpd3 (Chen et al. 1999). These data indicate that altering chromatin architecture may be an important mechanism for the regulation of target gene expression by the Wg and Wnt signaling pathways.
Eukaryotic organisms have evolved a number of multiprotein complexes that remodel chromatin structure to regulate gene expression, including the SWI/SNF and RSC complexes in yeast (for review, see Kadonaga 1998; Kingston and Narlikar 1999). Homologs of the SWI/SNF and RSC complexes have been identified in other organisms, including the hBRM and BRG1 complexes in humans (Wang et al. 1996) and the Brahma (Brm) complex in Drosophila (Papoulas et al. 1998). A purified yeast SWI/SNF or related human complex can catalyze an ATP-dependent reversible reaction that alters the structure of nucleosomal DNA, rendering it more accessible to transcription-factor binding (Cote et al. 1994; Kwon et al. 1994). Whereas chromatin remodeling by these complexes is generally thought to promote transcription of target genes, an increasing body of evidence suggests that they are also required for the direct repression of a subset of genes (Trouche et al. 1997; Holstege et al. 1998; Moreira and Holmberg 1999; Sudarsanam et al. 2000).
We have shown that the trithorax group gene osa encodes an ARID domain protein that is a component of Brm chromatin-remodeling complexes (Treisman et al. 1997; Collins et al. 1999). It has also been demonstrated that a human homolog of Osa, p270, is a component of the BRG1 complex (Dallas et al. 1998, 2000). Osa and Brm complexes have been shown to regulate the expression of a wide variety of genes including the segmentation gene even-skipped (Treisman et al. 1997) and the homeotic gene Antennapedia (Vasquez et al. 1999). Brm complexes have also been shown to be required for activation of expression by the trithorax group protein Zeste (Kal et al. 2000). Interestingly, loss of osa function in embryos and imaginal discs often induces phenotypes similar to those caused by ectopic activation of the wg pathway, although wg itself is not ectopically expressed (Treisman et al. 1997). This prompted us to suggest that osa functions to antagonize wg signaling (Treisman et al. 1997); however, it was not known whether this antagonism is direct or the result of the regulation of the expression of other components of the wg pathway by osa.
We show here that loss of osa function induces ectopic expression of Wg target genes and that overexpression of osa can repress the endogenous expression of the same genes. Furthermore, the lack of an effect of Osa on Arm up-regulation, the activity of an Osa-repressor domain fusion, and the specificity with which osa affects the Wg response of a minimal enhancer suggest that osa is required to directly repress the expression of these genes. Interestingly, loss of function of other components of the Brm complex also induces ectopic expression of Wg target genes. Thus, the repression of Wg target genes in vivo requires chromatin remodeling mediated by Osa-containing Brm complexes. These data provide further evidence that regulation of chromatin structure is an important mechanism for the control of target gene expression by the Wg and Wnt pathways.
Results
Osa is required for the repression of a Wg target gene
We have previously reported that loss of osa can induce phenotypes similar to those caused by ectopic wg expression (Treisman et al. 1997). Conversely, overexpression of full-length, wild-type Osa (UAS-Osa) results in dominant, gain-of-function phenotypes that often resemble those caused by loss of wg function (Collins et al. 1999; data not shown). However, osa appears to be epistatic to wg, and loss of osa function does not induce ectopic expression of wg (Treisman et al. 1997). Therefore, the wg gain-of-function phenotypes caused by osa loss of function are likely to result from de-repression of downstream target genes of Wg. To investigate this, we examined the regulation of nubbin (nub). nub encodes a POU domain protein that is required for the growth and patterning of the wing and is expressed throughout the wing primordium (or wing pouch) in third-instar wing discs (Fig. 1B; Ng et al. 1995, 1996; Cifuentes and Garcia-Bellido 1997). wg signaling is both necessary and sufficient for the expression of nub, as ectopic expression of wg (Ng et al. 1996) or ectopic activation of the wg pathway (Fig. 1D,K) can induce ectopic expression of nub, whereas blocking transmission of the wg signal in the wing pouch represses the endogenous expression of nub (Fig. 1F,H).
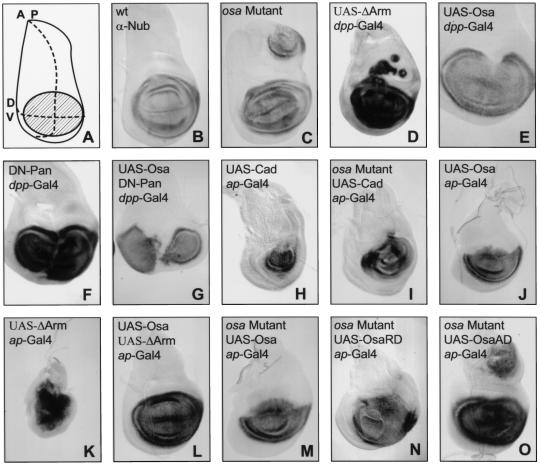
Figure 1.
Osa regulates the expression of a wg-dependent gene. (A) shows a diagram of the third-instar wing disc indicating the anterior/posterior (AP) and dorsoventral (DV) compartment boundaries. The shaded region represents the wing pouch, while the rest of the disc gives rise to the notum. The remaining panels show third-instar wing discs stained with anti-Nubbin antibody from wildtype (B), osaeld308/osa4h(C), UAS-Δarm/dpp-Gal4 (D), UAS-osad3/+; dpp-Gal4/+ (E), UAS-dTCFΔN1/+; dpp-Gal4/+ (F), UAS-dTCFΔN1, UAS-osad3/+; dpp-Gal4/+ (G), UAS-cadh7/ap-GAL4 (H), osaeld308/osa4h; UAS-cadh7/ap-GAL4 (I), UAS-osad3/+; ap-GAL4/+ (J), UAS-ΔArm/ap-GAL4 (K), UAS-osad3/+; UAS-ΔArm/ap-GAL4 (L), osaeld308/osa4h; UAS-osas2/ap-GAL4 (M), osaeld308, UAS-osaRD11c/osa4h; ap-GAL4/+ (N), and osaeld308/osa4h; UAS-osaAD8c/ap-GAL4 (O) larvae. osa mutant wing discs have ectopic nub expression (C) similar to that caused by ectopic activation of the wg pathway (D). Overexpression of osa along the anterior/posterior boundary causes a reduction in nub expression (E) similar to that caused by blocking the transmission of the wg signal (F). UAS-Osa and dominant negative Pan act synergistically to strongly reduce nub expression (G). The expression of UAS-Cad in the dorsal compartment results in the loss of nub expression in the dorsal wing pouch (H), and this expression is partially rescued in wing discs mutant for osa (I). Furthermore, the ectopic nub expression induced in osa mutant discs (B) is also rescued by the expression of UAS-Cad (I). Overexpression of osa in the dorsal compartment almost eliminates nub expression (J), whereas expression of activated Arm causes nub to be expressed in nearly all of the wing disc (K). More normal nub expression is restored when UAS-Osa and activated Arm are coexpressed (L). The ectopic nub expression induced in osa mutant wing discs (B) is rescued by expression in the dorsal compartment of UAS-Osa (M) or UAS-OsaRD (N) but not by UAS-OsaAD (O).
We found that nub was ectopically expressed in wing discs that were transheterozygous for null and hypomorphic alleles of osa (osaeld308 and osa4H, respectively; Fig. 1C). Similar ectopic nub expression was caused by ectopic activation of the wg pathway by an activated form of Armadillo (ΔArm; Zecca et al. 1996; Fig. 1D). Conversely, the endogenous expression of nub was reduced along the anterior/posterior (A/P) boundary when UAS-Osa was expressed there with a decapentaplegic (dpp)–Gal4 driver (Fig. 1E). A similar loss of nub expression was caused by the expression of a dominant negative form of Pangolin (Pan) that can no longer bind Arm to activate gene expression (DN-Pan; van de Wetering et al. 1997; Fig. 1F). When Osa and DN-Pan were coexpressed with dpp–Gal4, they acted synergistically to cause a severe reduction in nub expression (Fig. 1G).
In addition to its role in transmitting the wg signal, Arm binds directly to cadherins and is required for the formation of adherens junctions (Cox et al. 1996). Over-expression of Drosophila E-Cadherin (DE-Cadherin) can sequester Arm at the plasma membrane and prevent it from participating in Wg signaling; this results in the induction of wg-like phenotypes (Sanson et al. 1996). When DE-Cadherin (UAS-Cad) was overexpressed in the dorsal compartment of the wing disc with an apterous (ap)-Gal4 driver, dorsal expression of nub was lost and the growth of the wing pouch was reduced (Fig. 1H). Reduction of osa function in discs expressing UAS-Cad restored more normal nub expression and growth (Fig. 1I). Furthermore, the ectopic nub expression normally seen in osaeld308/osa4H discs (Fig. 1C) was suppressed by the expression of UAS-Cad in the dorsal compartment (Fig. 1I). Thus, the level of nub expression is determined by the relative levels of Arm and Osa when either of these levels is reduced. To increase the levels, we expressed UAS-Osa with ap-Gal4, causing a strong reduction of nub expression in the dorsal wing pouch (Fig. 1J). Expression of ΔArm with the same Gal4 driver caused nub to be expressed in almost the entire wing disc (Fig. 1K). The normal domain of nub expression was restored when UAS-Osa and ΔArm were coexpressed (Fig. 1L).
Taken together, these data demonstrate that Osa is required for the repression of a wg-dependent gene in vivo. Alterations in the dosage of osa can modulate the expression of wg-dependent genes even in the presence of an activated form of Arm or a dominant negative form of Pan, suggesting that Osa does not act upstream of Arm. Alterations in the level of active Pan/Arm complexes can also modulate nub expression in osa mutants; thus, lack of osa does not make Wg target genes entirely independent of Arm.
We have shown previously that the ARID DNA-binding domain of Osa fused to the repressor domain of Engrailed (UAS-OsaRD) or the activation domain of VP-16 (UAS-OsaAD) can target these domains to genes normally regulated by osa in vivo (Collins et al. 1999). The ectopic expression of nub in osaeld308/osa4H wing discs could be prevented by expression of either UAS-Osa or UAS-OsaRD with ap-GAL4 (Fig. 1M,N) but not by expression of UAS-OsaAD (Fig. 1O). This suggests that Osa functions as a repressor of transcription in the regulation of Wg target genes.
Osa function is independent of the up-regulation of cytosolic Arm
To test whether Osa was acting directly on Wg target genes or regulating the expression of some other gene that interacts with the wg pathway, we sought to determine at what level in the wg pathway Osa acts. In third-instar wing discs, wg is expressed in a narrow stripe of cells that straddle the dorsal/ventral (DV) boundary of the wing pouch and directs growth and patterning of the wing blade with respect to the DV axis (Neumann and Cohen 1997). Cells adjacent to the DV boundary respond to the wg signal by posttranscriptionally up-regulating cytosolic Arm (Fig 2A,C; Riggleman et al. 1990; Peifer et al. 1994). Arm then translocates to the nucleus and binds to Pan (Orsulic and Peifer 1996; Brunner et al. 1997; van de Wetering et al. 1997) to activate the expression of downstream target genes such as Distal-less (Dll; Fig. 2B,C; Zecca et al. 1996; Neumann and Cohen 1997).
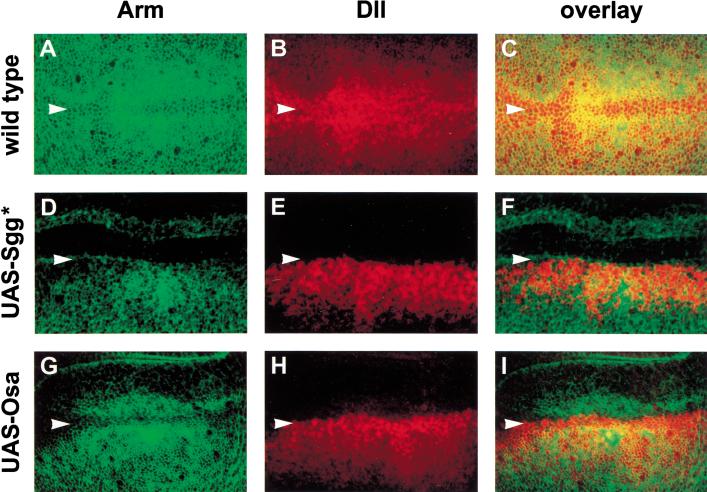
Figure 2.
Osa repression of Wg target genes is independent of Armadillo up-regulation. Confocal images of wing imaginal discs stained with anti-Arm (green) and anti-β-galactosidase to reflect Dll-lacZ expression (red) from Dll-lacZ/+ (A–C), UAS-sgga7, Dll-lacZ/ap-GAL4 (D–F), and UAS-osad3/+; Dll-lacZ/ap-GAL4 (G–I) third-instar larvae. wg expressed at the dorsal/ventral boundary of the wing pouch (indicated by arrowheads) signals the up-regulation of Arm in adjacent cells (A,C), which activates the expression of Dll (B,C). Expression of activated Sgg (UAS-Sgg*) in the dorsal compartment blocks the up-regulation of Arm (D,F) and as a result, these cells fail to express Dll (E,F). Expression of UAS-Osa in the dorsal compartment also represses the expression of Dll (H,I). However, these cells still respond to the Wg signal by up-regulating Arm (G,I).
When an activated form of the protein kinase Sgg that constitutively targets Arm for degradation (UAS-Sgg*; Hazelett et al. 1998) was expressed in the dorsal compartment using the ap-Gal4 driver, Arm was not up-regulated (Fig. 2D,F) and Dll was not expressed (Fig. 2E,F). Expression of UAS-Osa in the dorsal compartment similarly prevented the expression of Dll on the dorsal side of the DV boundary (Fig. 2H,I). However, these cells still responded to the Wg signal by up-regulating cytosolic Arm (Fig. 2G,I). Therefore, Osa represses Wg target genes without affecting the up-regulation of Arm. This places the activity of Osa in the nucleus and argues that Osa may directly repress the expression of Wg target genes.
Osa specifically represses the Wg response of a defined enhancer
To test the requirements for Osa to repress the expression of Wg target genes, we examined the expression of a lacZ reporter gene driven by a well-characterized wg-responsive enhancer. The midgut enhancer (UbxB) of the Ultrabithorax (Ubx) promoter drives lacZ expression in the embryonic midgut in a pattern that is dependent on both wg and decapentaplegic (dpp; Thuringer et al. 1993). In wild-type embryos UbxB–lacZ is expressed primarily in parasegment (ps) 6,7, and 8 with weaker expression in ps 3 (Fig. 3A; Thuringer et al. 1993). This expression was de-repressed in embryos lacking the maternal contribution of osa, such that the expression of lacZ expanded anteriorly as far as ps 3 (Fig. 3B). Similarly expanded expression was induced by ectopic expression of wg in the mesoderm using 24B-Gal4 (Fig. 3C; Thuringer et al. 1993). Conversely, expression of UAS-Osa (Fig. 3D) or UAS-DN-Pan (Fig. 3I) in the mesoderm repressed the expression of UbxB–lacZ. However, neither wg nor dpp was ectopically expressed in the midgut in embryos lacking maternal osa (data not shown).
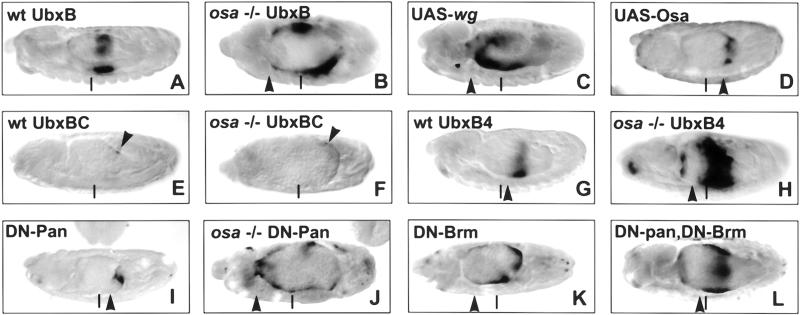
Figure 3.
Osa specifically represses a wg-responsive enhancer. Photomicrographs of stage-14 embryos stained with anti-β-Gal antibody. All embryos carry 24B-GAL4; (A–D,I–L) have UbxB-lacZ; (E,F) have UbxBC-lacZ; (G,H) have UbxB4-lacZ; (C) has UAS-wg; (D) has UAS-osa; (I,J,L) have UAS-dTCFΔN; and (K,L) have UAS-brmK804R. Embryos derived from osaeld308 germ-line clones are shown in B,F,H, and K. In wild-type embryos, UbxB-lacZ is expressed in the embryonic midgut in parasegments (ps) 6, 7, and 8 (A). In embryos lacking the maternal contribution of osa, UbxB-lacZ expression is de-repressed and expands anteriorly to ps 3 (B); similarly expanded expression is induced by ectopic expression of wg in the mesoderm (C). Conversely, overexpression of osa causes a dramatic reduction in UbxB-lacZ expression (D). A mutation in the Dpp response element in UbxB (UbxBC) causes a strong reduction of lacZ expression (E), and this expression is unchanged in embryos derived from osa mutant germ-line clones (F). A mutation in one of the Wg response elements in UbxB (UbxB4) also causes a reduction in the expression of lacZ (G). However, the expression of UbxB4-lacZ is de-repressed in embryos derived from osa mutant germ-line clones. Expression of a dominant negative form of Pan (DN-Pan) causes a reduction in UbxB-lacZ expression (I), whereas expression is expanded in embryos lacking maternal osa that also express DN-Pan (K). Expression of a dominant negative form of Brm also causes an expansion in UbxB-lacZ expression (K) and rescues the loss of expression caused by DN-Pan (L). Arrowheads indicate the position of the anterior extent of UbxB-lacZ expression. Vertical bars indicate 50% embryo length and mark the approximate position of the anterior extent of UbxB-lacZ expression in a wild-type embryo. Embryos derived from osa mutant germ-line clones have additional abnormalities, and constrictions fail to form in the midgut.
When the dpp response element in UbxB is mutated (UbxBC; Eresh et al. 1997), the expression of the lacZ reporter is severely reduced; only weak levels of lacZ expression are detectable in ps 8 (Fig. 3E; Eresh et al. 1997). Expression of UbxBC–lacZ was unchanged in the absence of maternal osa, suggesting that the dpp response element is still required for the expression of the reporter construct in the absence of Osa. When one of the two wg response elements in UbxB is mutated (UbxB4; Riese et al. 1997; Yu et al. 1998), the expression of lacZ is reduced in wild-type embryos (Fig. 3G; Riese et al. 1997). However, removal of maternal osa allowed an expansion of UbxB4–lacZ expression (Fig. 3H). This suggests that lack of osa can compensate for a reduction in the responsiveness of the promoter to Wg but not to Dpp. Furthermore, the expression of wild-type UbxB–lacZ was also de-repressed in embryos lacking maternal osa even in the presence of DN-Pan (Fig. 3J). These data argue that Osa functions specifically to repress the activation of the UbxB enhancer by the Wg pathway.
brahma and moira are also required to repress Wg target genes
Osa functions as a component of Brm chromatin-remodeling complexes (Collins et al. 1999) and might be acting through the Brm complex to repress Wg target genes. We therefore tested other components of the Brm complex for genetic interactions with the wg pathway.
Blocking Wg signaling at the wing margin by expressing UAS-Sgg* with vg-Gal4 caused a reduction in wing growth and a loss of the wing margin (Fig. 4A,B). These phenotypes were strongly enhanced in flies heterozygous for wg (Fig. 4C) or that coexpressed UAS-Osa (Fig. 4D) and were suppressed in flies heterozygous for axin (a negative regulator of Wg signaling; Hamada et al. 1999) or osa (Fig. 4E,F). The effects of UAS-Sgg* expression were also suppressed by the loss of one copy of brm or moira (mor), which encodes an essential component of the Brm complex (Crosby et al. 1999), or by coexpression of a dominant negative form of Brm (DN-Brm; Elfring et al. 1998; Fig. 4H,I; data not shown). In contrast, two other trithorax group genes, trithorax (trx) and absent, small, or homeotic discs 2 (ash2), which encode components of other nuclear complexes thought to regulate chromatin structure (Mazo et al. 1990; Adamson and Shearn 1996; Papoulas et al. 1998), failed to modify the UAS-Sgg* phenotype (data not shown).
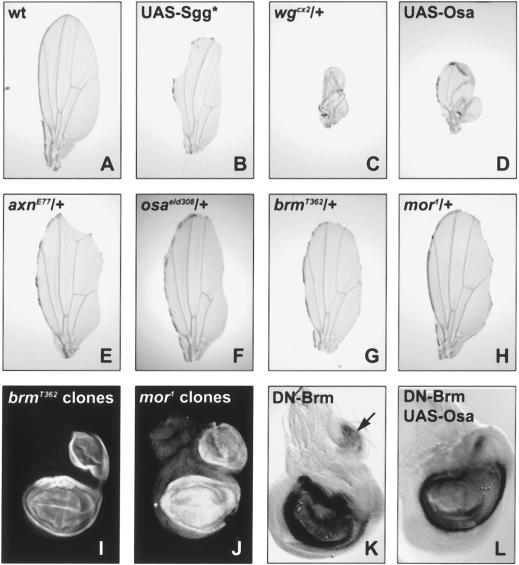
Figure 4.
Brm and Mor are required for the repression of Wg target genes. Photomicrographs of adult wings (A–H) and third-instar wing discs stained with anti-Nub antibody (I–L) from wild-type flies (A), UAS-sgga11/vgGal4 (B), UAS-Sgga11, wgcx2/vg-Gal4 (C), UAS-Osad3/+; UAS-Sgga11/vg-Gal4 (D), axnE77/+; UAS-Sgga11/vg-Gal4 (E), osaeld308/+; UAS-Sgga11/vg-Gal4 (F), brmT362/+; UAS-Sgga11/vg-Gal4 (G), mor1/+; UAS-sgga11/vg-Gal4 (H), FRT80, brmT362/FRT80, M(3)67C; ap-Gal4, UAS-FLP/+ (I), FRT82, mor1/FRT82, M(3)96C; ap-Gal4, UAS-FLP/+ (J), UAS-brmK804R/+; ap-Gal4/+ (K), and UAS-osad3/UAS-brmK804R; ap-Gal4/+ (L). Expression of an activated form of Sgg along the developing wing margin with the vg-Gal4 driver causes a reduction in the growth of the wing and a loss of the wing margin (B). These phenotypes are enhanced in flies that are heterozygous for wg (C) or that coexpress UAS-Osa (D) and are suppressed in flies heterozygous for axn (E) or osa (F). The growth and loss of margin phenotypes are also suppressed in flies heterozygous for brm and mor (G,H, respectively). nub is ectopically expressed in wing discs with clones of brm and mor that occupy a large part of the dorsal compartment (I,J, respectively). Expression of dominant negative Brm in the dorsal compartment induces ectopic expression of nub (arrow in K). Coexpression of dominant negative Brm also rescues the loss of endogenous expression of nub caused by expression of UAS-Osa alone (cf. L with Figs. 1J or 5A).
This demonstrates that there is a specific genetic interaction between the wg pathway and components of Brm complexes and suggests that these complexes are required for the repression of Wg target genes. Indeed, we found that the wg-dependent gene nub was ectopically expressed in wing discs that contained large clones of cells mutant for brm or mor (Fig. 4I,J) or that expressed DN-Brm in the dorsal compartment (Fig. 4K). Furthermore, the loss of nub expression caused by expression of UAS-Osa with ap-Gal4 was rescued by coexpression of DN-Brm (cf. Fig. 4L with Figs. 1J and 5A), indicating that Brm activity is required for the repression of Wg target genes by Osa. The Wg-dependent UbxB–lacZ reporter was also de-repressed in embryos that expressed DN-Brm (Fig. 3K), and coexpression of DN-Brm could rescue the loss of UbxB–lacZ expression caused by DN-Pan (Fig. 3L). These results suggest that Osa acts through the Brm chromatin-remodeling complex to prevent the expression of Wg target genes.
Figure 5.
Osa cooperates with Gro and Rpd3 to repress Wg target genes and is required for the Wg-dependent repression of dpp. Photomicrographs of third-instar wing discs stained with anti-Nub antibody (A–C) from larvae of genotype UAS-osad3/+; ap-Gal4/+ (A), UAS-osas2/ap-GAL4; rpd304556/rpd304556 (B), and osaeld308/groE48 (C). (D) osaeld308/groE48 adult. The loss of nub expression in the dorsal wing pouch caused by the expression of UAS-Osa with ap-Gal4 (A) is partially rescued in larvae homozygous for a hypomorphic allele of rpd3 (B). Flies that are transheterozygous for osa and gro often have ectopic expression of nub in wing discs (C) and notum-to-wing transformation in adults (arrow in D). (E–G) show leg discs stained for dpp-lacZ expression from larvae of genotype dpp-lacZ/+ (E), UAS-osad3/+; dpp-lacZ/Dll-Gal4 (F), osaeld308/osa4h; dpp-lacZ/+ (G). (H) adult leg from FRT82, osaeld616/FRT82, P(y+); hsFLP122/+ fly. dpp is expressed along the anterior/posterior boundary in the dorsal compartment of the leg disc (E). The expression of UAS-Osa represses the endogenous expression of dpp (F), whereas dpp is ectopically expressed in osa mutant leg discs (G). Clones of cells mutant for osa (marked with yellow) generated in leg discs can induce leg duplications in the ventral compartment (H). The black arrow indicates the yellow, osa mutant cells, and the red arrow points to transverse bristle rows, normally found ventrally, within the duplication.
osa interacts genetically with gro and rpd3
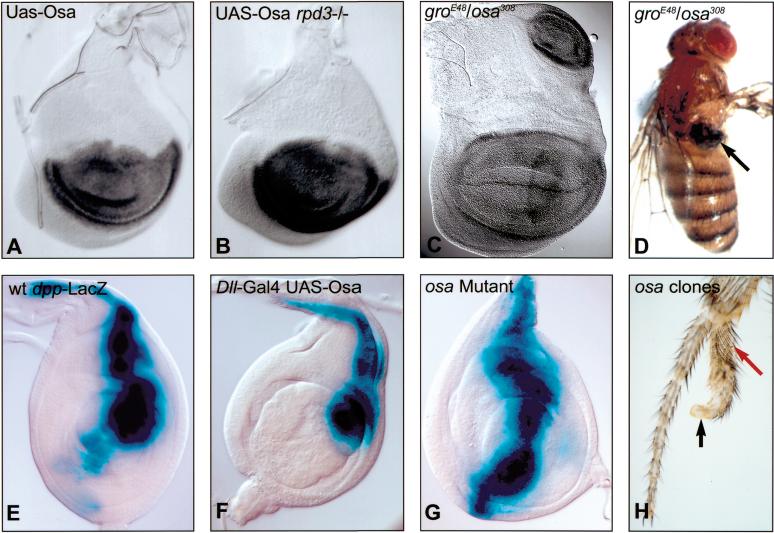
In addition to transducing the wg signal in a complex with Arm (Brunner et al. 1997; van de Wetering et al. 1997), Pan is also required for the active repression of Wg target genes in the absence of the Wg signal (Cavallo et al. 1998). This repression requires the association of Pan with the corepressor Groucho (Gro; Cavallo et al. 1998). Gro functionally interacts with the histone deacetylase Rpd3, and this interaction is important for at least some of the repressive activity of Gro (Chen et al. 1999). Thus, both Osa-containing Brm complexes and Pan/Gro/Rpd3 complexes repress the expression of Wg target genes and probably mediate this repression by altering the local chromatin architecture at the promoters of these genes. Consistent with this, we found that reduction of gro or rpd3 dosage reduced the ability of Osa to repress nub. The loss of nub expression caused by expression of UAS-Osa with ap-Gal4 (Fig. 5A) was significantly rescued in wing discs homozygous for a hypomorphic allele of rpd3 (rpd304556; Fig. 6B). Also, larvae transheterozygous for osaeld308 and groE48 often ectopically expressed nub in the wing disc (Fig. 5C), and 40% (n = 253) of transheterozygous adults had notum-to-wing transformations (Fig. 5D). These phenotypes were not seen when osa or gro single mutants were crossed to wild-type flies.
Figure 6.
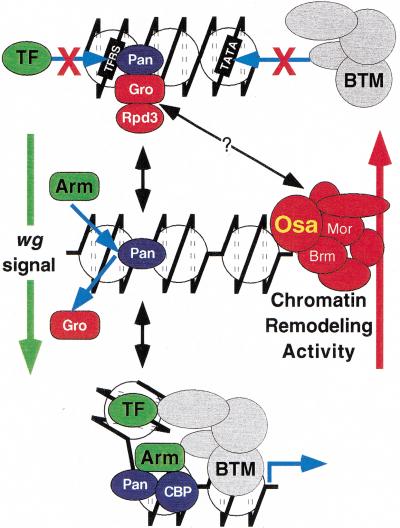
Model for the regulation of gene expression by the Wg pathway. See text for details. TF indicates a transcription factor that requires Wingless signaling for access to its binding site (TFBS). BTM indicates components of the basal transcriptional machinery.
Osa represses the expression of a gene that is repressed by Wg
Whereas many of the genes regulated by the wg pathway require wg for their expression, several genes appear to be repressed by high levels of wg signaling (Johnston and Schubiger 1996; Theisen et al. 1996; Cadigan et al. 1998; Yu et al. 1998; Payre et al. 1999). To determine the effect of Osa on the expression of genes that are normally repressed by wg, we examined the expression of dpp in leg discs with altered dosage of osa.
In third-instar leg discs, wg and dpp are expressed along the A/P boundary in the ventral and dorsal compartment, respectively, and mutually antagonize each other's expression (Brook and Cohen 1996; Jiang and Struhl 1996; Johnston and Schubiger 1996; Theisen et al. 1996). We found that dpp expression was repressed when UAS-Osa was expressed in a broad central domain of the leg disc with a Dll-Gal4 driver (Fig. 5F) and that dpp was ectopically expressed in the ventral compartment in osaeld308/osa4H leg discs (Fig. 5G). Clones of cells mutant for osa could also induce leg duplications in the ventral compartment of the leg (Fig. 5H), consistent with the ectopic expression of dpp (Brook and Cohen 1996; Jiang and Struhl 1996). Thus, in addition to repressing the expression of genes that are normally activated by the wg signal, Osa is also required for the repression of least one of the genes that are repressed by wg.
Discussion
Osa directly represses Wg target genes
We show here that the loss of osa function in the wing disc results in the ectopic expression of wg-dependent genes and that overexpression of osa can block the endogenous expression of these genes. Several lines of evidence suggest that Osa directly represses the expression of these genes. Firstly, up-regulation and nuclear accumulation of Arm is both necessary and sufficient for the response of a target gene to the Wg signal (for review, see Cadigan and Nusse 1997). Altering the dosage of osa modulates the expression of a Wg target gene without affecting the up-regulation of Arm. This indicates that Osa does not act upstream of Arm and that Osa activity is required in the nucleus for the repression of Wg target genes. In addition, Osa appears to act as a repressor rather than activating the expression of another repressor, as replacement of most of the protein with an exogenous repressor domain preserves its function in Wg target-gene regulation.
Second, increasing osa dosage suppresses the ectopic expression of Wg target genes induced when cytosolicnuclear Arm is maintained at artificially high levels, and reducing osa dosage restores Wg target gene expression when Arm is sequestered at the plasma membrane or when a form of Pan that is unable to bind to Arm is overexpressed. Therefore, the expression of a Wg target gene is determined by the relative levels of Osa and activating Pan/Arm complexes.
Finally, the UbxB enhancer contains two Wg response elements that are required for full expression of a reporter gene (Yu et al. 1998). A mutation in one of the Wg response elements prevents the recruitment of Arm to that site by Pan and results in a reduction in reporter gene expression (Riese et al. 1997). Loss of osa function can compensate for the reduced responsiveness of the enhancer to the Wg signal, allowing it to be more broadly expressed. This suggests that endogenous Osa is able to repress an enhancer lacking this Pan-binding site. However, the remaining Pan-binding site may be necessary for Osa to act on the enhancer; ectopic Osa can only repress the expression of a Wg response element from the optomotor-blind gene (K. Hofmeyer and G. Pflugfelder, pers. comm.) when its Pan-binding sites are intact (data not shown).
The most likely explanation of these data is that Osa functions to directly repress Wg target gene expression, with such target genes being defined by their inclusion of a Pan-binding site. Osa function is not exclusive to the Wg signaling pathway; Osa also functions as a promoter specific activator of Antennapedia expression and as a coactivator for Zeste and likely represses E2F-mediated gene expression (Staehling-Hampton et al. 1999; Vasquez et al. 1999; Kal et al. 2000). Furthermore, the expression of even-skipped is perturbed in embryos lacking maternal osa, a phenotype that precedes the expression of wg in the embryo (Treisman et al. 1997). However, the strong correlation of the expression of Wg target genes with the level of Osa suggests that counteracting Osa activity is an important function of the Wg pathway.
Brm complex chromatin remodeling activity is required to repress Wg target genes
Osa functions as a component of Brm chromatin-remodeling complexes (Collins et al. 1999). These complexes and closely related complexes in other species such as the yeast SWI/SNF complex catalyze an ATP-dependent alteration in the structure of nucleosomal DNA that can run in either direction to render the DNA either more or less accessible to binding by transcription factors (Cote et al. 1994; Kwon et al. 1994). Whereas chromatin-remodeling complexes are generally thought to promote gene expression, recent reports have demonstrated that they are also required for the repression of some genes. Genome-wide analysis shows that more genes have elevated than reduced expression in a swi2 mutant yeast strain, and some of these genes are directly repressed by SWI/SNF (Holstege et al. 1998; Sudarsanam et al. 2000). The hBRM complex in humans has been shown to cooperate with the retinoblastoma protein (Rb) to repress E2F-1-mediated activation (Trouche et al. 1997). Furthermore, brm, mor, and osa were identified as enhancers of an E2F gain-of-function phenotype, suggesting that Brm complexes also repress E2F activation in Drosophila (Staehling-Hampton et al. 1999).
Because we had found that Osa can antagonize Brm complex function in some tissues (Collins et al. 1999), it was possible that Brm complex activity could be required for the expression of Wg target genes and that Osa might be a negative regulator of Brm complex function. However, our findings that the effects of blocking the wg pathway at the wing margin can be suppressed by reducing the dosage of brm or mor and that nub and UbxB–lacZ are ectopically expressed when brm or mor function is lost suggest that Brm complexes are required for the repression, rather than the activation, of Wg target genes. Furthermore, expression of a form of Brm that has a mutation in its ATP-binding site also induces ectopic expression of nub and UbxB–lacZ and can rescue the loss of nub expression caused by overexpression of Osa. Because the ATPase activity of Brm is required for the chromatin-remodeling activity of the Brm complex (Elfring et al. 1998), this suggests that chromatin remodeling by the Brm complex is necessary for Osa to repress the expression of Wg target genes.
Osa cooperates with Pan and Gro to repress Wg target gene expression
In addition to activating gene expression by recruiting Arm to the promoters of wg-responsive genes, Pan also represses these same genes (Riese et al. 1997; Yang et al. 2000) by recruiting the transcriptional corepressor Gro (Cavallo et al. 1998). Interestingly, Gro has been shown to interact with the N-terminal tail of histone H3 and with the histone deacetylase Rpd3, and it has therefore been proposed that Gro mediates repression by altering chromatin structure (Palaparti et al. 1997; Chen et al. 1999). Consistent with this, we found a strong genetic interaction between osa and gro that suggests that their activities in repressing Wg target genes are closely related. Although it has not previously been reported that Rpd3 functions in the repression of wg target genes, we have shown that reducing the function of rpd3 can partly rescue the loss of nub expression caused by the overexpression of Osa. Rpd3 is therefore important for the repression of Wg target genes; testing whether it is essential awaits the isolation of null alleles.
The loss of either osa or gro leads to ectopic expression of Wg target genes; thus, the activity of one is not sufficient to repress the expression of these genes without the activity of the other. Osa and Gro may, therefore, be mediating the same repressive event rather than acting in parallel. Interestingly, Zhang et al. (2000) have reported that human SWI/SNF forms a repressor complex with Rb and the histone deacetylase HDAC. This complex interacts with the cyclin E promoter through the binding of Rb to E2F-1 and represses E2F-1 activation of cyclin E expression. This suggests the intriguing possibility that Osa and the Brm complex function in a larger repressor complex containing Gro and Rpd3 and that this complex is recruited to Wg target genes though the binding of Gro to Pan. However, Gro acts as a corepressor for a large number of transcription factors (for review, see Fisher and Caudy 1998), and Osa cannot be required for all repression mediated by Gro because loss of osa does not result in neurogenic phenotypes like those caused by the loss of gro (Treisman et al. 1997). Further research is needed to determine if Gro and/or Rpd3 can directly interact with components of the Brm complex and, if so, what determines the specificity of this interaction.
The mechanism by which Wg signaling leads to the active repression of genes such as dpp is not fully understood, although it is counteracted by Sgg (Jiang and Struhl 1996). However, our observation that dpp expression is repressed by Osa suggests that other factors may allow Wg signaling to reinforce repressive chromatin modeling by the Brahma complex on such promoters.
Model for the regulation of gene expression by the Wg pathway
The requirement of chromatin remodeling complexes for the repression of Wg target-gene expression clearly demonstrates that regulating chromatin architecture is important for the repression of these genes. However, it is becoming increasingly evident that altering the chromatin conformation at the promoters of Wg target genes is also important for the activation of these genes.
Pan and its vertebrate homologs Lef-1 and TCF belong to the high-mobility group, or HMG, family of proteins. The HMG domain is known to induce a sharp bend in DNA, and this DNA-bending activity has been shown to be important for the activation of gene expression by HMG box transcription factors (for review, see Bustin 1999). It is thought that DNA bending promotes activation by bringing distantly spaced transcription factors into proximity, thus promoting the formation of higher-order activation complexes. Interestingly, it has been reported that Lef-1 bound to the vertebrate homolog of Arm, β-catenin, can induce a sharper bend in DNA than Lef-1 alone (Behrens et al. 1996).
Hecht et al. (2000) have reported that β-cat binds to the histone acetyltransferase (HAT) proteins p300 and CBP and that this interaction can promote β-cat-dependent gene expression. They also report that the HAT activity of CBP is dispensable for this activation. However, these experiments were performed using the transient transfection of an artificial β-cat-responsive reporter gene that may not reflect the true requirement for CBP HAT activity for the expression of an endogenous gene in the context of chromosomal DNA. Alternatively, p300 may promote expression by recruiting other HATs or components of the basal transcriptional machinery (for review, see Grant and Berger 1999).
Drosophila CBP (dCBP) has also been shown to interact with Wg signaling; dCBP appears to negatively regulate Wg signaling by acetylating Pan and disrupting its association with Arm (Waltzer and Bienz 1998). Thus, CBP and dCBP may play opposing roles in Wnt and Wg signaling, respectively. However, in the case of the Interferon beta (IFNβ) enhanceosome complex, CBP HAT activity is not only required for activation by the complex (Merika et al. 1998) but also promotes the dissociation of the complex through the acetylation of the HMG I(Y) component of IFNβ (Munshi et al. 1998). Similarly, dCBP could promote expression of Wg target genes through acetylation of the core histones and later repress expression by acetylating Pan. While the full extent of the role of dCBP remains to be determined, the requirement of chromatin remodeling (and the apparent requirement of Rpd3) for the repression of Wg target gene expression makes it likely that Arm activates target-gene expression by recruiting the HAT activity of dCBP.
Our current model for the regulation of gene expression by components of the Wg pathway is depicted in Figure 6. The chromatin remodeling activity of the OsaBrm complex is required to maintain the chromatin at the promoters of wg-responsive genes in a repressive conformation. This would prohibit the association of other transcription factors with their binding sites and prevent the recruitment of components of the basal transcription machinery. Osa/Brm complexes may be recruited to Wg-responsive genes through an association with Pan/Gro/Rpd3 complexes. In response to the Wg signal, Arm is stabilized and accumulates in the cytosol. This accumulation of cytosolic Arm permits Arm to translocate to the nucleus and displace Gro from Pan and, in so doing, relieve the repression mediated by Gro, Rpd3, and Osa/Brm complexes. Arm may also promote a more open chromatin conformation by recruiting the HAT activity of dCBP, thus permitting the association of other transcription factors with their binding sites. Also, the stimulation of the DNA-bending activity of Pan by Arm may bring distantly spaced transcription factors into juxtaposition to promote the activation of gene expression.
In the absence of osa, the chromatin is maintained in a more open and less repressive conformation. This would permit other transcription factors to interact with their binding sites at lower concentrations than would otherwise be possible. Under these conditions, the low levels of Arm that are always present in the cell may be sufficient to promote the activation of gene expression without the Wg signal.
Materials and methods
Genetics
Alleles used were ash21, rpd304556, wgCX2, osaeld308, osaeld616, groE48, mor1, brm2 (Flybase), brmT362, brmT808 (J.E. Treisman, unpubl.), axnE77 (gift of J. Lee, New York University), and osa4H (gift of T. Lebestky, UCLA). The reporters were Dll01092 (Flybase), dpp-lacZ (Blackman et al. 1991), UbxB–lacZ (Thuringer et al. 1993), UbxB4–lacZ (Riese et al. 1997), and UbxBC–lacZ (Eresh et al. 1997) Gal4 driver lines used were how24B-Gal4, apmd544, Dllmd23 (Flybase), dpp-Gal4, ey-Gal4 (Hazelett et al. 1998), and vg-Gal4 (Simmonds et al. 1995). Transgenic lines used were UAS-brmK804R (Elfring et al. 1998), UAS-Cad (Sanson et al. 1996), UAS-dTCFΔN (van de Wetering et al. 1997), UAS-FLP (Duffy et al. 1998), UAS-fluΔArm (Zecca et al. 1996), UAS-osa, UAS-osaRD, UAS-osaAD (Collins et al. 1999), UAS-SggS9A (Hazelett et al. 1998), and UAS-wg (Azpiazu et al. 1996). To make mutant clones of brm and mor, flies of genotype FRT-80, brmT362/TM6B were crossed to flies of genotype hs-FLP122; FRT-80, M(3)67C/TM6B and flies of genotype FRT-82, mor1/TM6B were crossed to flies of genotype hs-FLP122; FRT-82, M(3)96C/TM6B, respectively. Larvae were heat shocked for 1 h at 38.5°C during first and second instars to induce expression of hs-FLP. To make germ-line clones mutant for osa, FRT-82, osaeld308/TM6B females were crossed to hsFLP122/Y; FRT82, ovoD/TM3 males, and larvae were heat shocked for 1 h at 38.5°C during first and second instars. The resulting females with germ-line clones were crossed to 24B-GAL4, UbxB(BC, B4) males. The controls for genetic interactions with UAS-sgg* were the FRT80 and FRT82 chromosomes, on which the other mutations were generated or onto which they were recombined.
Immunohistochemistry
Wing imaginal discs and embryos were stained as described by Treisman et al. (1997). The antibody dilutions were mouse anti-Nub (Ng et al. 1996) 1:5, mouse anti-Arm (Peifer et al. 1994) 1:1, and rabbit anti-β-Gal (Cappel) 1:500. Leg discs from late third-instar larvae were dissected into PBS and fixed for 10 min in 1% glutaraldehyde in PBS and washed twice with PBS. β-Gal activity was detected by incubating the discs in X-gal staining buffer. Embryos and adult wings were mounted in Canada balsam: methyl salicylate (2:1).
Acknowledgments
We thank Spyros Artavanis-Tsakonas, Mariann Bienz, Steve Cohen, Manfred Frasch, Kerstin Hofmeyer, Tim Lebestky, Jeff Lee, Armen Manoukian, Mark Peifer, Allen Shearn, Gary Struhl, John Tamkun, Jean-Paul Vincent, and the Bloomington Drosophila stock center for fly stocks and reagents. We are grateful to Ruth Lehmann and Michelle Starz-Gaiano for help with confocal microscopy and to Ulrike Heberlein for assistance with the analysis of osa mutant clones in the leg. We thank Ian Oliver, Aude Benlali, and Zara Martirosyan for technical assistance and the members of the Treisman and Lehmann labs for stimulating discussions. The manuscript was improved by the thoughtful comments of Jeff Lee, Corinne Zaffran, Michelle Starz-Gaiano, and Helen Sink. This work was supported by NIH grant GM56131.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL treisman@saturn.med.nyu.edu; FAX (212) 263-7760.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.854300.
References
- Adamson AL, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Lawrence PA, Vincent J-P, Frasch M. Segmentation and specification of the Drosophila mesoderm. Genes & Dev. 1996;10:3183–3194. doi: 10.1101/gad.10.24.3183. [DOI] [PubMed] [Google Scholar]
- Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of β-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh J-C, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Bhat KM. Frizzled and Frizzled 2 play a partially redundant role in Wingless signaling and have similar requirements to Wingless in neurogenesis. Cell. 1998;95:1027–1036. doi: 10.1016/s0092-8674(00)81726-2. [DOI] [PubMed] [Google Scholar]
- Blackman RK, Sanicola M, Raftery LA, Gillevet T, Gelbart WM. An extensive 3‘ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-β family in Drosophila. Development. 1991;111:657–666. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D. A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes & Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook WJ, Cohen SM. Antagonistic interactions between wingless and decapentaplegic responsible for dorsal-ventral pattern in the Drosophila leg. Science. 1996;273:1373–1377. doi: 10.1126/science.273.5280.1373. [DOI] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homolog that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93:767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes FJ, Garcia-Bellido A. Proximo-distal specification in the wing disc of Drosophila by the nubbin gene. Proc Natl Acad Sci. 1997;94:11405–11410. doi: 10.1073/pnas.94.21.11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RT, Furukawa T, Tanese N, Treisman JE. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 1999;18:7029–7040. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Cox RT, Kirkpatrick C, Peifer M. Armadillo is required for adherens junction assembly, cell polarity, and morphogenesis during Drosophila embryogenesis. J Cell Biol. 1996;134:133–148. doi: 10.1083/jcb.134.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby MA, Miller C, Alon T, Watson KL, Verrijzer CP, Goldman-Levi R, Zak NB. The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol. 1999;19:1159–1170. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas PB, Cheney IW, Liao D-W, Bowrin V, Byam W, Pacchione S, Kobayashi R, Yaciiuk P, Moran E. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas PB, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol. 2000;20:3137–3146. doi: 10.1128/mcb.20.9.3137-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR, Daubresse G, DePace A, et al. Genetic analysis of brahma: The Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eresh S, Riese J, Jackson DB, Bohmann D, Bienz M. A CREB-binding site as a target for decapentaplegic signalling during Drosophila endoderm induction. EMBO J. 1997;16:2014–2022. doi: 10.1093/emboj/16.8.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci. 1997;94:12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Berger SL. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–177. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- Hamada F, Tomoyasu Y, Takatsu Y, Nakamura M, Nagai S, Suzuki A, Fujita F, Shibuya H, Toyoshima K, Ueno N, et al. Negative regulation of Wingless signaling by D-axin, a Drosophila homolog of axin. Science. 1999;283:1739–1742. doi: 10.1126/science.283.5408.1739. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Hecht A, Litterst CM, Huber O, Kemler R. Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Complementary and mutually exclusive activities of decapentaplegic and wingless organize axial patterning during Drosophila leg development. Cell. 1996;86:401–410. doi: 10.1016/s0092-8674(00)80113-0. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Schubiger G. Ectopic expression of wingless in imaginal discs interferes with decapentaplegic expression and alters cell determination. Development. 1996;122:3519–3529. doi: 10.1242/dev.122.11.3519. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kal AJ, Mahmoudi T, Zak NB, Verrijzer CP. The Drosophila Brahma complex is an essential coactivator for the trithorax group protein Zeste. Genes & Dev. 2000;14:1058–1071. [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon RT. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in β-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Dierick HA, Kirkpatrick C, Moline MM, Baas A, Peifer M, Bejsovec A. Drosophila APC2 is a cytoskeletally-associated protein that regulates Wingless signaling in the embryonic epidermis. J Cell Biol. 1999;146:1303–1318. doi: 10.1083/jcb.146.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN β enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Moreira JM, Holmberg S. Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J. 1999;18:2836–2844. doi: 10.1093/emboj/18.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN β expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Distinct mitogenic and cell specification functions of wingless in different regions of the wing. Development. 1996;122:1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- ————— Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- Ng M, Diaz-Benjumea FJ, Cohen SM. nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development. 1995;121:589–599. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- Ng M, Diaz-Benjumea FJ, Vincent J-P, Wu J, Cohen SM. Specification of the wing by localized expression of wingless protein. Nature. 1996;381:316–318. doi: 10.1038/381316a0. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Peifer M. An in vivo structure-function study of Armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for Wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- Payre F, Vincent A, Carreno S. ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature. 1999;400:271–275. doi: 10.1038/22330. [DOI] [PubMed] [Google Scholar]
- Peifer M, Sweeton D, Casey M, Wieschaus E. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- Prieve MG, Waterman ML. Nuclear localization and formation of β-catenin-lymphoid enhancer factor 1 complexes are not sufficient for activation of gene expression. Mol Cell Biol. 1999;19:4503–4515. doi: 10.1128/mcb.19.6.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–788. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- Riggleman B, Schedl P, Wieschaus E. Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3b to the APC–β-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- Ruel L, Stambolic V, Ali A, Manoukian AS, Woodgett JR. Regulation of the protein kinase activity of Shaggy(Zeste-white3) by components of the wingless pathway in Drosophila cells and embryos. J Biol Chem. 1999;274:21790–21796. doi: 10.1074/jbc.274.31.21790. [DOI] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent JP. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Chou TB, Perrimon N. wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Brook WJ, Cohen SM, Bell JB. Distinguishable functions for engrailed and invected in anterior–posterior patterning in the Drosophila wing. Nature. 1995;376:424–427. doi: 10.1038/376424a0. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Ciampa PJ, Brook A, Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen H, Haerry TE, O’Connor MB, Marsh JL. Developmental territories created by mutual antagonism between Wingless and Decapentaplegic. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- Thuringer F, Cohen SM, Bienz M. Dissection of an indirect autoregulatory response of a homeotic Drosophila gene. EMBO J. 1993;12:2419–2430. doi: 10.1002/j.1460-2075.1993.tb05896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Luk A, Rubin GM, Heberlein U. eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes & Dev. 1997;11:1949–1962. doi: 10.1101/gad.11.15.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation funcitons of E2F1. Proc Natl Acad Sci. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ympa A, Hursh D, Jones T, Bejsovec A, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–800. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Vazquez M, Moore L, Kennison JA. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the Brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Willert K, Logan CY, Arora A, Fish M, Nusse R. A Drosophila Axin homolog, Daxin, inhibits Wnt signaling. Development. 1999;126:4165–4173. doi: 10.1242/dev.126.18.4165. [DOI] [PubMed] [Google Scholar]
- Yang X, van Beest M, Clevers H, Jones T, Hursh DA, Mortin MA. decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development. 2000;127:3695–3702. doi: 10.1242/dev.127.17.3695. [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes & Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- Yu X, Riese J, Eresh S, Bienz M. Transcriptional repression due to high levels of Wingless signalling. EMBO J. 1998;17:7021–7032. doi: 10.1093/emboj/17.23.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a Wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, III, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]