Abstract
Coliphage N4-coded, virion-encapsidated RNA polymerase (vRNAP) is able to bind to and transcribe promoter-containing double-stranded DNAs when the template is supercoiled and Escherichia coli single-stranded DNA-binding protein (Eco SSB) is present. We report that vRNAP–promoter recognition and activity on these templates require specific sequences and a hairpin structure on the template strand. Hairpin extrusion, induced by Mg(II) and physiological superhelical density, is essential to provide the correct DNA structure for polymerase recognition, as mutant promoters that do not form hairpins show reduced in vitro activity. Therefore, a supercoil-induced DNA structural transition regulates N4 vRNAP transcription. Eco SSB activates transcription at physiological superhelical densities by stabilizing the template-strand hairpin. Specific sequences at the promoters are conserved to provide proper contacts for vRNAP, to support hairpin extrusion, or both. We propose a model for in vivo utilization of the vRNAP promoters, and discuss the roles of DNA supercoiling and Eco SSB in promoter activation.
Keywords: DNA structure, supercoiling, single-stranded DNA-binding protein, promoter activation
Unlike other bacteriophages, N4 does not require the host RNA polymerase (RNAP) for the expression of its early genes. A phage-coded, virion-encapsidated RNA polymerase (N4 vRNAP) is injected into the host cell along with the phage genome to catalyze early transcription (Falco et al. 1977, 1980). N4 vRNAP is a sequence-specific, single-stranded DNA-binding protein. In vitro, it utilizes the promoter-containing template strand of DNA accurately and efficiently in the absence of the nontemplate strand (Glucksmann et al. 1992). N4 vRNAP activity on double-stranded templates requires both DNA supercoiling and Escherichia coli single-stranded DNA-binding protein (Eco SSB) (Falco et al. 1978; Markiewicz et al. 1992).
The consensus sequence of the three N4 early promoters (P1, P2, and P3) extends from positions −18 to +1 and contains a set of inverted repeats centered at position −12 and composed of conserved and nonconserved bases (Haynes and Rothman-Denes 1985). Analysis of the activity of promoter mutants present on single-stranded templates demonstrated that some, but not all, conserved sequences are required for vRNAP transcription (Glucksmann et al. 1992). In addition, the effect of mutations in the nonconserved positions of the inverted repeats indicated that the integrity of these repeats is essential for promoter activity. These results suggested that a 5- to 7-bp stem, 3-base loop hairpin is required for N4 vRNAP–promoter recognition (Glucksmann et al. 1992). To reconcile the peculiar template specificity of N4 vRNAP with the double-stranded nature of the N4 genome, we proposed a model for the interaction of vRNAP with its promoters on double-stranded DNA (Glucksmann et al. 1992). Hairpin extrusion is facilitated by negative supercoiling of the template generated by E. coli DNA gyrase. Subsequently, Eco SSB binds and stabilizes the hairpin to yield an active promoter conformation. This structure and the conserved sequences enable vRNAP to bind and initiate transcription.
Recently, we have shown that hairpin extrusion at the N4 vRNAP promoters occurs at physiological superhelical density in a Mg(II)-dependent manner (Dai et al. 1997). Besides the integrity of the inverted repeats, specific sequences at certain conserved positions encompassing the repeats are critical for extrusion of the promoter hairpins (Dai et al. 1997, 1998). Analysis of the in vivo activity of a mutant promoter that failed to extrude in vitro, but is utilized by vRNAP on single-stranded templates, indicated that hairpin extrusion is required for promoter activity in vivo (Dai et al. 1997). To determine the role of hairpin extrusion in vRNAP promoter activity, we analyzed a collection of mutant promoters present on supercoiled, double-stranded templates for their in vitro transcriptional activity. We show that the ability of a promoter to extrude a hairpin affects its in vitro activity. Comparison of promoter activities on supercoiled and single-stranded templates enabled us to identify the role of individual bases in polymerase contacts and/or hairpin extrusion. From these analyses, we conclude that conserved bases of the inverted repeats serve collectively two functions: Some positions provide critical direct contacts for vRNAP recognition and binding, other positions fulfill the sequence requirements essential for hairpin extrusion, and yet others are required for both functions. Furthermore, these studies have provided new insights into the effect of DNA supercoiling and of Eco SSB on promoter activation.
Results
Transcriptional activity of wild-type promoters requires supercoiling and Eco SSB at low superhelical densities
N4 DNA circles (2.2 kb) of varying superhelical densities, containing N4 vRNAP wild-type promoter P1 followed by its natural terminator t1 and N4 vRNAP wild-type promoter P2 followed by its natural N4 terminator t2 (Fig. 1A), were generated and prepared as described (Miller et al. 1996). In vitro transcription assays carried out in the absence or presence of Eco SSB indicated that promoters present on these circles are transcriptionally active (Fig. 1B). Two major RNA species with apparent sizes of 550 and 1100 bases were synthesized. The 1100-base RNA initiated at P1 and terminated at t2, whereas the 550-base RNA initiated at P2 and terminated at t2 (Markiewicz et al. 1992). As observed previously, promoter P2 is stronger than promoter P1 and terminator t2 is more efficient than t1 (Markiewicz et al. 1992). Whereas Eco SSB is essential for transcription from P1 and P2 at −σ < 0.071 (Fig. 1B), it was not required for, but still activated N4 vRNAP transcription at high superhelical densities (Fig. 1B, cf. left and right panels). Figure 1C shows the effect of increasing Eco SSB/DNA ratios on transcription at three superhelical densities. Eco SSB activated transcription markedly from promoter P1 at high superhelical density, whereas it had only a small effect on transcription from promoter P2 under these conditions (Fig. 1C, right panel). These results indicate that the functions of Eco SSB and DNA supercoiling might partially overlap.
Figure 1.
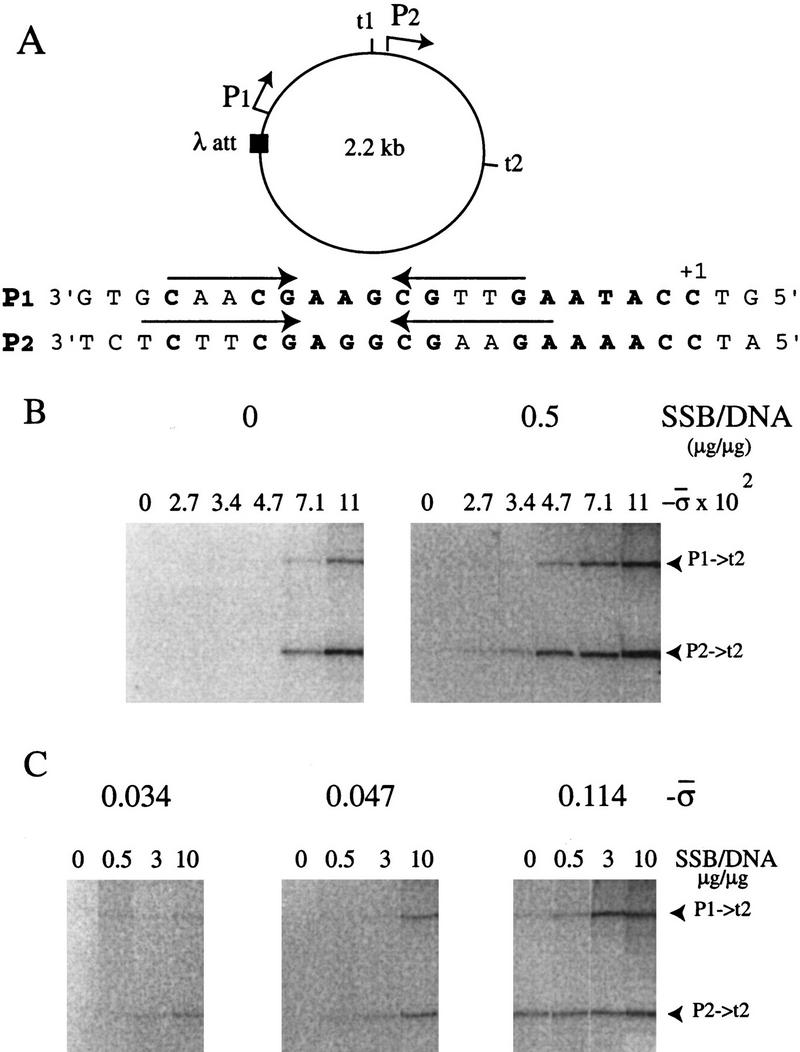
Dependence of promoter activity on superhelical density and Eco SSB. (A) Diagram of the 2.2-kb DNA circles used for transcriptional studies. The sequence of the template strand of promoters P1 and P2 is shown below. (B) Transcription on templates of different superhelical densities in the absence (left) and presence (right) of Eco SSB. (C) Eco SSB activation of transcription on templates of different superhelical densities. RNAs synthesized in in vitro transcription assays using topoisomers of N4 early promoter-containing DNA circles as templates were analyzed on denaturing gels. The two major transcripts, initiated from P1 or P2 and terminated at the strong terminator t2, are marked at right.
To assess the effect of base changes on promoter activity, the in vitro transcriptional activity of mutant promoters was determined using circles of physiological (σ = −0.034) or high (σ = −0.114) superhelical densities in the presence of Eco SSB [Eco SSB:DNA = 1:1 (wt/wt)]. To examine the intrinsic effect of promoter mutations on transcriptional activity, assays on highly supercoiled (σ = −0.114) templates were performed in the absence of Eco SSB, because at high superhelical densities, N4 vRNAP transcription no longer requires the presence of Eco SSB (Fig. 1B). Comparison of the activity of mutant promoters in the absence and presence of Eco SSB allowed us to examine the role of Eco SSB in activation. Promoter P2 remained unchanged and served as a control. The amount of transcripts initiated from promoters P1 and P2 was quantitated. The activity of mutant promoters (P → t2) was normalized against the activity of promoter P2 (P2 → t2) in each experiment. A summary of the activity of each promoter mutation relative to wild-type promoter P1 and measured under different conditions is presented in Tables 1 and 2. In all cases, sequences corresponding to the template strand of the promoter (3′ → 5′) are presented because no determinants of recognition are present in the complementary strand (Glucksmann et al. 1992). Names of mutant promoters designate the base change on the template strand of promoter P1 and its position with respect to +1.
Table 1.
Transcriptional activity of promoters containing mutations at the inverted repeats
| Activity
|
|||||||
|---|---|---|---|---|---|---|---|
| circles (−ς)
|
Extrusion
|
||||||
| Promoter
|
Sequence
|
ssDNA −SSB
|
0.114
|
0.034
|
−ς
|
||
| (−SSB)
|
(+SSB)
|
(+SSB)
|
0.034
|
0.114
|
|||
| P1 [A-17, A-16, A-12, T-8, T-7] |  |
1.0 | 1.0 | 1.0 | 1.0 | + | + |
| [T-17, T-16] | 0.2 | N.D. | N.D. | 0.2 | − | − | |
| [T-17, T-16, A-8, A-7] | 0.6 | N.D. | N.D. | 1.1 | + | + | |
| [T-17, T-16, G-12, A-8, A-7] | 5.2 | 3.2 | 2.3 | 1.6 | + | + | |
| [T-17, T-16, G-12] | N.D. | 0.3 | 0.4 | 0.3 | − | − | |
| 4STEM | 0.7 | 0.9 | N.D. | 1.0 | +/− | + | |
| 3STEM | 0.6 | 0.2 | N.D. | 0.3 | − | − | |
| [T-17, T-16, G-12, A-8, A-7, C-5] | 5.2 | 6.4 | 4.6 | 1.4 | + | + | |
| [G-17, G-12, C-7] | 2.6 | 8.6 | 4.6 | 1.9 | − | + | |
| [G-17, C-16, G-8, C-7] | 0.2 | 0.4 | 0.8 | 1.3 | − | +/− | |
| [G-15, C-9] | 0 | 0 | 0 | 0 | + | + | |
| [A-18, T-17, T-16, G-12, A-8, A-7, T-6] | 1.5 | 1.3 | 2.8 | 1.8 | + | + | |
| [C-14, G-10] | 0.4 | 0 | 0 | 0 | − | − | |
| [A-14, T-10] | 1.0 | 0.5 | N.D. | 0.5 | − | + | |
| [T-14, A-10] | 0.6 | 0 | N.D. | 0 | − | − | |
The sequence of the template strand of each promoter from −20 to −4 is shown. Position −12 corresponding to the center of the hairpin loop is indicated. The location of the inverted repeats is indicated by arrows above the sequence. Base changes with respect to promoter P1 are in boldface type. The amount of transcripts synthesized in vitro from the mutant promoters was quantitated and normalized against the amount of transcript synthesized from wild-type promoter P2, present in the same DNA template. Numbers are relative to the activity of promoter P1 under each condition tested. When present, Eco SSB was added at a Eco SSB/DNA ratio of 1:1 (wt/wt). (N.D.) Not determined. Hairpin extrusion at physiological and high superhelical densities (in the absence of Eco SSB) is indicated in the last two columns.
Table 2.
Transcriptional activity of promoters containing mutations in the loop of the promoter hairpin
| Activity
|
|||||||
|---|---|---|---|---|---|---|---|
| circles (−ς)
|
Extrusion
|
||||||
| Promoter
|
Sequence
|
ssDNA −SSB
|
0.114
|
0.034
|
−ς
|
||
| (−SSB)
|
(+SSB)
|
(+SSB)
|
0.034
|
0.114
|
|||
| P1 |  |
1.0 | 1.0 | 1.0 | 1.0 | + | + |
| [T-11] | 0.2 | 0 | 0 | 0 | − | − | |
| [T-12] | 0.5 | 0.3 | 0.2 | 0.2 | + | + | |
| [C-12] | 0.3 | 0.4 | N.D. | 0.3 | − | + | |
| [G-13, A-11] | 0.1 | 0 | 0 | 0 | − | − | |
| [C-14, G-13, A-11, G-10] | 0 | 0 | N.D. | 0 | − | − | |
| [T-17, T-16, G-12, A-8, A-7] | 5.2 | 3.2 | 2.3 | 1.6 | + | + | |
| [T-17, T-16, G-13, G-12, A-8, A-7] | 4.7 | 0.3 | 0.4 | 0.3 | − | − | |
| [T-17, T-16, T-13, G-12, A-8, A-7] | 5.2 | N.D. | N.D. | 0.4 | − | − | |
| [T-17, T-16, C-13, G-12, A-8, A-7] | 3.8 | 0.3 | 0.2 | 0.4 | − | − | |
For details, see Table 1 footnote.
Hairpin extrusion is required for in vitro N4 vRNAP promoter activity on supercoiled templates
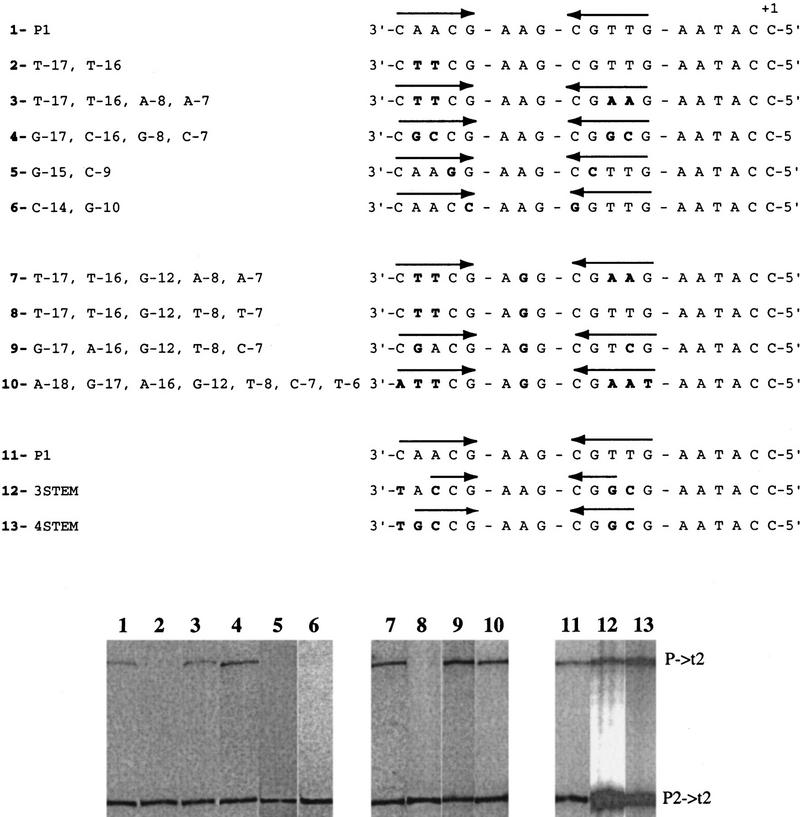
Figure 2 shows the results of transcription assays performed on a number of promoters with base changes in the inverted repeats, on circles of physiological superhelical density in the presence of Eco SSB.
Figure 2.
Electrophoretic analysis of RNAs synthesized from promoters containing mutations at the inverted repeats. In vitro transcription assays were carried out on wild-type or mutant promoter-containing DNAs at physiological superhelical density in the presence of Eco SSB at a 1:1 Eco SSB/DNA ratio (wt/wt) . The sequence from −18 to +1 of the template strand of the promoters is presented above the results of the transcription assays. In each case, sequence changes from promoter P1 are presented in boldface type. The location of the inverted repeats is indicated by arrows above the sequence.
Mutant promoters containing intact inverted repeats with base changes at the nonconserved positions were tested. Promoter [T-17, T-16, A-8, A-7] extruded hairpins as well as the wild-type promoter (Dai et al. 1998) and directed efficient transcription on single-stranded as well as supercoiled, Eco SSB-activated templates (Fig. 2, lane 3; Table 1). Therefore, these positions are not required for N4 vRNAP–promoter recognition. Promoters [G-17, C-16, G-8, C-7] and [G-17, A-16, G-12, T-8, C-7] have hairpin stems with 5- and 4-G:C bp, respectively. Hairpin extrusion at these promoters occurred at high (−0.114), but not at physiological superhelical densities (Dai et al. 1998). Surprisingly, both promoters exhibited higher than wild-type activity in the presence of Eco SSB at physiological superhelical density (Fig. 2, cf. lanes 4 and 1, and lanes 9 and 7; Table 1), suggesting that Eco SSB facilitates formation of the hairpin under these conditions.
We have observed previously that the length of the inverted repeats influences hairpin extrusion (Dai et al. 1998). Promoter 3STEM, with a 3-base stem hairpin, failed to extrude at both low and high superhelical densities, but still showed 30% of wild-type transcriptional activity at physiological superhelical density in the presence of Eco SSB (Fig. 2, lane 12; Table 1). Promoter 4STEM, with a 4-base stem hairpin, extruded inefficiently at low superhelical density but was as active as the wild-type control (Fig. 2, lane 13; Table 1). Both promoters were active when present on single-stranded templates (60%–70% of wild type). These results suggest that a 3- to 4-bp hairpin stem is sufficient to support activity and that the presence of Eco SSB must facilitate the formation of, or stabilize the template hairpin at physiological superhelical density. Promoters with disrupted inverted repeats caused by base changes at the nonconserved positions (−17, −16, −8, and −7) were also examined. These are [T-17, T-16] (Fig. 2, lane 2), a derivative of promoter P1 (Fig. 2, lane 1), and [T-17, T-16, G-12, T-8, T-7] (Fig. 2, lane 8), a derivative of [T-17, T-16, G-12, A-8, A-7] (Fig. 2, lane 7). These promoters displayed reduced activity on single-stranded as well as Eco SSB-activated, supercoiled templates (Table 1; Glucksmann et al. 1992) .
We also tested promoters containing mutations at the conserved positions (−18, −15, −14, −10, −9, and −6) of the inverted repeats. Promoter [A-18, T-17, T-16, G-12, A-8, A-7, T-6] extruded hairpins (Dai et al. 1998) and directed transcription on single-stranded (Table 1) and supercoiled Eco SSB-activated templates (Fig. 2, cf. lanes 10 and 7; Table 1) indicating that these two positions (C-18 and G-6) are not required for N4 vRNAP–promoter recognition. Although hairpin extrusion at promoter [G-15, C-9] occurs normally (Dai et al. 1998), this promoter was inactive on both single-stranded (Table 1) and supercoiled Eco SSB-activated templates (Fig. 2, cf. lanes 5 and 1; Table 1). Therefore, these positions are likely to be involved in direct contacts with the vRNAP.
Conserved positions −14 and −10 constitute the loop-closing base pair of the hairpin. A 3′G:C5′ [G-14, C-10] base pair is present in all three N4 vRNAP promoters (Haynes and Rothman-Denes 1985). On single-stranded templates, a promoter with a 3′A:T5′ [A-14, T-10] loop-closing base pair displayed wild-type activity, whereas promoters [C-14, G-10] and [T-14, A-10] had 40% and 60% of wild-type activity, respectively (Table 1), indicating a preference for 3′ Pu:Py5′ at these positions for N4 vRNAP contacts. We have shown previously that a 3′G:C5′ closing base pair is essential for hairpin extrusion at physiological superhelical density; when a 3′A:T5′ hairpin loop-closing base pair was present at that position, extrusion was observed only at high superhelical densities. No extrusion was detected, even at σ = −0.114 with a 3′ Py:Pu 5′ closing base pair (Dai et al. 1998). Accordingly, a template with a 3′G:C5′ loop-closing base pair gave maximal activity when tested at physiological superhelical density (Table 1). Templates with 3′Py:Pu5′ ([C-14, G-10] or [T-14, A-10]) were inactive at low or high superhelical densities both in the absence or presence of Eco SSB, once again confirming that hairpin extrusion is essential for vRNAP–promoter recognition (Table 1). Promoter [A-14, T-10] was active at high superhelical densities in the absence of Eco SSB (Table 1), in agreement with its ability to extrude a hairpin under these conditions. This promoter showed 50% of wild-type activity at physiological superhelical density in the presence of Eco SSB (Table 1), indicating that Eco SSB must facilitate hairpin extrusion, or stabilize the template-strand hairpin.
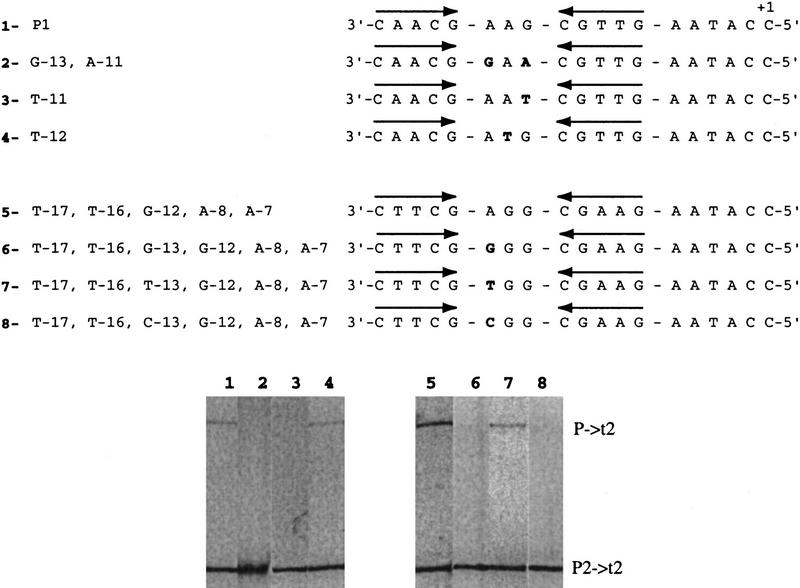
Role of bases at the hairpin loop in promoter activity on supercoiled templates
The N4 vRNA polymerase promoters contain the sequence 3′G[G/A]A5′ separating the inverted repeats. On all templates tested, a marked preference for purines (G > A > Py) at position −12 (center of the loop) was observed (Fig. 2, cf. lanes 3 [T-17, T-16, A-8, A-7] and 7 [T-17, T-16, G-12, A-8, A-7]; Table 1; Fig. 3, cf. lanes 1 [P1] and 4 [T-12]; Table 2, cf. P1, [T-12], and [C-12]). We conclude that the base identity at this position or, alternatively, a more rigid conformation of the hairpin loop generated by a pyrimidine at the center of the loop is responsible for the observed reduction in activity (M. Kloster and L. Rothman-Denes, unpubl.). Although a C-12 hairpin is as stable as a wild-type hairpin (Dai et al. 1997), we were unable to detect hairpin extrusion at promoter [C-12] at physiological superhelical densities (Dai et al. 1998). The ability of Eco SSB to activate this promoter at physiological superhelical density indicates that Eco SSB must facilitate hairpin extrusion.
Figure 3.
Electrophoretic analysis of RNAs synthesized from promoters containing mutations in the loop of the promoter hairpin. In vitro transcription assays were carried out on wild-type or mutant promoter-containing DNAs at physiological superhelical density in the presence of Eco SSB at a 1:1 Eco SSB/DNA ratio (wt/wt) . The sequence from −18 to +1 of the template strand of the promoters is presented above the results of the transcription assays. In each case, sequence changes from promoter P1 ( 2–4) and from promoter [T-17, T-16, G-12, A-8, A-7] (6–8) are presented in boldface type. The location of the inverted repeats is indicated by arrows above the sequence.
Replacing the G at position −11 with a T [T-11] resulted in an inactive promoter on single-stranded as well as supercoiled templates (Table 2). This substitution also abolished extrusion (Dai et al. 1998). Therefore, this position is required for both extrusion and contacting the polymerase.
Surprisingly, although position −13 is conserved in all three N4 vRNAP promoters (A-13), changes to any other base do not reduce promoter activity on single-stranded templates (Table 2). We have shown that an adenosine at this position is critical for hairpin extrusion at all superhelical densities (Dai et al. 1998). Accordingly, promoters that carry other bases at this position (G-13, T-13, and C-13) showed reduced activity on supercoiled templates (Fig. 3, lanes 6–8; Table 2).
Minimal requirements for vRNAP promoter recognition
The studies presented above indicate that specific sequences (3′-G-AXG-C 5′, where X = A, G, or T) and a minimal 4 bp hairpin stem, are required for hairpin extrusion, whereas C-15, G-11, G/A-12, and G-9 are required for N4 vRNAP–promoter recognition. Although downstream of the hairpin, all virion RNAP promoters contain the sequence 3′-AAXAC-5′ from positions −5 to −1 (Haynes and Rothman-Denes 1985), base changes in these sequences did not affect hairpin extrusion (Dai et al. 1998), promoter activity on single-stranded templates (Glucksmann et al. 1992), or Eco SSB-dependent or independent promoter activity on supercoiled templates (not shown). The possible function of these conserved sequences remains to be elucidated. A C at the +1 site, however, is required for transcription initiation as vRNAP initiates transcription solely with GTP (Glucksmann et al. 1992).
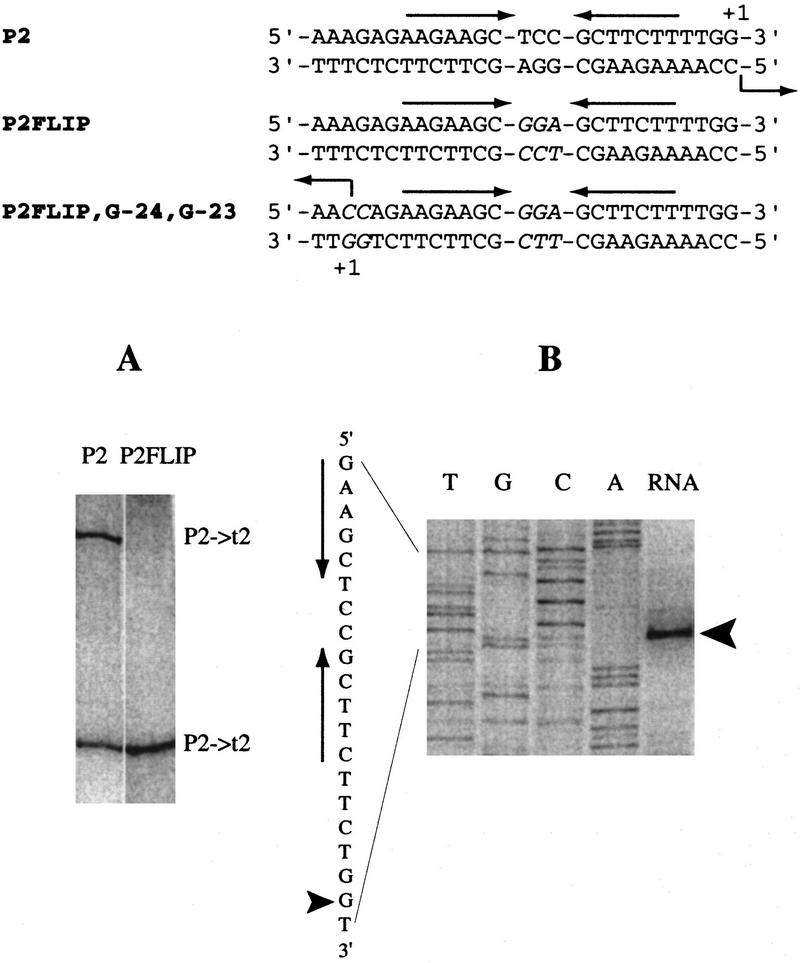
A mutant promoter (P2FLIP) in which the loop sequences of the template and nontemplate strand hairpins were exchanged, is inactive (Fig. 4A), although this mutant promoter extrudes hairpins (Dai et al. 1997). In P2FLIP, the nontemplate strand hairpin contains loop bases that are important for recognition. To test whether these sequences are sufficient for productive transcription, we engineered promoter [P2FLIP, G-24, G-23] that contains mutations to C at positions −23 and −24. Transcription in the opposite direction, using the original nontemplate strand as the template strand and initiating specifically at mutated position −24, was observed (Fig. 4B). These results demonstrate that, once the hairpin extrudes, the only sequences required for specific vRNAP–promoter recognition and transcription are a hairpin with 3′-C Pu-A [A/G] G-Py G-5′ (where Pu:Py) and a C at the +1 site.
Figure 4.
Transcription from the mutant promoters in which the bases at the loop are exchanged between template and nontemplate strands. In vitro transcription assays were carried out on supercoiled templates containing wild-type promoter P2 (cloned in place of P1) or mutant promoters P2FLIP (A) and P2FLIP, G-24, G-23 (B) at a 1:1 Eco SSB/DNA ratio (wt/wt). The sequences of the promoters are presented above the results, and the sequence of promoter P2FLIP, G-24, G-23 is also shown next to the DNA sequencing ladders (B). The location of the inverted repeats is indicated by arrows above or next to the sequence. Base changes are in italics. In A, radioactive RNAs were synthesized and visualized directly on a denaturing gel. In B, RNAs were subjected to primer extension analysis (see Materials and Methods), and the extension products are shown next to the sequencing ladder obtained on the DNA template using the same primer. The arrowheads indicate the major primer extension product and the corresponding site of transcription initiation at the mutant promoter.
Discussion
Site-specific mutagenesis was undertaken to define minimal promoter sequences required for vRNAP activity on both single-stranded and supercoiled templates. The results of these analyses indicate that conserved sequences at the promoters serve two roles: (1) to allow the formation of a DNA hairpin required for N4 vRNAP–promoter recognition; and (2) to provide specific contacts between promoter DNA and N4 vRNAP.
Role of promoter sequences in N4 vRNAP–promoter recognition
Transcription assays performed on single-stranded templates point to the role of specific bases in direct interactions with vRNAP. Results from these assays indicate that very few conserved sequences at the promoter are required for N4 vRNAP–promoter recognition: C-15, G/A-12, G-11, and G-9 (Tables 1 and 2; Fig. 5A). In addition, a purine at position −14 and a pyrimidine at position −10 are preferred. A C at position +1, although not required for vRNAP binding to the promoter, is required for transcription initiation since N4 vRNAP requires a G as the initiating nucleotide (Glucksmann et al. 1992). Transcription from the promoter [P2FLIP, G-24, G-23] in the opposite direction demonstrates that the only sequences required for specific vRNAP transcription are those encompassing the hairpin and the +1 site (Fig. 4). Footprinting experiments on single-stranded DNA templates indicated simultaneous N4 vRNAP occupancy of the hairpin sequences and the +1 site (M.A. Glucksmann-Kuis and L. Rothman-Denes, unpubl.).
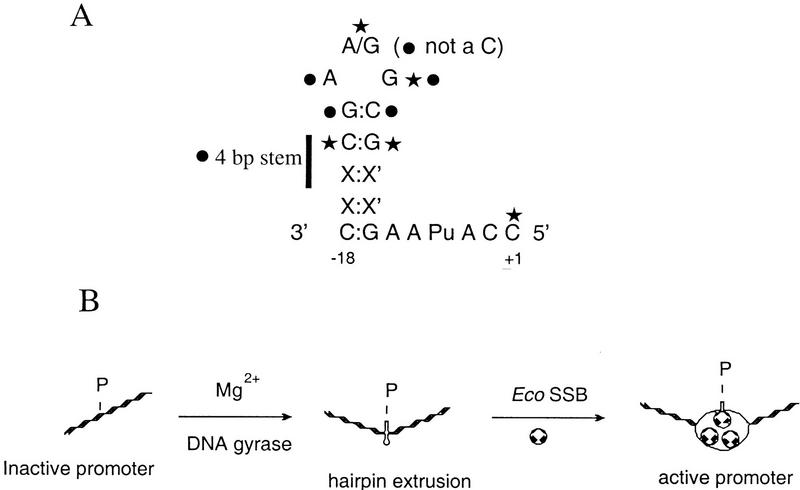
Figure 5.
(A) Summary of the sequence requirements at the N4 early promoters for hairpin extrusion (circle) and/or vRNAP contacts (star). The sequence of the template strand is shown. (B) Model for the N4 vRNAP–promoter utilization pathway. (P) Promoter. Top strand in active promoter indicates the template strand. The stoichiometry of Eco SSB in the activated promoter complex is unknown.
Role of promoter sequences in the supercoil-driven formation of the hairpin required for N4 vRNAP–promoter recognition
Extrusion of the promoter hairpins, which involves breaking the interstrand H-bonds to form intrastrand H-bonds at the hairpin stem, is a prerequisite for N4 vRNAP–promoter recognition on supercoiled templates. Cruciform formation results in the generation of an appropriate structure, the hairpin on the template strand, for N4 vRNAP binding. Results of hairpin extrusion assays indicate that specific sequences (G-14, A-13, A/G/T-12, G-11, and C-10) and at least a 4-bp hairpin stem are required for extrusion at physiological superhelical densities (Dai et al. 1998) (Fig. 5A). A-13 as well as G-11 and the loop-closing base pair (G-14, C-10) are essential for the formation of an unusually stable template-strand hairpin (Hirao et al. 1994; Chou et al. 1996; Dai et al. 1997). This hairpin drives extrusion from supercoiled, double-stranded DNA (Dai et al. 1998). Interestingly, the fact that mutation of A-13 does not affect promoter activity on ssDNA (Table 2) suggests that the unusually stable hairpin structure on the template strand is not essential for vRNAP recognition; it is just essential for supercoil-driven extrusion (M. Kloster and L. Rothman-Denes, unpubl.). Other conserved sequences (G-14, G/A-12, G-11, C-10) are required for both hairpin extrusion and contacting vRNAP (Fig. 5A). Finally, specific sequences at positions −15 (C) and −9 (G) are involved solely in N4 vRNAP contacts.
In general, mutant promoters that do not show detectable hairpin extrusion are transcriptionally inactive or less active than the wild-type promoter when present on supercoiled templates. However, mutations that affect hairpin extrusion still support some activity on single-stranded and on supercoiled templates ([T-17, T-16], [T-17, T-16, G-12], [A-14, T-10], and [G-17, G-12, C-7]; Table). It is possible that mutant promoters with 2- or 3-bp hairpin stems ([T-17, T-16], [T-17, T-16, G-12], and 3STEM) might form very small hairpins on single-stranded DNA. According to Hirao and colleagues, the oligonucleotide d[GC-GAA-GC] forms a stable hairpin structure with a Tm of 76.5°C in 0.1 m NaCl solution (Hirao et al. 1992). On supercoiled templates, the rate of N4 vRNAP binding to a promoter might be faster than the reaction rate of promoter sequences to the structural probes we used to detect hairpin extrusion. Alternatively, N4 vRNAP binding might stabilize the hairpin conformation through a protein-induced conformational switch under circumstances that usually do not allow hairpin formation.
Role of Eco SSB in N4 vRNAP–promoter recognition
Eco SSB is a specific activator of N4 vRNAP on single-stranded and supercoiled DNA templates. Other single-stranded DNA binding proteins cannot substitute for Eco SSB. Results of footprinting experiments on single-stranded templates indicate that this specificity results from Eco SSB’s ability to stabilize the template-strand hairpin, whereas the nontemplate strand hairpin is destabilized; other single-stranded DNA binding proteins destabilize the template-strand hairpin (Glucksmann-Kuis et al. 1996). However, three lines of evidence indicate that Eco SSB plays additional roles in vRNAP promoter activation.
First, promoters that contain inverted repeats but do not extrude hairpins at physiological superhelical densities (4STEM and [A-14, T-10]) are activated by Eco SSB. What is the basis for Eco SSB activation? We suggest that in these cases, although the extruded hairpin conformation is not stable, the presence of Eco SSB stabilizes this conformation by invading through the complementary strand to yield an ‘active promoter’ (see Fig. 5B).
Second, promoters [G-17, C-16, G-8, C-7] and [G-17, G-12, C-7], which contain four or five G:C bp in the hairpin stem, displayed reduced activity on single-stranded templates in the absence of Eco SSB (Table 1); when Eco SSB was present, these promoters were active at wild-type or even higher than wild-type levels (Table 1). Previous results showed that N4 vRNAP binds efficiently to promoter [G-17, C-16, G-8, C-7] (Glucksmann et al. 1992). Affinity-labeling experiments indicated that this mutant promoter does not support the formation of the first phosphodiester bond and is limited in promoter clearance (Glucksmann et al. 1992). Taken together, these results suggest that Eco SSB might facilitate promoter clearance by either interacting directly with vRNAP or by destabilizing the promoter hairpin to disrupt initial contacts.
Finally, the observation that N4 vRNAP is able to initiate transcription from promoters present on highly supercoiled templates in the absence of Eco SSB suggests that the functions of supercoiling and Eco SSB partially overlap. No transcription was detected from a heteroduplex template composed of a wild-type promoter template strand and a nontemplate strand from which the inverted repeat sequences were deleted, that is, one containing the looped-out sequence of the template strand inverted repeats (M.A. Glucksmann, E. Davydova, and L. Rothman-Denes, unpubl.). This result indicates that N4 vRNAP requires a single-stranded DNA region, in addition to specific sequences and a hairpin structure, for binding and transcription. How is a single-stranded region generated at the promoter? We propose that Eco SSB binding on templates of physiological superhelical density creates or stabilizes a large enough single-stranded region at the promoter for N4 vRNAP to initiate transcription. Whereas chemical and nuclease probes did not detect single-stranded bases immediately flanking the promoter hairpin on highly supercoiled circles (Dai et al. 1998), N4 vRNAP may bind to the hairpin and induce a DNA conformational change involving strand opening, which is facilitated at high superhelical densities, resulting in stable and productive vRNAP–promoter association.
A model for vRNAP–promoter recognition
In Figure 5B, we present a revised model for the N4 vRNAP–promoter recognition pathway. In essence, negative supercoiling and Mg(II) facilitate the formation of a cruciform, composed of two hairpins with different loop conformations and a four-way junction, at the N4 early promoters (Dai et al. 1997). The unusual conformation of the template-strand hairpin drives hairpin extrusion and determines its unusual interactions with Eco SSB; the template strand hairpin is preserved by Eco SSB for vRNAP recognition, whereas the nontemplate strand hairpin is disrupted (Glucksmann-Kuis et al. 1996). In addition, Eco SSB binding provides single-strandedness surrounding the hairpin. The template-strand hairpin and specific sequences within the hairpin are the two determinants for vRNAP recognition. These determinants are reminiscent of elements involved in RNA–protein interactions (Nagai 1992). At this point, we do not have any information on how Eco SSB invades the promoter region or on its stoichiometry in the activated promoter. Footprinting experiments on supercoiled templates are underway to study the interactions of Eco SSB with the two promoter strands.
The results presented in this paper indicate that N4 vRNAP is a sequence- as well as a structure-specific DNA-binding protein. Maximum vRNAP activity on its promoters on double-stranded templates is achieved by the optimization of at least three distinct but related processes: (1) formation of the required hairpin; (2) direct contacts of the polymerase with specific sequences in the hairpin and around the +1 site; and (3) efficient melting of the hairpin afterwards. Our results suggest strongly that a delicate balance must be maintained between these three processes through the identity of specific bases at the promoter. The promoter sequences have to yield a hairpin that is stable enough for extrusion to occur, not too stable to allow vRNAP to undergo promoter clearance, and yet still provide the correct interacting surface for the polymerase to make the desired contacts. Moreover, supercoiling and Eco SSB affect at least two of the three processes, adding yet another level of regulation. This work presents the first example of transcriptional regulation through changes in DNA secondary structure. Hairpin extrusion is essential for providing the proper DNA architecture required for assembly of the N4 vRNAP transcription machinery. Specific sequences at the loop of the template-strand hairpin and at the +1 site are required for N4 vRNAP recognition; the formation of a hairpin may help orient these loop bases with respect to +1 and allow interaction of N4 vRNAP with both sequences.
DNA structural transitions increase the repertoire of protein–DNA recognition elements beyond just DNA sequence, and may be a general mechanism used in other transcription systems. Indeed, McMurray and colleagues have found that two precisely arranged cAMP-responsive elements (CREs), present at the cAMP-inducible enhancer of the human proenkephalin gene, are bound by a single CRE-binding protein (Spiro et al. 1993, 1995). Several lines of evidence indicate that, in vitro, both strands of these sequences form stable hairpins (McMurray et al. 1991, 1994; Gacy and McMurray 1994). Therefore, they have proposed that formation of a cruciform structure might play a role in transcriptional regulation of the proenkephalin gene. Moreover, Levens and colleagues have shown that heterogeneous nuclear ribonucleoprotein K (hnRNP K), which binds to a specific, single-stranded sequence upstream of the human c-myc gene in vitro (Tomogawa and Levens 1995) activates transcription in vivo on circular, but not on linear templates, suggesting that hnRNP K recognizes a single-stranded region generated by negative supercoiling in circular plasmids (Tomogawa and Levens 1996).
Materials and methods
Preparation of DNA circles containing wild-type or mutant N4 early promoters
Strategies for site-specific mutagenesis to generate mutant promoters and the subsequent cloning of these promoters into circle-producing plasmids are described elsewhere (Dai et al. 1998). The generation and purification of circles containing the wild-type or mutant promoters and the generation of topoisomers of different superhelical densities were as described previously (Miller et al. 1996).
Purification of N4 virion RNA polymerase
Virion RNA polymerase was purified from CsCl-banded phage as described by Falco et al. (1980), with minor modifications (Miller et al. 1996).
Transcriptional activity of mutant promoters
Standard transcription reaction conditions were used (Haynes and Rothman-Denes 1985), at DNA template excess, with modifications. Reactions contained 10 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm ATP, 1 mm GTP, 1 mm UTP (or CTP), 0.1 mm CTP (or UTP), and 2–10 μCi of [α-32P]CTP (or [α-32P]UTP). RNasin (1 U/μl) was included in most reactions. Runoff transcription reactions contained 5 μg of BamHI-restricted single-stranded M13mp7 DNA, or heat-denatured, BamHI-restricted circle DNAs carrying wild-type or mutant promoters. When supercoiled circles were used as templates, 0.5 μg were used per 100-μl reaction volume, and Eco SSB was added to obtain the desired SSB/DNA ratios. The reactions were terminated by adding EDTA (5 mm final concentration) and tRNA to 100 μg/ml, followed by phenol extraction. The samples were ethanol-precipitated, resuspended in loading buffer [80% formamide, 50 mm Tris-HCl (pH 8.0), 20 mm EDTA, and 0.5% each of bromophenol blue and xylene cyanol], and run on 8% polyacrylamide/7 m urea gels. Gels were dried, exposed to X-ray film, and the transcripts quantitated either on a Molecular Dynamics PhosphorImager or by densitometer tracing of the autoradiograms to determine the relative activity of the mutant promoters.
RNA primer extension analysis
Standard in vitro transcription reactions were carried out in a volume of 300 μl in the presence of unlabeled rNTPs. The reactions were phenol-extracted, the DNA was ethanol-precipitated and, after a 70% ethanol wash, the pellets were resuspended in 75 μl of 10 mm Tris-HCl (pH 8.0), 0.1 mm EDTA. An equal volume of 10 mm Tris-HCl (pH 8.0), 20 mm MgCl2, 1 mm CaCl2 was then added, followed by addition of 12 units of RNase-free DNase I and 60 units of RNasin. The mixtures were incubated at 30°C for 30 min. After the addition of 6 μl of 0.5 m EDTA, samples were boiled for 3 min, phenol extracted, and ethanol precipitated in the presence of 0.3 m of sodium acetate (pH 5.0) and 0.2% SDS. Oligonucleotides used were: d[CATGCAGGTCGACTCTAGAGGATCCGTC], which hybridizes to the bottom strand (the template strand in the wild-type promoter) and d[GGCATGCAAGCTTTGTATAAAAAAGATGATACC], which hybridizes to the top strand (the nontemplate strand in the wild-type promoter). The primer labeling, hybridization to RNA, and extension reactions were carried out as described (Ausubel et al. 1994). The final extension products were resuspended in 4 μl of 10 mm Tris-HCl (pH 8.0), 1 mm EDTA (TE). Formamide (95%)/dye mix was then added (4 μl), and the samples were boiled for 3 min and loaded onto 8% polyacrylamide/7 m urea gels in TBE buffer. Primers used to synthesize RNA were used to generate the corresponding DNA sequence. Gels were dried and exposed to X-ray film.
Acknowledgments
We thank Jim Miller for excellent technical assistance and the members of the laboratory for comments on the manuscript. This work was supported by National Institutes of Health grant RO1 AI12575 to L.B.R.-D.; X.D. was partially supported by U.S. Public Health Service grant GM08369.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lbrd@midway.uchicago.edu; FAX (773) 702-3172.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Preparation and analysis of RNA. In: Janssen K, editor. Current protocols in molecular biology. New York, NY: John Wiley and Sons; 1994. pp. 4.8.1–4.8.5. [Google Scholar]
- Chou S-H, Zhu L, Reid BR. On the relative ability of centromeric GNA triplets to form hairpins vs self-paired duplexes. J Mol Biol. 1996;259:445–457. doi: 10.1006/jmbi.1996.0331. [DOI] [PubMed] [Google Scholar]
- Dai, X., M. Kloster, and L.B. Rothman-Denes. 1998. Sequence-dependent extrusion of a small DNA hairpin at the N4 virion RNA polymerase promoters. J. Mol. Biol. (in press). [DOI] [PubMed]
- Dai X, Greizerstein M, Nadas-Chinni K, Rothman-Denes L B. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc Natl Acad Sci. 1997;94:2174–2179. doi: 10.1073/pnas.94.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco SC, VanderLaan K, Rothman-Denes LB. Virion-associated RNA polymerase required for bacteriophage N4 development. Proc Nat Acad Sci. 1977;74:520–523. doi: 10.1073/pnas.74.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco SC, Zivin R, Rothman-Denes LB. Novel template requirements of N4 virion RNA polymerase. Proc Natl Acad Sci. 1978;75:3220–3224. doi: 10.1073/pnas.75.7.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco SC, Zehring W, Rothman-Denes LB. DNA-dependent RNA polymerase from bacteriophage N4 virions. Purification and characterization. J Biol Chem. 1980;255:4339–4347. [PubMed] [Google Scholar]
- Gacy AM, McMurray CT. Hairpin formation within the human enkephalin enhancer region. I. Kinetic analysis. Biochemistry. 1994;33:11951–11959. doi: 10.1021/bi00205a034. [DOI] [PubMed] [Google Scholar]
- Glucksmann MA, Markiewicz P, Malone C, Rothman-Denes LB. Specific sequences and a hairpin structure in the template strand are required for N4 virion RNA polymerase promoter recognition. Cell. 1992;70:491–500. doi: 10.1016/0092-8674(92)90173-a. [DOI] [PubMed] [Google Scholar]
- Glucksmann-Kuis MA, Dai X, Markiewicz P, Rothman-Denes LB. E. coli SSB activation of N4 virion RNA polymerase: Specific activation of an essential DNA hairpin required for promoter recognition. Cell. 1996;84:147–154. doi: 10.1016/s0092-8674(00)81001-6. [DOI] [PubMed] [Google Scholar]
- Haynes LL, Rothman-Denes LB. N4 virion RNA polymerase sites of transcription initiation. Cell. 1985;41:597–605. doi: 10.1016/s0092-8674(85)80032-5. [DOI] [PubMed] [Google Scholar]
- Hirao I, Nishimura Y, Tagawa Y-I, Watanabe K, Miura K. Extraordinarily stable mini-hairpins: electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 1992;20:3891–3896. doi: 10.1093/nar/20.15.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao I, Kawai G, Yoshizawa S, Nishimura Y, Ishido Y, Watanabe K, Miura K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: An extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994;22:576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz P, Malone C, Chase JW, Rothman-Denes LB. E. coli single-stranded DNA binding protein is a supercoiled-template dependent transcriptional activator of N4 virion RNA polymerase. Genes & Dev. 1992;6:2010–2019. doi: 10.1101/gad.6.10.2010. [DOI] [PubMed] [Google Scholar]
- McMurray CT, Wilson WD, Douglass JO. Hairpin formation within the enhancer region of the human enkephalin gene. Proc Natl Acad Sci. 1991;88:666–670. doi: 10.1073/pnas.88.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT, Juranic N, Chandrasekaran S, Macura S, Li Y, Jones RL, Wilson WD. Hairpin formation within the human enkephalin enhancer region. 2. Structural studies. Biochemistry. 1994;33:11960–11970. doi: 10.1021/bi00205a035. [DOI] [PubMed] [Google Scholar]
- Miller A, Dai X, Choi M-Y, Glucksmann-Kuis A, Rothman-Denes LB. Single-stranded DNA binding proteins as transcriptional activators. Methods Enzymol. 1996;174:9–20. doi: 10.1016/s0076-6879(96)74004-1. [DOI] [PubMed] [Google Scholar]
- Nagai K. RNA-protein interactions. Curr Opin Struct Biol. 1992;2:131–137. [Google Scholar]
- Spiro C, Richards JP, Chandrasekaran S, Brennan RG, McMurray CT. Secondary structure creates mismatched base pairs required for high affinity binding of cAMP response-binding protein to the human enkephalin enhancer. Proc Natl Acad Sci. 1993;90:4606–4610. doi: 10.1073/pnas.90.10.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro C, Bazett-Jones DP, Wu X, McMurray CT. DNA structure determines protein binding and transcriptional efficiency of the proenkephalin cAMP-responsive enhancer. J Biol Chem. 1995;270:27702–27710. doi: 10.1074/jbc.270.46.27702. [DOI] [PubMed] [Google Scholar]
- Tomogawa T, Levens D. Heterogeneous nuclear ribonucleprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- ————— Activating transcription from single-stranded DNA. Proc Natl Acad Sci. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]