Abstract
The RING-H2 finger protein Rbx1 is a subunit of the related SCF (Skp1–Cdc53/Cul1–F-box protein) and von Hippel-Lindau (VHL) tumor suppressor (elongin BC–Cul2–VHL) E3 ubiquitin ligase complexes, where it functions as a component of Cdc53/Rbx1 and Cul2/Rbx1 modules that activate ubiquitination of target proteins by the E2 ubiquitin-conjugating enzymes Cdc34 and Ubc5. Here we demonstrate that the Cdc53/Rbx1 and Cul2/Rbx1 modules also activate conjugation of the ubiquitin-like protein Rub1 to Cdc53 and Cul2 by the dedicated E2 Rub1 conjugating enzyme Ubc12. Our findings identify Rbx1 as a common component of enzyme systems responsible for ubiquitin and Rub1 modification of target proteins.
Keywords: Rbx1, ROC1, Hrt1, SCF, vonHippel-Lindau, ubiquitin ligase, cullin, Rub1, NEDD8
The RING-H2 finger protein Rbx1 (also known as ROC1 and Hrt1) (Kamura et al. 1999; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999; Tan et al. 1999) is a subunit of the multiprotein SCF (Skp1–Cdc53/Cul1–F-box protein) (Bai et al. 1996; Feldman et al. 1997; Skowyra et al. 1997; Lyapina et al. 1998; Patton et al. 1998a,b) and von Hippel-Lindau (VHL) tumor suppressor (elongin BC–Cul2–VHL) (Lisztwan et al. 1999) ubiquitin ligase complexes (Iwai et al. 1999). Studies on the mechanism of action of Rbx1 suggest that it functions as a component of Cdc53(Cul1)/Rbx1 and Cul2/Rbx1 modules that recruit and activate the E2 ubiquitin-conjugating enzymes Cdc34 and Ubc5 to ubiquitinate target proteins (Seol et al. 1999; Skowyra et al. 1999).
Cullin proteins, including the Cdc53(Cul1) and Cul2 subunits of SCF and VHL ubiquitin ligases, are targets for modification by the ubiquitin-like protein Rub1 (also known as NEDD8) (Hochstrasser 1998; Lammer et al. 1998; Liakopoulos et al. 1998, 1999; Wada et al. 1999). Rub1 is conjugated to a single site in the carboxyl termini of cullins by the concerted action of the dedicated Rub1 E1-activating and E2-conjugating enzymes Uba3/Ula1 and Ubc12 (Hochstrasser 1995; Lammer et al. 1998; Liakopoulos et al. 1998; Gong and Yeh 1999). Although the role of Rub1 in regulation of SCF and VHL ubiquitin ligase activities is not yet clear, a variety of evidence suggests that Rub1 modification of Cdc53 and Cul2 may be needed for optimal assembly or function of the E3 complex SCFCdc4 in yeast (Lammer et al. 1998), for regulation of auxin signalling in plants (del Pozo et al. 1998), and for VHL tumor suppressor activity (Liakopoulos et al. 1999).
In the course of experiments investigating the role of Rbx1 in SCF and VHL ubiquitin ligase activity, we discovered that Rbx1 is needed not only for efficient ubiquitination of multiple substrates by the E2 ubiquitin-conjugating enzymes Cdc34 and Ubc5, but also for Rub1 modification of the cullins Cdc53 and Cul2 by the dedicated E2 Rub1-conjugating enzyme Ubc12. Below we present these findings, which bring to light an expanded role for Rbx1 in cellular regulation.
Results and Discussion
Conjugation of Rub1 to Cdc53 depends on Rbx1
In experiments investigating functional interactions between Rbx1 and Cdc53, we noticed that expression of Cdc53 in Sf21 insect cells resulted in appearance in immunoblots of an ∼90 kD Cdc53 species, whereas coexpression of Cdc53 and mammalian Rbx1 (mRbx1) resulted in appearance of an additional, higher molecular mass species resembling Rub1-modified Cdc53 (Lammer et al. 1998; Liakopoulos et al. 1998) (Fig. 1A). The ubiquitin-like Rub1 proteins and the enzymes responsible for their conjugation to target proteins are very highly conserved across species. Notably, human Rub1 is conjugated to Saccharomyces cerevisiae Cdc53 in yeast and will satisfy the requirement for functional Rub1 in a cdc34-1 mutant yeast strain (Liakopoulos et al. 1999). Taken together, these observations raised the possibility that Rbx1 might promote conjugation of endogenous insect cell Rub1 to Cdc53 when Rbx1 and Cdc53 are coexpressed in insect cells. We therefore investigated the possibility that Rbx1 might be required for efficient conjugation of Rub1 to cullins.
Figure 1.
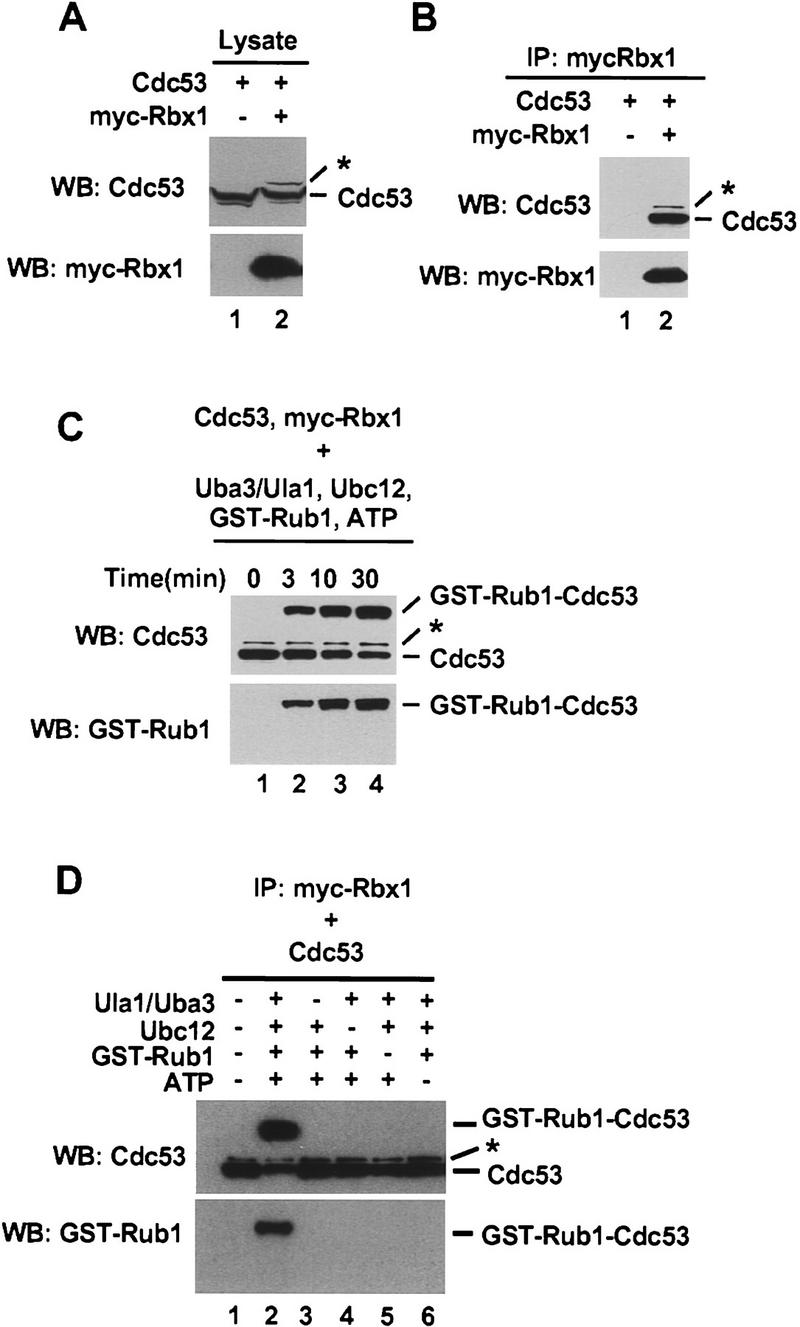
Conjugation of Rub1 to Cdc53 in vitro. (A) Total cell lysate (20 μg) from Sf21 cells infected with the indicated baculoviruses was immunoblotted with anti-Cdc53(yC-17) or anti-myc antibodies. (B) Total cell lysate (200 μg) from baculovirus-infected Sf21 cells was immunoprecipitated with 2 μg of anti-myc antibody and then immunoblotted with anti-Cdc53(yC-17) or anti-myc antibodies. (C) Total cell lysate (800 μg) from baculovirus-infected Sf21 cells was immunoprecipitated with 8 μg of anti-myc antibody. After immunoprecipitation, protein A Sepharose beads were divided into four tubes and and assayed for Rub1 conjugation activity at 25°C for the indicated times. Reaction products were immunoblotted with anti-Cdc53(yC-17) or anti-GST antibodies. (D) Total baculovirus-infected total cell lysate (1200 μg) was immunoprecipitated with 12 μg of anti-myc antibody. After immunoprecipitation, protein A Sepharose beads were divided into six tubes and assayed for Rub1 conjugation activity incubated at 25°C for 30 min in the presence or absence of Uba3/Ula1, Ubc12, GST–Rub1, and ATP as indicated and immunoblotted with anti–Cdc53(yC-17) or anti-GST antibodies. The asterisks (*) indicate a higher molecular mass Cdc53 species that is most likely conjugated to endogenous Rub1.
To accomplish this, we assembled an in vitro Rub1 modification system composed of purified recombinant S. cerevisiae Rub1-activating enzyme Uba3/Ula1 and S. cerevisiae Rub1-conjugating enzyme Ubc12. Myc–mRbx1 and Cdc53 were coexpressed in Sf21 cells. Myc–mRbx1/Cdc53 complexes were purified by immunoprecipitation with anti-myc antibodies (Fig. 1B) and added to reaction mixtures containing Uba3/Ula1, Ubc12, GST–Rub1, and ATP. The products of reactions were analyzed for the presence of GST–Rub1–Cdc53 conjugates by immunoblotting with anti-Cdc53 or anti-GST antibodies. As shown in Figure 1C, we observed time-dependent conversion of the input Cdc53 into a more slowly migrating species. This species was recognized by both anti-Cdc53 (top) and anti-GST antibodies (bottom), indicating that it corresponds to Cdc53 conjugated to GST–Rub1. Formation of GST–Rub1–Cdc53 conjugates was strongly dependent on Uba3/Ula1, Ubc12, and ATP (Fig. 1D).
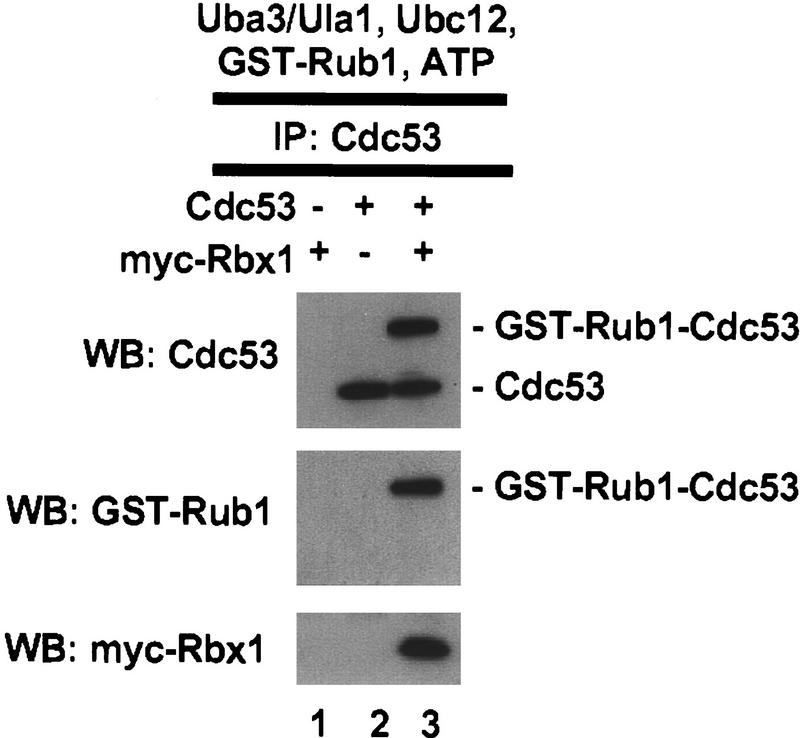
To determine whether formation of the GST–Rub1–Cdc53 conjugate depends on Rbx1, Cdc53 was expressed in Sf21 cells with or without coexpressed myc–mRbx1. Cdc53 or Cdc53/myc–mRbx1 complexes were purified by immunoprecipitation with anti-Cdc53 antibodies and added to reaction mixtures containing Uba3/Ula1, Ubc12, GST–Rub1, and ATP. As shown in Figure 2, formation of GST–Rub1–Cdc53 conjugates was strongly dependent on Rbx1; no detectable GST–Rub1–Cdc53 conjugates were formed in the absence of Rbx1, whereas ∼50% of the input Cdc53 was converted to GST–Rub1–Cdc53 conjugates in the presence of Rbx1 during the course of the reaction.
Figure 2.
Conjugation of Rub1 to Cdc53 in vitro depends on Rbx1. Total cell lyaste (200 μg) from Sf21 cells infected with the indicated baculoviruses was immunoprecipitated with 2 μg of anti-Cdc53(yN-18) antibody. Immunoprecipitates were assayed for Rub1 conjugation activity at 25°C for 30 min, and reaction products were analyzed by immunoblotting with anti-Cdc53(yC-17), anti-GST, or anti-myc antibodies.
Rbx1 activities depend most strongly on predicted zinc binding residues in the conserved RING-H2 domain
The RBX1 gene is highly conserved from yeast to mammals (Fig. 3A) and is essential for viability of yeast. Rbx1 deletion strains can be rescued by expression of either yeast or mammalian Rbx1. To investigate the relationship between mRbx1 activities in Rub1 modification of Cdc53 and in SCF-dependent ubiquitination, we constructed a series of mRbx1 mutants containing point mutations of conserved cysteine and/or histidine RING-H2 finger residues (Fig. 3A). Each mRbx1 mutant was then assayed for its ability (1) to rescue the rbx1 deletion strain, (2) to activate Rub1 modification of Cdc53, and (3) to support ubiquitination of the G1 cyclin Cln2 by the SCFGrr1 ubiquitin ligase complex.
Figure 3.
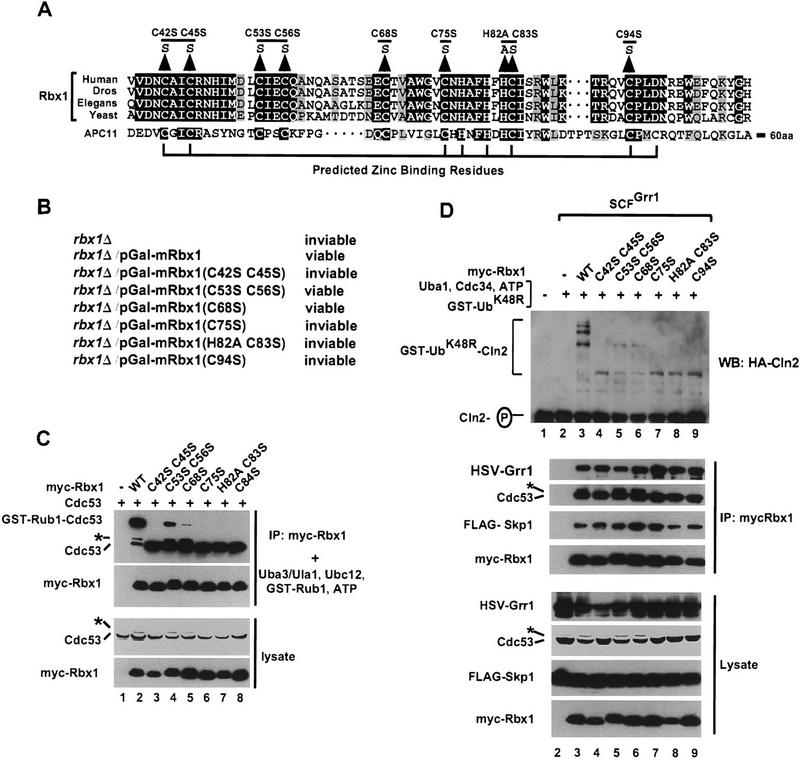
Mutation of predicted zinc-binding residues of Rbx1 is lethal in S. cerevisiae and interferes with Rbx1 activity in Rub1 conjugation and in ubiqutination of Cln2 by SCFGrr1. (A) Alignment of Rbx1 from human, Drosophila melanogaster (Dros), C. elegans (Elegans), or S. cerevisiae (Yeast) with APC11 from S. cerevisiae. (B) Effects of Rbx1 mutations on S. cerevisiae viability. (C) Cell lysate (200 μg) from Sf21 cells infected with the indicated baculoviruses was immunoprecipitated with 2 μg of anti-myc antibodies. The immunoprecipitates were assayed at 25°C for 50 min for Rub1 conjugation activity and reaction products were immunoblotted with anti-Cdc53(yC-17) or anti-myc antibodies (top panels). Cell lysate protein (20 μg) from the same cells was immunoblotted with anti-Cdc53(yC-17) or anti-GST antibodies (bottom panels). (D) Total cell lyaste (400 μg) from Sf21 cells infected with the indicated baculoviruses was immunoprecipitated with 4 μg of anti-myc antibody. One-half of the immunoprecipitate was immunoblotted with anti-HSV, anti-Cdc53(yC-17), anti-FLAG, or anti-myc antibodies (middle panels). The remaining immunoprecipitate was assayed for SCFGrr1 activity at 25°C for 60 min as described in Materials and Methods. Reaction products were immunoblotted with anti-HA antibody (top panel). The same cell lysates (20 μg) were immunoblotted with anti-HSV, anti–Cdc53(yC-17), anti-FLAG, or anti-myc antibodies (bottom panels).
The rbx1 deletion strain was rescued by expression of wild type mRbx1 and mRbx1 mutants mRbx1(C53S C56S) or mRbx1(C68S), but not by expression of mRbx1 mutants mRbx1(C42S C45S), mRbx1(C75S), mRbx1(H82A C83A), or mRbx1(C94S). Notably, all mRbx1 mutants that failed to rescue the rbx1 deletion strain contained mutations of cysteine or histidine residues that are predicted to be directly involved in zinc binding (Seol et al. 1999) and that are conserved between Rbx1 and the RING-H2 finger proteins Hrd1, Rad18, and Ubr1 (Skowyra et al. 1999), which interact with E2 ubiquitin-conjugating enzymes and function in ubiquitination of 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase, in DNA repair, and in amino-end rule protein degradation, respectively. In contrast, the two mRbx1 mutants that rescued the rbx1 deletion strain contained mutations of cysteine residues that are not predicted to be involved in zinc binding (Seol et al. 1999) and that are not conserved in Hrd1, Rad18, and Ubr1 (Skowyra et al. 1999).
To compare the activities of wild-type mRbx1 and Rbx1 mutants in Rub1 modification of Cdc53 in vitro, wild-type myc–mRbx1 and myc–mRbx1 mutants were coexpressed in Sf21 cells with Cdc53. Myc–mRbx1/Cdc53 complexes were purified by immunoprecipitation with anti-myc antibodies and added to reaction mixtures containing Uba3/Ula1, Ubc12, GST-Rub1, and ATP. As shown in Figure 3C, the abilities of mRbx1 RING-H2 finger mutants to support Rub1 modification of Cdc53 and ubiquitination of Cln2 by the SCFGrr1 ubiquitin ligase complex correlated with their abilities to rescue the rbx1 deletion strain. mRbx1 mutants mRbx1(C53S C56S) and mRbx1(C68S), which were the only mRbx1 mutants that rescued the rbx1 deletion strain, were also the only mutants capable of promoting formation of detectable GST–Rub1–Cdc53 conjugates in vitro, although they supported formation of significantly reduced levels of GST–Rub1–Cdc53 conjugates than wild type mRbx1. Interestingly, these two mRbx1 mutants were the only mutants that supported formation in Sf21 cells of the presumptive Rub1–Cdc53 conjugate indicated by the asterisks in Figure 3, C and D. mRbx1 mutants mRbx1(C42S C45S), mRbx1(C75S), mRbx1(H82A C83A), and mRbx1(C94S), which could not rescue the rbx1 deletion strain, did not support detectable Rub1 modification of Cdc53. As shown in Figure 3C, wild-type mRbx1 and mRbx1 mutants were expressed to similar levels in Sf21 cells and immunoprecipitated similar amounts of Cdc53, indicating that they were all capable of interacting with Cdc53 under the conditions of our assays.
To compare the activities of wild-type mRbx1 and mRbx1 mutants in ubiquitination of Cln2 by the SCFGrr1 ubiquitin ligase complex, wild-type myc–mRbx1 and myc–mRbx1 mutants were coexpressed in Sf21 cells with Cdc53, Skp1, and Grr1. SCFGrr1 complexes were purified by immunoprecipitation with anti-myc antibodies and added to reaction mixtures containing purified, recombinant S. cerevisiae E1 ubiquitin activating enzyme Uba1, S. cerevisiae E2 ubiquitin-conjugating enzyme Cdc34, GST–ubiquitinK48R (GST–UbK48R), ATP, and phosphorylated Cln2. As shown in Figure 3D (top), the relative activities of mRbx1 mutants in ubiquitination of Cln2 paralleled their activities in Rub1 modification of Cdc53. mRbx1 mutants mRbx1(C53S C56S) and mRbx1(C68S), which were the only mRbx1 mutants that rescued the rbx1 deletion strain and supported formation of detectable GST–Rub1–Cdc53 conjugates, were most active in ubiquitination of Cln2, although they were much less active than wild-type mRbx1. mRbx1 mutants mRbx1(C42S C45S), mRbx1(C75S), mRbx1(H82A C83A), and mRbx1(C94S), which neither rescued the rbx1 deletion strain nor supported detectable Rub1 modification of Cdc53, were less active than mRbx1 mutants mRbx1(C53S C68S) and mRbx1(C68S) in ubiquitination of Cln2. As shown in Figure 3D (middle and bottom), wild-type mRbx1 and mRbx1 mutants were expressed to similar levels in Sf21 cells and immunoprecipitated similar amounts of SCF subunits, indicating that they were all capable of interacting with the SCFGrr1 complex under the conditions of our assays. These observations are consistent with the findings of Ohta et al. (1999), who reported that Rbx1(C53A, 56A) and Rbx1(C75A, H77A) exhibit substantially reduced ability to support formation of polyubiquitin in vitro.
Conjugation of Rub1 to Cul2 depends on Rbx1
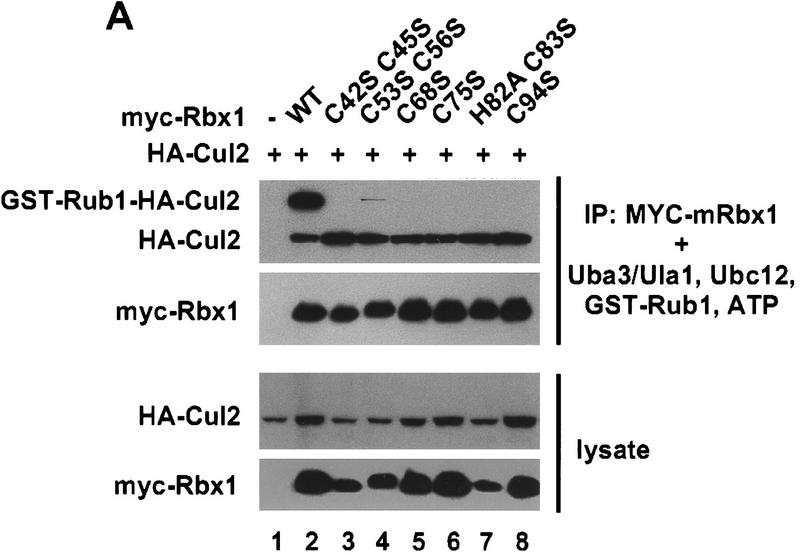
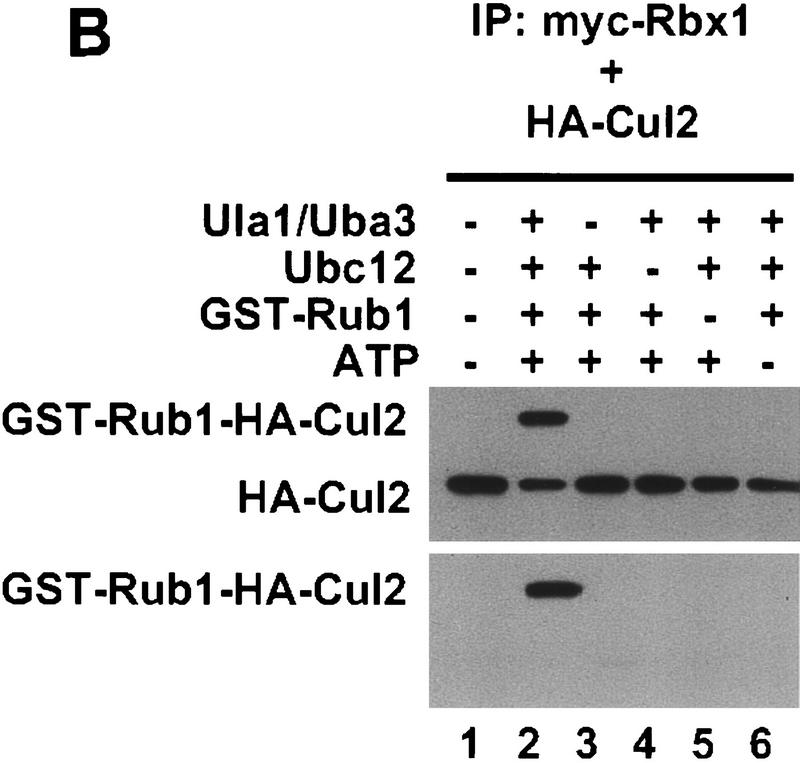
To determine whether Rub1 modification of Cul2 depends on Rbx1, wild-type myc–mRbx1 and myc–mRbx1 mutants were coexpressed in Sf21 cells with Cul2. Myc–mRbx1/Cul2 complexes were purified by immunoprecipitation with anti-myc antibodies and added to reaction mixtures containing Uba3/Ula1, Ubc12, GST–Rub1, and ATP. As shown in Figure 4A (lane 2), >70% of the input Cul2 was converted to GST–Rub1–Cul2 conjugates in the presence of wild type Rbx1. Formation of GST–Rub1–Cul2 conjugates was strongly dependent on Uba3/Ula1, Ubc12, and ATP (Fig. 4B). In addition, formation of GST–Rub1–Cul2 conjugates was strongly dependent on mRbx1, as the efficiency of Rub1 conjugation was substantially reduced with mRbx1(C53S C56S) and undetectable with the other mutants, even though wild-type Rbx1 and Rbx1 mutants were expressed to similar levels in Sf21 cells and immunoprecipitated similar amounts of Cul2 (Fig. 4A).
Figure 4.
Conjugation of Rub1 to Cul2 depends on Rbx1. (A) Cell lysate (20 μg) from the indicated baculovirus-infected Sf21 cells was immunoblotted with anti-HA or anti-myc antibodies. Total cell lyaste protein (200 μg) from baculovirus-infected Sf21 cells was immunoprecipitated with 2 μg of anti-myc antibody. The beads were incubated with 30 ng of Ula1, 20 ng of Uba3, 100 ng of Ubc12, 1 μg of GST–Rub1, and 1.5 mm ATP at 25°C for 50 min and immunoblotted with ant-HA or anti-GST antibodies. (B) After immunoprecipitation with 12 μg of anti-myc antibody from 1.2 mg of baculovirus infected Sf21 cell lysate, the beads were divided into six tubes and incubated at 25°C for 30 min in the presence or absence of Uba3/Ubc12, GST–Rub1, and ATP as indicated and immunoblotted with anti-HA or anti-GST antibodies.
Rbx1, an activator of ubiquitination by SCF and VHL ubiquitin ligases and Rub1 modification of cullins
In summary, in this report we present evidence that the evolutionarily conserved RING-H2 finger protein Rbx1 activates Rub1 modification of the cullins Cdc53 and Cul2 by the dedicated E1 Rub1-activating and E2 Rub1-conjugating enzymes Uba3/Ula1 and Ubc12. Rbx1 (also known as ROC1 and Hrt1) was initially identified as a subunit of the multiprotein SCF (Kamura et al. 1999; Ohta et al. 1999; Seol et al. 1999; Skowyra et al. 1999; Tan et al. 1999) and VHL tumor suppressor (elongin BC–Cul2–VHL) ubiquitin ligase complexes (Kamura et al. 1999), where it functions as a component of Cdc53(Cul1)/Rbx1 and Cul2/Rbx1 modules that recruit and activate the E2 ubiquitin conjugating enzymes Cdc34 and Ubc5 to ubiquitinate target proteins (Iwai et al. 1999; Seol et al. 1999; Skowyra et al. 1999).
The mechanism by which Rbx1 activates Rub1 modification of cullins remains unclear. Notably, we observed that Rbx1 RING-H2 finger mutations have similar effects on Cln2 ubiquitination by the SCFGrr1 complex and Rub1 conjugation of Cdc53 and Cul2, consistent with the model that Rbx1, as a component of Cdc53/Rbx1 modules or Cul2/Rbx1 modules, recruits and activates not only the E2 ubiquitin-conjugating enzymes Cdc34 and Ubc5, but also the E2 Rub1-conjugating enzyme Ubc12. Alternatively, the interaction of Rbx1 with Cdc53(Cul1) or Cul2 may induce a conformational change that allows access of the charged E2 Ubc12 to the site of Rub1 modification in the cullin. Regardless of the exact mechanism of Rbx1 action in these processes, our findings bring to light an expanded role for Rbx1 in regulation of SCF and VHL ubiquitin ligases and identify Rbx1 as a common component of enzyme systems responsible for ubiquitin and Rub1 modification of target proteins.
Materials and methods
Antibodies
Anti-HSV antibody was from Novagen. Anti–Cdc53 (yN-18 and yC-17) antibodies were from Santa Cruz Biotechnology, Inc. Anti-Flag antibody was from Sigma. Anti-HA (12CA5) and anti-Myc (9E10) antibodies were from Boehringer Mannheim. Anti-GST (4C10) antibody was from Covance.
Expression of recombinant proteins in Escherichia coli
S. cerevisiae Uba1 was subcloned into pET23 B (Novagen) with an amino-terminal Myc tag and a carboxy-terminal 6-histidine tag. S. cerevisiae Cdc34 was subcloned into pRSET B (Invitrogen) with an amino-terminal 6-histidine tag. S. cerevisiae Ubc12 was subcloned into pRSET B with an amino-terminal 6-histidine tag and a carboxy-terminal Flag tag. Mammalian ubiquitin (K48R) and S. cerevisiae Rub1 lacking the carboxy-terminal asparagine residue were subcloned into pGEX4T-2 (Pharmacia). Proteins were expressed in E. coli BL21 (DE3) and purified by Ni2+-agarose or glutathione-Sepharose affinity chromatography. After dialysis against 40 mm HEPES-NaOH (pH 7.9), 60 mm potassium acetate, 2 mm DTT, 1 mm MgCl2, 0.5 mm EDTA (pH 7.9), and 10% (vol/vol) glycerol, proteins were stored at −80°C.
Expression of recombinant proteins in Sf21 insect cells
Mouse wild-type Rbx1 and Rbx1 mutants containing amino-terminal 6-histidine and myc tags, S. cerevisiae Grr1 containing amino-terminal 6-histidine and HSV tags, S. cerevisiae Cln2 containing amino-terminal 6-histidine and HA tags, S. cerevisiae Cdc28 containing amino-terminal 6-histidine and Myc tags, S. cerevisiae Cks1 containing amino-terminal 6-histidine and T7 tags, S. cerevisiae Ula1 containing amino-terminal 6-histidine and myc tags, and S. cerevisiae Uba3 containing amino-terminal 6-histidine and Flag tags were subcloned into pBacPAK8, and recombinant baculoviruses were generated with the BacPAK baculovirus expression system (Clontech). The baculovirus vectors encoding S. cerevisiae Cdc53 (Willems et al. 1996), S. cerevisiae Skp1 (Skowyra et al. 1997), human Cul2 (Kamura et al. 1999), and mouse Rbx1 (mRbx1) (Kamura et al. 1999) have been described. Sf21 cells were cultured in Sf-900 II SFM with 5% fetal calf serum at 27°C and infected with the indicated recombinant baculoviruses. Sixty hours after infection, cells were collected and lysed as described previously (Kamura et al. 1998).
In vitro Rub1 conjugation assay
Ula1 and Uba3 were coexpressed in Sf21cells and purified by Ni2+-agarose affinity chromatography. Ubc12 and GST–Rub1 were expressed in E. coli BL21 (DE3) and purified by Ni2+-agarose and glutathione-Sepharose affinity chromatography, respectively. Sf21 cells (1 × 106) infected with the indicated baculoviruses were lysed with 1 ml of ice-cold buffer containing 40 mm HEPES-NaOH (pH 7.9), 450 mm NaCl, 1 mm DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 5 μg/ml leupeptin, 5 μg/ml of antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin. After centrifugation at 10,000g for 20 min at 4°C, the supernatants were immunoprecipitated with 2 μg of anti-myc or anti-Cdc53(yN-18) antibody and 10 μl of protein A–Sepharose beads. The beads were then mixed with 30 ng of Ula1, 20 ng of Uba3, 100 ng Ubc12, and 1 μg of GST-Rub1 in a 15-μl reaction containing 40 mm HEPES-NaOH (pH 7.9), 60 mm postassium acetate, 2 mm DTT, 5 mm MgCl2, 0.5 mm EDTA (pH 7.9), 10% (vol/vol) glycerol, and 1.5 mm ATP. Reaction mixtures were incubated at 25°C for the times indicated in the figure legends.
In vitro Cln2 ubiqitination assay
Phosphorylated Cln2 was prepared by coinfecting Sf21 cells with baculoviruses encoding Cln2, Cdc28, and Cks1. After lysis of cells, Cln2/Cdc28/Cks1 complexes were purified by Ni2+-agarose affinity chromatography and dialysed against 40 mm HEPEs-NaOH (pH 7.9), 60 mm potassium acetate, 2 mm DTT, 5 mm MgCl2, 0.5 mm EDTA (pH 7.9), and 10% (vol/vol) glycerol. Cln2 was phosphorylated by incubating 50 μg of Cln2/Cdc28/Cks1 complexes in dialysis buffer containing 1.5 mm ATP at 25°C for 1 hr.
Uba1, Cdc34, and GST–ubiquitin (K48R) were expressed in E. coli BL21(DE3) cells and purified by Ni2+-agarose or glutathione-Sepharose affinity chromatography. Sf21 cells (1 × 106) infected with the indicated baculoviruses were lysed with 1 ml of ice-cold buffer containing 40 mm HEPES-NaOH (pH 7.9), 150 mm NaCl, 1 mm DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin. After centrifugation at 10,000g for 20 min at 4°C, the supernatants were immunoprecipitated with 2 μg of anti-myc antibody and 10 μl of protein A–Sepharose. The beads were mixed with 50 ng of Cln2 complex, 50 ng of Uba1, 100 ng of Cdc34, and 1 μg of GST–ubiquitinK48R in a 15-μl reaction containing 40 mm HEPES-NaOH (pH 7.9), 60 mm potassium acetate, 2 mm DTT, 5 mm MgCl2, 0.5 mm EDTA (pH 7.9), 10% (vol/vol) glycerol, and 1.5 mm ATP. Reaction mixtures were incubated for 60 min at 25°C.
Construction of yeast strains
The EcoRI–BamHI fragment containing the GAL1,10 promoter was excised from YEp352–GAL (Benton et al. 1990) and introduced into EcoRI–BamHI-digested TRP1 plasmid YEplac112 (Gietz and Sugino 1988) to create pMCB340. PCR fragments encoding wild-type and mutant RBX1 were introduced between the BamHI and HindIII sites of pMCB340 downstream of the GAL1,10 promoter. Yeast strain MCY571 (MATa his3Δ200 ura3 leu2 lys2 trp1 can1R cyh2R rbx1::HIS3/YEp352–GAL–RBX1) is a random spore derived from MCY557 (MATa/MATα his3Δ200/his3Δ200 ura3/ura3 leu2/leu2 lys2/lys2 trp1/trp1 can1R/can1R cyh2R/cyh2R RBX1/rbx1::HIS3/YEp352–GAL–mRBX1) (Kamura et al. 1999). MCY571 was transformed with wild-type or mutant versions of mRBX1 under control of the GAL1,10 promoter in the plasmid pMCB340. Transformants were selected on trp− uracil-plates containing 2% galactose. Transformants were replica plated twice to uracil− galactose plates to allow time for loss of the URA3 plasmid and then tested for growth on trp− galactose medium containing 5-FOA.
Acknowledgments
We thank W. Harper and S. Elledge for baculoviruses encoding Cdc53 and Skp1 and K. Jackson of the Molecular Biology Resource Center of the Oklahoma Center for Molecular Medicine for oligonucleotide synthesis. This work was supported in part by a grant (2 ROI GM41628) from the National Institute of General Medicine to R.C.C. J.W.C. is an Associate Investigator of the HHMI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL conawayj@omrf.ouhsc.edu; FAX (405) 271-1580.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Benton BM, Eng WK, Dunn JJ, Studier FW, Sternglanz R, Fisher PA. Signal-mediated import of bacteriophage T7 RNA polymerase into the Saccharomyces cerevisiae nucleus and specific transcription of target genes. Mol Cell Biol. 1990;10:353–360. doi: 10.1128/mcb.10.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiqutination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- ————— There's the Rub: A novel ubiquitin-like modification linked to cell cycle regulation. Genes & Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, Klausner RD, Pause A. Identification of the von Hippel-Lindau tumor suppressor protein as the substrate recognition subunit of an E3 ubiquitin ligase complex. Proc Natl Acad Sci. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Conaway RC, Conaway JW. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes & Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes & Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes & Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina SA, Correll CC, Kipreos ET, Deshaies RJ. Human CUL1 forms an evolutionarily conserved ubiqutin ligase complex (SCF) with SKP1 and an F-box protein. Proc Natl Acad Sci. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complex that regulate cell division and methionine biosynthesis in yeast. Genes & Dev. 1998a;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Biochem Sci. 1998b;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina SA, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ. Cdc53/cullin and the essential hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme cdc34. Genes & Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Wada H, Yeh ETH, Kamitami T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias M, Kobayashi R, Wittenburg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]