Abstract
Activated transcription by RNA polymerase II (Pol II) requires coactivators, one of which is the SRB/mediator. Whereas Srb4, an essential subunit of the SRB/mediator, is broadly required for Pol II transcription in yeast, we have shown that it is dispensable for the transcriptional activation of some genes. Here, we show that transcriptional activation by different natural activators, and by artificial recruitment of various transcription factors, have very different degrees of Srb4 independence. These data, and the analysis of an rgr1 mutant, point to an Rgr1 subcomplex of the SRB/mediator as the mechanistic route of activation by Srb4-independent activators in vivo.
Keywords: Srb4, mediator, copper inducibility, CUP1, Rgr1, coactivator
The eukaryotic genome contains thousands of genes that show varied and sophisticated regulation. For example, transcriptional activation by upstream activator proteins requires not only general transcription factors (GTFs), but also coactivators that include TFIID (Verrijzer and Tjian 1996), histone acetyltransferase complexes such as SAGA (Grant et al. 1998a), and the SRB/mediator (Bjorklund and Kim 1996). TFIID is composed of TATA-binding protein (TBP) and TBP-associated factors (TAFs) (Verrijzer and Tjian 1996). Although the model of the TAFs having a general coactivator function has been challenged (Moqtaderi et al. 1996; Walker et al. 1996; Oelgeschlager et al. 1998), the in vivo importance of the TFIID-specific TAFs for transcription of about one-sixth of all yeast genes and of some developmentally regulated genes in higher organisms has been clearly demonstrated (Walker et al. 1997; Holstege et al. 1998; Zhou et al. 1998).
The yeast SAGA complex contains Gcn5, the catalytic subunit of HAT activity, Ada proteins, Spt proteins, and several TAFs (for review, see Grant et al. 1998b). A mutant of GCN5 resulted in altered transcription of <5% of yeast genes, whereas a mutation in the SAGA subunit, Taf17 (which is also a subunit of TFIID), affected transcription of over two-thirds of yeast genes (Holstege et al. 1998).
The SRB/mediator complex appears to be a more generally required coactivator. Mediator binds tightly to the carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II), mediates activated transcription in vitro (Kim et al. 1994), and can physically interact with at least some activators (Hengartner et al. 1995; Koh et al. 1998). These results, coupled with the observation that artificial recruitment of mediator results in a robust transcriptional activation both in vitro (Gaudreau et al. 1998) and in vivo (Barberis et al. 1995), suggest that the SRB/mediator is a primary target of activators in vivo. The general requirement of the SRB/mediator for Pol II transcription in vivo was shown in studies with temperature-sensitive mutants of essential subunits of the SRB/mediator, such as Srb4 and Srb6 (Thompson and Young 1995; Holstege et al. 1998). However, these experiments did not distinguish whether Srb4 and Srb6 are required for basal transcription or the transmission of upstream activation signals to the core transcription machinery. We and others have shown that the Ace1-driven transcription of the CUP1 gene, and Hsf-driven transcription of heat shock genes, SSA4 and HSP82, can be activated independently of Srb4 or Srb6 in vivo, whereas activation by Gal4 is severely reduced (Lee and Lis 1998; McNeil et al. 1998). The unique nature of the upstream activators Ace1 and Hsf was further revealed by studies of taf17ts mutants. Taf17 is required for the transcription of many genes, but not the Ace1 and Hsf-regulated CUP1 and SSA4 genes (Apone et al. 1998; Moqtaderi et al. 1998). These observations support the hypothesis that different upstream activators activate transcription by different mechanisms.
In this work, we explore the mechanism of Srb4 (and Taf17)-independent CUP1 transcription by examining the differential effects of Srb4 or Taf17 on transcriptional activation by different upstream activation domains, and on the activation caused by artificially recruiting various subunits of the SRB/mediator to the CUP1 promoter.
Results and Discussion
We have reported previously that the induced transcription of CUP1 and SSA4 genes can occur even after Kin28, Srb4, or Srb6 have been inactivated (Lee and Lis 1998). These results suggested that the transcription of these genes can be activated independently of the essential TFIIH kinase and the SRB/mediator. More recently, using transcription run-on assays, we demonstrate that the Srb4-independent increase in CUP1 mRNA occurs at a step that leads to an increase in the density of transcription complexes on the gene (data on our web site). These results agree with McNeil et al. (1998) and extend their studies to provide a direct measurement of the recruitment of Pol II on the CUP1 gene after copper induction.
The molecular basis for the Srb4-independent transcription must be specified by features of the promoter. By testing a series of hybrid promoters that contain the upstream activating sequence (UAS) from the Srb4-independent CUP1 gene and the core promoter from an Srb4-dependent gene (ADH1 and GAL1), and vice versa, we determined that Srb4-independent transcription is specified by the UAS, not by the core promoter (data on our web site). The same conclusion was derived independently with a different set of hybrid promoters by McNeil et al. (1998).
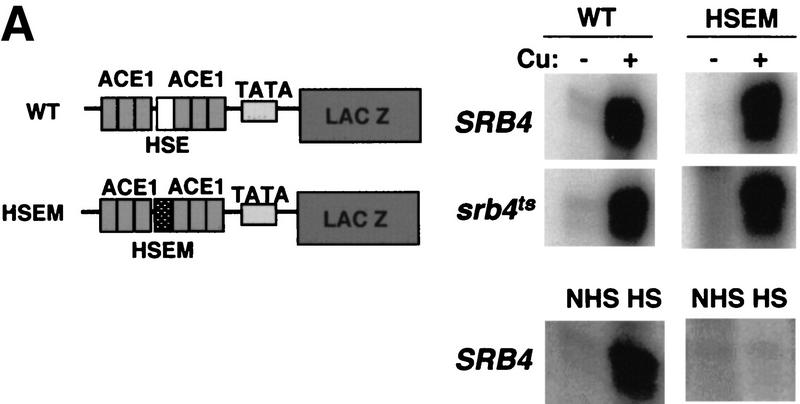
The UAS of the CUP1 gene contains known binding sites for two transcription factors, Ace1 (Evans et al. 1990) and Hsf (Tamai et al. 1994), that allow the CUP1 gene to be activated, respectively, by copper and by heat shock. One might assume that the Srb4-independent copper induction is mediated by the transcription factor Ace1 alone; however, Hsf shows some binding to its DNA sites in yeast even under non-heat shock conditions (Giardina and Lis 1995). Also, because transcription of other heat shock genes has been shown to be moderately heat inducible in yeast strains depleted of Srb4 (Lee and Lis 1998; McNeil et al. 1998), Hsf on its own could potentially specify the Srb4 independence. Therefore, we tested directly the role of the Hsf-binding site in the copper induction of the CUP1 gene, using a CUP1–LacZ reporter containing a point mutation in the HSE (HSEM) (Fig. 1A). The HSEM construct was defective in heat shock induction (Fig. 1A). However, in the srb4ts mutant, the HSEM construct was highly inducible by copper after Srb4 inactivation (Fig. 1A), demonstrating that the HSE is dispensable for CUP1 transcription in srb4ts cells, and that the Ace1 transcription factor is sufficient for Srb4-independent CUP1 activation by copper.
Figure 1.
Srb4 independence of the native CUP1 gene is activation domain specific. (A) S1 nuclease assays showing the heat shock element (HSE) is dispensable and Ace1-binding sites in the UAS of the CUP1 gene are sufficient for the Srb4-independent transcription. (Left) Reporter constructs used in the experiment. The binding sites for Ace1 (ACE1) and a single heat shock element (HSE), and TATA box (TATA) are shown. The HSE with a point mutation that destroys Hsf binding is designated as HSEM. (Top, right) The 30-min copper induction of wild-type (WT) and HSE mutant CUP1 promoter (HSEM) in SRB4 wild-type or srb4ts mutant. SRB4wt and srb4ts cells were grown until OD = 0.3–0.5 at 25°C, then cells were moved to a 37°C water bath and incubated for 1.5 hr; 8-ml cells were harvested (−Cu sample). Then, 1 mm CuSO4 was added and cells were incubated for 30 min at 37°C and again, 8-ml cultures were harvested (+Cu sample). (Bottom, right) The heat inducibility of each construct. Cells were grown to mid-log phase at 25°C and an aliquot was taken (NHS, non heat shock) or cells were transferred to a 39°C water bath for 15 min (HS, heat shock). RNAs were isolated and assayed by S1 nuclease protection. (B) Design of the experiment in part C. (C) Transcriptional activation of the CUP1 gene by Ace1-hybrid activators. Isogenic DLY981 (SRB4ace1Δ) and DLY982 (srb4tsace1Δ) strains are transformed with the following plasmids: pRS316 (vector), p316:ACE1 (ACE1), p316:ACVP (VP16), pAC–HSF (HSF), and pAC–GAL4 (GAL4). Cell growth and copper induction was done as in Fig. 1A. Fold induction of the endogenous CUP1 gene in wild-type and temperature-sensitive mutants are shown next to the S1 assay raw data. Standard deviation is shown for each value. Each value is an average of at least three experiments except for GAL4 (two).
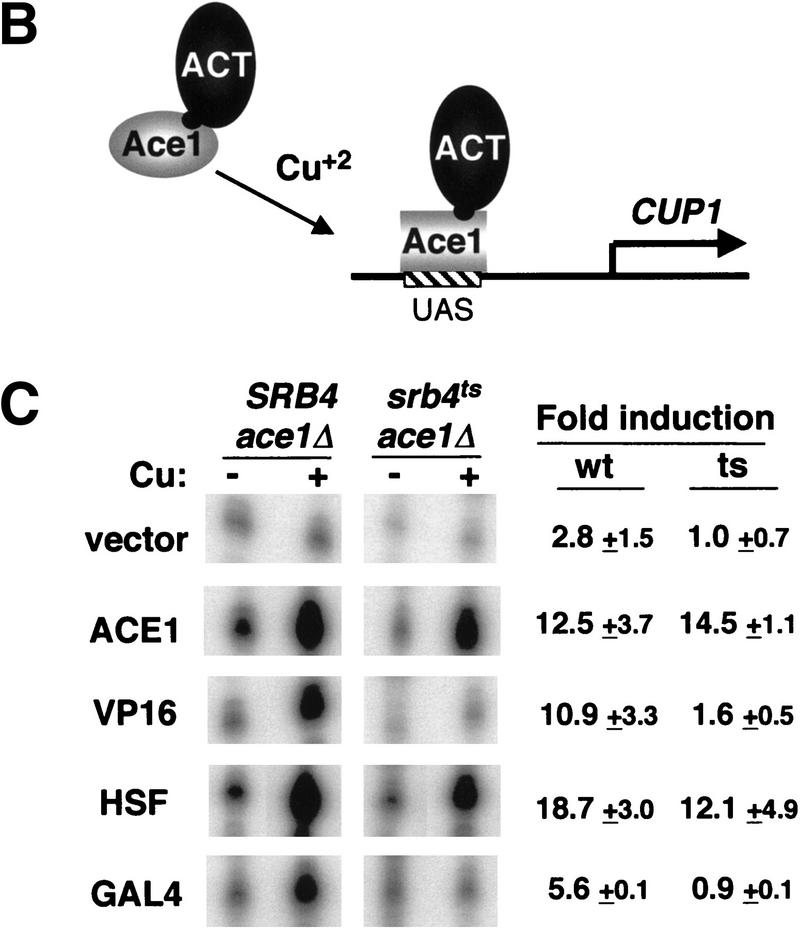
Is the activation domain of Ace1 qualitatively different from other activation domains that drive Srb4-dependent transcription? To answer this, we first deleted the endogenous ACE1 gene from SRB4 wild-type and srb4ts strains, and then transformed the resulting strains with plasmids expressing the Ace1 DNA-binding domain fused to various activation domains. The DNA-binding domain of Ace1 undergoes a conformational change on copper addition, allowing it to bind to the DNA sequence element within the CUP1 UAS (Furst et al. 1988). Therefore, the hybrid proteins should bind DNA in the same way wild-type Ace1 binds in response to copper (Fig. 1B). As expected, strong induction of the CUP1 gene is observed in both SRB4 ace1Δ and srb4ts ace1Δ strains expressing the wild-type Ace1 (Fig. 1C, ACE1). In contrast, whereas the Ace1–VP16 hybrid activated the CUP1 gene in the SRB4 strain, it failed to activate in the srb4ts strain at the nonpermissive temperature (Fig. 1C, VP16). Ace1–VP16 activated transcription normally in the srb4ts strain at the permissive temperature (data not shown). The Ace1–Gal4 construct, which contains the carboxy-terminal activation domain of Gal4, is also very sensitive to the inactivation of Srb4 (Fig. 1C, GAL4). These data clearly show that the Ace1 activation domain operates in a mechanistically distinct manner from VP16 and Gal4 activation domains in vivo.
It should be noted that although the ratio between uninduced and the Ace1-induced CUP1 gene transcription was similar in the SRB4 wild type and mutant, the uninduced level of CUP1 mRNA was two- to threefold less in the srb4ts mutant even at the permissive temperature. This lowered, uninduced level of CUP1 mRNA in the srb4ts mutant is likely to be a secondary effect, because the temperature shift to inactivate Srb4 did not further reduce the CUP1 mRNA level (data not shown). Additionally, this lowered uninduced level may be why the CUP1 gene was scored as Srb4 dependent in another study (Holstege et al. 1998). Nonetheless, it is clear that the CUP1 gene shows the normal fold inducibility in yeast that has an inactivated Srb4 protein.
Because some heat shock genes can be activated after inactivating Srb4, we anticipated that a fusion between the DNA-binding domain of Ace1 and the carboxy-terminal activation domain of Hsf (Ace1–Hsf) might activate the CUP1 gene transcription in the srb4ts mutant. Ace1–Hsf activated transcription in the srb4ts mutant (Fig. 1C, HSF). These results reinforce the proposal that the Srb4 requirement in transcription activation depends on the nature of the particular activation domain.
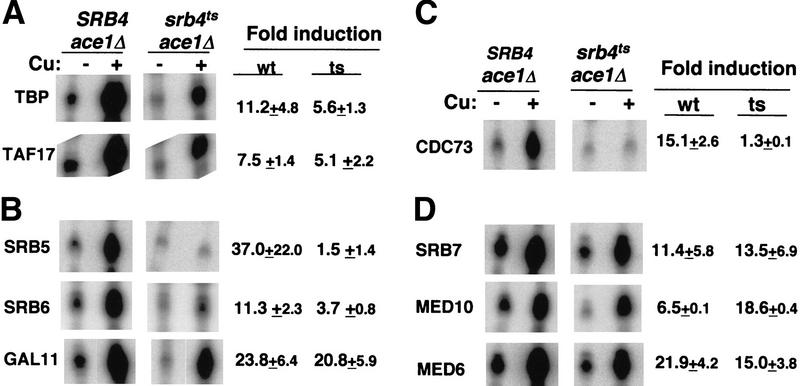
Activation domains may exert their effect on transcription through interactions with coactivators or components of the general transcription machinery. The artificial recruitment of these target proteins to the promoter, for example, via their fusion to a LexA DNA-binding domain, leads directly to high-level expression of promoters that have LexA sequence elements in the absence of any natural activator (Barberis et al. 1995; Gaudreau et al. 1998, and references therein). We reasoned that the particular target contacted by the Ace1 activation domain should activate transcription in an Srb4-independent manner, when it is directly recruited to a promoter. We first tested TBP and TAF17. Recruiting either of these could conceivably provide a platform for the assembly of a preinitiation complex with core Pol II (i.e., Pol II lacking Srb4/mediator). Tethering TBP or TAF17 to the Ace1 DNA-binding domain resulted in the copper-induced activation of the CUP1 reporter gene in the SRB4 wild type (Fig. 2A). In the srb4ts mutant, the fold induction by these hybrid proteins reached 50%–70% of that observed in wild type (Fig. 2A). Therefore, activation by recruiting TBP or Taf17 is partially independent of Srb4. Recent genetic studies have shown that the loss-of-function mutations in the negative regulators of TBP, such as NC2 and MOT complexes, can suppress the temperature-sensitive phenotype of the srb4ts mutant (Lee et al. 1998). This suggests that an activation mechanism that utilizes the recruitment and/or stabilization of TBP to the promoter may be independent of Srb4. It is possible that the artificial recruitment of TBP mimics such a situation at least partially. TAF17 is a subunit of both TFIID and SAGA complex. Therefore, the artificial recruitment of TAF17 may result in recruitment of either TFIID or SAGA, or both, and these complexes may function in a manner that is partially independent of Srb4.
Figure 2.
Transcriptional activation by artificial recruitment in the srb4ts mutant. DLY981 (SRB4ace1Δ) and DLY982 (srb4tsace1Δ) strains were transformed with following plasmids: pAC–TBP (TBP), pAC–TAF17 (TAF17), pAC–SRB5 (SRB5), pAC–SRB6 (SRB6), pAC–GAL11 (GAL11), pAC–CDC73 (CDC73), pAC–SRB7 (SRB7), pAC–MED10 (MED10), and pAC–MED6 (MED6). Cell growth and copper induction was done as in Fig. 1A. Fold induction of the endogenous CUP1 gene in wild-type and temperature-sensitive mutants were shown next to the S1 assay raw data. Standard deviation is shown for each value. Each value is an average of at least three experiments, except for CDC73 and MED10 (two). pAC–SRB5 and pAC–CDC73 activated CUP1 gene transcription normally in the srb4ts strain at the permissive temperature (data not shown). The TAF17 samples were run at the edge of a smiling gel. (A) Transcriptional activation by artificial recruitment of TBP and TAF17. (B) Activation by recruiting different subunits of SRB/mediator have different dependence on Srb4. (C) Activation by the artificial recruitment of Cdc73 is Srb4 dependent. (D) Subunits of the SMCC-like yeast Rgr1 subcomplex mediate Srb4-independent transcription.
We then tested the ability of artificially recruited subunits of SRB/mediator to activate transcription. It was proposed that the inactivation of the Srb4 subunit would result in an inactive SRB/mediator-holoenzyme (Thompson and Young 1995). Consistent with this hypothesis, the recruitment of either Srb5 or Srb6, two subunits of SRB/mediator, gave either very little activation (Fig. 2B, Srb5) or severely reduced activation (Fig. 2B, Srb6) in the srb4ts mutant. Surprisingly, however, the recruitment of Gal11, another subunit of SRB/mediator, resulted in a robust activation in the srb4ts mutant at the nonpermissive temperature (Fig. 2B, Gal11). One explanation for this result is that there exists another Gal11-containing complex, and activation through the recruitment of this complex is Srb4 independent. Recent work has suggested that there exists another form of RNA Pol II holoenzyme, which includes factors such as Cdc73, Paf1, and Gal11, but is devoid of SRBs (Shi et al. 1997). If the recruitment of Gal11 also recruits the Cdc73/Paf1/Gal11 holoenzyme, the transcription by this holoenzyme should be independent of Srb4. However, transcription activation by the Ace1–Cdc73 construct, which provides another way of recruiting the Cdc73/Paf1/Gal11 holoenzyme, is completely dependent on Srb4 (Fig. 2C). Therefore, the recruitment of the Cdc73/Paf1/Gal11 holoenzyme cannot explain the Srb4 independence of the Ace1–Gal11 activator. This result also suggests that the function of Cdc73/Paf1/Gal11 holoenzyme requires functional Srb4.
Another explanation of the Srb4-independent activation by Ace1–Gal11 derives from the observation that the SRB/mediator complex consists of two biochemically separable modules, an Srb4 subcomplex and an Rgr1 subcomplex that contains Gal11 (Lee and Kim 1998). The mediator lacking the Srb4 subcomplex may function to activate transcription when recruited to a promoter. More recently, Gu et al. (1999) purified a mammalian complex that consists of several homologs of yeast SRB/mediator subunits such as Rgr1, Srb7, Med6, and Med10, but is devoid of the Srb4 subcomplex. This complex, named SMCC, contains several homologs of the subunits of the Rgr1 subcomplex. This complex is identical to the human thyroid hormone receptor-associated TRAP complex (Ito et al. 1999). Considering the similarity between SMCC and the Rgr1 subcomplex, we speculate that the yeast Rgr1 subcomplex might also be able to act as a coactivator on its own. Recruiting Gal11 may then recruit the Rgr1 subcomplex, thereby activating transcription independently of functional Srb4, or even a functional Srb4 subcomplex. We tested the possibility by tethering another component of the Rgr1 subcomplex to the promoter to see if this results in an Srb4-independent activation. Ace1–Srb7 activated transcription in the srb4ts mutant to a level comparable with wild type (Fig. 2D, Srb7). Artificial recruitment of Med10 or Med6, both present in the SMCC complex (Gu et al. 1999), and at least one (Med10) proposed to be present in the yeast Rgr1 subcomplex (Han et al. 1999), also resulted in strong activation in the srb4ts mutant. These results support the idea that the transcriptional activation through a SMCC-like Rgr1 subcomplex is Srb4 independent in vivo.
Further support for the role of an Rgr1 subcomplex in the Srb4-independent activation of CUP1 comes from the analysis of the rgr1-Δ2 mutant, a strain that grows slowly and is temperature sensitive (Jiang et al. 1995). The mediator complex isolated from this strain is missing not only a portion of Rgr1 but also several subunits of the Rgr1 subcomplex including Sin4p, Gal11p, Med2, and Hrs1p (Li et al. 1995). Figure 3 shows that the CUP1 gene is poorly inducible by copper in this mutant line, providing additional support for the role of an Rgr1-containing complex in the Ace1 activation of CUP1.
Figure 3.
CUP1 induction is compromised in an rgr1 mutant strain. The copper inducibility of the CUP1 gene was measured by S1 nuclease protection assays in the rgr1-Δ2 yeast mutant and the isogenic RGR1 control. The average quantified fold inducibility and standard deviations for three experiments are shown.
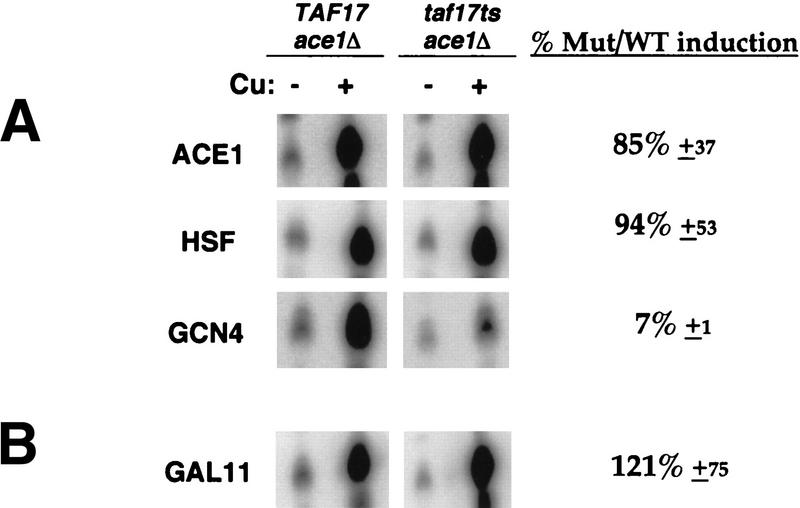
Finally, we applied our hybrid activators to investigate the role of another broadly required coactivator, Taf17, in the activation of CUP1. It has been shown that, whereas Taf17 is required for most Pol II transcription in vivo (Apone et al. 1998; Michel et al. 1998; Moqtaderi et al. 1998), transcriptional activation of CUP1 and SSA4 in taf17ts mutants is normal. Also, like the Srb4-independent CUP1 activation (McNeil et al. 1998; this work), the UASs of CUP1 and SSA4 promoters specify the Taf17 independence, suggesting a mechanism of activation by Ace1 and Hsf that is different from other gene-specific activators, such as Gcn4 (Moqtaderi et al. 1998). Here, we show that the activation domains of Ace1 and Hsf resulted in normal activation of the CUP1 gene in a taf17ts mutant when fused to the Ace1 DNA-binding domain (Fig. 4A, ACE1 and HSF). In contrast, the activation domain of Gcn4 was defective in activating transcription in the taf17ts mutant (Fig. 4A, GCN4). The basal level of CUP1 was not affected in this taf17ts mutant (unlike the srb4ts mutant). Therefore, the absolute levels after copper induction are similar between the wild type and taf17ts for both Ace1 and Hsf-mediated activation of the CUP1 gene and are reported in Figure 4 quantitatively as percent-induced level in mutant relative to wild type.
Figure 4.
Transcriptional activation in a taf17ts mutant. DLY983 (TAF17ace1Δ) and DLY984 (taf17tsace1Δ) strains were transformed with following plasmids: p316:ACE1 (ACE1), pAC–HSF (HSF), pAC–GCN4 (GCN4), and pAC–GAL11 (GAL11). Cell growth, copper induction, and S1 assays of CUP1 RNA were done as in Fig. 1A. The average induced levels in mutant relative to wild type are shown as percentages with standard deviations from three experiments (except two for HSF). (A) Activation domain of Ace1 and HSF, but not Gcn4, can activate Taf17-independent transcription. (B) Activation by artificial recruitment of Gal11 is Taf17 independent.
If the activation pathway through the Rgr1 subcomplex is utilized by Ace1 and/or Hsf, then it too should be independent of not only Srb4 but also Taf17. To test this hypothesis, we measured whether the Ace1–Gal11 can also activate transcription in a taf17ts mutant. As shown in Figure 4B, the recruitment of Gal11 resulted in strong activation in both wild type and mutant. Therefore, the transcriptional activation by Ace1–Gal11, like native Ace1 activation, is independent of both Srb4 and Taf17.
How do cells with inactive Srb4 still maintain the ability to mediate activation by Ace1 and Hsf? It was proposed that the activation domains of Ace1 or Hsf have the ability to interact with any of a number of targets including Srb4 (Moqtaderi et al. 1998). Alternatively, it is possible that distinct activation pathways exist in vivo, and some pathways, which can be utilized by Ace1 and Hsf, are independent of Srb4 and Taf17, but others, which are utilized by other activators such as Gal4, are not.
Our result that Ace1–Med6 can activate transcription in the srb4ts mutant indicates that Med6 behaves like other members of the Rgr1 subcomplex of the mediator. Yet, biochemical data show yeast Med6 is more tightly associated with the Srb4 subcomplex than with the Rgr1 subcomplex (Lee and Kim 1998). In contrast, hMed6 is present in a distinct coactivator complex, SMCC (Gu et al. 1999), along with human homologs of Rgr1, Srb7, and Med10. The coactivator activity of SMCC lacks any homologs of the yeast Srb4 subcomplex. Could an SMCC-like complex exist in yeast? The Rgr1 subcomplex is, to date, the best candidate for a yeast SMCC homolog on the basis of the similarity of subunit composition and the ability to mediate Srb4-independent activation. Our results do not distinguish whether the activation pathway we utilized by artificial recruitment of Gal11/Med6/Med10/Srb7 is through the Rgr1 subcomplex within the SRB/mediator, or through an SMCC-like complex (or a free Rgr1 subcomplex) that exists separately from the Srb4 subcomplex in yeast. Nonetheless, each of the tested yeast SRB/mediator subunits that have homologs in the human SMCC succeeded in mediating Srb4-independent activated transcription, whereas those subunits that fail are not found in the SMCC. These results are compatible with the existence of a functional SMCC-like complex in yeast.
The Rgr1 subcomplex has characteristics of an activation target for Ace1 and Hsf, in that the activation produced by recruiting components of this subcomplex is, like the activation produced by Ace1 and HSF, independent of Srb4 or Taf17. This finding and our observation that a mutation that disrupts the Rgr1 protein and its interaction with components of the Rgr1 complex dramatically reduces the activation of CUP1 strongly implicate an Rgr1 subcomplex (or a yeast SMCC) as the target of this activation.
In the broader perspective, our results show that by studying the coactivator requirements of natural activators and comparing them with the coactivator requirements of activation by artificial recruitment, one can gain insight into the activation mechanism of the natural activators in vivo. We propose that our strategy, applied with many different transcription factor mutants that already exist (e.g., Holstege et al. 1998), will help identify the coactivators and general transcription machinery required by particular upstream activators in vivo. Our results also strongly support the view that, in vivo, no single unified pathway of transcriptional activation exists. Rather, it is likely that many different activators target different coactivators (or GTFs), that is, different activation pathways, to achieve gene-specific activation. This view is consistent with the large number and complexity of transcriptional coactivators and the numerous interactions of activator proteins with coactivators and GTFs identified so far. The use of multiple pathways could accommodate the specificity of activation of many thousands of genes responding to a broad spectrum of signals.
Materials and methods
Construction of strains and plasmids
DLY981 (SRB4wt, ace1Δ:hisG) and DLY982 (srb4ts, ace1Δ:hisG) were derived from CTY233 (SRB4wt) and CTY271 (srb4-138), respectively (Thompson and Young 1995). To generate DLY981 and 982, CTY233 and CTY271 strains were transformed with XbaI–AflII-digested pACE1Δ:HISG.URA3 (Pena et al. 1998) and Ura+ colonies were scored. Ura+ transformants were then streaked on SCUra plates with 500 μm CuSO4. Those transformants that did not grow on CuSO4 plates were then grown overnight in YPD and plated on 5-FOA plates to lose the URA3 gene. Again, the sensitivity on CuSO4 was checked after 5-FOA selection. pACE1Δ:HISG.URA3 is a gift from Dennis Thiele (University of Michigan Medical School, Ann Arbor). DLY983 (TAF17 wild-type, ace1Δ:hisG) and DLY984 (taf17ts, ace1Δ:hisG) were derived from YSB380 (TAF17 wild-type) and YSB463 (taf17ts), respectively (Michel et al. 1998), by the same procedure as above. The taf17 mutant we have used is the allele (taf17-1) that was shown to dramatically reduce the global transcription on temperature shift. DY881(RGR1 wild-type) and DY2587(rgr1-Δ2) strains were gifts from David Stillman (University of Utah Health Science Center, Salt Lake City) (Jiang et al. 1995).
The HSEM construct, which is a 2-μ-based CUP1–LacZ reporter with a point mutation in the heat shock element within the CUP1 promoter, is a gift from Dennis Thiele (Tamai et al. 1994).
Details of construction of plasmids for run-on transcription and ACE1-hybrid activators can be found on the Lis laboratory website http://www.mbg.cornell.edu/lis/lis.html.
Run-on transcription assay
Run-on transcription assays were done with a modified procedure from Elion and Warner (1986). The detailed description of the run-on method can be found on the Lis laboratory website.
S1 nuclease assay
S1 assay was done as described (Ausubel et al. 1992), except 0.1 pmole of oligos were used per reaction. The sequences of oligos used can be found on the Lis laboratory website.
Acknowledgments
We thank Drs. Dennis Thiele, David Stillman, Rick Young, and Steve Buratowski for strains and constructs, Dr. Young-joon Kim and members of the Kim laboratory for discussion and sharing data before publication. D.-k.L. thanks Dr. Sang Jun Han for his suggestion to test the activation by Ace1–Cdc73. We also thank Drs. Jeff Roberts, Eric Alani, Erik Andrulis, Ernie Guzman, Paul Mason, and Hua Shi for critically reading the manuscript, and the members of Lis laboratory for help and suggestions. This work was supported by National Institutes of Health grant GM25232 to J.T.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jtl10@cornell.edu; FAX (607) 255-2428.
References
- Apone LM, Virbasius CA, Holstege FC, Wang J, Young RA, Green MR. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene Publishing Associates/Wiley Interscience; 1992. [Google Scholar]
- Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- Bjorklund S, Kim YJ. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- Elion EA, Warner JR. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CF, Engelke DR, Thiele DJ. Ace1 transcription factor produced in Escherichia coli binds multiple regions within yeast metallothionein upstream activation sequences. Mol Cell Biol. 1990;10:426–429. doi: 10.1128/mcb.10.1.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst P, Hu S, Hackett R, Hamer D. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988;55:705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- Gaudreau L, Adam M, Ptashne M. Activation of transcription in vitro by recruitment of the yeast RNA polymerase II holoenzyme. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- Giardina C, Lis JT. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998a;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998b;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- Han SJ, Lee YC, Gim BS, Ryu G-H, Park SJ, Lane WS, Kim Y-J. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner CJ, Thompson CM, Zhang J, Chao DM, Liao S-M, Koleske AJ, Okamura S, Young RA. Association of an activator with an RNA polymerase II holoenzyme. Genes & Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Dohrmann PR, Stillman DJ. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- Lee TI, Wyrick JJ, Koh SS, Jennings EG, Gadbois EL, Young RA. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Kim YJ. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bjorklund S, Jiang YW, Kim YJ, Lane WS, Stillman DJ, Kornberg RD. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes & Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Bai Y, Poon D, Weil PA, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager T, Tao Y, Kang YK, Roeder RG. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- Pena MM, Koch KA, Thiele DJ. Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2514–2523. doi: 10.1128/mcb.18.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai KT, Liu X, Silar P, Sosinowski T, Thiele DJ. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- Walker SS, Reese JC, Apone LM, Green MR. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- Walker SS, Shen WC, Reese JC, Apone LM, Green MR. Yeast TAF-II145 required for transcription of G1-S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zwicker J, Szymanski P, Levine M, Tjian R. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc Natl Acad Sci. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]