Abstract
Chromatin is actively restructured by a group of proteins that belong to the family of ATP-dependent DNA translocases. These chromatin remodelers can assemble, relocate or remove nucleosomes, the fundamental building blocks of chromatin. The family of ATP-dependent chromatin remodelers has many properties in common, but there are also important differences that may account for their varying roles in the cell. Some of the important characteristics of these complexes have begun to be revealed such as their interactions with chromatin and their mechanism of operation. The different domains of chromatin remodelers are discussed in terms of their targets and functional roles in mobilizing nucleosomes. The techniques that have driven these findings are discussed and how these have helped develop the current models for how nucleosomes are remodeled.
Keywords: ATP-dependent chromatin remodeling, DNA helicase, ISWI, SWI/SNF, nucleosome spacing, nucleosome disassembly
1. Introduction
A fundamental property of ATP-dependent remodelers is that they generally mobilize nucleosomes. If all ATP-dependent remodelers have the same basic enzymatic property of mobilizing nucleosome then why are there so many different kinds of remodelers? In Saccharomyces cerevisiae there is a minimum of nine or more different ATP-dependent chromatin remodelers and in humans the lowest estimate is well over 30 different complexes[1]. While significant progress has been made in identifying the roles of various remodelers in transcription activation/repression, DNA repair and replication, stem cell self renewal, and cell development; there is less understanding as to how the enzymatic activities of these complexes differ from one another. Some of the complexity could be due to alternative ways of recruiting complexes to distinct genomic sites. It seems unlikely that recruitment would be the main reason leading to the diversity of remodelers, because only a small subset of the total number of subunits would need to be varied to accommodate different modes of recruitment. Instead remodelers differ from each other in the underlying core subunits including the catalytic subunit and the remodeler specific accessory subunits thus creating a broad repertoire of remodelers. It seems likely that there are many important functional differences other than recruitment in the family of ATP-dependent chromatin remodelers. The significance of such variation in the composition of ATP-dependent chromatin remodelers will be uncovered by acquiring a more comprehensive understanding of their biochemical properties.
One key difference between remodelers is their interactions with nucleosomes. For convenience we divide the types of interactions into three categories based on the part of the nucleosome involved. Nucleosome mobilization at the very basic level is the interaction of the remodeler with nucleosomal DNA. There are at least two types of protein domains that bind to nucleosomal DNA and the first is the DNA translocase domain. Recent data show that the DNA translocase can engage nucleosomes in at least two distinct ways depending on the remodeler (SWI/SNF versus ISW2). These differences may be a reflection of one remodeler being more disruptive of nucleosome structure than the other. Other protein domains also interact with nucleosomal DNA and may facilitate in mobilizing nucleosomes and stabilizing remodeler binding to nucleosomes. Another part of the nucleosome that remodelers engage is the open face of the histone octamer not occluded by DNA. The extent of interactions between the remodeler and histone proteins seems to be quite varied with some remodelers making extensive interactions with histones while others do not. The interactions of these domains with histones and nucleosomal DNA are likely to have important roles in promoting nucleosome movement that work together with the DNA translocase domain to remodel chromatin. The third region that remodelers interact with is extranucleosomal or linker DNA. The requirement for linker DNA length can be quite varied, while some remodelers require particular lengths of linker DNA and others do not. An important outcome of being linker DNA length dependent is that remodelers move nucleosomes so they all have a particular linker DNA length in order to uniformly space nucleosomes. These effects appear to be mediated through protein-DNA interactions of the catalytic and/or accessory subunits with linker DNA. Finding how each remodeler interacts with nucleosomes and how they differ from one to the other provides us with the appropriate starting point to understand how they also differ in their mechanism of remodeling nucleosomes.
So far there is a minimum of two modes of remodeling with the outcome being either spaced or disassembled nucleosomes. Different models for how spacing of nucleosomes occurs will be reviewed. The important differences that cause one remodeler to disassemble and the other to space nucleosomes will be discussed. Each remodeler uses the same basic DNA translocase activity in different contexts to space or disassemble nucleosomes. The different experimental techniques that have been used to investigate the mechanisms of remodeling will be described. The advantages and limitations of techniques using either fluorescence or photo crosslinking are given along with that of doing bulk versus single molecule type experiments.
2. Functional Components of an ATP-dependent Remodeler
2.1 Binding to nucleosomal DNA
DNA helicases and translocases are divided into 6 superfamilies with the two largest groups being SF1 and SF2. Nucleosome remodelers belong to the SF2 superfamily of DNA translocases and unlike other members of this family generally do not have any helicase activity. It has been difficult to determine the structure of the ATPase domains of any SWI/SNF or ISWI remodeler, but crystal structures have been obtained of related complexes. The structure of the ATPase domain of Rad54 from Zebrafish and Sulfolobus Solfataricus has been solved [2,3]. The structure of Rad54 from Sulfolobus Solfataricus bound to DNA highlighted that it binds DNA much like other known DNA and RNA helicases. Recently the structure of the ATPase domain of the yeast Chd1 protein with tandem chromodomains has been solved [4]. It is informative to compare these structures to see both their similarities and their differences. Both of these DNA translocases have the same two lobe structure generally characteristic of DNA translocases as shown in Figure 1A and are referred to as lobes 1 and 2. In the case of Chd1 the orientation of the two lobes is different than that observed in Rad54 and indeed varies also from other SF2 translocases. The conformation of the two lobes in Chd1 suggests they are in an inactive state and would likely not be able to bind DNA because the two lobes are too far apart. Although not shown the tandem chromodomains binding across the two lobes would also interfere with DNA binding. When aligning lobe 1 in Rad54 with lobe 1 of Chd1 it is remarkable as to the extent of structure homology and conservation that is observed (Figure 1B). The same is true for lobe 2 when lobe 2 from Rad54 and Chd1 are aligned independently of the rest of the protein as shown in Figure 1C. The structural similarity seen between Rad54 and Chd1 is expected to be true for the other remodelers based on their respective sequence homology (Figure 1A).
Figure 1.
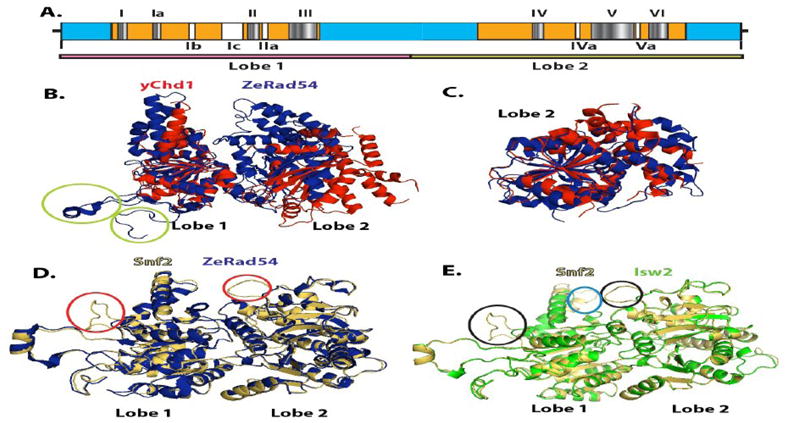
The structure of the ATPase domain from several chromatin remodelers.
(A) The ATPase domain of chromatin remodelers have 12 sequence motifs that are conserved and are number I through VI. The motif Ib is specific to the SWI/SNF subfamily of remodelers. Motifs I–III are all in lobe I, while motifs IV–VI are in lobe 2. (B) The X-ray crystal structure of the ATPase domain from Rad 54 (Zebrafish) and Chd1 (Saccharomyces cerevisiae) are shown in blue and red, respectively, and are overlaid to show their structural homology. The two regions of Rad54 that are not observed in Chd1 are highlighted by green circles. The orientation of the two lobes is offset and Chd1 as seen by lobe 2 in both structure not overlapping well in (B). Strong structural similarity is seen when lobe 2 from Rad54 and Chd1 are independently aligned to each other as shown in (C).
While it is extremely difficult to verify whether the ATPase domains within these different remodelers do indeed work differently given the size and complexity of these complexes, there are data that show such is the case for SWI/SNF and ISW2. The approach has been to use two different techniques to map the interactions of the catalytic subunit with DNA and to selectively focus on the ATPase domain while part of the complete complex. The first technique is to photochemically crosslink the proteins bound to nucleosomal DNA and find the part of the protein covalently linked to DNA. A protein footprinting technique was also used to map Snf2 interactions with free DNA and with nucleosomes in which Fe-EDTA is conjugated to either DNA or histones. Snf2 was crosslinked the same distance from the dyad axis as for Isw2, but was to an entirely different part of the DNA translocase which suggests they bind differently to the same part of the nucleosome[5]. SWI/SNF and ISW2 have been shown to translocate on nucleosomal DNA at this same position and in the same direction. Extensive probing of the interactions of the catalytic subunit by DNA crosslinking showed that Isw2 binds selectively to the face of DNA pointing away from the histone octamer, while Snf2 is preferentially associated inside the DNA gyre towards the histone octamer (data not shown).
Another piece of evidence for the differential interactions of the ATPase domains of ISW2 and SWI/SNF is also seen in ISW2 requiring the histone H4 tail for nucleosome remodeling while SWI/SNF does not. When the N-terminal tail of histone H4 is removed the interactions of the Isw2 ATPase domain with nucleosomal DNA are severely reduced as observed by both DNA footprinting and site-directed DNA crosslinking[6]. The reduced binding of the ATPase domain is functionally relevant as nucleosomes missing histone H4 tails are remodeled much less efficiently than intact nucleosomes. The H4 tail is therefore required to stabilize the interactions of the ATPase domain. SWI/SNF on the other hand does not have H4 tail dependence for its remodeling activity and points to the binding of its ATPase domain being different than that of ISW2.
Besides the DNA translocase binding to nucleosomal DNA there may be other parts of the remodeler that also bind to different sections of nucleosomal DNA. ISW2 is an excellent example in which the auxiliary subunit Itc1 binds to nucleosomal DNA in combination with the catalytic subunit Isw2 (Figure 2A). Itc1 was observed to be efficiently crosslinked to DNA just inside the nucleosome at 60–62 and 66–68 bp from the dyad axis in addition to Isw2[7]. The region of Isw2 crosslinked to nucleosomal DNA 60 and 62 bp from the dyad axis was found by peptide mapping to be in the C-terminal half of Isw2 encompassing the HAND domain [5]. The structure of the C-terminal end of ISWI from D. melanogaster including the HAND domain has been previously solved [8]. The HAND and SANT domains are closely associated together and have been suggested to work together as a functional unit. They are connected through a rigid alpha helical, rod-like structure to another domain called SLIDE that has DNA binding activity. Based on location it is attractive to suggest that the HAND and SANT domains could be involved in “pushing” DNA into the entry site of the nucleosome to facilitate moving DNA through and around the nucleosome[5]. In earlier reports it was indicated that Itc1 also binds at the SHL2 site along with Isw2 [7], but in later experiments we have found that Isw2 alone is crosslinked at this DNA site and not Itc1 (data not shown). The interactions of Itc1 with nucleosomal DNA are thus restricted in the core nucleosomes to only the entry site.
Figure 2.
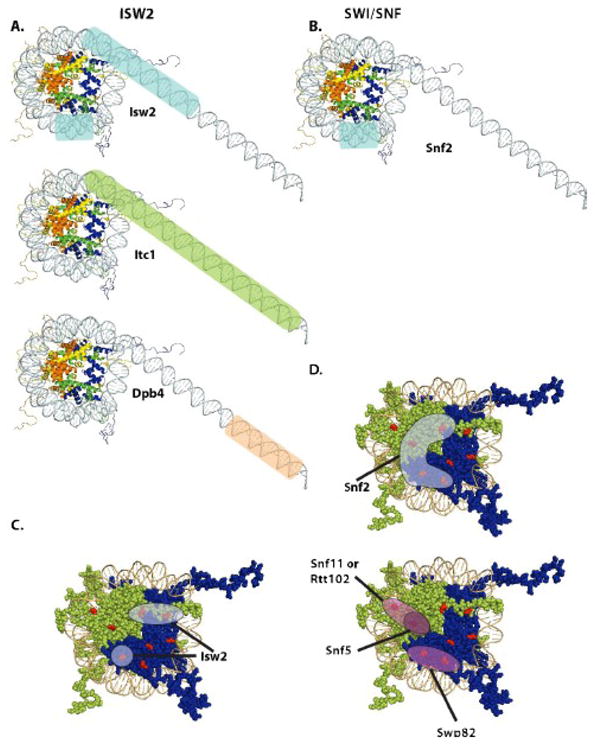
The contacts of SWI/SNF and ISW2 with histones and nucleosomal DNA were determined by site-directed crosslinking.
DNA crosslinking was used to find which subunits of ISW2 (A) and SWI/SNF (B) was associated with different regions of the nucleosomal DNA. Areas highlighted in light blue are those contacted by the catalytic subunits Isw2 and Snf2. The regions contacted by accessory subunits of ISW2 are highlighted in light green (Itc1) and tan (Dpb4). Contact with the globular portion of the histone proteins was mapped by histone crosslinking for ISW2 (C) and SWI/SNF (D). The regions where photoreactive probes were placed are shown in red and the H2A and H2B proteins are in green with H3 and H4 in blue. Those sites that crosslink to Isw2 and Snf2 are highlighted with light blue, while those to the accessory subunits of Snf2; namely Snf5, Swp82 and Snf11/Rtt102 are highlighted in purple.
Although SWI/SNF has a greater number of protein subunits, it appears to have less interactions with nucleosomal DNA than does ISW2 as reflected by the number of subunits and the range of sites that are crosslinked (compare Figure 2A and 2B). Snf2 is the only subunit of SWI/SNF that is appreciably crosslinked to any nucleosomal site [9]. Additional DNA crosslinking experiments in which the photoreactive group is placed in the phosphate backbone also showed photocrosslining of only Snf2 (data not shown). Snf2 was shown to be localized to a short ~10 bp stretch of DNA 2 helical turns from the dyad axis[9]. Only the DNA translocase domain of Snf2 is bound to nucleosomal DNA in the initial complex formed when SWI/SNF is recruited to nucleosomes. In some ways this is not entirely unexpected since SWI/SNF can push nucleosomes off DNA till the end of DNA is reached by the DNA translocase, an unraveling of 50 bps of nucleosomal DNA[10]. These observations indicate that any contacts with nucleosomal DNA that might occur between the DNA translocase at SHL2 and the entry site of the nucleosome would not be required for nucleosome movement.
Other protein interactions with nucleosomal DNA likely occur after remodeling is started and are part of the remodeling reaction. These kinds of changes in remodeler binding have been best shown for ISW2. In these experiments DNA footprinting and site-directed DNA crosslinking were used to track interactions with nucleosomal DNA[11]. Different stages in remodeling were examined by using non-hydrolysable ATP analogs and DNA gaps to look at conformational changes due to ATP binding and hydrolysis. The DNA gaps do not interfere with ATP hydrolysis, but instead block translocation and prevent nucleosomes from being moved [12,13,14,15]. These data are discussed in more detail in Section 3.2.3. These additional interactions of ISW2 with nucleosomal DNA cause the complex to bind nucleosomes tighter and form a template-committed complex thereby making ISW2 more processive. These transitions do not occur with ATP binding alone, but require ATP hydrolysis. Presumably the interactions of other remodelers with nucleosomal DNA may expanded during remodeling in order to promote nucleosome movement, but as of yet have not been characterized.
2.2 Binding to histones
These remodelers recognize the histones in nucleosomes as well as the nucleosomal DNA. Most of this type of work has focused on histone tails rather than the globular regions since the tails are key targets for post-translational modifications. Our focus is on the proteins and protein domains that recognize both modified and unmodified histone tails. The ATPase domain of ISW2 binds to the unmodified histone H4 N-terminal tail. The evidence that supports this is that deletion of the H4 tail and not other histone tails causes a strong reduction of the ATPase domain binding to SHL2[6]. This is consistent with other data that has shown the H4 tail is associated with nucleosomal DNA at SHL2[17,18]. Although it is not yet clear how Isw2 binds to the H4 tail, there is a suggestion that a cluster of acidic amino acid residues conserved only in ISWI proteins and predicted to be clustered together on the surface of the ATPase domain might be involved [5]. The H4 tail is also required for other ISWI chromatin remodelers such as CHRAC/ACF and NoRC[19,20,21]. The residues in the H4 tail that are required for ISWI remodeling are residues R17H18R19 at the base of the H4 tail[22]. As expected for interactions between the H4 tail and ISW2 being ionic, acetylation of lysine 16 in H4 disrupts the ability of ACF to remodel nucleosomes[22,23]. In these examples histone acetylation antagonizes binding of the remodeler, while in other remodelers it has the opposite effect. In another example an unmodified N-terminal tail of histone H2A has a negative effect on INO80 remodeling [24].
The bromodomain binds to acetylated lysines and is one of the signature motifs of the SWI/SNF family of remodelers. One copy of the bromodomain is found in Snf2 and Sth1, the catalytic subunits of yeast SWI/SNF and RSC. Other subunits within the RSC complex also contain bromodomains such as the Rsc4 subunit and subunits of mammalian SWI/SNF contain multiple copies. The bromodomain is not reserved just for ATP-dependent remodelers, but is frequently found in histone acetyltransferases as well. The other myriad of modified histone tail recognition domains such as the PHD finger and the chromodomain that recognize methylated lysines are also a common feature of nucleosome remodelers. One example of this is the PHD finger in the BPTF subunit of the NURF complex, an ISWI type of remodeler. BPTF recognizes lysine 4 in histone H3 when it is methylated and helps recruit the NURF complex to methylated chromatin[25,26]. Clearly the ability to recognize histone modifications is an integral part of ATP-dependent chromatin that may either target the complexes to particular genomic sites and/or modulate their activity.
Less well known is how remodelers recognize the globular portion of the histone proteins. The variable activity of remodelers with nucleosomes containing different histone variants implies that there may be structural features in the globular portion of the histones that are required by the remodelers. One feature of the globular histone region that has drawn a lot of attention is the acidic docking domain formed by histones H2A and H2B. H2A has the largest number of histone variants among all the core histone proteins. The H2A variants H2A.Bbd and H2AL which lack the acidic docking domain have been shown to interfere with SWI/SNF and RSC remodeling[27,28,29]. These data suggest that indeed the SWI/SNF and RSC complexes might bind to the H2A/H2B docking domain. Other data suggest that H2AZ, the variant of H2A that has an enhanced acidic docking domain, could form chromatin regions that are preferred for recruiting SWI/SNF as in the case of the Gal 1 promoter[30]. More work is needed to understand the important structural features that these remodelers recognize within the globular histone region of the nucleosome.
There are histone modifications that occur in the globular region which could also demarcate remodeler binding sites. Acetylation of lysine 56 in histone H3 is in the globular region and is in the alpha helix that extends to the entry site of the nucleosome. Although at first it was suggested that acetylated lysine 56 positively regulates SWI/SNF, careful biochemical experiments have shown that this modification has no significant effects on SWI/SNF or RSC remodeling[31,32]. As of yet there is no data to support modifications of the globular histone regions being recognized by remodelers.
An approach that has been used to examine the interactions of remodelers with the globular region of the histone proteins has been protein crosslinking. Since nucleosomes can be reconstituted with recombinant histones and single cysteines engineered into the nucleosome surface, it has been possible to incorporate photoreactive probes to specific sites in the globular region. The probes are designed with the feature that a radiolabel is transferred to the target protein after crosslinking. The subunits and domains bound across the exposed surface of the globular histone region are mapped by varying the location of the photoreactive probe. Several of the subunits of SWI/SNF, including the Snf2 catalytic subunit, have been shown to be bound across a large section of the histone octamer surface[9]. A total of eight different residues within the histone octamer were modified spanning the H3/H4 tetramer and H2A/H2B dimer surfaces. Many of the crosslinks to SWI/SNF were in the H2A/H2B region and within the acidic docking domain. The Snf5 subunit was found to be selectively associated only near the acidic domain and nowhere else on the nucleosome (Figure 2D). Snf5 was not seen to be bound close to nucleosomal DNA and appears to make only discrete contacts with nucleosomes through the H2A/H2B dimer. Snf2 covers about half of the face of the histone octamer and spans from the tetramer into the dimer as shown by crosslinking. The interactions of SWI/SNF appear to be more extensive with the histone octamer face than with nucleosomal DNA as shown by protein-protein and DNA-protein crosslinking. The ISW2 complex is different and only makes limited contacts with the histone octamer (Figure 2C). ISW2 crosslinking is restricted to the histone octamer near SHL2 and the entry site of the nucleosome (data not shown). Both of these sites are proximal to where ISW2 is bound to nucleosomal DNA. Some key differences then between SWI/SNF and ISW2 is that SWI/SNF has more extensive interactions with the histone octamer face while ISW2 has more interactions with nucleosomal DNA.
2.3 Binding to linker DNA
A key feature that sets apart chromatin remodelers is their requirement for linker DNA. Initially studies with nucleosome remodelers paid little attention to whether the length of linker DNA was an important factor or not. After a while it became apparent that not only did some remodelers require a certain length of linker DNA for binding, but also for their nucleosome mobilizing activity. The class of remodelers that were first found to have a strong dependence on linker DNA length was the ISWI subfamily[7,12]. In retrospect this does not seem to be surprising given that some of these complexes have been shown to arrange nucleosomes with uniform length of linker DNA between adjacent nucleosomes referred to as nucleosome spacing[33]. Further characterization has shown that ISW2 causes nucleosome positions to shift at the 5′ and 3′ ends of genes [34,35].
Both ISW2 and INO80 from Saccharomyces cerevisiae require a minimum of ~20 bp of extranucleosomal DNA to bind nucleosomes efficiently[7,24] and more than 30 bp for efficient binding of Isw1a and Chd1 [36,37]. However, not all remodeling complexes need extranucleosomal DNA to bind well. RSC bound to nucleosomes with 0, 7, and 20 bp of linker DNA with comparable affinities as shown by gel shift assays [15]. The yeast ISW1b complex does not require extranucleosomal DNA to bind efficiently to nucleosomes either, but its affinity is generally less for nucleosomes than for ISW1a and ISW2 [37]. The ISW1b binding data has been fraught with the fundamental problem that that the ISW1b-nucleosome complex is not stable to gel shift assay conditions and therefore these measurements are likely to be less reliable. Other data suggest that ISW1b does bind to extranucleosomal DNA, because exonuclease footprinting of ISW1b-nucleosome complexes shows that ISW1b binds to 13–19 bp of extranucleosomal DNA[36]. However ISW1b moves nucleosomes slightly off the edge of DNA by 5–11 bp which suggest it may not have the extranucleosomal DNA length dependence of ISW1a or ISW2.
Besides extranucleosomal DNA being involved in recruiting remodelers, it can be required for the remodeler’s enzymatic activity. Often the length of extranucleosomal DNA needed for optimal nucleosome movement is longer than that required for binding. ISW2 needs almost 70 bp of extranucleosomal DNA for optimal movement; whereas, INO80 needs a minimum of 53 bp of linker DNA for nucleosome movement [7,24] and are significantly longer than that required for efficient binding. A similar linker DNA requirement was also observed for nucleosome movement by Chd1 [36,37]. ISW1a is in a class by itself because it has a more complicated extranucleosomal DNA length requirement. Rather than sensing only one arm of linker DNA at a time like ISW2, ISW1a can adjust its activity depending on the length of both linker DNA present in a single nucleosome. If there is extranucleosomal DNA of 33 bp or longer at any one entry site then ISW1a moves nucleosomes towards the center of DNA. When there is 33 bp of extranucleosoml DNA at both entry sites ISW1a does not move nucleosomes or appreciably hydrolyze ATP, even though it binds quite well. When the lengths of the two arms of linker DNA become disproportionate and one of them is longer than 33 bp, ISW1a is again able to move nucleosomes in the direction of the longer linker DNA. It is clear that other remodelers such as SWI/SNF do not depend on linker DNA for mobilizing nucleosomes [38].
The interactions of ISW2 and ISW1a with nucleosomal DNA change with linker DNA length as shown by DNA footprinting. ISW2 binds to extranucleosomal DNA and the entry site regardless of extranucleosomal DNA length, but ISW2 binding to SHL2 is lost when extranucleosomal DNA is reduced from 35 to 20 bp[6]. Stable binding of the ATPase domain is therefore dependent on extranucleosomal DNA length and implies that there is communication between the ATPase domain and the part of ISW2 bound to linker DNA. ISW1a interactions with SHL2 also changes with linker DNA length like ISW2. SHL2 is protected by ISW1a when there is only one linker DNA, but loses this protection when there are two linker DNA of 33 bp each[36]. Human ACF complex containing ISWI has also been shown to move nucleosomes in a linker DNA dependent manner. The rate of nucleosome remodeling and ATP turn over for ACF increases as extranucleosomal DNA is lengthened[39]. ACF has been proposed to bind nucleosomes as a dimer, presumably with one ACF bound to each linker DNA and the one bound to the longer linker DNA is the one actively mobilizing nucleosomes [39,40,41,42].
It has been important to identify the protein subunits and domains of these remodeling complexes that interact with extranucleosomal DNA. These subunits and domain are probably integral parts of the machinery that help modulate the binding and activity of the ATPase domain. Several subunits of the remodeling complex interact with linker DNA and recruit the remodeler to target sites to properly orient the remodeler on nucleosomes. In ISW2 the accessory subunit Itc1 extensively interacts with at least 53 bp of extranucleosomal DNA starting from the entry site (Figure 2A and reference [7]). The accessory subunit Ioc3 of ISW1a crosslinks to linker DNA 85 to 92 bp from the dyad axis[36]. The activity of human Snf2h is regulated by a variety of accessory subunits found in the hACF, CHRAC, RSF and WICH complexes. Snf2h alone requires extranucleosomal DNA for optimal nucleosome mobilization, but its dependence on extranucleosomal DNA is even more enhanced when associated with hACF1[43]. The effect of the accessory subunit on the basic remodeling activity of hSnf2h changes with different accessory subunits as seen by comparing the remodeling activities of hACF, RSF, and WICH which all contain the same hSnf2h catalytic subunit[44]. The differential effects of accessory subunits on ISWI activity is seen when comparing the remodeling activities of ISW1a to ISW1b which have the same catalytic subunit Isw1. Besides accessory subunits contacting linker DNA, the catalytic subunit also makes key contacts with extranucleosomal DNA. Isw2 interacts with ~19 bp of extranucleosomal DNA starting at the entry site [7]. Peptide mapping of crosslinked subunits revealed that the SLIDE domain of Isw2 interacts with linker DNA 19 bp from the entry site [5]. X-ray crystallography studies of dISWI C-terminus shows that SLIDE domain resembles the structure of the DNA binding domain of the c-Myb transcription factor that contains three alpha helical regions [8].
3. Mechanism of Chromatin remodeling
Chromatin remodeling complexes utilize the energy derived from ATP hydrolysis to disrupt histone-DNA contacts of nucleosomes and alter nucleosome positions on DNA. The outcome of this alteration could be equal spacing of nucleosomes, repositioning or disassembling nucleosomes. How different remodelers, with significantly similar ATPase domains, achieve such diverse tasks is currently unresolved. Recent reports with different biochemical and biophysical approaches have found some of the changes that occur during remodeling and the forces required to move nucleosomes. These techniques will be reviewed first before discussing some of the prevailing models for remodeling nucleosomes.
3.1 Approaches
3.1.1 Tracking nucleosome movement and changes in histone-DNA interactions
There have been several innovative approaches that have been used to investigate the mechanism of nucleosome remodeling. These methods have taken advantage of reconstituting nucleosome with recombinant histones to engineer into nucleosomes at well defined locations either photoreactive or fluorescent type probes. These kinds of experiments either examine histone-DNA or histone-remodeler interactions. One type of site-directed crosslinking is designed with histone modification sites close to nucleosomal DNA such that the photoreactive group preferentially crosslinks to nucleosomal DNA[7,10,45,46]. The location of the crosslinked site in DNA is readily determined because the photoadduct formed with DNA is generally more sensitive to cleavage under alkaline conditions than unmodified DNA. This is done by radiolabeling one 5′ of DNA and analyzing the cleaved products on a DNA sequencing gel along with an appropriate DNA sequencing ladder. A strength of this approach is the one-to-one correlation of a specific histone residue (i.e. residue 53 of histone H2B) being in close proximity to a particular bp in DNA. Another strength is the multiple histone positions that can be examined to provide a more complete view of the interactions of DNA with histone octamer. There are four different histone positions that have been tested which are histone H2B (residue 53), H2A (residue 45), H3 (residue 120) and H4 (residue 47). Often site-directed mapping of histone-DNA interactions have been used in a static format where remodeling is either in an arrested or steady state, but can also be used to study the dynamics of remodeling[11,12,13,38]. It is usually necessary to slow the remodeling reaction by reducing the concentration of ATP when using site-directed crosslinking. Although this technique provides a direct read-out of nucleosome movement, it does considerably more because it highlights multiple positions throughout the nucleosome. For example one can determine after the nucleosome has been moved if the canonical nucleosome structure has been maintained in terms of the wrapping of DNA on the histone octamer. A fundamental difference between ISW2 and SWI/SNF has been revealed by measuring the length of DNA between different points on the octamer before and after remodeling. While ISW2 maintains the canonical nucleosome structure after the nucleosome has moved, nucleosomes remodeled by SWI/SNF have changed more than just its position on DNA [10,46]. One of these changes is the increase in the length of DNA between two points on the histone octamer and is evidence for residual DNA bulges on the surface of the nucleosome. Another difference is the disruption of histone-DNA contacts at residue 45 of histone H2A after remodeling.
The other approach for tracking nucleosome movement is placing a donor and acceptor fluorescent group in the histone octamer and DNA, respectively, close to the entry site of the nucleosome [39,41,47,48]. The distance between the histone and DNA sites is measured in terms of fluorescence resonance energy transferred or FRET [47]. A key advantage of an approach like this is that it rapidly measures changes in nucleosome position of single nucleosome molecules and provides a more comprehensive view of the remodeling dynamics[47]. The location of the donor and acceptor can make a large difference and is impossible to predict apriori which pairs will function well and which will not. The other limitation so far has been that it has not been possible to probe the movement of DNA inside the nucleosome and is restricted to examining only the rate at which DNA exits the nucleosome. Recently single molecule (sm)FRET was used to study the dynamic movement of nucleosomes by human ACF. ACF with ATP moved end-positioned nucleosomes first ~ 7 bp from its original translational position and then in steps of 3–4 bp [47]. Binding of ACF was also stimulated by ATP as shown using fluorescently labeled ACF. The rate at which ACF moved nucleosomes was 2 bp per second and the total distance nucleosomes moved before ACF was released averaged about 200 bp in a bi-directional manner.
These approaches provide more information about nucleosome movement than the often used method of restriction endonuclease accessibility assay. The basic principle of the accessibility assay is to monitor nucleosome movement or changes in nucleosome structure by finding if a particular DNA site can be cleaved by its cognate restriction endonuclease. The particular DNA site often starts out within the nucleosome and as such is protected, but once the nucleosome has moved sufficiently becomes exposed and is cleaved. The problems with this approach are the important factors of the intrinsic kinetics of the restriction enzyme itself and that once it becomes exposed it is insensitive to how far the nucleosome has moved in total. Another factor that can also complicate this analysis is that the nucleosome translational position does not have to be necessarily changed for the remodeler to be able to make a site accessible as transient bulges on the nucleosome may also be cut by restriction endonucleases.
Many of these techniques measure only the average behavior of the molecules in the sample and not the distribution of molecules with their differing behavior. Sometimes because only class average characteristics are observed it can lead to particular details being missed or overlooked that reveal important mechanistic information. Besides smFRET, DNA methylation protection assays or MAP-IT can be used to examine nucleosome position in individual molecules and has been used to examine the nucleosome remodeling products of SWI/SNF [38,49]. MAP-IT was particularly useful for tracking nucleosome movement in short arrays where there are likely to be many different patterns of nucleosome movement which could not be properly identified by bulk measurements. The progressive movement of one nucleosome toward an adjacent nucleosome by SWI/SNF was observed by MAP-IT. Within the population of nucleosomes there were species that either had the one nucleosome moved a short distance or others that were moved to the extent that it was significantly invading the space of the adjoining nucleosome. These assays were also adept at showing that nucleosome movement by SWI/SNF was dictated by Gal4-VP16 recruitment to the immediately adjoining nucleosome.
Another single molecule approach used to find the rate of DNA translocation, processivity, and the force generated by the remodeler involves magnetic and optical tweezers (or trap). The magnetic trap approach immobilizes one end of DNA onto the glass surface of a microscope flow chamber and the other end is attached to a small (~1μM) paramagnetic bead. The bead can be twisted or pulled to supercoil or stretch the DNA. In an optical trap, DNA is attached at one end to polystyrene beads with the other end attached to the microscope flow chamber. Video microscopy is used to record changes in the position of the tethered bead and from that the DNA length, horizontal motion of the bead, and stretching force on DNA can be calculated. Approaches involving magnetic tweezers have been extensively used to study coiling and uncoiling in DNA [50,51]. Recently magnetic trap experiments were used to study the interactions of both ATP-dependent RSC and SWI/SNF on DNA. Both enzymes translocate on DNA forming transient DNA loops and introduce negative supercoils into DNA in an ATP-dependent manner [52,53]. Under low stretching tension (0.3pN), RSC reversibly translocates on DNA surface at 200 bp/sec forming loops of 100 to 400 bp. A similar study using nucleosomal templates and the optical tweezer method found that under high stretching forces (≥1pN) RSC translocates on nucleosomes at ~13 bp per sec, forms loops varying in size from 20 to 1200 bp, and travels an average of ~ 105 bp before dissociating [54,55]. These studies provide important mechanistic details of how remodelers reposition nucleosomes on DNA. One limitation of these experiments has been their inability to detect translocations on DNA less than ~20 bp.
3.1.2 Tracking the interactions of remodelers with nucleosomes
DNA footprinting has been used to map the interactions of remodeler with nucleosomal DNA. The high reactivity, lack of base pair specificity and smaller size of hydroxyl radical makes it an excellent choice for DNA footprinting of protein-DNA complexes [56]. Footprinting is completed in seconds, only tight interactions are observed with single bp resolution, and can be done at different time intervals after addition of ATP. An inherent problem with this approach is that nucleosome mobilization is not a uniform or well synchronized process thus giving a heterogeneous mixture of products that makes the analysis difficult. One way to avoid these problems is to arrest remodeling at specific stages. Gaps in nucleosomal DNA can block translocation of the remodeler and depending on their location can be used to move nucleosomes to any defined position on DNA. The intermediates of ISW2 remodeling have been investigated in this manner and have significantly helped identify nucleosomal DNA interactions that occur as a result of remodeling [11].
Complementary to footprinting is finding the protein(s) involved in contacting nucleosomal DNA or histones by site-directed crosslinking. Photoreactive groups can be placed at specific nucleotide bases or amino acids in order to find the targets bound to extranucleosomal or nucleosomal DNA or to histone proteins. In difficult cases where DNA footprinting may be problematic, site-directed DNA crosslinking can provide something similar to a footprint with a systematic scanning of protein-DNA interactions. Photo crosslinking is complete in a matter of seconds making this considerably slower than fluorescent based techniques, but nonetheless if done in a quench flow format key intermediates in the remodeling reaction can be observed. There are two different methods for modifying DNA, one in which the crosslinker is incorporated in the phosphate backbone and the other to the nucleotide base. Each of these has different spatial constraints and therefore scans different parts of the DNA tertiary structure [7,9,16]. Since not all interactions are restricted to the DNA in nucleosomes, we have also designed a similar approach for site-specific attachment of photo crosslinkers to histone proteins[9]. The fundamental design is similar with attachment of a photo crosslinker to a cysteine and the transfer of radiolabel to the target crosslinked protein. Nucleosomes with modified octamers are bound to the remodelers and after crosslinking a radiolabel is transferred to the remodeler subunit. Structural and conformational changes in the remodeling complex as a result of ATP binding and hydrolysis can be detected by comparing the crosslinking pattern in the presence and absence of ATP and ATP analogs.
The location of the DNA translocase and direction in which it travels on nucleosomal or extranucleosomal DNA is determined by using DNA gaps to block translocation. Random one nucleotide gaps are created by incorporating ~1 dUTP per DNA by PCR. The gap is created by removal of deoyxuridine with uracil DNA glycosylase and endonuclease III. Nucleosomes reconstituted with DNA containing gaps at a variety of positions are remodeled to find out which gaps will prevent nucleosomes from moving. This is an effective way to scan a large number of DNA positions in one experiment to find the gaps that selectively block DNA translocase and thereby prevents nucleosomes from moving. Both ISWI and SWI/SNF were found using this technique to initiate remodeling from an internal site of the nucleosome about two helical turns from the dyad axis [13,57]. In order to further confirm these results other nucleosomes are made in which the DNA gap is placed at a position to block nucleosome movement or at position further away that will not block remodeling until the nucleosome has moved to the appropriate position on DNA. Gaps at one particular site are created by incorporating deoxyuridine in the oligonucleotide primers that are used to make the DNA and removed the same as before after incorporation into DNA. The number of uracil bases incorporated could be varied to create gaps of one or more nucleotides. Interference assays are also done with DNA nicks to find if DNA twist propagation is also required for nucleosome movement in addition to DNA translocation [14]. Randomly nicked DNA is created by digestion with an appropriate amount of DNaseI so as to create one nick per DNA.
3.2 Nucleosome mobilization and spacing: the two stories of ISW2 and ACF
3.2.1 Dimer versus monomer
There are two stories of the ISWI subfamily of remodelers with different outcomes and conclusions deriving from yeast ISW2 and human/Drosophila ACF. ISW2 and CHRAC/ACF are likely orthologs of each other given that they have similar subunit organization and functional activity. The difference between ACF and CHRAC is that CHRAC has two histone fold subunits which are absent in ACF. ISW2 also has two histone fold subunits like those found in CHRAC. The accessory subunit, ACF1 in human and Drosophila and Itc1 in yeast, has limited sequence homology that hints at them being similar. Both ISW2 and ACF move mononucleosomes to the center of DNA fragments and make discrete contacts near SHL2 [7,41]. What seems to set these two apart is that data supports recombinant ACF binding to nucleosomes as a dimer, while native ISW2 binds as a monomer. The evidence for an ACF dimer is that Snf2h, the catalytic subunit of ACF, alone exists as a monomer in solution but binds cooperatively as a dimer to nucleosomes as shown in a fluorescence based binding assays[41]. Gel shift assays with ISW2 and nucleosomes found that ISW2 does not bind cooperatively and has a Hill coefficient of only 1.1. In these assays the apparent affinity of Snf2h for nucleosomes was considerably lower than that for either ISW2 complex or the catalytic subunit Isw2 alone (630 nM vs. 11 and 54 nM, respectively) which may account for some of these underlying differences. Nucleosome mobilization assays with Snf2h or ACF shows cooperativity in the rate of nucleosome remodeling (Hill’s coefficient is ~ 1.8) consistent with the nucleosome binding data for Snf2h. Other data that points to ACF acting as a dimer is Snf2h can bind to both H4 tails as shown by electron paramagnetic resonance (EPR). Snf2h engages with only one H4 tail in presence of ADP, but engages both H4 tails in the presence of the ATP analog ADP·BeFx, representing the activated state. Of course the concern with this data is that the catalytic subunit alone will not behave the same as the complex with its accessory subunit. Supporting evidence for ACF is that ACF protects only one SHL2 on the linker DNA side of the nucleosome in presence of ADP, while both SHL2 positions were protected by ACF with ADP·BeFx thus reflecting that observed for Snf2h by EPR. These observations led the authors to propose that ACF binds nucleosome cooperatively and the ACF dimer actively engages both H4 tails when in the activated or transition state by binding ADP·BeFx. After hydrolysis of ATP one of the catalytic sites of the dimer stays associated with an H4 tail and the other releases. The ATPase domain bound to the longer linker DNA hydrolyzes ATP faster thus making it more prone to translocate along DNA than the other ATPase domain (Figure 3B). This difference in rates of the two bound translocase domains therefore account for its ability to sense linker DNA and to center nucleosomes on DNA or space nucleosomes [39], ACF samples linker DNA in this manner till it comes to equilibrium at midpoint and spaces nucleosomes evenly in this process.
Figure 3.
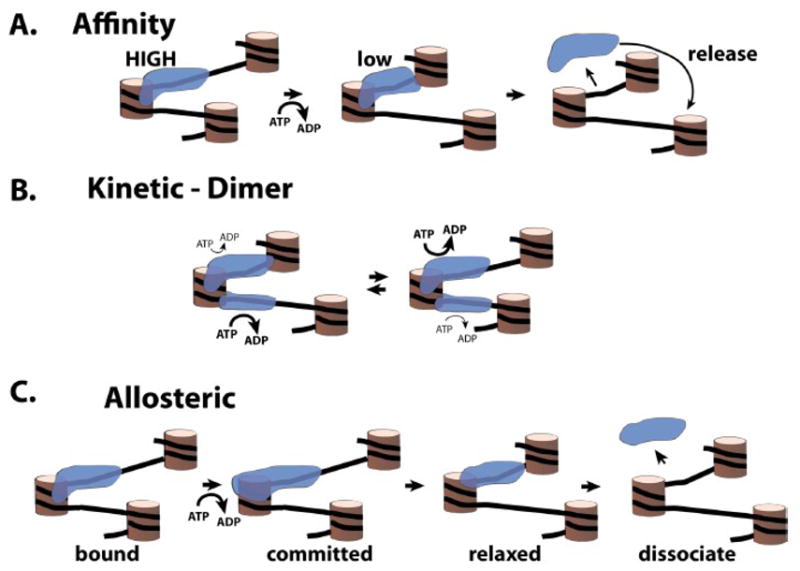
Three models for nucleosome spacing by ISWI.
(A). ISWI positions nucleosomes to particular DNA positions based on its affinity. ISWI has a higher affinity for those nucleosomes with longer linker DNA. During remodeling the linker DNA becomes shorter causing the affinity of ISWI to be reduced for the nucleosome. ISWI dissociates from the nucleosome due to the reduced affinity and searches out a new and higher affinity site. (B). Often a key part of the kinetic model is that ISWI binds to nucleosomes as a dimer with the rate of ATP hydrolysis varying from one monomer to the other based on the length of linker DNA length. When the linker DNA is longer the rate of ATP hydrolysis is increased (indicated by the thickness of the arrows and type size) and consequently the rate of nucleosome movement. The kinetic model has nucleosomes shuttling back and forth between the adjacent nucleosomes. (C). Three different conformational states may exist during ISWI remodeling. The first state is ISWI initially bound to nucleosomes without ATP. During ATP hydrolysis, ISWI makes additional interactions with nucleosomes that stabilizes the ISWI-nucleosome complex and makes it more processive (committed). When the linker DNA becomes too short ISWI undergoes a conformational change that reduces its interactions with the nucleosome (relaxed), causing ISWI to stop moving and dissociate from the nucleosome.
3.2.2 Why does ACF and ISW2 space nucleosomes?
The first model for how ISW2 is able to center nucleosomes on DNA and sense linker DNA for nucleosome spacing is based on the observation that the affinity of ISW2 for nucleosome is dependent on extranucleosomal DNA length (Figure 3A). The model is simply that ISW2 stops moving nucleosomes when it can no longer remain stably bound to nucleosomes because the length of extranucleosomal DNA to which ISW2 is bound becomes too short [7]. This effect would explain why when remodeling nucleosomes with ≤70 bp of extranucleosomal DNA, ISW2 and ACF tend to move nucleosomes generally well to one central position on DNA and when the extranucleosomal DNA is longer that the steady state position of remodeled nucleosomes is more dispersed. Similarly the reduced affinity of ACF and Chd1 for nucleosomes at particular positions dictated the end position to which nucleosomes were moved [58]. This model is attractive because it suggests that ISW2 could start with an array of nucleosomes in which there is some nucleosomes spaced far enough apart to present a high affinity site for ISW2 and serve as an entry point. Once ISW2 starts in this region it will continue to move nucleosomes potentially until the nucleosome spacing is such that ISW2 can no longer bind well to this region. ISW2 would consequentially move on to another region to continue the process elsewhere. This model is supported by a recent report with human Snf2H and Snf2L that utilizes the intrinsic fluorescence of these proteins [59].
The other model has more than one version, but the essence of it is that rather than just the binding affinity there is a conformational switch that occurs in the remodeler complex that is dependent on the length of linker DNA. The ACF dimer model discussed earlier is one version of this model in which both DNA translocases do not have the same conformation which accounts for one hydrolyzing ATP faster than the other (Figure 3B and reference [39]). Unfortunately, it has not been clear as to the nature of this conformational difference between the two translocase domains since both appear to bind H4 tail and nucleosomal DNA at SHL2 when ATP is bound. There is however more evidence for ISW2 having different conformational states that are dependent on extranucleosomal DNA length (Figure 3C). As extranucleosomal DNA length is shortened there is one key change in ISW2 interactions with nucleosomal or extranucleosomal DNA. DNA footprinting of ISW2-nucleosome complexes shows that the contact of Isw2 with SHL2 is highly dependent on extranucleosomal DNA and is lost when DNA is shortened from 35 to 20 bp[6]. This transition correlates well with a loss in nucleosome mobilizing activity observed for these same nucleosome constructs[12]. The other interactions of ISW2 with extranucleosomal DNA and just inside in the nucleosome near the entry site are all still present with only 20 bp of extranucleosomal DNA. These data point to communication between the proteins that interact with linker DNA and the ATPase domain being critical in this process of sensing linker DNA length. When linker DNA becomes too short during remodeling, the ATPase domain of ISW2 lifts off from nucleosomal DNA, stops translocating and moving nucleosomes in response to some allosteric switch from the protein(s) bound to linker DNA. The proteins or protein domains that might be involved in this communication are the accessory subunit Itc1 and those domains in the C-terminus of Isw2 that contact the linker DNA such as the SLIDE domain. Studies are ongoing to investigate the role of these in regulating the remodeling activity of ISW2.
While some may think it may have to be one or the other of these two basic models, it seems possible that both mechanisms can be at work in this process. If the conformational switch was only at work then ISW2 would likely get trapped on nucleosomes as an inactive complex once the linker DNA has been sufficiently shortened. After ISW2 move nucleosomes to the critical length of linker DNA, the ATPase domain disengages, and subsequently dissociates because of the reduced affinity of ISW2 for nucleosomes with shortened linker DNA. The dissociation of ISW2 would make this a much more catalytic process and cause it to have a fast turnover rate, all of which has been observed both in vitro and in vivo [7,59,60].
3.2.3 Template-commitment and intermediate states in ISW2 remodeling
While chromatin remodeling complexes bind and interact with specific nucleosomal regions, the molecular events that occur during remodeling are not well understood. Yeast ISW2 complex binds nucleosome at three distinct regions; linker DNA, the entry/exit site, and SHL2 site in the absence of ATP. Binding of ATP tightened ISW2 interactions at all three sites without increasing the overall binding affinity of the complex to nucleosome. Upon hydrolysis of ATP, ISW2 contacts increased to an entire DNA gyre from the entry exit site to SHL2 as observed by rapid hydroxyl radical DNA footprinting. At even the fastest time point it is difficult to get a homogenous population of nucleosome that have moved the same distance from the original location making it difficult to interpret the footprinting data. This problem was avoided using nucleosomes with gaps 24/25 or 35 bp from the dyad axis to block nucleosome movement and DNA translocation at different stages in remodeling [13]. Footprinting of arrested ISW2 produced similar results where an entire gyre of DNA was protected whether blocked at the very beginning or after a short movement of 10 bps. A 3–4 bp region four helical turns from the dyad axis on the opposite DNA gyre was found to be protected in the arrested complexes only in the presence of ATP. This protection moves 10 bp to a new site when nucleosomes were arrested using a gap at 35 bp from the dyad axis. It appears that ISW2 binds this 3–4 bp region in a skewed manner away from the other gyre of DNA located above where the DNA translocase is bound to the other DNA gyre. These ATP hydrolysis induced conformational changes in ISW2 cause it to form a complex that is resistant to competition by competitor DNA or nucleosomes and is referred to as template-committed complex[11]. A similar kind of template commitment by Drosophila ACF complex towards DNA was observed previously during nucleosome assembly [61].
The conformation of ISW2 changes with ATP hydrolysis as it extends its interactions across an entire gyre of nucleosomal DNA. The subunits of ISW2 involved in these additional interactions were investigated using site-directed DNA crosslinking. There was an increase in crosslinking of Isw2 17 to 18 bp from the dyad axis after remodeling was initiated with ATP and translocation blocked with gaps. DNA crosslinking revealed that histone fold subunit Dpb4 interacts with nucleosomal DNA 40 bp from the dyad axis, about 10–20 bp from where the DNA translocase is bound[11]. Movement of nucleosomes by 10 bp caused the crosslinking of Dpb4 to also shift by 10 bp and demonstrated that along with Isw2 the Dpb4 subunit provides additional interactions during ATP hydrolysis to help form a committed complex. During remodeling, Dpb4 tracks along nucleosomal DNA ~15 bp in front of the translocase in the same direction and may help in destabilizing the histone-DNA interaction to facilitate remodeling[11].
When considering template commitment and the role of linker DNA in regulating ISW2 remodeling activity, it is important to view these as interconnected. It is possible that in order for ISW2 to disengage its ATPase domain from SHL2 that it will need to convert from a template-committed complex to a non-committed or relaxed complex and that these are interconnected (Figure 3C). Once ISW2 starts moving nucleosomes it is stably bound as a template-committed complex, but as soon as it moves the nucleosome close to an adjoining nucleosome, the ATPase domain dissociates from SHL2. Dissociation of the ATPase domain could then signal to change from a template-committed to a non-committed complex preparing the way for the complex to be effectively released. It is however not certain as to the order in which these events occur. Evidence in support of this idea is that ISW2 is unable to form a template-committed complex in the absence of the histone H4 N-terminal tail even though ISW2 can still hydrolyze ATP and remodel nucleosomes albeit at a significantly slower rate[11].
In conclusion it seems that ISWI remodelers have developed intricate ways in which they sense linker DNA length and modulate their catalytic activity accordingly. There are already indications that these mechanisms will vary from one family member to another such as the differences between ISW2 and ISW1a. In time and with more effort we suspect that there will be other variations on this theme than those discussed here.
3.3 Nucleosome disassembly by SWI/SNF
3.3.1 Unraveling nucleosomes is a property unique to SWI/SNF
Do the fundamental difference of ISW2 and ISW1a remodeling being dependent on linker DNA while SWI/SNF is not reflect something important about their roles in the cell? Because SWI/SNF can move nucleosomes even when there is no linker DNA it has an intrinsic ability to more aggressively disrupt nucleosome structure than other remodelers that do require linker DNA. The simplest example of this is that SWI/SNF can unravel mononucleosomes or in other words move the DNA far enough on the histone octamer that it leaves a ~1/3 of the octamer free of DNA to which it is normally bound [10,62]. A direct outcome of this is that one of the H2A/H2B dimers could be more readily displaced [63]. This activity is not as prevalent as might be expected when one H2A/H2B dimer is no longer being contacted by DNA and occurs only with a minor fraction of the nucleosomes. The likely explanation for H2A/H2B dimer displacement not occurring is because SWI/SNF also actively places DNA back onto this exposed surface of the nucleosome and thereby stabilizes the association of the dimer with the rest of the octamer [10]. Another reason why SWI/SNF may be able to unravel nucleosomes and ISW2/1a cannot is the fundamental difference in how these complexes interact with nucleosomes. As described earlier SWI/SNF appears to have more extensive interactions with histones than ISW2 which could provide a strong anchor to nucleosomes that would be independent of nucleosomal DNA (Figure 2). Without this anchor it would be impossible for the DNA translocation domain alone bound to nucleosomal DNA to have sufficient force to unravel or mobilize nucleosomes. If there was no way to fix the ATPase domain in relation to the nucleosome then the ATPase domain would merely travel along nucleosomal DNA till it reaches the entry site without changing the nucleosome translational position.
3.3.2 Structural considerations of SWI/SNF
These histone interactions with SWI/SNF most likely occur with one face of the nucleosome. The data that support this idea are the cryo-electron microscopy of free SWI/SNF and the DNA footprint of nucleosomes bound to SWI/SNF. The 3-dimensional structure of SWI/SNF differs from that observed for RSC[9,64,65,66]. The distinctive feature of SWI/SNF is a large trough on its surface that is of the correct dimensions to accommodate a nucleosome (Figure 4). The trough has three walls; a high and low side walls and one back wall. The DNA footprint of nucleosomes bound to SWI/SNF recruited by Gal4-VP16 appears to coincide with the model of the nucleosome bound to the trough region of SWI/SNF. The dimensions of the trough are such that the nucleosome would of necessity have to be placed with the DNA gyres at the base of the trough and the DNA free region of the histone octamer facing the high and low walls of the trough. The DNA footprint suggests that the nucleosome is likely tilted towards the high wall such that the DNA gyre closest to the high wall is protected while the other DNA gyre would be more exposed. Based on this model the part of SWI/SNF contacting the open face of the histone octamer would be that in the high wall of the trough. Combining aspects of this model with DNA and histone crosslinking data one can make tentative assignments as to where the DNA translocase domain would be located as well as the Snf2, Snf5, Swp82, Swp29 and other subunits of SWI/SNF in the 3-D structure of SWI/SNF based on crosslinking data [9].
Figure 4.
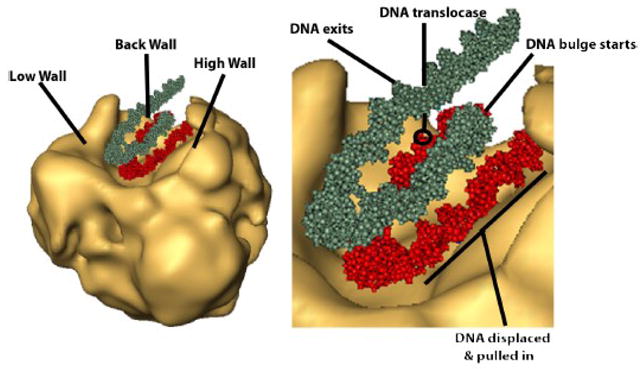
Structural model of the SWI/SNF-nucleosome complex.
Shown in gold is the structure of SWI/SNF as observed by cryo-electron microscopy and the nucleosome is fitted into the trough based on DNA footprinting and site-directed DNA and histone crosslinking data. Only the DNA of the nucleosome is shown for ease in pointing out key features in the structure and the proposed movements of the nucleosomal DNA. DNA in red is that part of the nucleosomal DNA in close contact with SWI/SNF while that in gray is not.
A benefit of this model is that it provides certain structural parameters to consider for how nucleosomal DNA moves around the histone octamer during remodeling. The model predicts that the DNA gyre proximal to the high wall is the DNA that is pulled into the nucleosome towards the dyad axis (Figure 4). This implies that the region which becomes “unraveled” is that part of the nucleosome associated with the high wall and therefore might be protected or shielded. Additional contacts with SWI/SNF might help to keep the H2A/H2B dimer associated with the nucleosome while DNA is being stripped off this region and even sterically prevent it from leaving. As DNA is pushed inside the nucleosome, DNA bulges might be created behind the DNA translocase as it moves and would be in the area not bound by SWI/SNF. The DNA bulge would continue to migrate around the nucleosome in the direction where there is less steric hindrance and away from the DNA translocase which would be proximal to the low wall. Changes in histone-DNA contacts at early stages in nucleosome movement catalyzed by SWI/SNF have been examined by site-directed crosslinking as described in section 3.1.1. The contacts of residue 53 of histone H2B with DNA is 54 bp from the dyad axis in nucleosomes. Initially the H2B contact with the DNA gyre closest to the high wall is lost upon SWI/SNF remodeling consistent with this DNA being pulled in and disrupting histone-DNA contacts near the entry site[13]. After a short period of time DNA is rebound to this surface, but has moved 52 bp from where it was initially bound. It appears that the H2B site is not bound to DNA for an appreciable length of time while remodeling is occurring and only when SWI/SNF reaches the DNA end and cannot translocate any further does DNA reassociate.
One other implication from the SWI/SNF structure and model is that SWI/SNF can only bind one nucleosome at a time. This idea has been proven to be correct by binding SWI/SNF to dinucleosomes. First, the affinity of SWI/SNF for dinucleosomes is not greater than that for mononucleosomes and argues against any cooperative binding effects. Second, when the two nucleosomes are spaced close together the affinity of SWI/SNF is greatly reduced for these dinucleosomes suggesting a steric hindrance problem. This last point is particularly important because it shows that SWI/SNF is not able to directly move nucleosomes close together or overlapping because there would be a steric clash in the active site of SWI/SNF. Some data however suggested that RSC could make overlapping nucleosomes when remodeling arrays[67]. Based on the structural constraints the overlapping nucleosomes are likely formed after the RSC is removed and the nucleosomes collapse into this type of structure. So what happens when SWI/SNF is remodeling one nucleosome and it comes into close proximity to an adjoining nucleosome? The answer to this question may really get more to the essence of how SWI/SNF functions inside the cell.
3.3.3 Nucleosome disassembly by SWI/SNF requires two adjoining nucleosomes
In a recent study SWI/SNF was found to displace nucleosomes only when using substrates that had at least two nucleosomes per DNA template[38]. These data showed that SWI/SNF systematically disassembles nucleosomes by first removing one histone H2A/H2B dimer followed by a slower step of the complete eviction of one nucleosome. What was surprising is that SWI/SNF did not disassemble nucleosomes when there were only one nucleosome per template and that in general there was always one nucleosome left behind after the others had been disassembled. This data reflected earlier results showing that nucleosomes were disassembled in vivo at the PHO5 promoter in a manner that also required SWI/SNF and left on the average one nucleosome remaining[68,69,70,71]. Mapping of nucleosome translational position with the techniques described earlier found that when SWI/SNF was recruited by Gal4 VP16 that it moved preferentially the most proximal nucleosome away from the Gal4 binding site and towards the adjacent nucleosome. As it did so the adjoining nucleosome became more destabilized as shown by DNA methylation protection assays or MAP-IT[38]. These data suggest that the nucleosome which is not being mobilized by SWI/SNF is the one that becomes displaced and that the nucleosome which is retained on DNA is the one bound to the active site of SWI/SNF.
So what causes the one nucleosome to be displaced from DNA? The characteristic loss of first one H2A/H2B dimer followed by the complete nucleosome is reminiscent of the action of RNA polymerase on nucleosomes[72,73]. SWI/SNF as a powerful DNA translocation machine is suggested to pull DNA from the edge of the adjoining nucleosome, first releasing one H2A/H2B dimer. As it continues to pull on the DNA and move it from the adjacent nucleosome it will eventually disrupt the H3/H4 tetramer interactions with DNA and completely disassemble an entire nucleosome. After removal, the other nucleosome is freely moved along DNA with no further reduction in nucleosomes bound to DNA unless it encounters another nucleosome. When SWI/SNF is removed at different stages in the process the nucleosomes can collapse into what appear to be overlapping nucleosomes, but it doesn’t seem likely that such is actually formed during the process with SWI/SNF still associated since it is not possible to have two nucleosomes simultaneously bound to the active site. These data provide a more complete view of the context of SWI/SNF in vivo in which it will encounter nucleosomal arrays and thereby help create semi-nucleosomal free regions. The ability to pull DNA off a nucleosome so that it can reel in the adjacent nucleosome close enough to the SWI/SNF to compete for binding to DNA is a feature unique to SWI/SNF and RSC that is not found in other remodelers thus far. These observations explain how not having linker DNA dependence can be beneficial in vivo for being able to remove nucleosomes and points to how SWI/SNF stands out in the cell from other remodelers.
Conclusions
We can see there are clear differences between the ISWI and SWI/SNF subfamilies of ATP-dependent chromatin remodeler that help explain their differing roles in the cell. Many of these differences were not fully appreciated until sufficient biochemical characterization had been done with these complexes. It is expected that more differences will be uncovered even now between members of the same subfamily such as ISW2 vs. ISW1a or SWI/SNF vs. RSC. This task becomes even more demanding when considering how many different forms of human SWI/SNF exist and the potential underlying properties of each that make them distinctive. There are other remodelers such as the CHD/Mi-2 and INO80/SWR1 subfamilies that will also add to the variety of operations and likely have key differences from those so far observed for the ISWI and SWI/SNF subfamilies. Only as we learn the intrinsic properties of each will we be able to better understand their roles in gene regulation, chromosome maintenance and repair, replication, and developmental control.
Research Highlights.
The large number of ATP-dependent chromatin remodelers is a reflection of their different modes of operation. It is important to understand how different each remodeler is from another in order to understand their in vivo roles.
Each remodeler has three ways of interacting with nucleosomes. We discuss how these interactions are different between SWI/SNF and ISW2 and the protein subunit/domains that are have been identified so far to be involved. These interactions heavily influence the efficiency of remodeling and the ultimate outcome of remodeling.
The models for how the ISWI remodelers “sense” linker DNA length and space nucleosomes is discussed. These are the affinity, kinetic, and allosteric models. We discuss the data supporting each model and the underlying similarities in these models.
The mechanism of SWI/SNF remodeling of nucleosomes is reviewed with a particular focus on its action in nucleosomal arrays and nucleosome disassembly.
The advantages and limitations of bulk and single molecule techniques used in studying the mechanism of chromatin remodeling are highlighted. We also compare those techniques that rely on photoreactive groups versus those with fluorescent reporters.
Acknowledgments
We would like to thank Jim Persinger and Rashmi Prasad for suggestions and critical reading of the manuscript. This work was supported by the National Institutes of Health (GM 48413 and GM70864).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 3.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 9.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-Nucleosome Complex. Mol Cell Biol. 2008 doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 11.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 14.Lorch Y, Davis B, Kornberg RD. Chromatin remodeling by DNA bending, not twisting. Proc Natl Acad Sci U S A. 2005;102:1329–1332. doi: 10.1073/pnas.0409413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 16.Dang W, Kagalwala MN, Bartholomew B. The Dpb4 subunit of ISW2 is anchored to extranucleosomal DNA. J Biol Chem. 2007;282:19418–19425. doi: 10.1074/jbc.M700640200. [DOI] [PubMed] [Google Scholar]
- 17.Ebralidse KK, Grachev SA, Mirzabekov AD. A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nature. 1988;331:365–367. doi: 10.1038/331365a0. [DOI] [PubMed] [Google Scholar]
- 18.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 19.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alenghat T, Yu J, Lazar MA. The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor. EMBO J. 2006;25:3966–3974. doi: 10.1038/sj.emboj.7601280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2002;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2010 doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 27.Syed SH, Boulard M, Shukla MS, Gautier T, Travers A, Bednar J, Faivre-Moskalenko C, Dimitrov S, Angelov D. The incorporation of the novel histone variant H2AL2 confers unusual structural and functional properties of the nucleosome. Nucleic Acids Res. 2009;37:4684–4695. doi: 10.1093/nar/gkp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyen CM, Montel F, Gautier T, Menoni H, Claudet C, Delacour-Larose M, Angelov D, Hamiche A, Bednar J, Faivre-Moskalenko C, Bouvet P, Dimitrov S. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shukla MS, Syed SH, Goutte-Gattat D, Richard JL, Montel F, Hamiche A, Travers A, Faivre-Moskalenko C, Bednar J, Hayes JJ, Angelov D, Dimitrov S. The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemieux K, Larochelle M, Gaudreau L. Variant histone H2A.Z, but not the HMG proteins Nhp6a/b, is essential for the recruitment of Swi/Snf, Mediator, and SAGA to the yeast GAL1 UAS(G) Biochem Biophys Res Commun. 2008;369:1103–1107. doi: 10.1016/j.bbrc.2008.02.144. [DOI] [PubMed] [Google Scholar]
- 31.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 35.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 36.Gangaraju VK, Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol. 2007;27:3217–3225. doi: 10.1128/MCB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 40.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzgerald DJ, DeLuca C, Berger I, Gaillard H, Sigrist R, Schimmele K, Richmond TJ. Reaction cycle of the yeast Isw2 chromatin remodeling complex. Embo J. 2004;23:3836–3843. doi: 10.1038/sj.emboj.7600364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 alters the remodeling strategy of SNF2h. J Biol Chem. 2006;281:28636–28647. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
- 44.He X, Fan HY, Garlick JD, Kingston RE. Diverse regulation of SNF2h chromatin remodeling by noncatalytic subunits. Biochemistry. 2008;47:7025–7033. doi: 10.1021/bi702304p. [DOI] [PubMed] [Google Scholar]
- 45.Kassabov SR, Bartholomew B. Site-directed histone-DNA contact mapping for analysis of nucleosome dynamics. Methods Enzymol. 2004;375:193–210. doi: 10.1016/s0076-6879(03)75013-7. [DOI] [PubMed] [Google Scholar]
- 46.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang JG, Narlikar GJ. FRET-based methods to study ATP-dependent changes in chromatin structure. Methods. 2007;41:291–295. doi: 10.1016/j.ymeth.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouazoune K, Miranda TB, Jones PA, Kingston RE. Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res. 2009;37:5279–5294. doi: 10.1093/nar/gkp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 51.Strick TR, Croquette V, Bensimon D. Homologous pairing in stretched supercoiled DNA. Proc Natl Acad Sci U S A. 1998;95:10579–10583. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lia G, Indrieri M, Owen-Hughes T, Finzi L, Podesta A, Milani P, Dunlap D. ATP-dependent looping of DNA by ISWI. J Biophotonics. 2008;1:280–286. doi: 10.1002/jbio.200810027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain SS, Tullius TD. Footprinting protein-DNA complexes using the hydroxyl radical. Nat Protoc. 2008;3:1092–1100. doi: 10.1038/nprot.2008.72. [DOI] [PubMed] [Google Scholar]
- 57.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 58.Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Langst G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci U S A. 2007;104:15635–15640. doi: 10.1073/pnas.0702430104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erdel F, Schubert T, Marth C, Langst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci U S A. 2010;107:19873–19878. doi: 10.1073/pnas.1003438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fyodorov DV, Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- 62.Flaus A, Owen-Hughes T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol Cell Biol. 2003;23:7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asturias FJ, Ezeokonkwo C, Kornberg RD, Lorch Y. Electron microscopic analysis of the RSC chromatin remodeling complex. Methods Enzymol. 2004;376:48–62. doi: 10.1016/S0076-6879(03)76004-2. [DOI] [PubMed] [Google Scholar]
- 65.Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci U S A. 2007;104:4913–4918. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 70.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 71.Korber P, Luckenbach T, Blaschke D, Horz W. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol Cell Biol. 2004;24:10965–10974. doi: 10.1128/MCB.24.24.10965-10974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem Sci. 2004;29:127–135. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 73.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]


