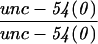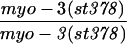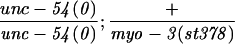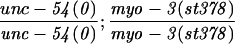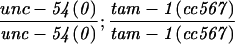Abstract
Context-dependent gene silencing is used by many organisms to stably modulate gene activity for large chromosomal regions. We have used tandem array transgenes as a model substrate in a screen for Caenorhabditis elegans mutants that affect context-dependent gene silencing in somatic tissues. This screen yielded multiple alleles of a previously uncharacterized gene, designated tam-1 (for tandem-array-modifier). Loss-of-function mutations in tam-1 led to a dramatic reduction in the activity of numerous highly repeated transgenes. These effects were apparently context dependent, as nonrepetitive transgenes retained activity in a tam-1 mutant background. In addition to the dramatic alterations in transgene activity, tam-1 mutants showed modest alterations in expression of a subset of endogenous cellular genes. These effects include genetic interactions that place tam-1 into a group called the class B synMuv genes (for a Synthetic Multivulva phenotype); this family plays a negative role in the regulation of RAS pathway activity in C. elegans. Loss-of-function mutants in other members of the class-B synMuv family, including lin-35, which encodes a protein similar to the tumor suppressor Rb, exhibit a hypersilencing in somatic transgenes similar to that of tam-1 mutants. Molecular analysis reveals that tam-1 encodes a broadly expressed nuclear protein with RING finger and B-box motifs.
Keywords: TAM-1, Chromatin Silencing, RING finger, C. elegans, Rb, RAS Pathway
Eukaryotes have evolved a number of concerted mechanisms that allow extended domains of individual chromosomes to be rendered inaccessible to the transcription apparatus. The use of such gene silencing mechanisms requires two different mechanistic operations. First, a cell must choose specific regions of the genome for silencing. Second, a silent state must be enforced. The most surprising feature of chromatin silencing (and the feature that distinguishes silencing from more localized modulation of gene expression) is the substantial distance at which silencing effects are seen. One striking example of this is the inactivation of the human X chromosome, which extends from a single locus (Xist) throughout a chromosome of 108 bases (Riggs and Porter 1996).
The silencing apparatus can apparently be recruited by a variety of distinct features at the DNA sequence level. In many cases, a specific sequence element in DNA can be identified as necessary and sufficient for silencing of a surrounding domain. Examples of silencing effects with a clear nucleation point include telomere and mating-type silencing in yeast (Grunstein 1998; Haber 1998). In these cases, binding of a multiprotein silencing complex to the cis-silencing site is thought to nucleate an extended change in local chromatin structure.
Heterochromatin in many organisms includes regions of highly repetitive structure, and extensive speculation has attempted to link this repetitive structure to silencing properties of heterochromatin (Henikoff 1998). A potentially related class of silencing effects has been inferred from studies with transgenic fungi (Selker 1997), plants (Assaad et al. 1993; Ye and Signer 1996) and animals (Garrick et al. 1998). In these cases, a sequence that can be active as a single-copy locus in the genome becomes partially or completely inactive when repeated. Strong effects generally require that the repeats are present at a single site in the genome, often as a tandem repeat. The identification of tandemly repeated sequences as appropriate for silencing could conceivably occur by two distinct mechanisms. In the first model, a weak cis-silencing element might be present in each member of the tandem repeat. In this case, the repetition would initiate silencing by juxtaposing several copies of the weak cis-silencing element. Alternatively, the organism could possess a mechanism allowing repeated structures to be recognized by virtue of repeated character and not of any specific sequence. The latter seems to be the situation in Neurospora (Selker et al. 1987; Cambareri et al. 1989; Selker 1997) and Ascobolus (Goyon and Faugeron 1989; Rossignol and Faugeron 1994), whereas the limited number of transgenic constructs tested for repeat-dependent silencing in the other systems leave both models open.
Despite the apparently conserved ability to recognize certain repeated sequences in DNA, there are differences between biological systems in the consequences of repetition. Ascobolus, Neurospora, mammals, and plants respond to repeated DNA structures by localized methylation of DNA (although Neurospora has the additional consequence of promoting point mutations in the DNA). Repeated structures in DNA in Drosophila produce variegated or mosaic expression without any methylation of DNA (Dorer and Henikoff 1994,1997).
In Caenorhabditis elegans, transgenes are generally carried as heritable extrachromosomal structures consisting of long tandem arrays with several hundred copies of the injected DNA (simple arrays) (Stinchcomb et al. 1985; Mello et al. 1991). Although expression (and phenotypic rescue) by such tandem arrays is readily observed, a difference in transcriptional activity compared with the endogenous locus is seen. The activity is frequently mosaic and expression levels (per copy) are frequently much lower than for the endogenous gene (Mello and Fire 1995). This difference is not a simple consequence of the extrachromosomal nature of the simple arrays; transgenes can be integrated into the chromosome by γ-irradiation producing stable transgenic lines, and these integrated arrays can still exhibit mosaic expression (e.g., Krause et al. 1994; J. Hsieh and A. Fire, unpubl.). Another factor that may affect the expression of simple arrays is the repetitive character inherent in the long tandem array. Consistent with this hypothesis, silencing of certain germ-line and somatic expression constructs can be relieved when the plasmid DNAs are cotransformed with an excess of C. elegans genomic DNA as carrier, producing a more complex array (Kelly et al. 1997; S. Xu, and A. Fire, unpubl.). To avoid specific assumptions about the role of particular sequences or general repeated structures in the silencing of tandem arrays in C. elegans, we will use the term context-dependent gene silencing. This paper describes the genetic identification and molecular characterization of one component that acts to attenuate the context-dependent silencing mechanism affecting tandem array transgenes in C. elegans.
Results
Isolation of modifiers of context-dependent gene expression
To identify factors responsible for context-dependent gene silencing, we screened for mutations that altered the activity of transgenes present in tandemly repeated arrays. We started with a stably integrated myo-3::gfp transgene expressing uniformly in all bodywall muscles (ccIs4251; Fire et al. 1998b). After mutagenesis with ethylmethanesulfonate (EMS), ∼4500 mutagenized gametes were screened. Four putative mutant lines were recovered in which most, if not all, the animals have reduced-GFP expression (Fig. 1A, B). These strains were outcrossed to determine whether the reduced-GFP expression resulted (1) from changes in the transgene array, or (2) from (potentially interesting) mutations in endogenous genes. One mutation appeared to be dominant and linked to the transgene array; this could have reflected a change in copy number or in the structure of the array and was not analyzed further.
Figure 1.

tam-1 effects on a repetitive myo-3::gfp transgene. An integrated myo-3::gfp transgene was examined in wild-type and tam-1(cc567) mutant backgrounds. (A) Wild-type and (B) tam-1(cc567) animals photographed under fluorescent illumination showing GFP in bodywall muscles (the array produces GFP in both nuclear and mitochondrial compartments). Scale bars, 55 μm. Quantitative measurements of GFP fluorescence in wild-type and tam-1(cc567) mutant backgrounds. Differences in fluorescence level were most striking in the central region of the animal; thus quantitations were carried out in bodywall muscle nuclei in this region. Eight animals were analyzed for each genetic background. Fluorescence was quantitated for six nuclei in each animal with a Nikon U-III multipoint sensor system. Linearity of fluorescence measurements in the range of assay was confirmed by a series of neutral density filters. (wild-type mean= 140.9, wild-type (s.e.m.)= 5.9; tam-1 mean = 7.2, tam-1 s.e.m. = 0.4). A second measure of tam-1 effects on the expression of a repeated transgene is observed with a lin-3 transgene (syIs1). The average induction of vulval precursor cells (VPCs) caused by the expression of syIs1 in wild type is 5.8 VPCs induced (n = 20), however, in tam-1(sy272), there were 3.2 VPCs induced (n = 1 0) (data not shown).
The three reduced-GFP expression strains were shown to contain chromosomal mutations (cc566, cc567, and cc587). All three were recessive and showed a temperature-sensitive effect with the decrease in myo-3::gfp expression (relative to the parent line), most pronounced at 25°C. At 16°C, the mutations had no observable effect on GFP levels. Because the mutant effects were temperature sensitive, all of the experiments were carried out at 25°C. Genetic mapping placed the three mutations on chromosome V. Complementation tests showed the three mutations to be allelic. The corresponding gene has been named tam-1 (tandem-array modifier). The magnitude of the tam-1(cc567) effect on the myo-3::gfp transgene was estimated by optical quantitation of fluorescence to be ∼18-fold (Fig. 1C).
The endogenous myo-3 gene is not significantly affected by tam-1
Alteration in the expression of the endogenous myo-3 locus in a tam-1 background was first tested genetically (Table 1). In an unc-54 null mutant background, loss of one copy of the endogenous myo-3 locus dramatically increases the severity of the muscle phenotype (Waterston 1989). unc-54; tam-1 double mutants do not show a more severe phenotype (compared with unc-54 alone), indicating that expression of the endogenous myo-3 locus in a tam-1 background is within twofold of wild type. Immunofluorescence experiments with a MYO-3 antibody similarly showed a lack of difference between and tam-1 mutant animals (data not shown).
Table 1.
Assays for tam-1 modulation of endogenous myo-3 function
| Genotype
|
Phenotype
|
Viability
|
Reference
|
|
|---|---|---|---|---|
|
paralyzed as adult | yes | Brenner (1974); Epstein et al. (1974) | |
|
L1 arrest | no | Waterston (1989) | |
|
paralyzed after hatch; reaches adulthood | yes | Waterson (1989) | |
|
paralyzed as embryo | no | Waterston (1989) | |
|
paralyzed as adult [looks like unc-54(0) alone] | yes | this work |
Transgene silencing in tam-1 mutants depends on chromosomal context
We took advantage of the ability to manipulate the transgene context as a means to investigate the apparent ability of tam-1 to distinguish between transgene arrays and endogenous loci. We used myo-3 as a target for these assays, as earlier experiments (above) had shown that myo-3::gfp transgenes were affected, whereas the endogenous locus was not. The original ccIs4251 transgene had been produced under a standard set of conditions that produce a highly repetitive tandem array carrying several hundred copies of the injected plasmid DNAs. We considered that differences between endogenous and transgene loci could have reflected (1) a difference in the genomic context of the endogenous gene and chromosomal locus, or (2) a difference between the gfp fusion and endogenous gene. To distinguish between these possibilities, we examined a novel type of transgene (Kelly et al. 1997) in which the incoming plasmid DNA was cotransformed with an excess of genomic DNA from C. elegans. The latter type of transgene (called a complex array) has a less repetitive structure, and might thus be a closer approximation to the native chromosomal context. Three new myo-3::gfp transgenes were produced in which the identical promoter was fused to gfp, but, in this case, was diluted 50- to 100-fold with fragmented genomic DNA. These complex arrays were strongly expressed and showed no difference in tam-1(+) versus tam-1(−) genetic backgrounds (Fig. 2A,B). Taken with the strong effect on simple arrays, these data indicate that tam-1 effects on transgene activity are context dependent.
Figure 2.
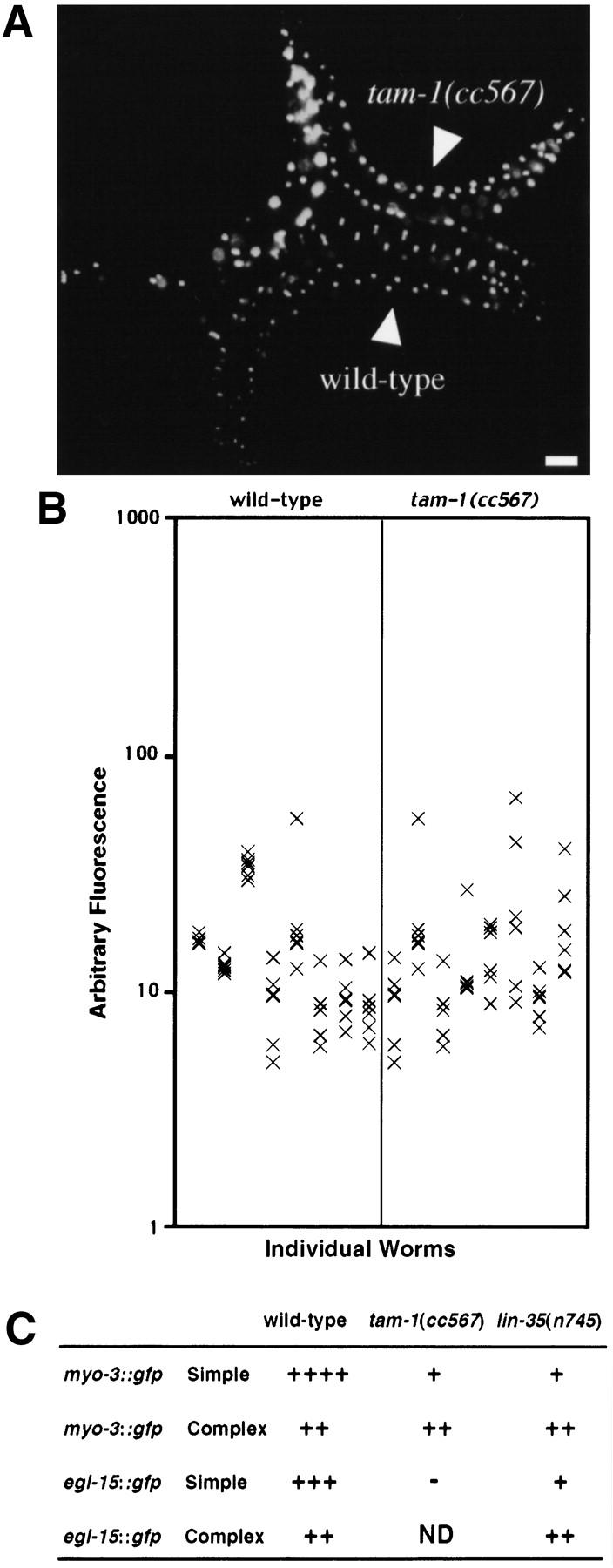
Assessment of tam-1 effects in a nonrepetitive context. A nonrepetitive transgene array (ccEx6172) was compared in wild-type and tam-1(cc567) mutant backgrounds. (A) Wild-type and tam-1(cc567) animals photographed under fluorescent illumination showing GFP in bodywall muscles (the array produces GFP in nuclei). Scale bar, 55 μm. (B) Quantitation of fluorescence was carried out in six nuclei in the central region of the body for each of eight animals as in Fig. 1B. (wild-type mean = 15.2, wild-type s.e.m. = 1.4; tam-1 mean = 15.6, tam-1 s.e.m. = 1.7). (C) Analysis of repetitive (simple) and nonrepetitive (complex) transgene activity in wild-type, tam-1(cc567) and lin-35(n745) backgrounds. Transgenes are as described in Materials and Methods.
tam-1 mutants have a silencing effect on many different repetitive transgenes
The overexpression of a repetitive lin-3 transgene (syIs1) results in a fully penetrant multivulva (Muv) phenotype and provides an alternative screen for decreased transgene activity (lin-3 encodes a diffusible signal for vulval induction, Horvitz and Sulston 1980; Hill and Sternberg 1992) . From a screen of 73,000 EMS-mutagenized gametes, 15 additional alleles of tam-1 were recovered. These mutants mapped to chromosome V and failure to complement tam-1(cc567) was observed. A representative allele of this group (sy272) also decreased the activity of the repetitive myo-3::gfp transgene, whereas a representative of the original group (cc567) showed the ability to decrease the activity of the lin-3 transgene. As with the effects on myo-3::gfp, the tam-1(sy272) effect on a lin-3 transgene was temperature dependent: strong suppression was seen at 25°C, whereas only limited suppression was observed at 16°C. The distinct expression patterns of lin-3 (somatic gonad) and myo-3 (bodywall muscle) suggested that tam-1 might have effects on gene expression in many or all tissues. To test this hyposthesis, the expression of additional repetitive transgenes was analyzed in tam-1 mutants (Table 2A). We observed a decrease in the expression of a number of diverse reporter constructs with different control signals and patterns of tissue specificity (including mesodermal, endodermal, and neuronal tissues). In addition, effects were seen with both lacZ and gfp transgenes.
Table 2.
tam-1 effects on reporter transgene activity
|
|
Construct
|
Relative intensities of reporters
|
||
|---|---|---|---|---|
| expression pattern
|
wild-type
|
tam-1
|
||
| A | myo-3::gfp | body-wall muscle vulval muscle | ++++ +++ | + + |
| unc-54::gfp | body-wall muscle | +++ | + | |
| hlh-1::gfp | body-wall muscle | ++ | − | |
| myo-2::gfp | pharyngeal muscle | ++++ | ++ | |
| F22B7.9::gfp | gut | ++ | − | |
| egl-15::gfp | vulval muscle | +++ | − | |
| gut | + | − | ||
| neurons near head | + | − | ||
| lin-3::lacZ | anchor cell | +++ | + | |
| gpa-1::lacZ | head neurons | +++ | + | |
| plasmid neurons | +++ | + | ||
| lin-11::lacZ | 2° vulval lineages | ++ | − | |
| VC motor neurons | +++ | − | ||
| ventral uterine π cells and their progeny | ++ | − | ||
| mab-5::gfp | posterior region neurons near tail | + + | −− | |
| B | let-858::gfp | all somatic nuclei | +++ | +++ |
| rps-5::gfp | all somatic nuclei | +++ | +++ | |
| glr-1::gfp | subset of neurons | +++ | +++ | |
| unc-119::gfp | many neurons | +++ | +++ | |
Expression of tandemly repeated transgenes in wild-type and tam-1 mutants. GFP transgenes were examined in tam-1(cc567), and lacZ transgenes were examined in tam-1(sy272). Number of plus signs (+) indicate relative intensity of GFP fluorescence or lacZ signal. (−) No observable signal. In cases where no expression was observed in tam-1 mutants, an outcross was performed to confirm the reappearance of reporter expression. (A) tam-1 effects were seen on a number of different reporter constructs. (B) Other transgenes showed little or no effect in tam-1 mutants. Additional myo-3::gfp and rps-5::gfp transgenes (independently derived) were tested in wild-type and tam-1(cc567) backgrounds and showed similar results (data not shown).
As shown in Table 2B, not all reporter transgenes were strongly affected by tam-1. No obvious characteristic was shared between the transgenes in which we saw no tam-1 effect. It is interesting, however, that several ubiquitously expressed transgenes (let-858, rps-5) appeared to be unaffected. Two neuronally expressed gfp fusions were also in this category. This may not be a universal property of neuronal expression: tam-1 had strong effects on expression of egl-15::gfp, lin-11::lacZ, gpa-1::lacZ, and mab-5::gfp in neuronal tissues.
tam-1 effects on endogenous genes
Although tam-1 mutants were fertile, they were not wild type. Brood sizes were considerably reduced. In three independent alleles (cc567, cc587, and sy272), average brood sizes at 25°C ranged from 55.8–105.1, compared with 204 for wild-type and 187 for the parental (gfp fusion) strain. tam-1 mutants also showed defects in X chromosome segregation, as evidenced by a high incidence of males (XO animals) derived from XX parents. Male frequencies for the three alleles varied between 0.5% and 8.6% compared with the <0.2% incidence of spontaneous males in wild-type animals and in the parent strain PD4251.
Sensitized genetic assays were used to test for tam-1 effects on several endogenous genes (Table 3). No effects (i.e., less than a twofold difference) was seen for unc-22 (data not shown), or for gain-of-function alleles of let-23 and let-60. In a number of other sensitized genetic assays, we found modest enhancement by tam-1, in each of these cases (lin-2, let-2, lin-3), a partial loss-of-function allele gave a more severe phenotype in a tam-1(−) genetic background. For example, tam-1 reduced vulval differentiation in a lin-3 hypomorphic strain e1417 to the level of the e1417/null hemizygote; thus, there was an approximately twofold effect. Similar arguments indicated that the magnitude of the tam-1 effect on lin-2 and let-2 activity was also approximately twofold.
Table 3.
tam-1 effects on endogenous gene activity
| Genotype
|
Phenotype
|
Statistical significance
|
|---|---|---|
| let-2(mn114) | sterile or semisterile adults (n = 8) | |
| let-2(mn114); tam-1(sy272) | L1 or L2 lethal (n > 20) | |
| lin-2(n768) | 3.25 VPCs induced (n = 26) | P < 0.0001 |
| lin-2(n768); tam-1(sy272) | 1.81 VPCs induced (n = 18) | |
| lin-3(e1417) | 0.66 VPCs induced (n = 102) | P < 0.0001 |
| lin-3(e1417); tam-1(sy272) | 0.29 VPCs induced (n = 92) | |
| let-23(sa62) | 3.42 VPCs induced (n = 6) | P = 0.17 |
| let-23(sa62); tam-1(sy272) | 4.4 VPCs induced (n = 4) | |
| let-60(n1046) | 3.97 VPCs induced (n = 16) | P = 0.0029 |
| let-60(n1046); tam-1(sy272) | 4.93 VPCs induced (n = 32) |
Phenotypes are described for a number (n) of animals for each of the genotypes. The state of vulval morphogenesis was assessed by counting induced vulval precursor cells (normally three in wild type). Statistically significant differences between wild-type and tam-1(sys272) were observed in lin-2(n768), let60(n1046), and lin-3(e1417) backgrounds. The average induction of VPCs for lin-3(e1417)/lin-3(n1059) is 0.34, n = 22 (P.W. Sternberg, unpubl.). For statistical analysis, a Fisher's exact test was performed using Instat (GraphPad Software).
tam-1 has properties of a class-B synthetic Muv gene
Two C. elegans genes were recently identified (lin-35 and lin-53) with a redundant role in the modulation of RAS signaling (Lu and Horvitz 1998). Mutations in these genes, like tam-1, are viable and have modest genetic interactions with extragenic mutations. lin-35 and lin-53 encode proteins similar to Rb and Rb-associated protein p48, respectively. lin-35 and lin-53 are members of a larger class of genes, called class-B synMuv genes, which are defined by the observation that null or hypomorphic mutations lead to little or no phenotypic effect unless a second mutation is present in a distinct set of genes termed the class-A synMuv genes (Horvitz and Sulston 1980; Ferguson and Horvitz 1989). Double mutants carrying a class-A and a class-B allele show phenotypes characteristic of hyperactivity of the RAS-signaling pathway (most notably a Muv phenotype). Numerous class-A and class-B synMuv genes have thus been identified by these studies, with the working model that these sets of genes define two redundant pathways inhibiting EGFR–RAS-signaling activity.
Given the fact that lin-35 and lin-53, two class-B SynMuv genes, encode factors shown to mediate chromatin repression in other systems (Lu and Horvitz 1998; Luo et al. 1998), we investigated the relationship of tam-1 to these groups of genes. Double mutants carrying a tam-1 mutation and a mutation in lin-15A (a class-A synMuv gene) showed a Muv phenotype at 25°C (Table 4; Fig. 3A). In contrast, tam-1(cc567); lin-15B(n744) double mutants did not have a Muv phenotype (Table 4). Two other class-A synMuv genes were analyzed for interactions with tam-1. Double mutants carrying tam-1(cc567) and the class-A synMuv mutation lin-38(n751) showed a synthetic multivulval phenotype, whereas no such phenotype was observed in tam-1(cc567); lin-8(n111). The latter result may indicate a functional division within the A class of synMuv genes, although it should be noted that differences in penetrance of the Muv phenotype in different A/B combinations have been observed previously (Ferguson and Horvitz 1989; Thomas and Horvitz 1999). In any case, it is clear from the interaction with lin-15A and lin-38 that tam-1 shares functional properties with the previously identified class-B synMuvs.
Table 4.
tam-1 has properties of a class B synMuv gene
| Genotype
|
Phenotype
|
|---|---|
| tam-1(cc567) | 3 VPCs induced (n = 15) |
| lin-15A(n767) | 3 VPCs induced (n = 15) |
| tam-1(cc567); lin-15A(n767) | 4.9 VPCs induced (n = 20) |
| tam-1(cc567); lin-15B(n744) | 2.9 VPCs induced (n = 15) |
The average induction of vulval precursor cells for each genotype is shown. All counts of VPC induction were performed with L4 staged animals at 25°C. At 20°C, the frequency of multivulva animals in tam-1(cc567); lin-15A(n767) double mutants decreased considerably.
Figure 3.
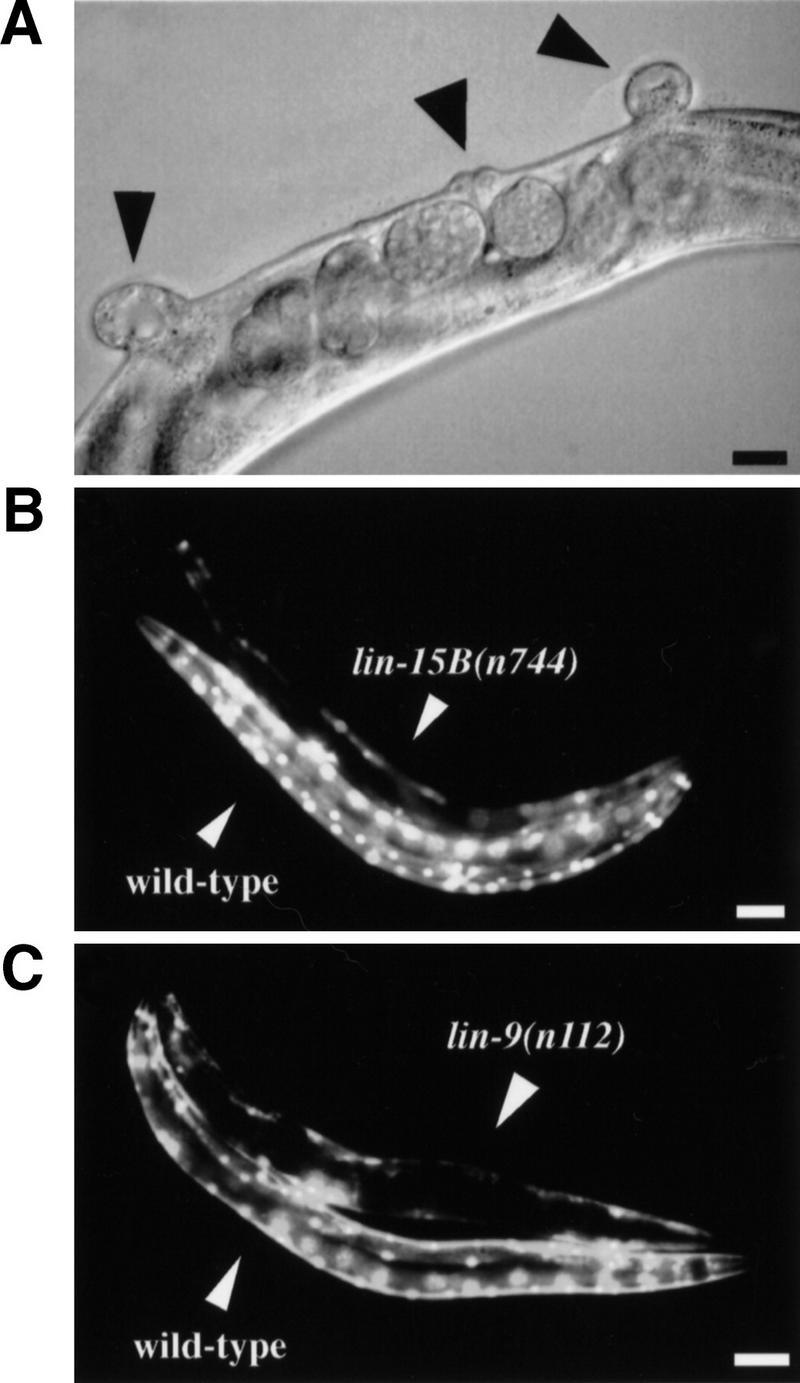
tam-1 has properties of a class-B synMuv gene. (A) lin-15A(n767); tam-1(cc567) double mutants are Muv (arrowheads indicate the position of vulvae). Scale bar, 27.5 μm. Other class-B synMuv mutants have decreased transgene expression. (B) The expression of ccIs4251(myo-3::gfp) is examined in wild-type and homozygous lin-15B(n744) animals. Scale bar, 87.5 μm. (C) The expression of the same transgene is also examined in wild-type and homozygous lin-9(n112) animals. Scale bar, 87.5 μm.
An additional connection between tam-1 and the class-B synMuv family comes from analysis of tandem array transgene activity in previously characterized class-B synMuv mutants. A mutation eliminating the RB homolog Lin-35 was tested by simple and complex arrays of myo-3::gfp and of egl-15::gfp (Fig. 2C). The two simple transgene arrays were both silenced in a lin-35(n745) background, whereas no effect of lin-35(n745) on either complex array was observed. Five additional class-B mutants (lin-9(n112), lin-15B(n744), lin-36(n766), lin-51(n770), and lin-52(n771)) were tested for silencing of a simple transgene array; all except for lin-36(n766) were found to produce a transgene-silencing phenotype similar to tam-1 (Fig. 3B,C; data not shown). The class-A synMuv mutations lin-8(n111), lin-15A(n767), and lin-38(n751) showed no transgene-silencing phenotype.
Molecular identification of tam-1
The tam-1 locus was mapped more precisely to a small region of chromosome V very close to the gene unc-46 (Fig. 4A). On the basis of the known position of unc-46 on the physical map, (E. Jorgensen, pers. comm.) we obtained cosmids that covered this region, injected pools of overlapping cosmids, and assayed for rescue of tam-1 effects on myo-3::gfp expression. We found that cosmid F26G5 rescued the decreased-GFP phenotypes of three tam-1 alleles (Fig. 4B). Testing of subcloned fragments from this cosmid led to the identification of an 8.6-kb fragment with rescuing activity (Fig. 4C). A single gene had been predicted in this region (C. elegans Sequencing Consortium 1998). When we created a frameshift in a unique AatII site within the coding region, the resulting fragment was incapable of rescuing tam-1(cc567). The mRNA structure predicted by the sequencing consortium (Fig. 4D) was correct, as determined by completing the sequence of a full-length cDNA clone obtained from Y. Kohara [pers. comm.; accession nos. C42160 (5′), C31634 (3′)]. Next, we identified the molecular lesions associated with three of the tam-1 alleles (Fig. 4E). All three alleles resulted in nonsense mutations, with cc567 containing the most amino-terminal stop codon.
Figure 4.
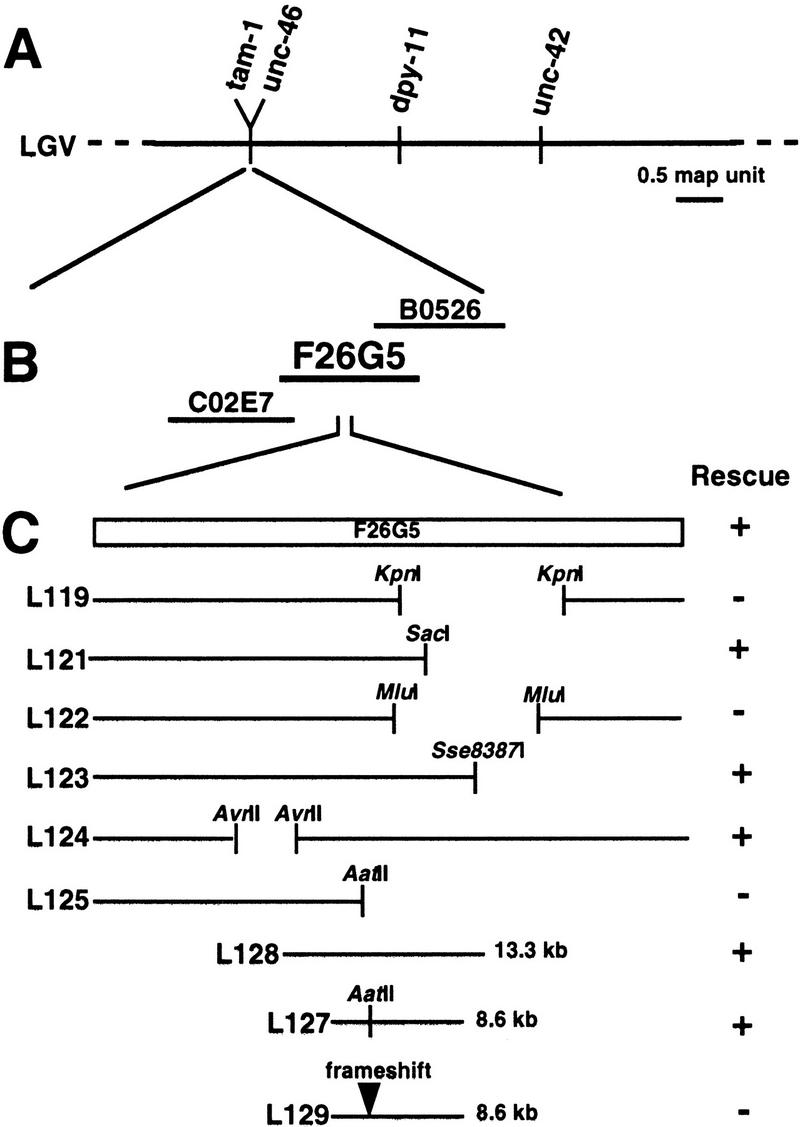

Molecular identification of tam-1. (A) Genetic mapping placed the tam-1 locus to the left of the gene unc-46. (B) Pools of cosmids were assayed for rescue of the reduced-transgene-expression phenotype of tam-1(cc567). Cosmid F26G5 was found to rescue the decreased-GFP phenotype of tam-1(cc567); ccIs4251(myo-3::gfp). Rescue was also observed for two other tam-1 alleles, cc587 and sy272. (C) Deletion analysis of cosmid F26G5 led to the identification of an 8.6-kb fragment (L127) with rescuing activity. Analysis of the sequence of this fragment had predicted a single coding region (C. elegans Sequencing Consortium 1998). Creation of a frameshift in a unique AatII site within this coding region abolished rescue. (D) A full-length cDNA was obtained from the Kohara laboratory. Sequencing of this cDNA confirmed that the tam-1 mRNA sequence contains seven exons and six introns. (E) Amino acid sequence of TAM-1. Underlined are the RING finger (solid line) and B-box (broken line). Sequencing of tam-1 alleles (cc567, sy272, and cc587) revealed in each case the conversion of a glutamine codon (CAA) to a nonsense codon (TAA). DNA from tam-1 mutants was amplified by PCR; this was done in several segments covering a region from 288-bp upstream of the ATG to 440-bp downstream of the deduced tam-1 translational stop. PCR products were directly sequenced, with mutations confirmed by sequencing products from at least two different PCR reactions. The three distinct mutations are indicated by arrowheads.
The predicted TAM-1 protein contains two cysteine-rich regions, a C3HC4 (RING finger) motif and a B-box motif. The RING finger and B-box motifs have been defined by primary sequence comparisons; although both domains may act as metal (zinc)-binding domains, there is not yet any basis to predict specific biochemical function (Saurin et al. 1996). C. elegans contains 90 RING finger proteins (C. elegans Sequencing Consortium 1998). Several Drosophila proteins with roles in global regulation of chromosome activity also contain RING finger motifs; these include the Posterior Sex Combs (PSC) and Suppressor Two of Zeste [Su(Z)2] proteins (which are involved in the maintenance of repressed transcriptional states by the polycomb family; Brunk et al. 1991), and MSL-2 (involved in sex-specific activation of the X chromosome in males; Zhou et al. 1995). Although these comparisons are intriguing, proteins containing RING finger and B-box motifs have been found in many additional biological processes (Fig. 5). The similarity of TAM-1 to other characterized RING finger and B-box proteins does not extend outside of these two domains.
Figure 5.

Sequence alignments of RING finger and B-box motifs of TAM-1 with related segments of other proteins. Conserved cysteine and histidine residues are underlined. A limited number of members of each family were chosen arbitrarily. GenBank contains >100 entries with RING finger motifs. Alignment of RING finger motifs: Numbering below each conserved cysteine refers to zinc binding ligands cysteine 1-cysteine 7 (Saurin et al. 1996) . Percent identity refers to just the RING finger domain. Alignment of B-box motifs: Sources for RING fingers and B-Box motifs used in this comparison are RAD18 (Jones et al. 1988); RPT-1 (Patarca et al. 1988); PSC, SU(Z)2 (Brunk et al. 1991); SS-A/Ro (Chan et al. 1991); XNF7 (Reddy et al. 1991); RAD16 (Bang et al. 1992); PML (Kastner et al. 1992); PAF-1 (Shimozawa et al. 1992); PwA33 (Bellini et al. 1993); MEL-18 (Ishida et al. 1993); PAR-2 (Levitan et al. 1994); BRCA1 (Miki et al. 1994); HT2A (Fridell et al. 1995); T18 (Miki et al. 1991; LeDoyerin et al. 1995); RFP (Cao et al. 1997); RING1 (Satijn et al. 1997); BMI-1 (Hemenway et al. 1998); and XL43, XL75 (Perrin and Lacroix 1998).
TAM-1 protein is present in nuclei throughout development
We raised polyclonal antibodies against two nonoverlapping protein fragments from the carboxyl terminus of TAM-1. The two antisera gave identical patterns, with staining in the nuclei of most, if not all, somatic cells in wild-type embryos (Fig. 6A,B). The nuclear distribution was uniform, with staining beginning at about the 20-cell stage and persisting throughout development. There was also nuclear staining (albeit somewhat fainter) in larval and adult cells and germ-line and oocyte nuclei (Fig. 6C,D). Antibody specificity was verified by examining null mutant embryos, larvae, and adults (no detectable staining by either antisera).
Figure 6.
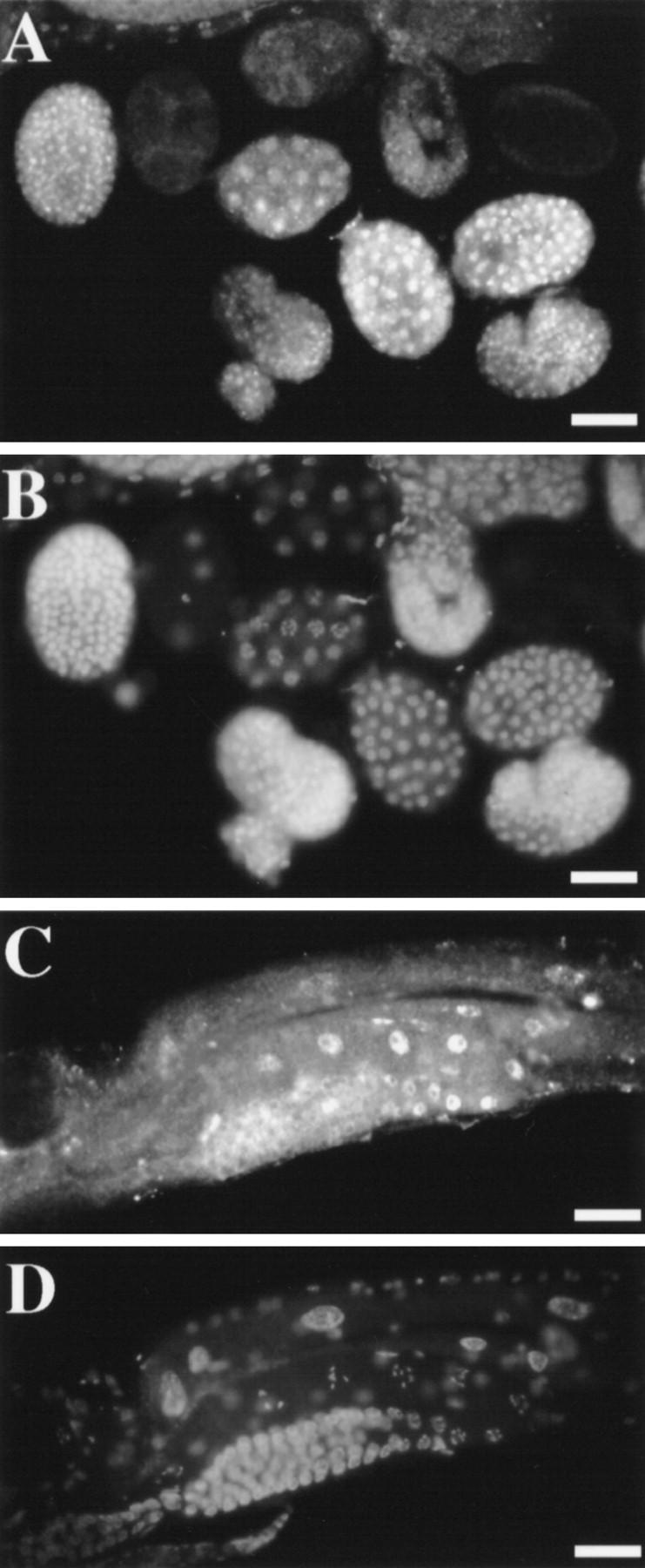
Immunofluorescence detection of TAM-1 protein in nuclei. (A) Embryo population and (C) an adult stained with mouse anti-TAM-1 and visualized with a secondary goat anti-Cy3 antibody. (B,D) Corresponding images of DNA localization, visualized with DAPI. Photos shown are from antisera to TAM-1 residues 594–712. Similar results were seen with antisera to TAM-1 residues 778–856. Signal in C has been augmented by electronic contrast enhancement. Scale bars, 27.5 μm.
tam-1 is not essential for viability
Several observations support the hypothesis that the viable chain-termination allele tam-1(cc567) is a null mutation: (1) tam-1(cc567) is predicted to result in translational arrest after synthesis of only 50 amino acids; (2) Hemizygous [tam-1(cc567)/nDf32] animals showed a phenotype indistinguishable from tam-1(cc567)/tam-1(cc567) (data not shown); (3) double-stranded RNA interference (Fire et al. 1998b) with a segment of tam-1 produced a nonlethal reduced-GFP phenotype identical to that of tam-1(cc567) (data not shown); (4) viable tam-1 alleles arose at a high frequency (2.2 × 10−4 after EMS mutagenesis) close to reported knockout frequencies for C. elegans (Brenner 1974); and (5) there was no detectable staining with either TAM-1 antibody in tam(cc567) (data not shown).
Discussion
tam-1 and context-dependent gene silencing
We have presented evidence that TAM-1, a novel RING finger and B-Box factor, plays a role in modulating context-dependent gene silencing. C. elegans transgenes are often subject to silencing when present in tandemly repeated array structures (simple arrays). This silencing is particularly evident in the germ line (Kelly et al. 1997), but can also be observed in the soma of C. elegans (Okkema et al. 1993; Krause et al. 1994). This silencing appears to be context dependent because it can be attenuated or abolished by placing the transgene in a less repetitive context (complex arrays).
In tam-1 null mutants, the silencing of transgenes present in tandemly repeated arrays was potentiated. Two comparisons suggest that tam-1 effects on gene expression are sensitive to chromosomal context. First, the dramatic reductions in activity seen with simple arrays (e.g., 18-fold for a canonical myo-3::gfp array) were not seen with the corresponding endogenous loci (e.g., no effect on the endogenous myo-3 locus in an assay in which a twofold effect would have been observed). Second, complex arrays are much less susceptible to silencing in a tam-1 null mutant background. This suggests that the silencing activity in tam-1 mutants has some degree of preference for repetitive transgene arrays. Hence, the wild-type TAM-1 product appears (directly or indirectly) to be attenuating context-dependent silencing.
Although tam-1 had no discernible effect on the endogenous myo-3 locus, a number of genes showed an apparent decrease in expression levels in a tam-1 mutant background. Although the mechanistic basis for these modest effects was not investigated further, one possibility is that all effects of tam-1 might reflect a direct potentiation of context-dependent silencing; in this model, the silencing mechanism might have drastic activity for some regions of chromosomes (e.g., regions that might behave similarly to highly repeated transgene arrays) with a less evident effect on other regions (e.g., subtle or indirect effects on genes in partially repetitive chromosomal areas). An additional possibility is that tam-1 might be a specific transcription factor regulating a small number of target genes, including one or more genes with a general role in context-dependent silencing. Clearly the above models are not mutually exclusive.
A connection between context-dependent gene silencing and control of the RAS pathway?
Genes of the class-B synMuv family have been shown to play a role in the modulation of EGFR–RAS pathway activity during vulval specification. This role is redundant in that loss-of-function mutants in class-B genes show vulval specification defects (apparent hyperactivity of the RAS pathway) only if a second mutation in a class-A synMuv gene is present (Horvitz and Sulston 1980; Ferguson and Horvitz 1989; Clark et al. 1994; Huang et al. 1994). Although much of the genetic analysis of class-B synMuv activity has involved analysis of vulval specification, mutants have also been reported to exhibit defects in growth and fertility, suggesting additional functions for these genes outside the vulva (Ferguson and Horvitz 1989).
Our work extends this analysis in two ways. First, TAM-1 was shown to have properties of a class-B synMuv factor and also to modulate transgene reporter activity in a wide variety of nonvulval tissues including mesoderm (pharyngeal muscles, body muscles, and somatic gonad), endoderm (gut) and at least a subset of neurons (from expression of egl-15, lin-11, gpa-1, and mab-5). Second, we have observed effects of previously characterized class-B synMuv genes (lin-9, lin-15B, lin-35, lin-51, and lin-52) on transgene activity levels in extensive subsets of mesoderm. Although not all class-B synMuv genes have been examined, and not all cell types have been included in the reporter expression patterns that were tested, it appears likely that this family of genes might act in most or all tissues of the animal. Consistent with this hypothesis, tam-1 and several other class-B synMuv genes encode proteins with broad expression patterns (Lu and Horvitz 1998; this work; L.Huang, J. DeModena, and P.W. Sternberg, unpubl.).
What are the natural targets of the class-B synMuv family? As noted above for tam-1, one possibility for the class-B SynMuv factors is that they form a relatively specific regulatory complex whose targets include both RAS pathway components and (independently) factors involved in the modulation of repetitive transgenes. Under these circumstances, there would be no direct link between RAS signaling and silencing of repetitive arrays. We favor the alternative hypothesis that some relationship exists between these two processes. In particular, our current working model is that the class-B synMuv factors modulate context-dependent silencing of a wide variety of biological targets; some of these targets may be endogenous genes that are present in areas of repetitive sequences or an otherwise inopportune genomic environment.
At some level, the effects of the class-B synMuv genes are likely to involve changes in the acetylation state of histones in chromatin. The strongest indication for this hypothesis comes from the ability to induce a class-B synMuv phenotype by selective disruption of histone deacetylase-1 (hda-1) function and from the existence of a complex containing LIN-35 (Rb), LIN-53 (p48), and HDA-1 (Lu and Horvitz 1998). It is not clear whether TAM-1 participates as a part of this complex, or whether the complex interacts directly with repetitive transgene arrays. The directionality of the class-B synMuv effect should be considered in evaluating the following: transgene expression is reduced in the absence of LIN-35 or TAM-1 function. Thus, if there were a direct interaction in wild-type animals, it would be (unexpectedly) a positive effect. In many cases, histone deacetylase complexes are thought to act as negative regulators of gene expression (Kuo and Allis 1998). An additional possibility is that the class-B synMuvs normally act to direct silencing factors to a defined set of targets not including tandem array transgenes; under these circumstances, class-B mutants might result in an excess of unused silencing factors and consequent hypersilencing of certain loci (including transgene arrays) with susceptibility to the process. A similar proposal involving differential partitioning of a limiting supply of generalized silencing factors was proposed by Grewal and Klar (1998) in their studies of chromosomal silencing in fission yeast.
Materials and methods
C. elegans strains and culture
Conditions for growth and maintenance were as described (Brenner 1974). Unless stated otherwise, experiments were conducted at 25°C. Wild-type N2 (var. Bristol) and the standard mutant strains listed below are as described by Brenner (1974), unless otherwise noted. The following genes and alleles were used: LG I, unc-54(e190), lin-35(n745) unc-13(e189) (Ferguson and Horvitz 1989); LG II, let-23(sa62) unc-4(e120) (Katz et al. 1996), lin-8(n111) dpy-10(e128) (Ferguson and Horvitz 1989), lin-38(n751) (Thomas and Horvitz 1999); LG III, lin-9(n112) (Ferguson and Horvitz 1989), lin-36(n766) unc-32(e189) (Thomas and Horvitz 1999), lin-51(n770) unc-32(e189) (Thomas and Horvitz 1999), lin-52(n771) unc-32(e189) (Thomas and Horvitz 1999); LG IV, dpy-20(e1282), lin-3(e1417) (Horvitz and Sulston 1980), let-60(n1046) unc-31(e169) (Ferguson and Horvitz 1985; Han and Sternberg 1990); LG V, tam-1(cc566, cc567, cc587, and sy272), unc-46(e177), dpy-11(e224), unc-42(e270), dpy-11(e224) nDf32/eT1 (Park and Horvitz 1986); and LG X, lin-2(n768), let-2(mn114) unc-3(e151), lin-15A(n767) (Ferguson and Horvitz 1985), lin-15B(n744) (Ferguson and Horvitz 1985).
Plasmid constructs used in transgenes
pSAK-2 (myo-3::Ngfp–lacZ) contains the myo-3 promoter driving gfp expression in all nonpharyngeal muscles; a nuclear localization signal (NLS) and lacZ sequences are appended to the reporter coding sequence, and lead to predominantly nuclear localization (Fire et al. 1998b).
pSAK-4 (myo-3::Mtgfp) contains the myo-3 promoter driving gfp expression in nonpharyngeal muscles; the GFP has a mitochondrial localization signal (Fire et al. 1998b).
pPD96.02 (unc-54::Ngfp–lacZ) contains the unc-54 promoter driving NLS::gfp::lacZ in nonpharyngeal muscles (Fire et al. 1998a).
pPD105.19 (unc-54::Mtgfp) contains the unc-54 promoter driving gfp expression with a mitochondrial localization signal in nonpharyngeal muscles (Fire et al. 1998a).
pPD93.92 (hlh-1::gfp) is a translational fusion containing the hlh-1 promoter and first exon driving gfp expression in myogenic precursor cells and bodywall muscles (derived from pPD37.48 (Krause et al. 1990) by substitution of gfp for lacZ).
pPD118.33 (myo-2::gfp) contains the myo-2 promoter driving gfp expression in pharyngeal muscle (S. Xu and A. Fire, unpubl.).
pPD103.68 (F22B7.9::gfp) is a translational fusion driving gfp in the gut (S. Xu and A. Fire, unpubl.).
pNH#300 (egl-15::gfp) contains the egl-15 promoter driving gfp expression in Vm1 vulval muscles (Harfe et al. 1998).
pRH9 (lin-3) is a lin-3 genomic construct (Hill and Sternberg 1992).
pCC2 (lin-3::lacZ) is a translational fusion of the lin-3 promoter and coding region before the first cytoplasmic domain driving lacZ expression in the anchor cell and many other tissues (Chang et al. 1999).
pjM1HG.11 (gpa-1::lacZ) contains the gpa-1 promoter driving lacZ expression in plasmid neurons, PHA and PHB (J. Mendel and P.W. Sternberg, unpubl.).
pGF66 (lin-11::lacZ) is a translational fusion that drives lacZ expression in descendants of vulval precursor cells (P5.pp and P7.pa), VC motor neurons, and in the ventral uterine p cells and their progeny (Freyd 1991).
pCH22 (mab-5::gfp) contains the mab-5 promoter driving gfp expression in cells of the posterior region (Hunter et al. 1999).
BK48 (let-858::gfp) contains a complete copy of the let-858 gene tagged by in-frame insertion of a gfp cassette within the coding region, yielding gfp expression in all somatic nuclei (Kelly et al. 1997).
pPD129.51 (rps-5::gfp) contains the promoter for the gene T05E11.1 (encoding ribosomal protein S5) driving gfp expression in all somatic nuclei (J. Fleenor and A. Fire, unpubl.).
pG75#KP6 (glr-1::gfp) contains the glr-1 promoter driving gfp expression in a subset of neurons (H. Hutter, pers. comm.).
pDP#MMUGD12 (unc-119::gfp) is a translational fusion containing the unc-119 promoter and unc-119 genomic region driving gfp expression throughout the nervous system (Maduro and Pilgrim 1995).
pRF4 (rol-6 [su1006]) carries a dominant allele of the gene rol-6 that leads to an easily identified rolling phenotype.
pMH86 (dpy-20[+]) carries a wild-type copy of the gene dpy-20 that provides a ready selection for transgene function in a dpy-20(e1282ts) background (Han et al. 1993; Clark et al. 1995).
Transgenic lines
In some cases (lines referred to as Is), the transgene arrays had been integrated by γ-ray mutagenesis. In all other lines (referred to as Ex), the transgene arrays remained extrachromosomal.
For each transgene, the comparison of wild-type with tam-1(−) and/or lin(−) backgrounds was made several generations after the strain had been established; at this point simple culture of the strains produced no evident instability in expression or transmission properties. Effects on transgene expression were observed in all animals from each affected population (>100 animals for each transgene).
Simple array lines were produced by injection of a mixture of the circular reporter plasmid and a circular selectable marker plasmid, with no other DNA added as carrier (Mello and Fire 1995).
ccIs4251
pSAK2(myo-3::Ngfp–lacZ), pSAK4(myo-3::Mtgfp), and pMH86 (Fire et al. 1998b). The multicopy tandem-array character of the ccIs4251 transgene was confirmed by Southern blot analysis (showing ∼100 copies each of pSAK2 and pSAK4; data not shown). ccIs4251 was obtained following treatment with γrays and shows some crossover suppression activity in the center of chromosome I.
ccEx6188
pSAK2(myo-3::Ngfp–lacZ) and pRF4 (this work). A simple extrachromosomal transgene with expression levels comparable with the integrated ccIs4251 array, ccEx6188 was used to assay mutations on chromosome I (e.g., lin-35) for silencing effects.
ccIs9385
pPD96.02(unc-54::Ngfp–lacZ), pPD105.19(unc-54:: Mtgfp), and pRF4 (Fire et al. 1998a).
ccIs7963
pPD93.92(hlh-1::gfp) and pRF4 (K. Dej and A. Fire, unpubl.).
ccIs4792
pPD95.29(pes-10::gfp), pPD118.33(myo-2::gfp), pPD103.68(F22B7.9::gfp), and pMH86 (M. Edgley, K. Liu, A. Fire, and D. Riddle, unpubl.)
ayIs2
pNH#300(egl-15::gfp) and pMH86 (Harfe et al. 1998).
syIs1
pRH9(lin-3) and an unc-31 subclone (selectable marker) (Hill and Sternberg 1992; this work; J. Liu, unpubl.).
syEx241
pCC2(lin-3::lacZ), pMH86, and pSK+ as carrier (Chang et al. 1999).
syIs19
cpjM1HG.11(gpa-1::lacZ) and pMH86 (J. Mendel and P.W. Sterberg, unpubl.).
nIs2
pGF66(lin-11::lacZ) and pGF60(lin-11 rescuing plasmid) (Freyd 1991).
muIs16
pCH22(mab-5::gfp) and pMH86 (Hunter et al. 1999).
ccEx7997
BK48(let-858::gfp) and pRF4 (Kelly et al. 1997).
ccEx8151
pPD129.51(rps-5::gfp) and pRF4 (J. Fleenor and A. Fire, unpubl.).
rhIs4
pG75#KP6(glr-1::gfp) and pMH86 (H. Hutter, pers. comm.).
edIs6
pDP#MMUGF12(unc-119::gfp) and pRF4 (Maduro and Pilgrim 1995).
Complex array lines were produced by injection of a mixture of the linear reporter plasmid, a linear selectable marker plasmid, and an excess of PvuII-fragmented genomic DNA from C. elegans as carrier. Plasmids were linearized with a unique blunt-cut enzyme in the vector backbone. Mass ratios (genomic DNA/plasmid DNA) were ∼50–100:1, with the concentration of plasmid ∼1–2 μg/ml.
ccEx6172
pSAK2(myo-3::Ngfp–lacZ) and pRF4, with excess genomic C. elegans DNA as carrier. Copy number of pSAK2 within the ccEx6172 transgene was confirmed to be 1–2 (Southern blot analysis; data not shown).
ccEx6176
pNH#300(egl-15::gfp) and pMH86 with excess genomic C. elegans DNA as carrier.
Genetic screens
Screen for decreased expression of myo-3::gfp
PD4251 animals carrying an integrated myo-3::gfp transgene expressing in bodywall and vulval muscles were mutagenized with EMS as described (Brenner 1974). Nonclonal F2 and F3 populations were observed under direct fluorescent illumination and animals with uniformly reduced GFP activity recovered for further analysis. We screened ∼9000 F2 animals and identified four independent mutations with distinctly decreased GFP activity. One of these mutations was subsequently shown to be tightly linked to the original transgene and was not further characterized. The other three mutations (cc566, cc567, and cc587) were all recessive and failed to complement, defining the tam-1 locus on chromosome V. We observed some differences in phenotype between alleles; cc566 and cc567 animals showed strongest reduction of myo-3::gfp activity in the center of the body, with lesser effects in head and tail regions. cc587 showed a less dramatic reduction in GFP with no evident differences between head, body, and tail.
Screen for decreased expression of a lin-3 transgene
Additional alleles of tam-1 were identified as part of an independent screen for reversion of a Muv defect caused by an integrated high-copy lin-3 transgene (syIs1). This screen yielded (1) several mutants corresponding to known components of the lin-3-signaling pathway (J. Liu and P.W. Sternberg, unpubl.) and (2) 15 alleles of tam-1 (sy270, sy272, sy273, sy342, sy366, sy372, sy394, sy395, sy398, sy399, sy401, sy402, sy410, sy411, and sy412).
Genetic and molecular analysis of tam-1
Mapping of tam-1 was carried out with allele cc567 by standard methods. Three-factor mapping was done with unc-46 dpy-11/tam-1; ccIs4251. 16/16 Dpy-non-Unc picked up tam-1, whereas 10/10 Unc-non-Dpy did not pick up tam-1; also 9/9 Dpy-non-Tam picked up unc-46, indicating that tam-1 is very close or to the left of unc-46. A genetic deficiency in this region, nDf32 failed to complement tam-1. unc-46 has thus been used as a linked marker in introducing tam-1 into various genetic backgrounds. unc-46 (without tam-1) has also been introduced into a variety of transgene-carrying strains and has shown no effect on transgene expression.
By use of the intensity of GFP fluorescence in adults carrying the ccIs4251 transgene as an assay, all tam-1 alleles tested (cc567, cc566, cc587, and sy272) were recessive and showed no evidence of maternal effect. This assay was used for all complementation and genetic rescue assays with tam-1.
Phenotypic rescue of tam-1 mutants was assayed in transgenic lines derived from microinjection of DNAs into homozygous tam-1 mutants (Mello and Fire 1995). Cosmid DNAs were injected at 10–20 μg/ml. To identify transgenic lines, plasmid pRF4, which contains the semidominant mutation rol-6 (su1006), was coinjected at 100 μg/ml (Mello et al. 1991). For each cosmid mixture, several independent transgenic lines were recovered and scored for restoration of gfp expression to levels comparable with those seen in the parental strain PD4251. A positive pool of five cosmids was successively divided, leading to the identification of a single rescuing cosmid, F26G5. F26G5 was shown to rescue three alleles of tam-1 (cc566, cc567, and sy272).
Protein localization
To produce antisera recognizing TAM-1, two different regions of the cDNA (Pro594–Glu712 and Ala778–Ser856) were cloned into GST gene fusion vectors (pGEX-2T) (Smith and Johnson 1988) to produce fusion proteins of 39 and 35 kD. SDS-polyacrylamide gel slices containing purified fusion proteins were prepared for the immunization of mice. Polyclonal mouse sera were collected and used for immunohistochemistry as described (Miller and Shakes 1995). Freeze-cracked adult C. elegans were freeze substituted in methanol (−20°C, 4 min) and fixed with 4% formaldehyde (23°C, 30 min).
Acknowledgments
We thank K. Liu, S. Parrish, B. Kelly, L. Timmons, J. Goodliffe, C. Lee, C. Ceol, E. Davison, B. Horvitz, M. Bellini, V. Guacci, P. McGee, E. Jorgensen, and A. Shearn for help and advice; M. Edgley, D. Riddle, H. Hutter, L. Huang, J. DeModena, S. Xu, J. Fleenor, J. Mendel, Y. Kohara, and K. Dej for useful materials; and R. Feldman for isolating several tam-1 alleles. This work was supported by U.S. Public Health Service grants GM37706 (A.F.); HD23690 (P.W.S.); T32GM07231 (J.H., S.K.); GM07616 (J.L., C.C.); and by the Carnegie Institution of Washington. P.W.S. is an investigator of the Howard Hughes Medical Institute. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is supported by National Institutes of Health's National Center for Research Resources.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: fire@mail1.ciwemb.edu; FAX (410) 243-6311.
References
- Assaad F, Tucker K, Signer E. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- Bang D, Verhage R, Goosen N, Brouwer J, van de Putte P. Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Lacroix J, Gall J. A putative zinc-binding protein on lampbrush chromosome loops. EMBO J. 1993;12:107–114. doi: 10.1002/j.1460-2075.1993.tb05636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk B, Martin E, Adler P. Drosophila genes Posterior Sex Combs and Suppressor Two of Zeste encode proteins with homology to the murine bmi-1 oncogene. Nature. 1991;353:351–353. doi: 10.1038/353351a0. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cambareri E, Jensen B, Schabtach E, Selker E. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Cao T, Borden K, Freemont P, Etkin L. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J Cell Sci. 1997;110:1563–1571. doi: 10.1242/jcs.110.14.1563. [DOI] [PubMed] [Google Scholar]
- Chan E, Hamel J, Buyon J, Tan E. Molecular definition and sequence motifs of the 52-kD component of human SS-A/Ro autoantigen. J Clin Invest. 1991;87:68–76. doi: 10.1172/JCI115003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Newman A, Sternberg P. Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia. Curr Biol. 1999;9:237–246. doi: 10.1016/s0960-9822(99)80112-2. [DOI] [PubMed] [Google Scholar]
- Clark S, Lu X, Horvitz H. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D, Suleman D, Beckenbach K, Gilchrist E, Baillie D. Molecular cloning and characterization of the dpy-20 gene of C. elegans. Mol Gen Genet. 1995;247:367–378. doi: 10.1007/BF00293205. [DOI] [PubMed] [Google Scholar]
- Dorer D, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- ————— Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H, Waterston R, Brenner S. A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. J Mol Biol. 1974;90:291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- Ferguson E, Horvitz H. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Kelly W, Hsu M, Xu S, Ahnn J, Harfe B, Kostas S, Hsieh J. The uses of green fluorescent protein in Caenorhabditis elegans. In: Chalfie M, Kane S, editors. Green fluorescent protein: Properties, applications, and protocols. New York, NY: Wiley-Liss, Inc.; 1998a. pp. 153–168. [Google Scholar]
- Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998b;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Freyd G. Department of biology. Cambridge, MA: Massachusetts Institute of Technology; 1991. Molecular analysis of the C. elegans cell lineage gene Lin-11. [Google Scholar]
- Fridell R, Harding L, Bogerd H, Cullen B. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin D, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Goyon C, Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989;7:2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S, Bonaduce M, Klar A. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: Regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Haber J. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- Han M, Sternberg P. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a RAS protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- Han M, Golden A, Han YM, Sternberg PW. C. elegans lin-45 raf gene participates in let-60 RAS-stimulated vulval differentiation. Nature. 1993;363:133–140. doi: 10.1038/363133a0. [DOI] [PubMed] [Google Scholar]
- Harfe B, Branda C, Krause M, Stern M, Fire A. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. 1998;125:2479–2488. doi: 10.1242/dev.125.13.2479. [DOI] [PubMed] [Google Scholar]
- Hemenway C, Halligan B, Levy L. The Bmi-1 oncoprotein interacts with dinG and MPh2: The role of RING finger domains. Oncogene. 1998;16:2541–2547. doi: 10.1038/sj.onc.1202042. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. BioEssays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Hill R, Sternberg P. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature. 1992;358:470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- Horvitz H, Sulston J. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Tzou P, Sternberg P. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Harris J, Maloof J, Kenyon C. Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit Wnt signaling. Development. 1999;126:805–814. doi: 10.1242/dev.126.4.805. [DOI] [PubMed] [Google Scholar]
- Ishida A, Asano H, Hasegawa M, Koseki H, Ono T, Yoshida M, Taniguchi M, Kanno M. Cloning and chromosome mapping of the human Mel-18 gene which encodes a DNA-binding protein with a new 'RING-finger' motif. Gene. 1993;129:249–255. doi: 10.1016/0378-1119(93)90275-8. [DOI] [PubMed] [Google Scholar]
- Jones J, Weber S, Prakash L. The Saccharomyces cerevisiae RAD18gene encodes a protein that contains potential zinc finger domains for nucleic acid binding and a putative nucleotide binding sequence. Nucleic Acids Res. 1988;16:7119–7131. doi: 10.1093/nar/16.14.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): Structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W, Lesa G, Yannoukakos D, Clandinin T, Schlessinger J, Sternberg P. A point mutation in the extracellular domain activates LET-23, the Caenorhabditis elegans epidermal growth factor receptor homolog. Mol Cell Biol. 1996;16:529–537. doi: 10.1128/mcb.16.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W, Xu S, Montgomery M, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Fire A, Harrison S, Priess J, Weintraub H. CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell. 1990;63:907–919. doi: 10.1016/0092-8674(90)90494-y. [DOI] [PubMed] [Google Scholar]
- Krause M, Harrison S, Xu S, Chen L, Fire A. Elements regulating cell- and stage-specific expression of the C. elegans MyoD family homolog hlh-1. Dev Biol. 1994;166:133–148. doi: 10.1006/dbio.1994.1302. [DOI] [PubMed] [Google Scholar]
- Kuo M, Allis C. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier J, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D, Boyd L, Mello C, Kemphues K, Stinchcomb D. par-2, a gene required for blastomere asymmetry in Caenorhabditis elegans, encodes zinc-finger and ATP-binding motifs. Proc Natl Acad Sci. 1994;91:6108–6112. doi: 10.1073/pnas.91.13.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Horvitz H. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Luo R, Postigo A, Dean D. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mello C, Kramer J, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Fleming T, Crescenzi M, Molloy C, Blam S, Reynolds S, Aaronson S. Development of a highly efficient expression cDNA cloning system: Application to oncogene isolation. Proc Natl Acad Sci. 1991;88:5167–5171. doi: 10.1073/pnas.88.12.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal P, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Miller D, Shakes D. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- Okkema P, Harrison S, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Horvitz H. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 1986;113:821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarca R, Freeman G, Schwartz J, Singh R, Kong Q, Murphy E, Anderson Y, Sheng F, Singh P, Johnson K, et al. rpt-1, an intracellular protein from helper/inducer T cells that regulates gene expression of interleukin 2 receptor and human immunodeficiency virus type 1. Proc Natl Acad Sci. 1988;85:2733–2737. doi: 10.1073/pnas.85.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin K, Lacroix J. XL43 and XL75: two novel RING finger-containing genes expressed during oogenesis and embryogenesis in Xenopus laevis. Gene. 1998;210:127–134. doi: 10.1016/s0378-1119(98)00052-3. [DOI] [PubMed] [Google Scholar]
- Reddy B, Kloc M, Etkin L. The cloning and characterization of a maternally expressed novel zinc finger nuclear phosphoprotein (xnf7) in Xenopus laevis. Dev Biol. 1991;148:107–116. doi: 10.1016/0012-1606(91)90321-s. [DOI] [PubMed] [Google Scholar]
- Riggs A, Porter T. X-Chromosome Inactivation and Epigenetic Mechanisms. In: Russo V, Martienssen R, Riggs A, editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 231–248. [Google Scholar]
- Rossignol J, Faugeron G. Gene inactivation triggered by recognition between DNA repeats. Experientia. 1994;50:307–317. doi: 10.1007/BF01924014. [DOI] [PubMed] [Google Scholar]
- Satijn D, Gunster M, van der Vlag J, Hamer K, Schul W, Alkema M, Saurin A, Freemont P, van Driel R, Otte A. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A, Borden K, Boddy M, Freemont P. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Selker E. Epigenetic phenomena in filamentous fungi: Useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- Selker E, Cambareri E, Jensen B, Haack K. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y. A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science. 1992;255:1132–1134. doi: 10.1126/science.1546315. [DOI] [PubMed] [Google Scholar]
- Smith D, Johnson K. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D, Shaw J, Carr S, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Horvitz HR. The C. elegans gene lin-36 acts cell autonomously in the lin-35 Rb pathway. Development. 1999;126:3449–3459. doi: 10.1242/dev.126.15.3449. [DOI] [PubMed] [Google Scholar]
- Waterston R. The minor myosin heavy chain, mhcA, of Caenorhabditis elegans is necessary for the initiation of thick filament assembly. EMBO J. 1989;8:3429–3436. doi: 10.1002/j.1460-2075.1989.tb08507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Signer E. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc Natl Acad Sci. 1996;93:10881–10886. doi: 10.1073/pnas.93.20.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yang Y, Scott M, Pannuti A, Fehr K, Eisen A, Koonin E, Fouts D, Wrightsman R, Manning J, et al. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 1995;14:2884–2895. doi: 10.1002/j.1460-2075.1995.tb07288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]