Abstract
The mechanisms regulating vertebrate heart and endoderm development have recently become the focus of intense study. Here we present evidence from both loss- and gain-of-function experiments that the zinc finger transcription factor Gata5 is an essential regulator of multiple aspects of heart and endoderm development. We demonstrate that zebrafish Gata5 is encoded by the faust locus. Analysis of faust mutants indicates that early in embryogenesis Gata5 is required for the production of normal numbers of developing myocardial precursors and the expression of normal levels of several myocardial genes including nkx2.5. Later, Gata5 is necessary for the elaboration of ventricular tissue. We further demonstrate that Gata5 is required for the migration of the cardiac primordia to the embryonic midline and for endodermal morphogenesis. Significantly, overexpression of gata5 induces the ectopic expression of several myocardial genes including nkx2.5 and can produce ectopic foci of beating myocardial tissue. Together, these results implicate zebrafish Gata5 in controlling the growth, morphogenesis, and differentiation of the heart and endoderm and indicate that Gata5 regulates the expression of the early myocardial gene nkx2.5.
Keywords: Gata, cardia bifida, nkx2.5, organogenesis, morphogenesis, liver
Fusion of the cardiac primordia is the first in a complex series of morphogenetic steps required to form a functional heart. The cardiac primordia are contained within the anterior lateral plate mesoderm (ALPM). During segmentation stages, the bilateral populations of ALPM migrate medially and fuse to form a cell layer that crosses the embryonic midline. As part of this process, the two cardiac primordia meet and fuse at the embryonic midline to form the definitive heart tube. In the absence of migration, the two cardiac primordia develop in their original lateral positions, resulting in a phenotype called cardia bifida. Six zebrafish mutations causing cardia bifida were identified in the large-scale screens for genes regulating embryonic development: faust (fau), casanova (cas), one-eyed pinhead (oep), bonnie and clyde, miles apart, and natter (Chen et al. 1996; Stainier et al. 1996).
Two of these mutations, oep and cas, also disrupt endoderm formation (Schier et al. 1997; Alexander et al. 1999), underscoring the developmental relationship between the heart and endoderm. Classical embryological experiments initially identified the ability of endoderm to induce rhythmic contraction in cardiac explants (Jacobson and Sater 1988). More recent experiments have confirmed that the endoderm is required for avian cardiac myofibrillogenesis (Gannon and Bader 1995). The endoderm is also implicated in the earlier process of precardiac mesoderm induction; coculture of Xenopus deep endoderm with the heart primordia enhances cardiogenesis (Nascone and Mercola 1995), and BMP2, which is secreted by the anterior endoderm, is sufficient to induce expression of myocardial genes in avian noncardiogenic mesoderm (Schultheiss et al. 1997).
Heart and endoderm development in the mouse both require the zinc finger transcription factor Gata4 (Kuo et al. 1997; Molkentin et al. 1997). Although 35% of gata4 null embryos arrest at the egg cylinder stage for unknown reasons, those that do gastrulate fail to form a fused heart or a foregut and develop partially outside the yolk sac and amnion. Gene expression within the precardiac mesoderm is not significantly affected, suggesting that cardiac differentiation is not the primary defect. In support of this conclusion, chimera analyses indicate that Gata4 function in the visceral and/or anterior definitive endoderm restores ventral closure and cardiac fusion to otherwise gata4 null embryos (Narita et al. 1997).
Gata4 is one of a family of six vertebrate Gata transcription factors, each of which contains two zinc fingers and binds to regulatory elements containing A/TGATAA/G or closely related sequences. The gata genes can be divided into two groups on the basis of sequence homology and expression pattern. The members of the first group, gata1, gata2, and gata3 have unique roles in hematopoiesis (for review, see Orkin and Zon 1997), whereas gata4, gata5, and gata6 are expressed in the ALPM and endoderm (for review, see Evans 1997). Studies of cis-regulatory elements have suggested that gata4, gata5, and gata6 have important roles in promoting the expression of both myocardial and endodermal genes. Transfection of heterologous cells with Gata factors activates transcription of myocardial genes including α-myosin heavy chain (α-MHC), cardiac troponin C, and B-type natriuretic peptide (BNP) (for review, see Evans 1997), as well as endodermal genes such as IFABP and HNF4 (Gao et al. 1998; Morrisey et al. 1998). Evidence that Gata factors directly activate the transcription of myocardial and endodermal genes has come from studies identifying functionally important Gata-binding sites in the regulatory regions of the myocardial genes α-MHC and BNP (for review, see Evans 1997) and the liver enhancer of serum albumin (Bossard and Zaret 1998). Recently, Gata-binding sites have been identified in two different regulatory elements of nkx2.5, one of the earliest known markers of the myocardial precursors (Searcy et al. 1998; Lien et al. 1999). Each of these elements activates expression in a manner similar to the endogenous nkx2.5 promoter and depends on Gata sites for activity.
Experiments in chick and Xenopus provide further in vivo evidence for the role of gata factors in heart development. Inhibition of Gata activity through the addition of three antisense oligonucleotides complementary to gata4, gata5, and gata6 produces variable abnormalities in avian cardiac morphogenesis (Jiang et al. 1998). Ectopic expression of gata4 and gata5 in Xenopus embryos leads to the premature expression of the myocardial genes α-cardiac actin and α-MHC (Jiang and Evans 1996). In contrast, sustaining the expression of Xenopus gata6 after it has normally declined inhibits expression of markers of terminal myocardial differentiation, perhaps by maintaining the precardiac mesoderm in an undifferentiated state (Gove et al. 1997).
gata genes also play evolutionarily conserved roles in endoderm development, as the Drosophila gata gene serpent and the Caenorhabditis elegans gata gene elt-2 are essential for differentiation of the embryonic gut (Rehorn et al. 1996; Fukushige et al. 1998). Another C. elegans gata gene, end-1, is implicated in the earlier process of endoderm specification (Zhu et al. 1997, 1998).
Here, we show that the zebrafish fau locus is essential for multiple aspects of heart and endoderm development and that fau encodes Gata5. Analysis of fau mutants reveals that Gata5 is required for the normal expression of nkx2.5 and for other aspects of myocardial differentiation. Mutations in gata5 also prevent the formation of normal amounts of cardiac and endodermal tissue and inhibit the normal morphogenesis of the heart, pharyngeal endoderm, and gut. Furthermore, we demonstrate that the overexpression of gata5 is sufficient to activate ectopic expression of nkx2.5 and other cardiac genes and to cause the formation of ectopic regions of rhythmically contracting tissue. Thus, our results establish Gata5 as a critical regulator of nkx2.5 expression and of multiple aspects of cardiac and endoderm development.
Results
fau mutants display defects in heart development
The zebrafish fau locus is defined by two alleles; fautm236a is a previously reported, ethylnitrosourea (ENU)-induced allele (Chen et al. 1996), whereas the second allele, faus26, arose spontaneously in an unrelated line. Both alleles segregate as fully penetrant, completely recessive mutations and cause equivalent phenotypes. The transheterozygous phenotype is also fully penetrant and is indistinguishable from the homozygous phenotypes.
Most conspicuously, fau mutations disrupt formation of the definitive heart tube, an early step in cardiac morphogenesis. Heart-tube formation depends upon the medial migration and fusion of the bilateral cardiac primordia (Stainier et al. 1993). In fau mutants this step of cardiac morphogenesis usually fails to occur, resulting in cardia bifida. However, 0%–45% of the fau mutant embryos in a given clutch produce fused, although morphologically abnormal, hearts. This variable expressivity is present in both alleles and also pertains to other aspects of the phenotype.
To examine when and how fau functions in heart development, we examined the expression of markers of the developing cardiac primordia. In fau mutants at 18.5 hr postfertilization (hpf), myocardial precursors expressing cardiac myosin light chain 2 (cmlc2) (Yelon et al. 1999) and endocardial precursors expressing tie2 (Lyons et al. 1998) fail to migrate to the embryonic midline (Fig. 1B,C), demonstrating that the morphogenesis of both tissues that comprise the zebrafish heart is affected. Development of more posterior lateral plate mesodermal derivatives, such as the fin bud and blood, are unaffected in fau mutants (data not shown). Therefore, fau is essential for the proper morphogenesis of both of the cellular components of the zebrafish heart tube but is not generally required for the development of lateral plate mesodermal derivatives.
Figure 1.
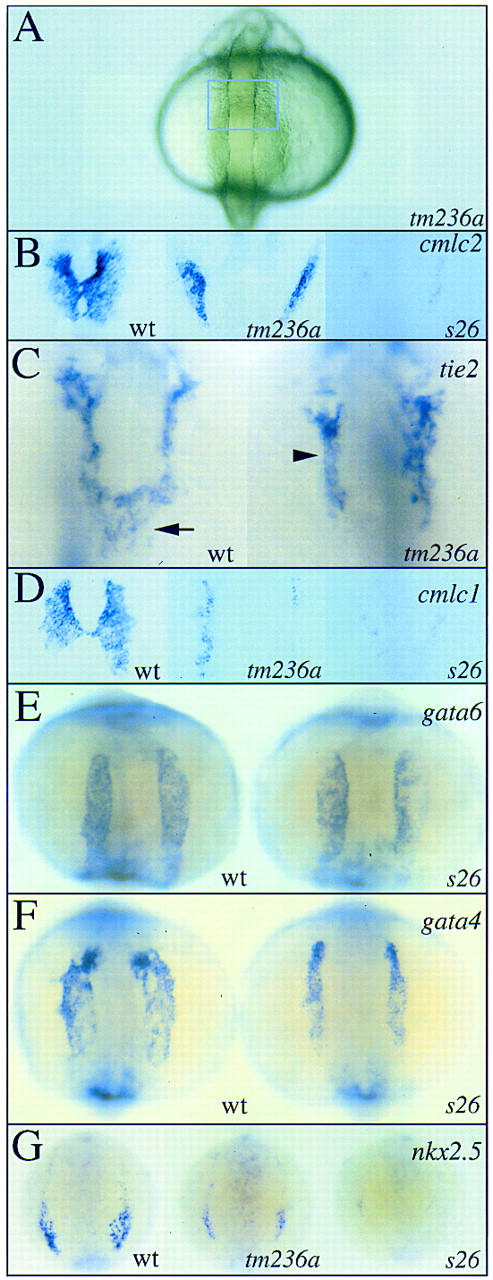
fau mutants display abnormal cardiac morphogenesis and differentiation. Expression of cmlc2 (B), tie2 (C), cmlc1 (D), gata6 (E), gata4 (F) and nkx2.5 (G) in wild-type embryos and fau mutant siblings at the 19-somite (18.5 hpf; A,B,C,D), 12-somite (15 hpf; E,F) and 6-somite (12 hpf; G) stages. All views are dorsal with anterior to the top. (A) At the 19-somite stage, both fautm236a and faus26 mutants are morphologically indistinguishable from wild-type siblings. The blue box approximately outlines the cardiogenic region, which lies ventral and just posterior to the midbrain–hindbrain boundary. B and D depict enlarged views of this region. (B) In wild-type embryos, the myocardial primordia express cmlc2 and have fused at the embryonic midline (left). In contrast, the myocardial primordia of fautm236a mutants do not migrate to the embryonic midline, resulting in cardia bifida (middle). The myocardial primordia of faus26 mutants also fail to migrate to the embryonic midline (right) and in addition display a severe reduction in the number of cells expressing cmlc2. (C) Formation of the endocardium (arrow) also depends on the migration of precursors from the ALPM to the embryonic midline. In fau mutants, endocardial precursors expressing tie2 are absent from the midline. tie2 expression in endothelial cells (arrowhead) reveals that more anterior vasculature forms normally in fau mutants. (D) cmlc1 is also expressed in the myocardial primordia. In the right fautm236a cardiac primordium (middle), <20 cells express cmlc1 whereas >120 cells express cmlc1 in each wild-type primordium (left). In contrast, fautm236a mutants and wild-type siblings contain approximately equal numbers of cmlc2-expressing cells (see B). (E) gata6 is widely expressed throughout the ALPM throughout somitogenesis. gata6 expression is not perturbed in fau mutants. (F) In contrast, ALPM expression of gata4 is slightly reduced in fau mutants. (G) nkx2.5 expression is also reduced in fau mutants. Although the number of cells expressing nkx2.5 is only moderately reduced in fautm236a mutants (middle), nkx2.5 expression is severely diminished in faus26mutants (right).
Mutations in fau lead to the abnormal expression of genes encoding myocardial sarcomere components. cardiac troponin T, tropomyosin, cmlc1, and cmlc2 are all expressed in fewer cells in fau mutants than in wild-type siblings (Fig. 1B,D; data not shown). Detailed examination of myocardial sarcomere gene expression in fautm236a mutants reveals a differential requirement for the fau gene product. For example, cmlc1 expression is more profoundly reduced in fautm236amutants than is the expression of cmlc2 (Fig. 1B,D, middle panels). This distinction is less apparent in faus26 mutants as too few cells express sarcomere genes in these mutants to allow easy comparison. The more severe defect in cmlc2 and cmlc1 expression observed in faus26 mutants than in fautm236a mutants indicates that the faus26 allele is the stronger of the two.
During earlier segmentation stages before the migration of the cardiac primordia to the midline, the ALPM exists as two parallel stripes of cells lateral to the head and anterior notochord. gata6, a gene expressed throughout the ALPM, is expressed normally in fau mutants (Fig. 1E). However, the expression of the related gene gata4 in the ALPM is slightly reduced (Fig. 1F). Together, these results indicate that fau is not required for the formation of the ALPM but is critical for the expression of cardiac genes within this tissue.
nkx2.5, an Nk homeobox gene required for heart development (for review, see Evans 1999), is also expressed in part of the ALPM from early in development. Expression of nkx2.5 is dramatically reduced in fau mutants (Fig. 1G), demonstrating that fau is a direct or indirect regulator of nkx2.5 expression and is required at a very early stage of ALPM differentiation. fautm236a mutants display less severely diminished nkx2.5 expression (Fig. 1G), further indicating that faus26 is the stronger allele.
Later in development, the zebrafish heart differentiates into an anterior ventricle and a posterior atrium. Immunofluorescent staining using monoclonal antibodies that recognize myosin heavy chain epitopes differentially expressed in ventricular and atrial tissues (Stainier and Fishman 1992) reveals that ventricular and atrial differentiation does not depend on cardiac fusion. For example, a robust amount of both ventricular and atrial tissue forms in the cardia bifida mutant cas (Fig. 2A,B). However, fautm236a myocardia, in addition to being smaller than cas myocardia, contain proportionally much less ventricular tissue (Fig. 2B,C). This defect is also observable at earlier stages, before the cardiac primordia normally fuse. At the 18-somite stage the expression of ventricular myosin heavy chain (vmhc), a marker of ventricular myocardium (Yelon et al. 1999), is dramatically reduced in fautm236a mutants (Fig. 2D). vmhc expression is more severely reduced, or even absent, in faus26 mutants (Fig. 2D).
Figure 2.
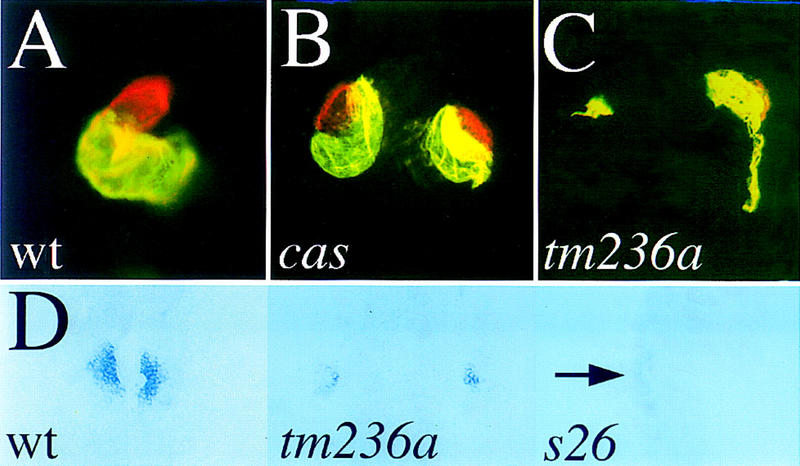
Formation of ventricular myocardial tissue is abnormal in fau mutants. Mutants (33 hpf) and wild-type siblings were stained with MF20 and S46 antibodies (A–C). Red fluorescence indicates MF20 (TRITC) staining of ventricular myocardium and yellow fluorescence indicates the overlap of MF20 (TRITC) and S46 (FITC) staining of atrial myocardium. A–C show ventral views with anterior to the top. D shows dorsal views of vmhc expression at the 18-somite stage (18 hpf). (A) Wild-type embryos develop a single, looped heart patterned into a ventricle (red) and atrium (yellow). (B) cas mutants display cardia bifida and form both ventricular and atrial tissue. (C) fau mutants also display cardia bifida but form less total myocardium and only a very small amount of ventricular tissue. Here, one myocardium (left) has not formed any ventricular tissue. (D) vmhc is expressed in the preventricular component of the developing myocardium. vmhc expression is dramatically reduced in fau myocardial primordia even prior to cardiac fusion. faus26 mutants display a variable loss of vmhc expression; (right) a faus26 mutant in which less than five cells weakly express vmhc in the left ALPM (arrow), whereas none do on the right.
fau mutants display defects in endoderm morphogenesis
As embryological experiments in chick and amphibia have suggested that the endoderm is a source of instructive signals for the precardiac mesoderm, we investigated the effects of the fau mutations on endoderm development. From 24 to 48 hpf, the zebrafish gut endoderm coalesces at the midline where it will form a tube, loop to the left, and differentiate into the liver, pancreas, swim bladder, and epithelium of the gastrointestinal tract (Warga and Nüsslein-Volhard 1999). The anterior gut endoderm expresses axial, a homolog of HNF3β (Strahle et al. 1996), whereas the posterior endoderm expresses fkd2, a homolog of HNF3γ (Odenthal and Nüsslein-Volhard 1998). In fau mutants, axial-expressing cells fail to coalesce at the midline, disrupting anterior gut tube morphogenesis (Fig. 3A, arrow). The posterior endoderm displays more variable defects that range from the near absence of gut to the failure of gut looping (Fig. 3B).
Figure 3.
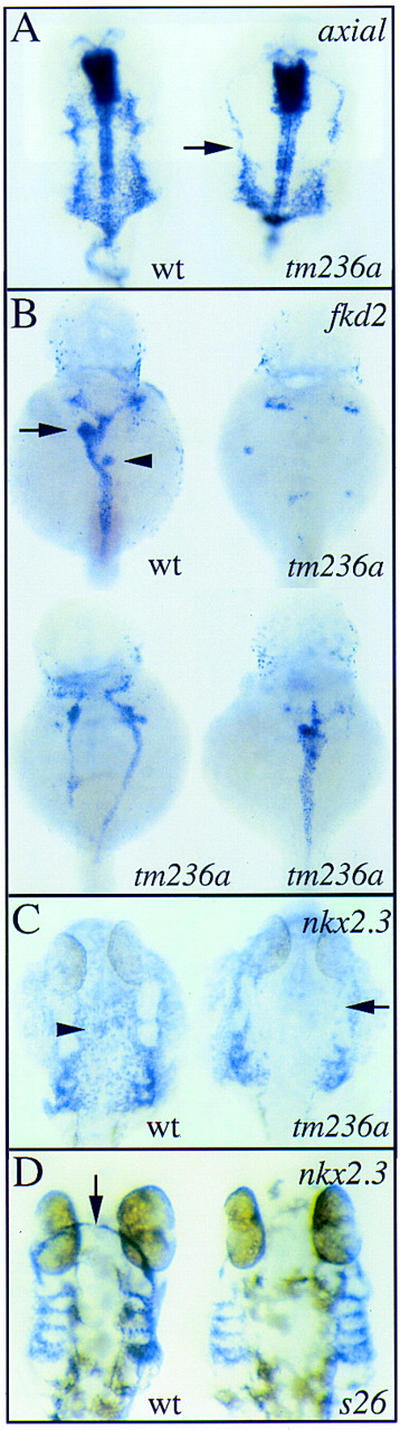
Endoderm development is abnormal in fau mutants. Expression of axial (A), fkd2 (B), and nkx2.3 (C,D) in wild-type embryos and fau mutant siblings at 44 hpf (A,B,D) and 28 hpf (C). All views are dorsal with anterior to the top. (A) axial is expressed in the anterior gut endoderm and the ventral neuroectoderm. At this stage, the anterior gut endoderm is coalescing at the embryonic midline in wild-type embryos. In fau mutants, gut endoderm is only present laterally (arrow). For purposes of orientation, the arrow is at the level of the midbrain–hindbrain boundary. (B) fkd2 is expressed throughout the pancreas (arrowhead), liver (arrow), and posterior gut tube in wild-type embryos. fau mutants display variable abnormalities in the morphogenesis and amount of fkd2-positive tissue. These defects range from near absence of fkd2-positive endoderm (top right) to formation of two lateral gut tubes (bottom left) to the absence of gut looping (bottom right). (C) Morphogenesis of the pharyngeal pouches is disrupted in fau mutants. The mediolateral distance between the pouches is greater in fau mutants and the anterior pouches (arrow) are disorganized. Also, midline nkx2.3-expressing cells (arrowhead) appear to be absent in fau mutants. (D) In faus26 mutants, the anterior domain of nkx2.3 expression (arrow) is absent.
fau mutations also disrupt the morphogenesis of the pharyngeal endoderm. In fau mutants, the pharyngeal pouches are separated by a larger mediolateral distance (Fig. 3C), a defect that is reminiscent of the inability of the gut endoderm and precardiac mesoderm to migrate to the embryonic midline. In addition, the anterior pharyngeal pouches are dysmorphic in fautm236a mutants (Fig. 3C, arrow), whereas the most anterior nkx2.3-expressing cells appear to be absent in faus26 mutants (Fig. 3D, arrow).
fau encodes Gata5
Initially, we mapped the fau locus 1.5 cM proximal to the simple sequence repeat marker z20895 on linkage group 23 (Shimoda et al. 1998). Using an enriched selection for amplified fragment length polymorphisms (AFLPs), we identified a number of markers located within a centimorgan of fau. In a parallel approach, we mapped gata5, a gene dynamically expressed in both the endoderm and cardiac mesoderm (see below), to the same genomic region using the Goodfellow radiation hybrid panel (Kwok et al. 1998). Linkage analysis using a restriction fragment length polymorphism (RFLP) within the gata5 gene demonstrated that the mutant gata5 allele segregated with the fautm236a phenotype in >300 meioses.
Sequencing of the fautm236a allele of gata5 revealed a G-to-A substitution at the first base of the third intron (Fig. 4A). This mutation disrupts the highly conserved GT dinucleotide necessary for recognition of the splice site by the U1 snRNP (Krawczak et al. 1992). Sequencing of gata5 cDNAs from fau mutants demonstrated that the fautm236a mutation leads to mis-splicing at flanking cryptic splice sites (Fig. 4B). The most frequently isolated cDNA results from the use of a downstream cryptic splice site and contains a 31-bp insertion predicted to create a translational frameshift (Fig. 4C,D). A less frequently isolated cDNA is produced by use of a cryptic splice site 30 bp upstream of the inactivated site (Fig. 4B). This mutant splice variant is predicted to produce a 10-amino-acid deletion in the second zinc finger (Fig. 4C,D).
Figure 4.
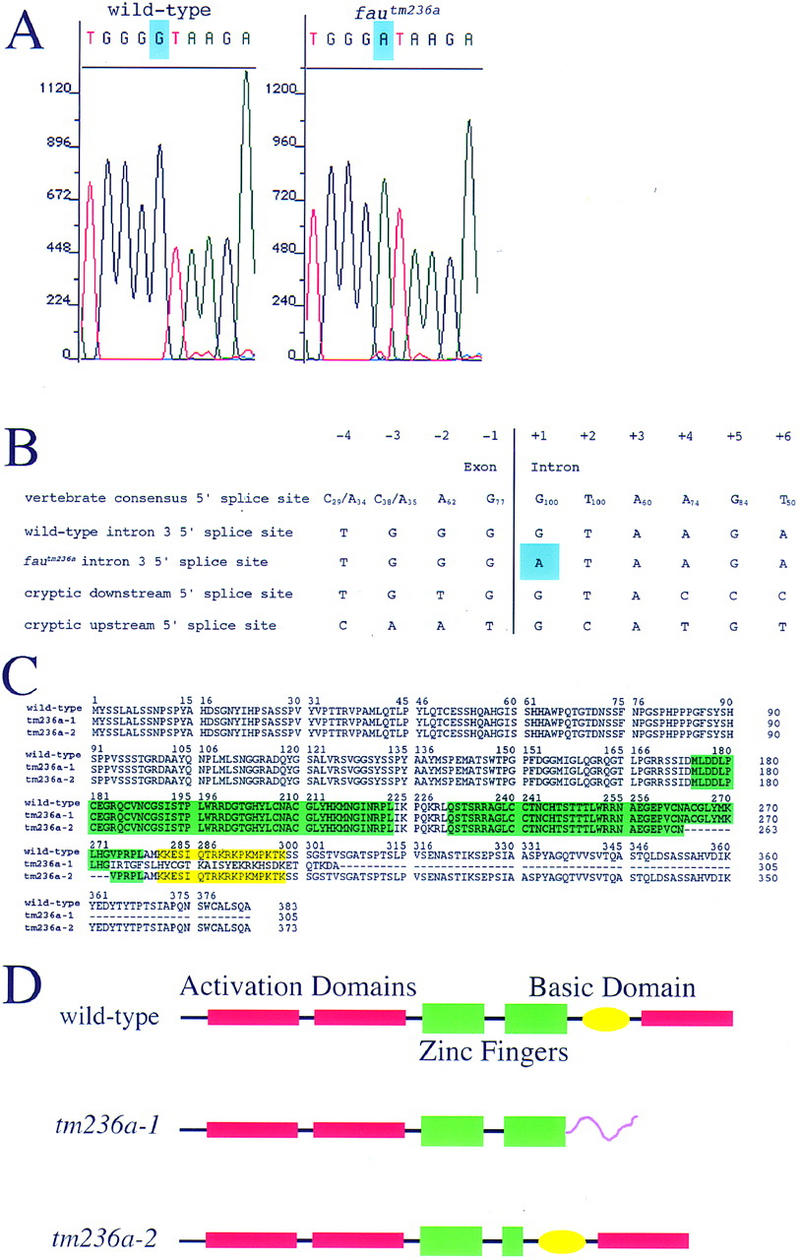
fau encodes zebrafish Gata5. (A) Sequencing of fautm236a and wild-type genomic DNA reveals a G-to-A substitution at the +1 position of the third intron of gata5 (blue). (B) Comparison of the vertebrate consensus 5′ splice site (Krawczak et al. 1992) with the wild-type gata5 intron 3 5′ splice site, the mutant sequence, the cryptic downstream 5′ splice site, and the cryptic upstream 5′ splice site. Subscripts indicate the frequency at which nucleotides occur in the consensus splice site. Blue shading marks the fautm236a mutation. (C) Comparison of the complete wild-type Gata5 protein sequence with the mutant sequences. tm236a-1 is the sequence of the protein predicted to result from use of the downstream cryptic splice site and was most frequently isolated from fautm236a mutants. tm236a-2 is the sequence of the protein predicted to result from use of the upstream cryptic splice site and was less frequently isolated from fautm236a mutants. The two zinc fingers are shaded green. The basic domain required for nuclear localization is shaded yellow. (D) Schematic of the expected effects of the fautm236a mutation on Gata5 structure. The purple carboxy-terminal tail of tm236a-1 represents the nonfunctional domain created by the frameshift mutation. The deletion of the carboxy-terminal portion of the second zinc finger of tm236a-2 is represented by the smaller size of the second green rectangle. The transcriptional activation domains depicted in red are predicted based on comparison of zebrafish Gata5 domains with identified murine Gata4 and Gata5 activation domains (Durocher et al. 1997; Morrisey et al. 1997b).
Structure/function studies have revealed that murine Gata5 contains two amino-terminal transcriptional activation domains, as well as a basic domain required for nuclear localization (Morrisey et al. 1997b). Murine Gata4 also contains a carboxy-terminal transcriptional activation domain required for synergistic activity with Nkx2.5 (Durocher et al. 1997; Lee et al. 1998; Sepulveda et al. 1998). As Gata5 shows a similar synergism with Nkx2.5 (Durocher et al. 1997), Gata5 may also contain a carboxy-terminal activation domain. Sequence comparison of murine Gata5 and zebrafish Gata5 suggests that the two proteins are similar in structure (Rodaway et al. 1999). The more prevalent 31-bp insertion caused by the fautm236a mutation disrupts the putative carboxy-terminal activation domain and the basic nuclear localization domain of zebrafish Gata5 (Fig. 4C,D). The rarer splicing event deletes a 30-bp region that encodes part of the carboxy-terminal zinc finger required for DNA binding and for interaction with the transcription factors NF-AT3 and Nkx2.5 (Durocher et al. 1997; Lee et al. 1998; Molkentin et al. 1998). Additionally, the carboxy-terminal zinc finger has also been implicated in mediating heterotypic interactions between Gata factors (Charron et al. 1999). Therefore, we suspect that both mutant forms of Gata5 retain little, if any, activity. However, wild-type message can be detected in fautm236a mutants (see Materials and Methods), indicating that the splicing machinery is still able to recognize the mutant splice site, albeit at a much reduced frequency. Taken together, these data indicate that fautm236a is a mutation in the gata5 gene that substantially reduces, but does not eliminate, the production of wild-type Gata5.
Examination of the faus26 gata5 cDNA sequence did not identify a mutation. As gata5 expression is dramatically diminished in faus26 mutants (see below), we hypothesize that the faus26 mutation disrupts the function of the gata5 promoter. We sequenced 800 bp upstream of the transcriptional start site and have identified a putative core promoter without successfully identifying the faus26 lesion (data not shown). Therefore, we speculate that the faus26 mutation disrupts a distal enhancer of the gata5 gene that reduces gata5 transcription to very low levels.
gata5 expression in wild-type and fau mutant embryos
In zebrafish, gata5 is first expressed at the dome stage (late blastula) in the most marginal cells (Fig. 5A, arrowhead), a population that includes the endodermal progenitors and some mesodermal progenitors (Warga and Nüsslein-Volhard 1999), and in the yolk syncytial layer (YSL), an extraembryonic tissue underlying the blastoderm that may be equivalent to the mouse visceral endoderm (Fig. 5A, arrow). During gastrulation, gata5-expressing cells in the hypoblast adopt one of two different morphologies. Predominantly dorsally, gata5-expressing cells adopt the large, flat morphology characteristic of endoderm (Fig. 5C, arrowhead) (Warga and Nüsslein-Volhard 1999). Further evidence that these large gata5-expressing cells are endodermal progenitors comes from the very similar distribution of these cells and the axial- and sox17-expressing endodermal progenitors (Alexander and Stainier 1999). In a more ventral domain reflecting the regional propensity to become heart (Stainier et al. 1993), other gata5-expressing cells adopt a rounder morphology characteristic of mesodermal progenitors (Fig. 5C, arrow). After completion of gastrulation, gata5 expression persists in the endoderm and ALPM (Fig. 5D). ALPM expression of gata5 is maintained throughout somitogenesis (10–24 hpf) whereas the level of endodermal expression diminishes (Fig. 5E). Later, from 20–22 hpf, cardiac expression of gata5 becomes predominantly endocardial (Fig. 5F, arrow), as has been described for murine gata5 (Morrisey et al. 1997a).
Figure 5.
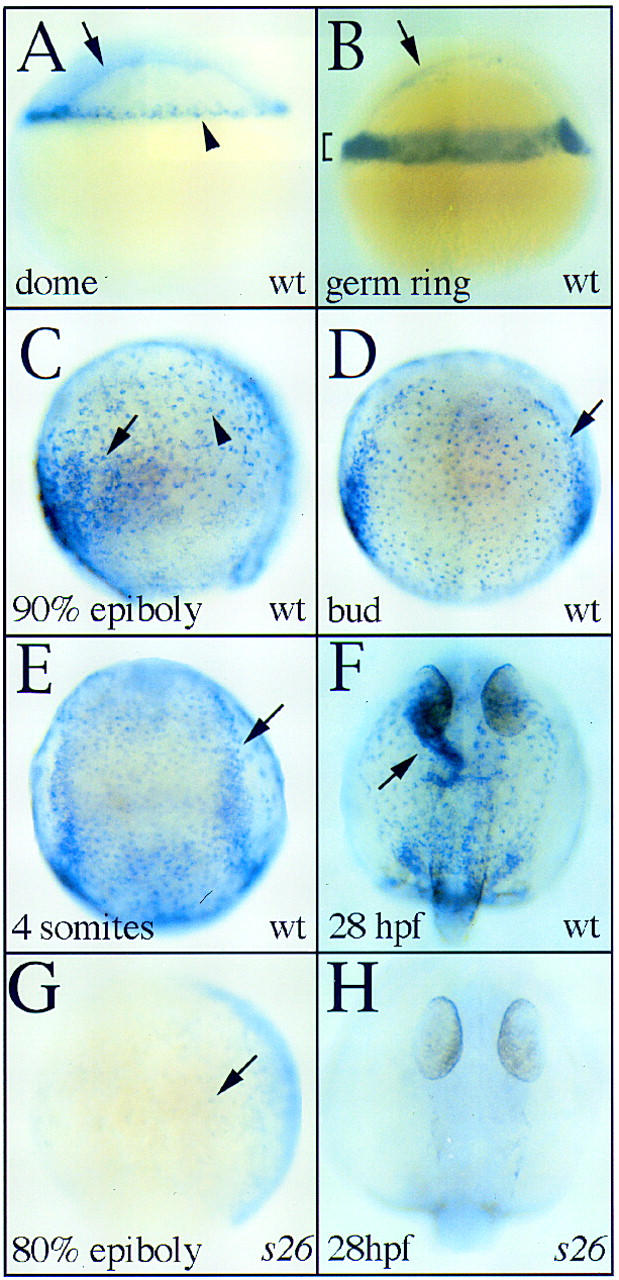
gata5 expression in zebrafish embryos at dome (4.3 hpf; A), germ ring (5.7 hpf; B), 90% epiboly (9 hpf; C), bud (10 hpf; D), 4-somite (11.3 hpf; E), 28 hpf (F,H), and 80% epiboly (8.3 hpf; G) stages. A,B,C, and G show lateral views with dorsal to the right. D,E,F, and H show dorsal views with anterior to the top. (A) At dome stage, gata5 is expressed in the yolk syncytial layer (arrow) and the most marginal tiers of the blastoderm (arrowhead). (B) During early gastrulation, gata5 expression is maintained in the marginal zone (bracket) and yolk syncytial layer (arrow). (C) Late in gastrulation, gata5 is expressed by cells of two different morphologies. Predominantly dorsally, endodermal cells displaying a characteristically large and flat morphology (arrowhead) express gata5. Ventrally, hypoblast cells with a rounder, more mesodermal appearance (arrow) also express gata5. (D) After the completion of gastrulation, gata5 is expressed in the endoderm (stippled staining) and ALPM (arrow). (E) During early somitogenesis, both the endoderm and lateral plate mesoderm (arrow) continue to express gata5. (F) At 28 hpf, gata5 is expressed most strongly in the endocardium (arrow) and weakly in the surrounding noncardiogenic ALPM. (G) faus26 mutants display markedly reduced gata5 expression. At 80% epiboly, gata5 mRNA is undetectable in the YSL and observed only faintly (arrow) in the blastoderm of faus26 mutants. (H) gata5 expression is undetectable in faus26 mutants at 28 hpf.
Expression of gata5 in fautm236a mutants and wild-type siblings is indistinguishable until morphogenetic defects become apparent. In contrast, faus26 mutants display markedly decreased gata5 expression throughout development and in both the YSL and embryonic tissues (Fig. 5G,H). Very low levels of gata5 expression can be detected in faus26 mutants by in situ hybridization (Fig. 5G, arrow) and RT–PCR (data not shown), indicating that although expression is strikingly diminished, the faus26 mutation does not completely eliminate the production of gata5 message.
Overexpression of wild-type gata5 rescues heart fusion in fautm236a mutants
To confirm that the fau mutant phenotype is due to the absence of Gata5 activity, we injected mRNA encoding wild-type Gata5 into fautm236a mutants at the 1–4 cell stage. Although this approach leads to widespread misexpression, injection of 20 pg of gata5 mRNA restores cardiac fusion in fautm236a mutants (Fig. 6A; Table 1), confirming that the fau phenotype is due to a loss-of-function mutation in gata5. gata5-injected fau mutants are not rescued to viability as gata5 misexpression causes variable abnormalities in morphogenesis, including anterior truncations and compression of the anterior–posterior axis. Injection of tm236a-1, a gata5 mRNA containing the same insertion and frameshift as most commonly caused by the fautm236a mutation, does not rescue cardiac fusion (Table 1). Moreover, tm236a-1 overexpression does not disrupt morphogenesis, indicating that it has little if any biological activity.
Figure 6.
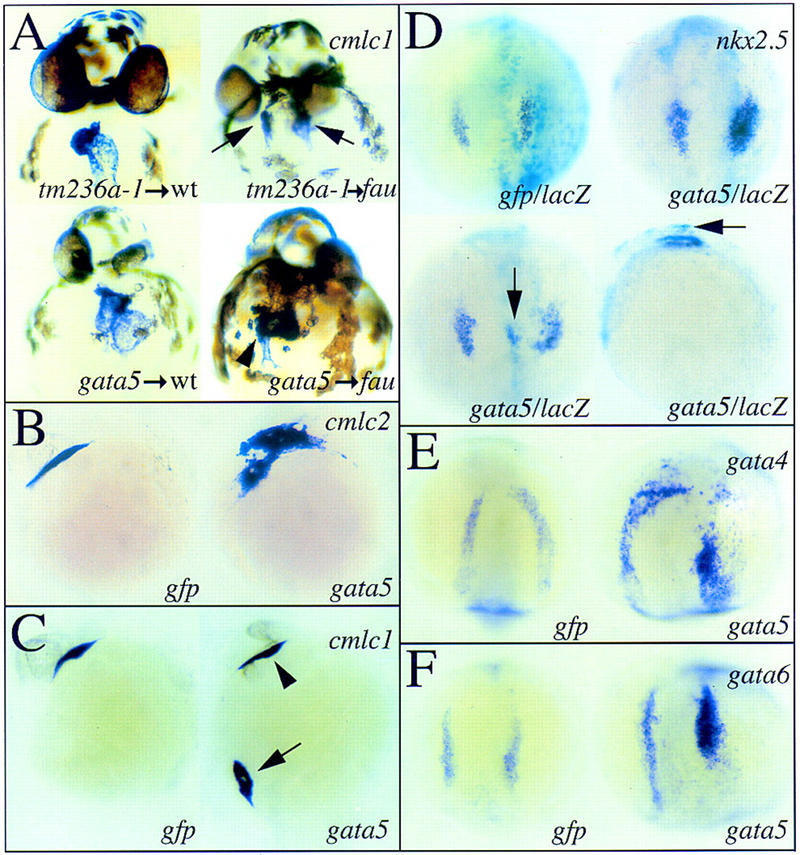
Overexpression of gata5 rescues cardiac fusion in fautm236a mutants and leads to ectopic expression of myocardial genes. Expression of cmlc1 (A,C), cmlc2 (B), nkx2.5 (D), gata4 (E), and gata6 (F) in wild-type embryos and fau mutant siblings at 36 hpf (A), 25 hpf (B,C), 5-somite stage (11.7 hpf; D), and 12-somite stage (14.5 hpf; E,F). Views are anterior (A), lateral [B,C,D (bottom right) ], and dorsal [D (top, and bottom left), E,F]. (A) Injection of 20 pg of tm236a-1 mRNA has no demonstrable effect on development (top left) and does not restore cardiac fusion in fautm236a mutants (top right, arrows). In contrast, injection of an equivalent amount of wild-type gata5 mRNA causes variable defects in head and body morphology (bottom left) and can restore cardiac fusion in fautm236a mutants (bottom right, arrowhead). (B) Injection of a higher dose of gata5 mRNA (50 pg) can cause expanded expression of cmlc2 in a domain contiguous to the ALPM. (C) gata5 overexpression can also cause ectopic expression of cmlc1. Here, gata5 overexpression has led to ectopic cmlc1 expression in cells overlying the ventral side of the yolk (arrow). A heart tube has also formed in the expected location (arrowhead). (D) Injection of 50 pg of gata5 mRNA can increase the number of cells expressing nkx2.5 (top right) and cause ectopic nkx2.5 expression (bottom). The bottom panels show dorsal and lateral views of the same embryo, which exhibits a dorsal midline region of ectopic nkx2.5 expression (arrows). β-Galactosidase staining produces a turquoise color marking the distribution of coinjected lacZ mRNA. gata5 overexpression can also lead to increased expression of gata4 (E) and gata6 (F).
Table 1.
Overexpression of wild-type gata5 restores cardiac fusion in fautm236a mutants
| Injected mRNA
|
Number of fautm236amutants (by genotype)
|
Number with cardia bifida
|
|---|---|---|
| gata5 | 39 | 9 (23%) |
| tm236a-1 | 25 | 19 (76%) |
| Uninjected | 35 | 23 (70%) |
The observed value of the test χ2(1), 17.33, indicates that the increased rate of cardiac fusion following overexpression of gata5 in fautm236amutants is statistically significant.
To test whether the defects in organogenesis exhibited by fau mutants are due to reduced Gata5 activity in the extraembryonic YSL, we injected gata5 mRNA directly into the YSL. Overexpression of gata5 mRNA in the YSL does not restore cardiac fusion in fautm236a mutants (14 cardia bifida mutants observed in a clutch of 60 YSL-injected embryos), suggesting that gata5 is required embryonically to promote heart fusion.
Ectopic expression of gata5 leads to the ectopic expression of myocardial genes
Injection of wild-type embryos with 50 pg of gata5 mRNA expands the ALPM expression of cmlc1, cmlc2, vmhc, and the MF20 and S46 myosin heavy chain proteins and leads to their ectopic expression (Fig. 6B,C; Table 2; data not shown). Occasionally, ectopic domains of myocardial gene expression both in the head and the tail were observed to contract rhythmically (Table 2). Control embryos injected with gfp mRNA did not manifest either expansion of myocardial gene expression or ectopic regions of beating tissue.
Table 2.
gata5 overexpression causes ectopic expression of myocardial genes and beating tissue
| Injected mRNA
|
Riboprobe
|
Expanded expression
|
Ectopic expression
|
|---|---|---|---|
| gata5 | cmlc1 | 32/95 | 10/95 |
| gata5 | cmlc2 | 20/64 | 7/64 |
| gata5 | vmhc | 13/22 | 4/22 |
| gata5 | nkx2.5 | 11/18 | 4/18 |
| gata5 | nkx2.5 | 0/6 | 0/6 |
| gata5 and lacZ | nkx2.5 | 84/237a | 47/237a |
| gfp and lacZ | nkx2.5 | 0/36 | 0/36 |
| gata5 | gata4 | 9/16 | 6/16 |
| gfp | gata4 | 0/6 | 0/6 |
| gata5 | gata6 | 14/22 | 10/22 |
| gfp | gata6 | 0/5 | 0/5 |
| Ectopic beating tissue | |||
| gata5 | 4/52 | ||
| gfp | 0/7 |
Embryos were scored as displaying expanded expression if expression was noted in cells adjacent to the normal domain [see Fig. 6B,D (upper right), E,F for examples]. Embryos were scored as displaying ectopic expression or ectopic beating tissue if tissue was noted dorsally or posteriorly within the embryo in a domain discontinuous with the ALPM or heart [see Fig. 6C,D (bottom) for examples].
Of the gata5-overexpressing embryos exhibiting expanded or ectopic nkx2.5 expression, 84/84 and 46/47 of the embryos, respectively, displayed increased expression within a domain of β-galactosidase activity.
Because nkx2.5 is downregulated in fau mutants, we investigated whether gata5 overexpression was sufficient to induce the expression of zebrafish nkx2.5 or other myocardial regulatory genes. Notably, we found that injection of 50 pg of gata5 mRNA expands the domain of nkx2.5, gata4, and gata6 expression within the ALPM (Fig. 6D–F; Table 2) and causes them to be ectopically expressed (Fig. 6D; Table 2). Coinjection of lacZ and gata5 mRNA allowed us to correlate the embryonic distribution of the exogenous RNA with the increased expression of nkx2.5. In all cases but one (n = 131), regions of expanded or ectopic nkx2.5 expression overlapped with the distribution of exogenous gata5 mRNA as indicated by β-galactosidase activity (Fig. 6D; Table 2), suggesting that Gata5 autonomously induces the expression of myocardial genes.
Surprisingly, the ectopic expression of these genes does not appear to be limited to the mesodermal germ layer. Ectopic expression of nkx2.5, gata4, and gata6 was observed in dorsal regions of the embryo normally fated to form neuroectoderm (Fig. 6D; data not shown). Neuroectodermal expression of nkx2.5 and gata6 is never observed during normal zebrafish embryogenesis.
Human gata5 maps to 20q13.2–q13.3
Mutations in human NKX2-5 can cause congenital cardiac septation defects (Schott et al. 1998). As Gata4 and Gata5 physically interact and synergize with Nkx2.5 (Durocher et al. 1997; Lee et al. 1998; Sepulveda et al. 1998), and as zebrafish Gata5 is essential for the correct expression of nkx2.5, it is possible that mutations in gata genes also cause human congenital heart defects. In support of this hypothesis, gata4 haploinsufficiency has been associated with congenital heart malformations (Pehlivan et al. 1999). Human gata4 and gata6 map to chromosome 8p23 (Pehlivan et al. 1999) and 18q11.1–q11.2 (Suzuki et al. 1996), respectively. Using the Stanford G3 radiation hybrid panel, we mapped human gata5 to chromosome 20q13.2–q13.3. The study of appropriate pedigrees should determine whether gata5 also plays an important role in human congenital heart disease.
Discussion
gata5 is required for cardiac morphogenesis and differentiation, as well as the production of normal amounts of myocardial tissue
In this study we establish that the fau locus encodes zebrafish Gata5, an evolutionarily conserved zinc finger transcription factor, and that mutations in gata5 profoundly disrupt cardiac development. Most conspicuously, diminished Gata5 activity prevents the precardiac mesoderm from migrating to and fusing at the embryonic midline, resulting in cardia bifida.
Reduced Gata5 activity also disrupts the expression of several genes in the ALPM, including nkx2.5. The defect in nkx2.5 expression is observable early in somitogenesis indicating that Gata5 is required very early for proper myocardial differentiation. As previous studies have demonstrated that two murine nkx2.5 regulatory elements contain essential Gata-binding sites (Searcy et al. 1998; Lien et al. 1999), Gata5 may be a direct endogenous regulator of nkx2.5. Consistent with this hypothesis, gata5 is expressed during blastula stages in a domain that includes the myocardial precursors, whereas we first observe nkx2.5 expression in a subset of gata5-expressing cells only after the completion of gastrulation (data not shown). Similar relative timing of expression of these genes is seen during chick development (T. Evans, pers. comm.).
Gata5 is capable of activating transcription from a wide range of myocardial promoters in vitro (Charron and Nemer 1999). Our results suggest that the same is true in vivo; gata5 mutants display marked defects in the expression of many genes encoding components of the myocardial sarcomere (e.g., cmlc1, cmlc2, vmhc, cardiac troponin T, tropomyosin). However, it is not clear at this point which of these genes are direct targets of Gata5 and which require intermediate Gata5-dependent regulators. In fact, there may be no clear division between these cases as Gata5 may act both directly and indirectly on a single promoter. For example, Gata5 may participate in the induction of nkx2.5 and also bind cooperatively with Nkx2.5 to regulatory elements of a wide range of myocardial genes. It is also interesting to note the different requirements various sarcomeric protein genes have for Gata5. For example, although the expression patterns of cmlc1 and cmlc2 are indistinguishable in wild-type embryos, cmlc1 expression is more severely reduced than cmlc2 expression in fau mutants. Perhaps these differences reflect different affinities of Gata5 for the respective cis regulatory elements.
In addition to exhibiting defects in myocardial differentiation, the hearts of fau mutants at 33 hpf are very small in comparison to those of wild-type siblings and other cardia bifida mutants such as cas. Similarly, the number of developing myocardial precursors is reduced in fau mutants throughout earlier segmentation stages. This deficit does not seem to reflect problems in ALPM specification or maintenance as gata6 expression demonstrates that the ALPM is present and morphologically normal in gata5 mutants. Instead, Gata5 appears to play a role in the induction, differentiation, and/or proliferation of the myocardial precursors within the ALPM.
Differentiation of the myocardium into atrial and ventricular components is also defective in fau mutants. fau myocardia preferentially form atrial tissue, indicating that gata5 is required for ventricular induction or differentiation and that the two chambers of the heart have different requirements for gata5. However, zebrafish gata5 does not appear to be differentially expressed in the two chambers of the heart. Although we have not examined the distribution of Gata5 protein, the expression pattern of the gata5 gene suggests that it is not directly involved in patterning the cardiac chambers. Instead, the defect in ventricular development may be secondary to the reduced amount of myocardial tissue produced in gata5 mutants if embryos must elaborate a threshold amount of myocardial tissue to sustain ventricular induction or differentiation. Alternatively, Gata5 may act with an unidentified ventricle-specific factor to promote ventricular development.
Taken together, these results demonstrate that Gata5 has a critical role in multiple aspects of zebrafish heart development including the expression of nkx2.5 and other myocardial genes, the production of a normal amount of myocardial precursors, the morphogenesis of both the myocardial and endocardial components of the heart, and the differentiation of the ventricular chamber.
gata5 is required for endoderm morphogenesis
Previous expression studies have suggested a role for Gata factors in the early development of the embryonic endoderm (Laverriere et al. 1994; Morrisey et al. 1997a; Bossard and Zaret 1998). Here, we have confirmed that zebrafish gata5 is essential for the formation of normal amounts and morphogenesis of the endoderm. Although gut and pharyngeal endoderm lineages separate early during zebrafish development and have distinct genetic requirements (Alexander 1999), gata5 is necessary for the morphogenesis of both tissues. Interestingly, Gata5 is required for the movement toward the midline of both the heart primordia and the gut and pharyngeal endoderm, suggesting a causative link between these defects or a common role of Gata5 in promoting the medial migration of several tissues.
gata5 overexpression causes the ectopic expression of myocardial genes
We show that overexpression of zebrafish gata5 leads to the expanded or ectopic expression of cmlc1, cmlc2, vmhc, gata4, gata6, nkx2.5, and the MF20 and S46 myosin heavy chain proteins, indicating that zebrafish Gata5 can act as a positive regulator of many myocardial genes, even in noncardiogenic tissues. This is the first demonstration of the ability of a Gata factor to enhance cardiogenesis in the context of a developing embryo. In contrast, overexpression of gata4 and gata5 in Xenopus does not affect the spatial pattern of myocardial gene expression or heart morphogenesis (Jiang and Evans 1996). The apparently different capacities of Xenopus and zebrafish gata5 may reflect species-dependent differences in the plasticity of cell fate or differences in the activities of the two homologs. In fact, zebrafish Gata5 function may be more similar to that of murine Gata4, which can also potentiate cardiogenesis, at least in the P19 cell line (Grepin et al. 1997).
The ability of Gata5 to induce ectopic myocardial gene expression is not entirely unexpected, given that Gata5 regulates nkx2.5 expression and overexpression of nkx2.5 can induce some ectopic myocardial gene expression (Chen and Fishman 1996). It will be interesting to determine whether Nkx2.5 is an obligatory effector of Gata5 function in myocardial cells. Another intriguing issue concerns whether Gata5, when forcibly expressed in noncardiogenic tissues, simply activates the transcription of a subset of myocardial genes or whether Gata5 can reprogram an otherwise noncardiac cell to become completely myocardial in character. The unique ability of gata5 overexpression to produce ectopic beating tissue suggests that, at least in some contexts, these cells exhibit myocardial behavior. Therefore, both phenotypic analysis of fau mutants and gain-of-function experiments indicate that Gata5 plays an essential role in the induction, differentiation, and/or proliferation of the myocardial precursors.
Although the normal expression of gata5 in fautm236a mutants suggests that Gata5 does not regulate its own expression, fau mutants exhibit modestly decreased expression of gata4, and overexpression of gata5 is able to induce the ectopic transcription of gata4. Therefore, Gata5 appears to positively regulate gata4, through which Gata5 may mediate some of its effects on cardiac differentiation. Although no defects in gata6 expression were noted in fau mutants, gata5 overexpression can cause ectopic gata6 expression, suggesting that Gata5 may have a redundant role in regulating gata6. Perhaps embryos mutant for both gata4 and gata5 will reveal a nonessential role for Gata5 in gata6 gene regulation. Previous work on mouse gata4 and gata6 mutants has demonstrated that Gata4 negatively regulates gata6 in the ALPM whereas Gata6 positively regulates gata4 in extraembryonic tissues (Kuo et al. 1997; Molkentin et al. 1997; Morrisey et al. 1998). Thus, complex interregulation by Gata4, Gata5, and Gata6 may have an important role in controlling the early events of cardiac and endodermal development.
gata5 may act both tissue autonomously and nonautonomously in heart development
Previous studies in both mouse and zebrafish have demonstrated that cardia bifida can be secondary to defects in endoderm development (Narita et al. 1997; Peyrieras et al. 1998). This work leads us to hypothesize that the cardia bifida in fau mutants is also secondary to their severe endodermal defects. Moreover, the presence of endodermal defects in the cardia bifida mutants fau, oep, and cas (Schier et al. 1997; Alexander et al. 1999) suggests that zebrafish cardia bifida may be a general consequence of defects in endodermal morphogenesis or differentiation. Conclusive experimental tests of the autonomy of the morphogenetic defects in fau mutants will require analyses of embryos in which interacting tissues are of distinct genotypes, the production of which is contingent on techniques not yet developed in zebrafish.
Although the absence of cardiac fusion in fau mutants may be secondary to endoderm defects, other cardiac abnormalities may be due to reduced Gata5 activity in the ALPM. Despite the complete and early absence of gut endoderm in cas mutants, cas myocardia exhibit no defect in gene expression (Alexander et al. 1999), are larger than fau myocardia, and contain a normal ratio of atrial to ventricular tissue. Thus, fau mutants display defects in gene expression, cardiac growth, and ventricular development beyond what is attributable to the loss of endoderm, implicating ALPM gata5 expression in these roles. The observation that gata5 overexpression autonomously promotes the ectopic expression of nkx2.5 further suggests that Gata5 acts within cells of the ALPM to regulate myocardial differentiation. We therefore propose that the defects in fau cardiac morphogenesis are secondary to the reduction of Gata5 activity in the endoderm, whereas the additional defects in cardiac growth and differentiation are due to the reduction of Gata5 function in the ALPM.
Zebrafish Gata5 and murine Gata4 may function similarly during organogenesis
Although mouse gata5 null mutants have not been extensively described, they are viable (Morrisey et al. 1998), suggesting that some of the functions performed by zebrafish Gata5 may be fulfilled by Gata4 or Gata6 in the mouse. Unfortunately, mouse embryos deficient for Gata4 (Kuo et al. 1997; Molkentin et al. 1997) or Gata6 (Morrisey et al. 1998; Koutsourakis et al. 1999) arrest at embrionic day (E) 7.0–9.5 and E 5.5-7.5, respectively, impeding the analysis of later developmental events such as cardiac chamber formation and the development of endoderm-derived organs. However, gata4 null mice and gata5 mutant zebrafish both display cardia bifida, prompting a comparison. gata4 null mice do not undergo ventral closure, which has led to the idea that cardia bifida in gata4 null mice is a direct consequence of the defect in ventral folding. Our study, indicating that zebrafish cardia bifida can result from gata-dependent defects in the migration of the cardiac primordia to the embryonic midline, suggests an alternative interpretation. In zebrafish, the cardiac precursors migrate on the yolk syncytial layer to reach the embryonic midline (Stainier et al. 1993). Perhaps murine precardiac mesoderm reaches the embryonic midline similarly, by actively migrating on the underlying visceral endoderm and not through ventral folding of the body wall. In support of this hypothesis, in the mouse the midline cardiogenic plate appears to form before the beginning of ventral folding (Plate 8d in Kaufman 1992; Saga et al. 1999). Therefore, cardia bifida in gata4 null mice may stem from a defect in the migration of the precardiac mesoderm to the embryonic midline in much the same way as cardia bifida arises in mice lacking MesP1 (Saga et al. 1999). We propose that mouse gata4 is required for two early and distinct morphogenetic processes: (1) the initial migration of the bilateral cardiac precursors to the embryonic midline where they form the cardiogenic plate, and (2) the subsequent ventral folding required for foregut and ventral body-wall morphogenesis. Determining whether cardiac fusion is truly independent of ventral closure will require the identification of mouse mutations that inhibit ventral closure without affecting cardiac fusion.
In summary, we have shown that gata5 mutants show reduced expression of nkx2.5 and that gata5 overexpression leads to ectopic nkx2.5 expression. These data together with the demonstrated presence of Gata-binding sites in nkx2.5 regulatory elements indicate that gata5 functions upstream of nkx2.5 to initiate myocardial differentiation. Further studies will be required to identify additional regulators of heart and endoderm development that function downstream of or in combination with Gata5.
Materials and methods
Zebrafish strains and maintenance
Zebrafish were maintained and staged as described (Westerfield 1995).
Whole-mount in situ hybridization and immunohistochemistry
We performed in situ hybridization as described (Alexander et al. 1998); in certain cases, embryos older than 28 hpf were incubated in 0.003% phenylthiourea to inhibit pigmentation. The gata4 (GenBank accession no. AF191577) and gata6 (GenBank accession no. AF191578; previously called gata5 in Pack et al. 1996) genes were isolated in Leonard Zon's laboratory (Harvard Medical School, Boston, MA; unpubl.). Full-length cDNA was used as the template for the gata6 riboprobe, but the gata4 riboprobe was generated from a truncated cDNA comprised of the first 954 bp. The gata5 riboprobe is described below. The cmlc1 gene (GenBank accession no. AF119163) was isolated as described (Yelon et al. 1999). As fau mutants younger than 24 hpf could not be phenotypically distinguished from wild-type siblings, earlier stage embryos were genotyped post in situ hybridization to confirm that any defects in gene expression segregated with the fau mutation. Briefly, this entailed serial rehydration with PBS + 0.1% Tween, proteinase K digestion at 55°C overnight, and PCR genotyping with a tightly linked marker.
Immunohistochemistry using the antibodies MF20 and S46 (generous gift of F. Stockdale, Stanford University, Palo Alto, CA) was performed as described (Alexander et al. 1998). MF20 was obtained from the Developmental Studies Hybridoma Bank, maintained by the Department of Biological Studies, University of Iowa, Ames under contract NO1-HD-2-3144 from the National Institute of Child Health and Development (NICHD).
β-Galactosidase staining was performed as described (Rodaway et al. 1999).
Genetic mapping
The mapping strain was constructed by crossing a fau Tü male heterozygote to a WIK female. Genomic DNA extraction from zebrafish embryos was performed as described (Westerfield 1995), except that proteinase K digestion was done for 2–4 hr. By using only those embryos recombinant at simple sequence length polymorphisms flanking fau in a search for AFLPs, we successfully restricted our identified markers to those that mapped within a useful genetic distance of fau. Otherwise, AFLP selection was performed as described (Vos et al. 1995). An AFLP marker linked to the fau mutation (primers: 5′-AAGAGAGCGAGCACTGCTACTAC-3′ and 5′-CAGCTTTCCCATATTCTGGC-3′) and an amplicon from the gata5 3′ UTR (primers: 5′-TGATCCGAGACCTCCAC-3′ and 5′-CTCCCGATCATTGGAAC-3′) were placed on the Goodfellow zebrafish radiation hybrid panel (Research Genetics). These markers are separated by 14 cR (lod score 18.96). Human gata5 was mapped using primers designed from the sequence of EST W00677 (primers: 5′-AGACCACCCACTGAGTCACC-3′ and 5′-GGAGTCCCTTGCTGTACGTG-3′) on the Stanford G3 radiation hybrid panel (Research Genetics). Human gata5 is linked to marker SHGC-57473 with a lod score of 18.28.
PAC library screen and linkage analysis
A PAC library was screened by PCR as pools of clones using primers that amplify the gata5 3′ UTR. Internal PAC sequencing was performed using the primer 5′-ACACTGTGGAGGAGAAACGCA-3′. A single-strand conformational polymorphism identified at one end of a PAC clone containing gata5 was linked to the fautm236a mutation in all of 272 gynogenetic haploid embryos examined. Additionally, a BsmFI RFLP at position 380 in the gata5 gene was linked to fautm236a in all of 160 diploid embryos examined. The 400-bp region containing the RFLP was amplified by PCR (primers: 5′-GTACCAGCCATGCTCCAGAC-3′ and 5′-CTCCTTCCCGGTAGAGTTCC-3′). The faus26 mutation was linked to SSRP z4003 in all of 40 gynogenetic haploid embryos examined. Amplification of SSRPs flanking gata5 from faus26 genomic DNA failed to reveal evidence of a deletion.
Isolation of gata5 cDNA
Total RNA was isolated using Trizol (Gibco BRL). Reverse transcription was performed using Superscript II (Gibco BRL). The gata5 ORF was amplified by PCR (primers: 5′-GGACGTTGACAAGGTTTTTACTAG-3′ and 5′-GTCTGTCTACATTTGTGTAAAAGTCC-3′). The 1330-bp product was cloned into the pGEM-T vector (Promega) and sequenced using an ABI 377 DNA sequencer. We sequenced 20 independent clones of mutant-derived and 8 independent clones of wild-type-derived gata5 cDNA (one of which was used as the template for the gata5 riboprobe). Among the mutant-derived clones, the 31-bp insertion was represented by 17 clones, the 30-bp deletion was represented by 2 clones, and the wild-type cDNA was detected once. PCR analysis using a primer bridging the wild-type splice site (primer: 5′-GCTTCATGGGGTACCAAGG-3′) confirmed that wild-type gata5 message is detectable in fautm236a mutants.
Although the amount of gata5 mRNA is profoundly diminished in faus26 mutants, low levels were detectable by RT–PCR performed as above. Sequencing of the subcloned RT–PCR products indicated that faus26 is not a mutation in the gata5 coding sequence or UTRs.
gata5 gene structure
PCR of genomic amplicons and subsequent sequencing revealed that the first intron (723 bp) is present 3′ of cDNA nucleotide 602, the second intron (1207 bp) is present 3′ of cDNA nucleotide 771, the third intron (1.6 kb) is present 3′ of cDNA nucleotide 901, and the fourth intron (343 bp) is present 3′ of cDNA nucleotide 986. Additional internal PAC sequencing upstream of the transcriptional start site using the primer 5′-GGCTAGTTAAAAACCTTGTCAACG-3′ revealed a putative promoter sequence containing a consensus TATA box and several potential Gata-binding elements. Sequencing of this region in faus26 mutants failed to identify a mutation.
gata5 mRNA injection
Full-length, capped mRNA was generated from pCS2:gata5, pCS2:tm236-1:MT, and pGST:GFP using the SP6 mMESSAGE mMACHINE system (Ambion). pCS2:tm236-1:MT was created by subcloning the coding sequence of the gata5 cDNA species most frequently isolated from fautm236a mutants into pCS2:MT. For embryonic overexpression, 20 or 50 pg of mRNA was microinjected at the 1–4 cell stage. For autonomy studies, 100 pg of lacZ mRNA was coinjected. To restrict overexpression to the YSL, mRNA was microinjected directly into the YSL at the 512-cell stage. Injected embryos were raised to 11–36 hpf for in situ hybridization or to 30 hpf for immunohistochemical analysis. After phenotypic analysis, embryos were genotyped as described above.
Acknowledgments
We dedicate this paper to the memory of Nigel Holder. We thank Sarah Howard, Ken Nakashima, Alex Navarro, and Elliot Perens for invaluable assistance; members of the Stainier laboratory, Brian Black, Todd Evans, Richard Harvey, Ira Herskowitz, and Patrick Tam for helpful discussions and/or comments; Joseph DeYoung and Travis Taylor for sequencing; Len Zon for providing gata4 and gata6; and Frank Stockdale for generously providing the S46 antibody. J.F.R. and J.A. are members of the Medical Scientist Training Program of the National Institutes of Health. D.Y. is an Amgen fellow of the Life Sciences Research Foundation. This work was supported by the American Heart Association (J.A. and D.Y.R.S.), the Medical Research Council and Wellcome Trust (A.R., N.H., and R.P.), the Packard Foundation and the Life and Health Insurance Medical Research Fund (D.Y.R.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL didier_stainier@biochem.ucsf.edu; FAX (415) 476-3892.
References
- Alexander J. “Gut feelings: Molecular and genetic analysis of endoderm development in zebrafish”, Ph.D. Thesis. San FranciscoSan Francisco, CA: University of California; 1999. [Google Scholar]
- Alexander J, Stainier DYR. A molecular pathway underlying endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DYR, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Alexander J, Rothenberg M, Henry GL, Stainier DYR. casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-N, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- Chen J-N, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM. Vertebrate tinman homologues and cardiac differentiation. Semin Cell Dev Biol. 1999;10:73–83. doi: 10.1006/scdb.1999.0282. [DOI] [PubMed] [Google Scholar]
- Evans T. Regulation of cardiac gene expression by GATA-4/5/6. Trends Cardiovasc Med. 1997;7:75–83. doi: 10.1016/S1050-1738(97)00010-8. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Gannon M, Bader D. Initiation of cardiac differentiation occurs in the absence of anterior endoderm. Development. 1995;121:2439–2450. doi: 10.1242/dev.121.8.2439. [DOI] [PubMed] [Google Scholar]
- Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gove C, Walmsley M, Nijjar S, Bertwistle D, Guille M, Partington G, Bomford A, Patient R. Over-expression of GATA-6 in Xenopus embryos blocks differentiation of heart precursors [published erratum appears in EMBO J. 1997 16: 1806–1807] EMBO J. 1997;16:355–368. doi: 10.1093/emboj/16.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grepin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124:2387–2395. doi: 10.1242/dev.124.12.2387. [DOI] [PubMed] [Google Scholar]
- Jacobson AG, Sater AK. Features of embryonic induction. Development. 1988;104:341–359. doi: 10.1242/dev.104.3.341. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Evans T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev Biol. 1996;174:258–270. doi: 10.1006/dbio.1996.0071. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Tarzami S, Burch JB, Evans T. Common role for each of the cGATA-4/5/6 genes in the regulation of cardiac morphogenesis. Dev Genet. 1998;22:263–277. doi: 10.1002/(SICI)1520-6408(1998)22:3<263::AID-DVG8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The atlas of mouse development. London, UK: Academic Press; 1992. [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes & Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Kwok C, Korn RM, Davis ME, Burt DW, Critcher R, McCarthy L, Paw BH, Zon LI, Goodfellow PN, Schmitt K. Characterization of whole genome radiation hybrid mapping resources for non-mammalian vertebrates. Nucleic Acids Res. 1998;26:3562–3566. doi: 10.1093/nar/26.15.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JB, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese RJ, Markham BE, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development. 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- Lyons MS, Bell B, Stainier DYR, Peters KG. Isolation of the zebrafish homologues for the tie-1 and tie-2 endothelium-specific receptor tyrosine kinases. Dev Dyn. 1998;212:133–140. doi: 10.1002/(SICI)1097-0177(199805)212:1<133::AID-AJA12>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes & Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. GATA-5: A transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997a;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Parmacek MS. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J Biol Chem. 1997b;272:8515–8524. doi: 10.1074/jbc.272.13.8515. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes & Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev Biol. 1997;189:270–274. doi: 10.1006/dbio.1997.8684. [DOI] [PubMed] [Google Scholar]
- Nascone N, Mercola M. An inductive role for the endoderm in Xenopus cardiogenesis. Development. 1995;121:515–523. doi: 10.1242/dev.121.2.515. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nússlein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Genetics of erythropoiesis: Induced mutations in mice and zebrafish. Annu Rev Genet. 1997;31:33–60. doi: 10.1146/annurev.genet.31.1.33. [DOI] [PubMed] [Google Scholar]
- Pack M, Solnica-Krezel L, Malicki J, Neuhauss SC, Schier AF, Stemple DL, Driever W, Fishman MC. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83:201–206. [PubMed] [Google Scholar]
- Peyrieras N, Strahle U, Rosa F. Conversion of zebrafish blastomeres to an endodermal fate by TGFβ-related signaling. Curr Biol. 1998;8:783–786. doi: 10.1016/s0960-9822(98)70303-3. [DOI] [PubMed] [Google Scholar]
- Rehorn KP, Thelen H, Michelson AM, Reuter R. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development. 1996;122:4023–4031. doi: 10.1242/dev.122.12.4023. [DOI] [PubMed] [Google Scholar]
- Rodaway A, Takeda H, Koshida S, Broadbent J, Price B, Smith JC, Patient R, Holder N. Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGFβ family signals and discrimination of mesoderm and endoderm by FGF. Development. 1999;126:3067–3078. doi: 10.1242/dev.126.14.3067. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed p inhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes & Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- Searcy RD, Vincent EB, Liberatore CM, Yutzey KE. A GATA-dependent nkx-2.5 regulatory element activates early cardiac gene expression in transgenic mice. Development. 1998;125:4461–4470. doi: 10.1242/dev.125.22.4461. [DOI] [PubMed] [Google Scholar]
- Sepulveda JL, Belaguli N, Nigam V, Chen CY, Nemer M, Schwartz RJ. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: Role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda N, Knapik E, Ziniti J, Kaplan S, Sim C, Yamada E, Fishman MC. Zebrafish development and genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. Survey of CA repeats in the zebrafish genome; p. 274. [Google Scholar]
- Stainier DYR, Fishman MC. Patterning the zebrafish heart tube: Acquisition of anteroposterior polarity. Dev Biol. 1992;153:91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- Stainier DYR, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development. 1993;119:31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- Stainier DYR, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- Strahle U, Blader P, Ingham PW. Expression of axial and sonic hedgehog in wildtype and midline defective zebrafish embryos. Int J Dev Biol. 1996;40:929–940. [PubMed] [Google Scholar]
- Suzuki E, Evans T, Lowry J, Truong L, Bell DW, Testa JR, Walsh K. The human GATA-6 gene: Structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics. 1996;38:283–290. doi: 10.1006/geno.1996.0630. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Nüsslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Yelon D, Horne SA, Stainier DYR. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes & Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes & Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]


