Abstract
Homologous recombination is a central pathway to maintain genomic stability and is involved in the repair of DNA damage and replication fork support, as well as accurate chromosome segregation during meiosis. Rad54 is a dsDNA-dependent ATPase of the Snf2/Swi2 family of SF2 helicases, although Rad54 lacks classical helicase activity and cannot carry out the strand displacement reactions typical for DNA helicases. Rad54 is a potent and processive motor protein that translocates on dsDNA, potentially executing several functions in recombinational DNA repair. Rad54 acts in concert with Rad51, the central protein of recombination that performs the key reactions of homology search and DNA strand invasion. Here, we will review the role of the Rad54 protein in homologous recombination with an emphasis on mechanistic studies with the yeast and human enzymes. We will discuss how these results relate to in vivo functions of Rad54 during homologous recombination in somatic cells and during meiosis.
1. Introduction
Homologous recombination (HR) is a complex mechanism to maintain genomic stability. This process uses template-dependent DNA synthesis to repair or tolerate DNA damage, such as DNA double-stranded breaks (DSB), single-stranded DNA gaps, and interstrand crosslinks [1]. HR is intimately linked to DNA replication, as initially elaborated in phage T4 [2], and the function of HR in the recovery of stalled and collapsed replication forks is critical to faithfully duplicate and segregate the genome to daughter cells [1, 3, 4]. Moreover, HR is essential for accurate chromosome segregation during the first meiotic division, also generating genetic diversity among the meiotic products in the process [5].
RAD54 is a member of the RAD52 epistasis group and a core component of the recombination machinery found in all eukaryotes and in some archaea, but not in bacteria [6–8]. The Rad54 protein belongs to a group of the Snf2/Swi2 family of SF2 helicases that share a prominent motor domain which powers ATP-dependent tracking on dsDNA but does not lead to strand separation like classical DNA helicases [9]. The Snf2/Swi2 family of proteins is represented by well-known ATP-dependent chromatin remodelers, such as Snf2/Swi2 or ISWI, but also other proteins that remodel non-chromatin protein-dsDNA complexes such as Mot1, which displaces the TATA-binding protein from DNA [10, 11] (see other reviews in this issue of BBA). The budding yeast Saccharomyces cerevisiae contains 17 Snf2 family proteins, of which 10 function in different aspects of the DNA damage response, including Rad54, Rdh54, Uls1, Rad5, Rad16, Rad26, Ino80, Snf2, Sth1, and Swr1 [7, 12].
This review will focus on yeast and human Rad54 proteins, their paralogs, and their function in HR. We will also discuss the yeast Uls1 protein, which appears to have a partially overlapping function with Rad54 and Rdh54. Excellent reviews are available with comprehensive discussions of the HR pathway [13–16], the Rad52 group proteins [17, 18], meiotic recombination [5, 19], and the Snf2/Swi2 family chromatin remodelers [12, 20–22]. We will emphasize the significant novel developments since our last review on Rad54 in 2006 [7] and refer the reader also to other reviews dedicated to the Rad54 protein [6, 8]. To distinguish between the yeast and human proteins, we will adhere to the following nomenclature: Saccharomyces cerevisiae Rad54 protein, ScRad54; human RAD54 protein, hRAD54; S. cerevisiae Rdh54 protein (also known as Tid1), ScRdh54; and human RAD54B protein hRAD54B. We will use the generic term Rad54 when we refer to both yeast and human Rad54.
2. Homologous recombination – A mechanistic overview
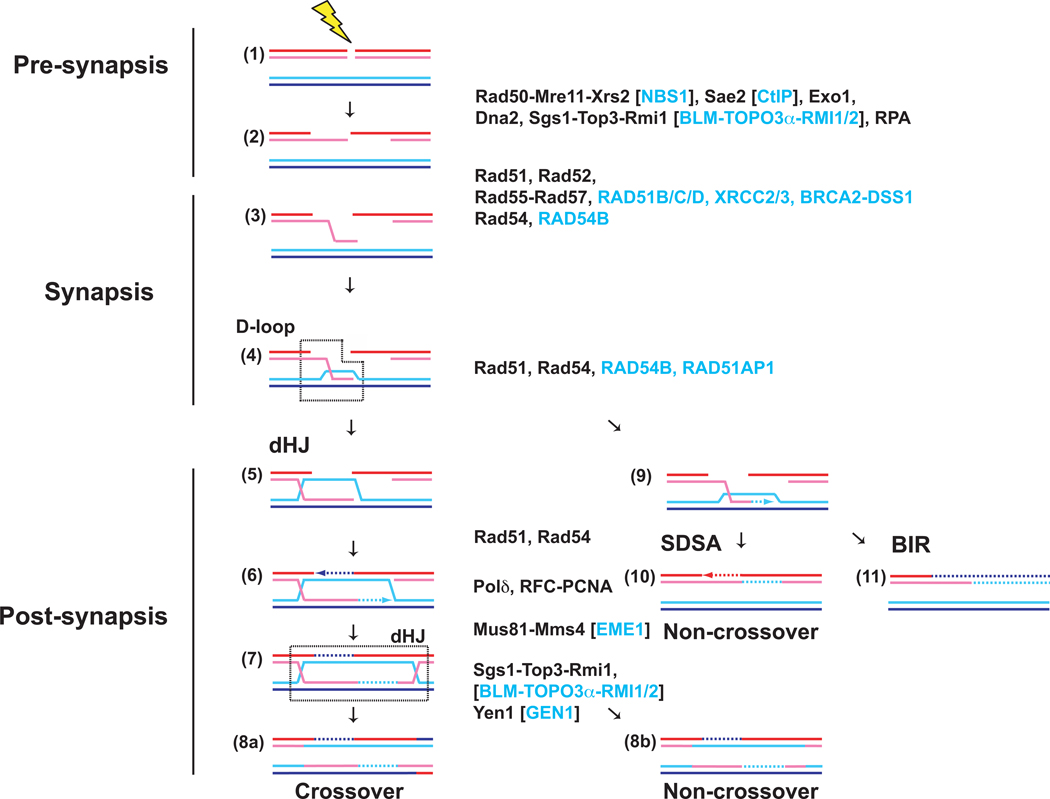
HR can be conceptually divided into three mechanistic stages: Pre-synapsis, synapsis, and post-synapsis (Fig. 1) [13–18]. During pre-synapsis, the DNA damage is processed to generate ssDNA, onto which Rad51 can assemble the pre-synaptic filament. The Rad51-ssDNA filament performs homology search and DNA strand invasion, the key steps during HR that define synapsis. During post-synapsis, HR is completed by several pathways that entail DNA synthesis and different modes to resolve recombination-mediated junction intermediates. The double Holliday Junction pathway (dHJ in Fig. 1) is of particular interest, because it can lead to crossovers, which are essential for meiotic recombination [5, 19]. In somatic (vegetative) cells, the predominant recombinational repair pathway appears to be Synthesis-Dependent Strand Annealing (SDSA in Fig. 1), as it inherently avoids crossover and thus the possibility of deleterious genome rearrangements. Lastly, Break-Induced Replication (BIR in Fig. 1) appears restricted to conditions of one-sided DSBs, where the second DSB end is missing, for example after breakage of the replication fork.
Figure 1. Double-strand break repair by homologous recombination.
Homologous recombination can be conceptually divided into three stages: Pre-synapsis (1–2), synapsis (3–4), and post-synapsis (5–11). The Saccharomyces cerevisiae proteins identified to function at the individual stages are listed in black, the human proteins with alternate nomenclature are listed in blue in square brackets and human proteins not found in yeast are listed in blue without brackets. Three different pathways emanate from the D-loop intermediate (4), the product of DNA strand invasion by the Rad51-ssDNA filament. The dHJ (double Holliday junction; steps 5–8) pathway engages both ends of the DSB to ultimately form a double Holliday junction intermediate (dHJ), which can be resolved into crossover and non-crossover products. SDSA (synthesis-dependent strand annealing; step 10) retracts the invading strand after DNA synthesis on the target duplex to anneal the newly synthesized strand with the single strand resulting from resection of the second end, leading to localized conversion without crossover. BIR (break-induced replication; step 11) was proposed to assemble a replication fork at the D-loop to copy the entire chromosome arm distal to the DSB site leading to long gene conversion events.
Recent genetic and biochemical work defined two independent DSB resection pathways in budding yeast controlled by Exo1 and Sgs1-Top3-Rmi1 with Dna2, respectively, which function after the initial processing by Rad50-Mre11-Xrs2 in conjunction with Sae2 (for review [23]). Similar pathways exist in humans (see Fig. 1). The resulting 3’ overhanging ssDNA is bound by the ssDNA binding protein, RPA, which removes potential secondary structure in the ssDNA. Since RPA has exceptionally high affinity to ssDNA, it interferes with binding of Rad51. This impediment is overcome by so-called mediator proteins, Rad52 and Rad55-Rad57 in yeast, which allow the formation of Rad51-ssDNA filaments on RPA coated ssDNA. In humans, the breast and ovarian cancer tumor suppressor BRCA2 and its associated factor DSS1 perform mediator function, and the RAD51 paralogs, RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 have been implicated as well [24–26]. The Rad51 filament performs homology search and DNA strand invasion. Extension of the invading 3-OH end marks the onset of post-synapsis. In yeast, the primary DNA polymerase for D-loop extension appears to be DNA Polymerase delta with its cofactors PCNA/RFC [27]. In vertebrates, genetic and biochemical data suggest an involvement of the translesion polymerase Pol eta [28, 29].
In the dHJ pathway, the displaced strand of the D-loop anneals to the second DSB end through the action of Rad52 protein [30], to form a junction intermediate that has the potential to mature into a dHJ upon ligation of all strand interruptions. Such junction intermediates, including also nicked and partial dHJs, can be cleaved to crossover or non-crossover products by specific endonucleases, such as Yen1 (human GEN1) or Mus81-Mms4 (human MUS81-EME1) [31, 32]. Alternatively, the two junctions of a dHJ can be migrated towards each other to result in a hemicatenane that can be dissolved into non-crossover products by the combined action of Sgs1 helicase with the type IA topoisomerase Topo3 and the specificity factor Rmi1 (human BLM-TOPOIIIα-RMI1/RMI2 [33]). In the second post-synaptic pathway, the DNA synthesis-extended strand of the D-loop can be extricated to anneal with the second DSB end, in a process called Synthesis-Dependent Strand Annealing (Fig. 1). Lastly, in the absence of a second DSB end, Break Induced Replication establishes a replication fork at the D-loop to initiate long-range DNA synthesis to copy the entire distal chromosome arm resulting in loss-of-heterozygosity [34]. In this review, we will use this mechanistic framework to discuss the functions of Rad54 and its paralogs during HR.
3. Genetic analysis of RAD54 in Saccharomyces cerevisiae and vertebrates
In eukaryotes, RAD54 is a highly conserved gene. There is only a single Snf2/Swi2-like protein in bacteria, RapA (HepA). However, RapA does not directly function in DNA repair but rather in transcription to release the nascent transcript from transcription complexes [35, 36]. A Rad54 homolog has been identified in the Archaeon Sulfolobus solfataricus, but Rad54 is absent in many other groups of Archaea [37, 38]. Biochemical analysis of SsoRad54 suggest that the archaeal protein has a comparable function to its eukaryotic homolog in stimulating the recombination activity of the cognate Rad51 homolog [37].
ScRAD54 was first discovered in screens isolating mutants that were sensitive to ionizing radiation (IR) [39–41]. In S. cerevisiae, rad54 is one of the most IR-sensitive single mutants along with rad51 and rad52. A single DSB induces significant lethality in rad54 cells [42]. Considering the relatively normal growth of rad54 (and rad51 and rad52) mutants compared to wild-type cells, the data suggest that spontaneous DSBs are relatively rare in yeast [43]. Similar to other mutants in the RAD52 epistasis group, rad54 mutants are hypersensitive not only to IR but also to crosslinking (e.g., mitomycin C, cis-platinum) or alkylating agents (e.g., methyl methanesulfonate), and a multitude of other agents that are capable of causing DSBs [17, 18]. Rad54-deficient S. cerevisiae cells are defective in intrachromosomal gene conversion as well as interchromosomal recombination [44]. In hetero-allele assays (HR between different alleles of one gene residing on two homologs in diploids cells) rad54 mutants displayed a similar defect than rad52 mutants [45]. The effect of the rad54 mutation on meiotic recombination is much milder than on mitotic recombination, which is likely due to partial redundancy in function between ScRad54 and ScRdh54 (see below) [44, 46]. The analysis of the Walker A box mutations (Rad54-K341R/A) established that ATP hydrolysis is essential for protein function in yeast and mice, as these catalytically defective mutants displayed phenotypes in DNA repair assays that were quantitatively identical to null mutations [47–49]. In general, the defects of ScRad54-deficient yeast cells in DNA repair are highly similar in extent to the defects in Rad51-deficient cells in yeast, whereas the meiotic phenotype of ScRad54-deficient yeast cells is much less pronounced than that of Rad51-deficient cells [5, 7, 17, 50].
Of particular importance regarding the in vivo function of ScRad54, are results from the analyses of double mutants, specifically the synthetic lethality of the srs2 rad54 double mutant. Srs2 is an anti-recombinase that dissociates Rad51 from ssDNA, antagonizing the assembly of the presynaptic filament [51, 52]. Importantly, the observed synthetic lethality in srs2 rad54 cells is suppressed by mutations in the RAD51, RAD52, RAD55, and RAD57 genes [53–55]. This implies that Rad51 filaments assembled in the srs2 rad54 double mutants lead to recombination-dependent toxic intermediates that cause the lethality. This strongly suggests that the critical function of ScRad54 is after the assembly of Rad51-ssDNA filaments.
Genetic analysis of RAD54 in mouse and chicken DT40 cells established the importance of RAD54 and its essential ATPase activity for recombination in vertebrates [49, 56, 57]. The function of hRAD54 in human cells is inferred from these results. The hRAD54 cDNA partially suppressed the MMS-sensitivity of rad54Δ yeast cells, suggesting remarkable conservation in protein function [58]. Mouse RAD54 knockout embryonic stem (ES) cells display a reduction in HR (measured as gene conversion and sister chromatid exchange), sensitivity to IR and interstrand crosslinking agents, and defects in recombination-mediated gene targeting [56, 59]. At the organismal level, however, RAD54 is not essential for embryonic development and Rad54-deficient mice do not display IR-sensitivity, but are hypersensitive to the interstrand crosslinking agent mitomycin C [56]. Double mutants of mouse RAD54 with defects in nonhomologous endjoining (NHEJ), an alternative pathway of DSB repair, caused by deletion of DNA dependent protein kinase (SCID) or DNA ligase 4 (LIG4), exhibit synergistic sensitivities to IR and interstrand crosslinking agents, showing competition of HR and NHEJ for DSB repair in vertebrates [60, 61]. RAD54-deficient mice do not display an obvious meiotic defect [56]. It is interesting to note that in mice, unlike mutations in RAD54, disruption of the RAD51 gene causes early embryonic lethality [62, 63]. This represents an interesting contrast to budding yeast, where rad51 and rad54 mutants are both viable with nearly identical phenotypes in terms of DNA damage sensitivities or mitotic recombination defects. This difference might be related to functional differences between the yeast and human Rad51 and Rad54 proteins, or possibly indicates the existence of an additional protein with a function similar to RAD54 in vertebrates. As the RAD54 RAD54B double mutant mouse is also viable [64], hRAD54B is not that candidate, but yet another third gene would have to be postulated. The genetic data, largely obtained in yeast and mouse, are critical to guide the interpretation of the biochemical results discussed below.
4. Biochemical and cytological analysis of Rad54 protein in Saccharomyces cerevisiae and vertebrates
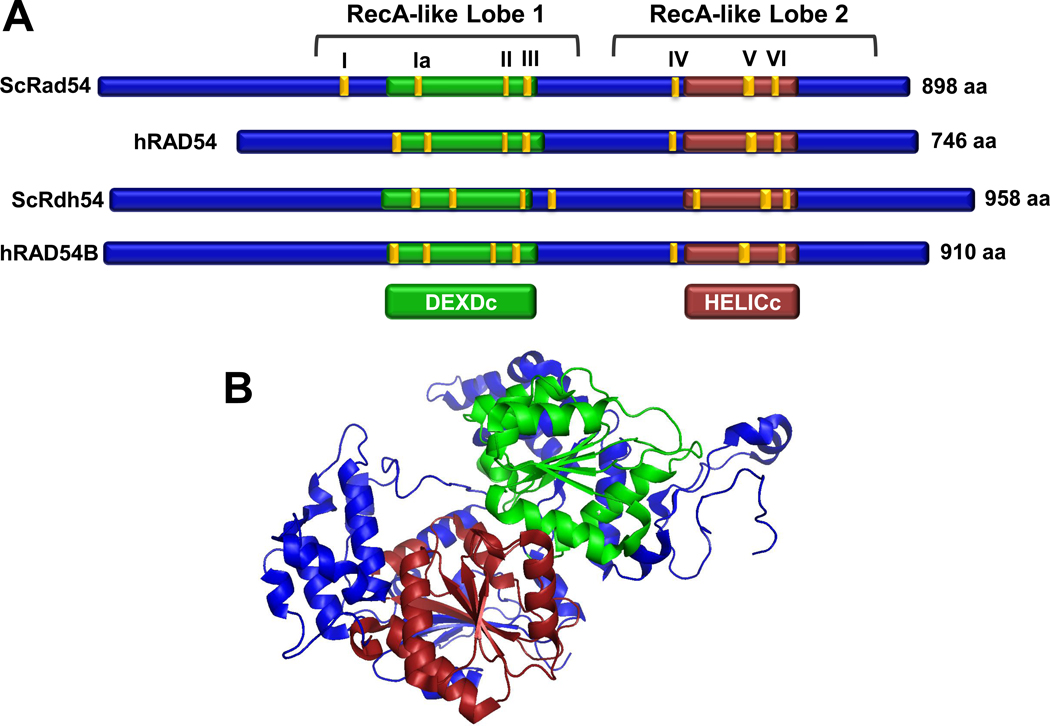
ScRad54 and hRAD54 are highly conserved proteins with 68% similarity and 48% identity [58, 65]. Sequence analysis identified Rad54 as a member of the SF2 superfamily of helicases with the classic seven “helicase” motifs: I, Ia, II, III, IV, V, and VI that define the ATP-dependent motor domain essential for translocation on DNA (Fig. 2) [9, 66]. More precisely, Rad54 falls in the Swi2/Snf2 group of ATP-dependent motor proteins that track along dsDNA but are unable to catalyze the strand displacement reactions typical for a DNA helicase [12]. The ATPase activity requires dsDNA and is not supported by ssDNA [67] (Table 1), whereas classical DNA helicases translocate on ssDNA and ssDNA stimulates their ATPase activity [66]. dsDNA tracking by Rad54 is associated with topological changes in the DNA (positive and negative supercoils in topologically constrained DNA molecules) that were documented biochemically and through the use of atomic force microscopy [68–72]. Translocation on dsDNA was directly visualized in single-molecule experiments with ScRad54, measuring a rate of ~300 bp/sec with an average travel distance of 11.5 kbp [73]. This high level of processivity suggests that Rad54 forms a multimeric assembly on the DNA, possibly a ring encircling the DNA. Indeed, there is biochemical and electron microscopic evidence for Rad54 multimerization on DNA [47, 68, 74], but the exact architecture of this assembly remains to be determined. The fundamental biochemical activity of Rad54 is its dsDNA-dependent ATPase which powers translocation on duplex DNA, and the ATPase activity is significantly stimulated by the cognate Rad51-DNA complex [69, 70, 75]. As discussed below, this biochemical finding strongly supports that the physical interaction between Rad54 and Rad51 [76–78] is of biological significance and that both proteins cooperate during HR.
Figure 2. Structure and architecture of Rad54 proteins and Rad54 paralogs.
(A) Schematic alignment of Rad54 proteins and Rad54 paralogs from Homo Sapiens and S. cerevisiae. The motor domains are outlined in green (DEXDc) and red (HELICc) [172], the 7 conserved ATPase motifs are highlighted in yellow. (B) X-ray crystal structure of the zebra fish Danio rerio Rad54 protein, an N-terminal truncation representing amino acids 91–738, is shown with the DEXDc and HELICc domains colored as in (A) (PDB code:1Z3I) [79].
Table 1.
Rad54 and its paralogs in budding yeast and humans.
| Protein | Organism | MW Length |
ATPase min−1 |
Protein interactions |
|---|---|---|---|---|
| ScRad54 | Saccharomyces cerevisiae | 101,753 Da 898 aa | 1,200 1 [75, 114] | Mus81-Mms4 [145] Rad51 [76, 78] Histone H3 [106] Mek1 [161] |
| hRAD54 | Homo sapiens | 84,352 Da 747 aa | 800 1 [67, 125] | RAD51 [77] MUS81-EME1 [147] |
| ScRdh54 | Saccharomyces cerevisiae | 107,946 Da 958 aa | 2,200 [165] | Dmc1 [88] Rad51 [165] |
| hRAD54B | Homo sapiens | 102,836 Da 910 aa | 69 [99] | DMC1 [169, 170] RAD51 [95, 99] |
The ATPase of ScRad54 and hRAD54 is given for linear dsDNA, the activity is enhanced on junction DNA substrates or substrates partially covered with the cognate Rad51 protein.
Two X-ray crystallographic structures for Rad54 proteins are available: the core (motor) domain of the zebra fish protein [79] (Fig. 2) and of the Sulfolobus enzyme, which has also been crystallized with duplex DNA [38]. Both proteins fold into two lobes (Fig. 2), each consisting of a RecA-like domain found in helicases [66] with clear topological and structural similarity to other SF2 helicases [80]. The two lobes contain specific insertions, only found in the Snf2/Swi2 group. The overall structural homology to other SF2 helicases supports an inchworm mechanism of translocation, tracking along the minor groove of the DNA [81]. The relative orientation of the two lobes in the two crystal structures of the zebra fish and Sulfolobus enzymes is significantly different [38, 79]. The lobe orientation of the DNA-bound form of the Sulfolobus Rad54 protein was very similar to the free form of the protein [38]. The significance of this difference is presently unclear, but may be related to the mechano-chemical cycle of the enzyme [82]. The specificity of Rad54 function is likely mediated by its protein interactions, in particular with Rad51. It will be of interest to obtain structural information for the Rad51 interaction domain of Rad54, which maps to the N-terminus of Rad54 protein [76, 83]. Unfortunately, this region is lacking in the present structures.
In vivo cytological analysis provides significant insights into the biological functions of Rad54. A comprehensive analysis of HR proteins in mitotically growing yeast cells after IR-induced DNA damage revealed that Rad54 forms DNA damage-induced foci, dependent on the presence of Rad51 protein [84]. Conversely, formation of DNA damage-induced Rad51 foci was independent of Rad54 [84], suggesting that Rad54 acts downstream of the formation of the Rad51 presynaptic filament. Analogous experiments in mice discovered that DNA damage-induced RAD51 foci are not as robust in RAD54-deficient cells compared to wild type [64, 71]. Further work with chromosomally integrated ATPase-deficient RAD54 mutants in mice showed that this function of RAD54 protein was independent of its ATPase activity [49]. Elegant real-time imaging and photo-bleaching experiments revealed additional facets of RAD54 protein function and dynamics in living cells [49]. Mouse cells expressing the RAD54 ATPase-deficient mutants (RAD54-K189A/R) show an increase in spontaneous RAD54 foci and RAD51 foci, whereas RAD54 null mutant cells have normal levels of spontaneous RAD51 foci. Interestingly, this increase was not accompanied by increased endogenous DNA damage, as indicated by normal levels of γ-H2AX, a sensitive cellular marker for DNA damage that can be repaired by HR. Although foci of DNA repair proteins appear as relatively stable structures, the residence time of individual proteins can vary greatly, from a few seconds for RAD54 to several minutes for RAD51 [85]. FRAP (fluorescence recovery after photobleaching) experiments revealed that the RAD54 ATPase activity is required for its dynamic association with DNA repair foci in mammalian cells [49]. Surprisingly, the ATPase activity of RAD54 was also required to release DNA repair foci after relocalization to the nuclear periphery [49]. The mechanisms and significance of intranuclear redistribution of DNA repair foci are not understood, but this phenomenon has also been observed in yeast [86]. The release of DNA repair foci from the periphery may reflect the completion of the repair event. These important in vivo observations are invaluable to interpret the biochemical data and establish the biological significance of the in vitro results.
5. Rad54 paralogs in Saccharomyces cerevisiae and humans
There are 17 Snf2/Swi2-like proteins in budding yeast [12], but ScRdh54 is the paralog that is evolutionarily most similar to Rad54 by functional and phylogenetic criteria, and we therefore treat ScRdh54 as the sole Rad54 paralog in the stricter sense. Both proteins appear to perform partially overlapping roles in HR as discussed in more detail below (Fig. 2). Similarly in humans, where several dozen Snf2/Swi2-like proteins exist, hRAD54B represents the only hRAD54 paralog clearly involved in the core mechanism of HR (Fig. 2) [12]. Other Snf2/Swi2-like proteins, such as Rad5 (human HLTF and SHPRH), Rad16, and Rad26 (human CS-B) are involved in different DNA repair pathways. In addition, the classical ATP-dependent chromatin remodeling factors and other Snf2/Swi2-like factors function in other aspects of the DNA damage response and transcriptional control (for review see [7]).
Budding yeast cells deficient for the Rad54 paralog ScRdh54 display very mild phenotypes compared to mutants in RAD54. Mutants in RDH54 exhibit no MMS or IR sensitivity, somewhat increased levels of intrachromosomal recombination compared to wild-type, and only very slightly reduced interchromosomal recombination [44]. Mitotic recombination is reduced in the diploid mutant strains but no reduction is observed in haploids [87]. The meiotic phenotype of rdh54 cells is also rather mild with 82% spore viability compared to 53% for rad54 cells. However, the rad54 rdh54 double mutant is completely defective in meiosis, suggesting that both proteins perform partially overlapping functions in meiosis [44]. It has been suggested that ScRdh54 preferentially functions in HR between homologs, possibly through its preferential interaction with Dmc1 (see Table 1), the meiosis-specific Rad51 paralog that is required for meiotic interhomolog recombination [5, 88], whereas Rad54 is envisioned to primarily function in DNA repair involving the sister chromatid through its association with Rad51 [44, 87, 89]. Further insights into the differential roles of ScRad54 and ScRdh54 will likely shed light on the mechanisms governing interhomolog events during meiotic recombination.
In somatic yeast cells, Rdh54 is recruited to a DSB by Rad51 and Rad52, as demonstrated in ChIP experiments [90]. This result is rather surprising, given that ScRdh54-deficient cells are not sensitive to this type of DNA damage. However, ScRdh54 has also been implicated in signaling from DSBs, specifically during adaptation from DSB-induced cell cycle arrest in cells with a non-repairable DSB [91]. While wild type cells eventually adapt after prolonged arrest and resume the cell cycle even in the presence of a DSB, rdh54 cells remain terminally arrested. It is possible that the localization of ScRdh54 and its recruitment by Rad51 and Rad52 is related to this signaling function [92]. The mechanisms involved remain to be determined. In vivo, ScRdh54 localizes to kinetochores, but the functional significance is unclear [84]. In cells lacking ScRdh54, ScRad54 appears to take the place of ScRdh54 at kinetochores [84], suggesting that this localization is not related to the ScRdh54 signaling function in adaption evident in the single mutant.
Biochemically, ScRdh54 is exceedingly similar to ScRad54, and it appears that their specificity is mediated by their differential affinities for Rad51, Dmc1, and possibly other proteins (Table 1). Like ScRad54, ScRdh54 is a dsDNA-specific ATPase, albeit with apparently lower specific activity (Table 1). ScRdh54 translocates on dsDNA, with lower velocity (84 bp/min) and processivity than ScRad54, inducing the topological changes identified to be associated with ScRad54 translocation [93, 94]. In summary, the genetic, biochemical, and cytological data suggest some functional overlap between ScRdh54 and ScRad54, particularly in meiosis, but there are clearly unique functions for both proteins, e.g. of ScRad54 in DNA repair in somatic cells and ScRdh54 in adaptation from DNA damage checkpoint-mediated cell cycle arrest.
Unlike the yeast Rad54 paralog, ScRdh54, the mammalian RAD54 paralog, hRAD54B, has a clear function in DNA repair. In human cells hRad54B colocalizes with hRad51 and hRad54 in the nucleus [95]. The availability of a human cell line (HCT116) knocked out for both copies of RAD54B directly demonstrated the relevance of hRad54B to HR, as these cells show reduced gene targeting frequencies and genomic instability [96, 97]. Importantly, hRAD54B-deficiency is synthetically lethal with depletion of the FEN1 nuclease, which is critical for lagging strand replication [97].mics the synthetic lethality of rad27 mutants (RAD27 encodes yeast FEN1) with any HR mutant in budding yeast [98]. The conclusions from the genetic studies in human cells are corroborated by results with RAD54B-deficient mouse ES cells, which display sensitivity to IR and interstrand crosslinking agents (mitomycin C) [64]. Interestingly, RAD54 RAD54B double mutant ES cells do not exhibit increased mitomycin C sensitivity, whereas the double mutant animal does, suggesting possible tissue-specific expression and function of both proteins [64]. Likewise, the yeast rad54 rdh54 double mutant is completely deficient in meiosis, whereas the RAD54 RAD54B double mutant mouse is fertile [44, 64]. The overall biochemical properties of hRAD54B are highly similar to hRAD54, although the ATPase activity appears to be surprisingly low (Table 1, see below) [64, 99, 100].
While it is tempting to draw the analogy that both yeast and vertebrates (human) contain one Rad54 protein (ScRad54, hRAD54) and one principal Rad54 paralog (ScRdh54, hRAD54B), the genetic data in yeast and mouse strongly suggest that hRAD54B is not the equivalent of ScRdh54. The phenotypes of the respective single mutants are too different, and the genetic analysis in mice documents clear differences to yeast.
6. Functions of Rad54 protein
Rad54 protein has been implicated in nearly all mechanistic stages of HR, except DNA end resection [49]. Here, we discuss the functions of Rad54 protein in the consecutive stages of HR (see Figure 1), followed by a brief summary of the roles of Rad54 paralogs in HR.
6.1. Chromatin remodeling
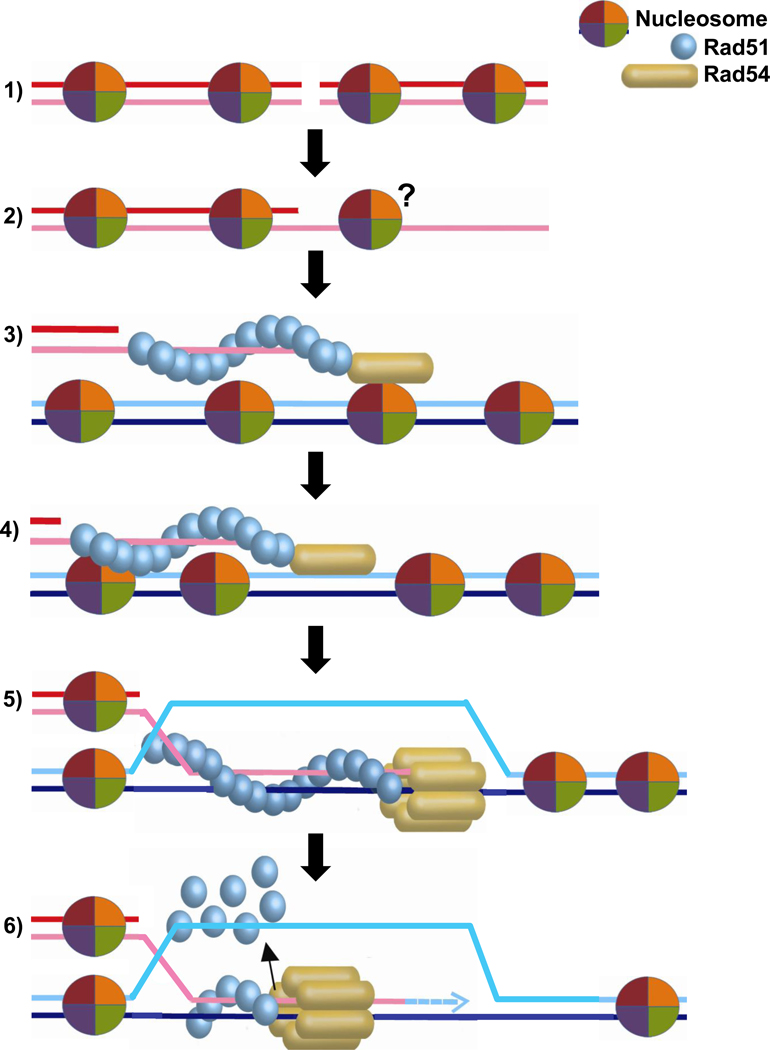
Snf2/Swi2 is the founding member of ATP-dependent chromatin remodeling factors that can slide or eject nucleosomes, as well as exchange constituent histones ([101]; also see other contributions in this issue of BBA). The homology between Rad54 and Snf2/Swi2 provided an immediate impetus to test whether Rad54 functions to relieve the repressive context of chromatin on HR, which could be relevant during DSB resection, Rad51 filament assembly, homology search, DNA strand invasion or later stages (Fig. 3). Biochemical experiments using purified S. cerevisiae, Drosophila melanogaster, and human Rad54 proteins on reconstituted chromatin templates demonstrated that Rad54 remodels nucleosomes in an ATP-dependent manner in vitro [72, 102–104]. The chromatin remodeling activity of Rad54 was stimulated by the Rad51 presynaptic filament [72, 102–104], which is known to stimulate the Rad54 ATPase activity from studies involving non-chromatinized substrates [6–8]. Rad54 enhanced DNA strand invasion by Rad51 on chromatin substrates [102, 104], but ScRad54 was not required for Rad51-mediated homology search on such substrates [105]. Rad54 is a potent and processive dsDNA motor protein [73] and may remodel nucleosome in a non-specific manner as it translocates on duplex DNA. Rad54-mediated nucleosome remodeling is relatively inefficient compared to established remodelers such as the Snf2 complex. Rad54 chromatin remodeling appeared limited to mononucleosomes and Rad54 did not affect nucleosome positioning in a nucleosomal array [72]. While the Rad54 ATPase activity is highly stimulated by the Rad51-dsDNA filament [75], a similar stimulation by nucleosomes has not been reported. Moreover, there is a fundamental difference between the architecture of Rad54, which may assemble in a homo-polymeric ring on DNA [47, 73, 74], and that of established ATP-dependent chromatin remodelers that occur in large multi-subunit assemblies [101]. However, a direct protein interaction has been described between ScRad54 and histone H3 [106], which is consistent with a specific function of Rad54 in chromatin remodeling.
Figure 3. Chromatin remodeling during HR.
Schematic representation of possible stages, where nucleosomes on chromosomal DNA (1) may exert an effect on HR, potentially requiring chromatin remodeling, including resection (2), homology search (3), DNA strand invasion (4), branch migration (5), and recombination-associated DNA synthesis (6). The question mark in (2) indicates the possibility that nucleosomes may be associated with ssDNA after resection. The architecture of the Rad54 protein (monomer, multimer) in association with the Rad51-ssDNA filament or duplex DNA is unknown. For convenience, we represent here and in subsequent figures Rad54 associated with the Rad51-ssDNA filament as a monomer. The active translocating motor on duplex DNA is depicted here and in subsequent figures as a hexameric ring encircling the DNA in an interpretation of the single molecule and electron microscopic studies discussed in the text based on the analogy of processive, homo-hexameric DNA helicases.
Does ScRad54 remodel chromatin during HR in vivo? At the moment this question cannot be answered definitively, but it can be narrowed down. In yeast, Rad51 filaments form in vivo and can perform successful homology search in vivo and in vitro with chromatinized substrates in the absence of ScRad54 [105, 107]. This suggests that Rad54 and its ATP-dependent chromatin remodeling activity are not essential for DSB resection, Rad51 presynaptic filament formation or homology search. Rad54 chromatin remodeling may still be relevant in later stages of HR, such as DNA strand invasion, branch migration, or during recombination-associated DNA synthesis. During in vivo recombinational repair in the MAT system using the HML locus as a donor site, changes in a positioned nucleosome and the accessibility by HO endonuclease dependent on ScRad54 have been noticed [108, 109]. However, it is presently not possible to distinguish a direct chromatin remodeling action by Rad54 from an indirect effect by DNA strand invasion, branch migration, or DNA synthesis. The occurrence of Rad54 in thermophilic Crenarchaea, such as Sulfolobus, shows that the appearance of Rad54 is not linked to the appearance of histones in evolution, as many members of this group of Archaea lack histones [110]. In summary, more work is needed to establish the specificity and biological significance of chromatin remodeling by Rad54.
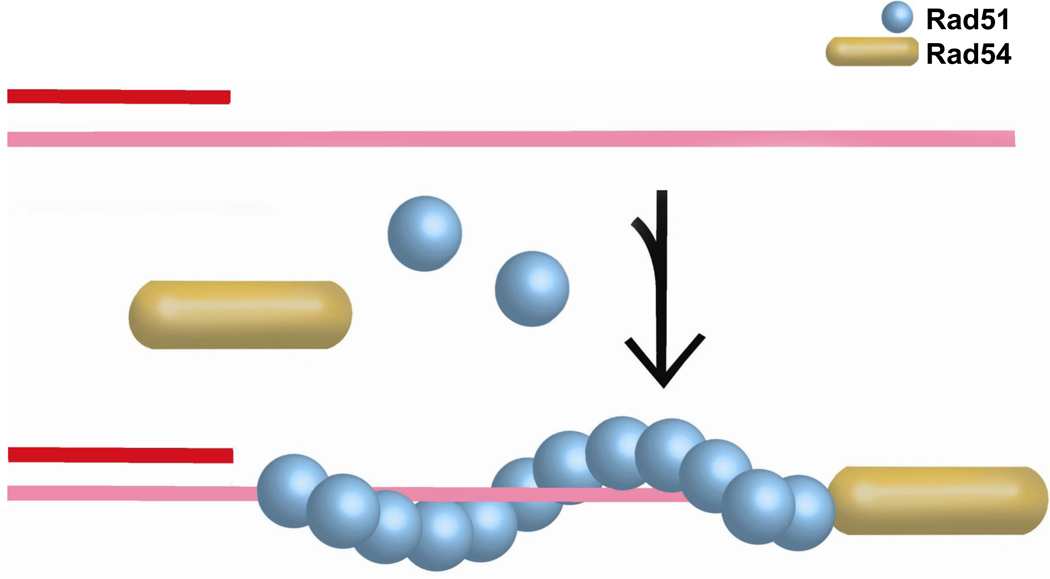
6.2. Rad51-ssDNA filament stability
The Rad51 presynaptic filament is the central HR intermediate. During its formation, mediator proteins help in the assembly of Rad51 filaments on RPA-covered ssDNA. These mediators may act either in the nucleation or stability of the Rad51-ssDNA filament (Fig. 4). Biochemical experiments uncovered a role of ScRad54 in stabilizing Rad51-ssDNA filaments [111]. This function was independent of ATPase activity, as also the ATPase-deficient ScRad54-K341R protein stabilized Rad51 filaments. The increase in stability is likely contributed by the additional protein-protein contacts promoted by the specific binding of Rad54 to Rad51. Stabilization of the Rad51-ssDNA filaments provides a satisfying mechanism for the observed mediator function of Rad54 in reconstituted in vitro recombination reactions [112].
Figure 4. Rad51 presynaptic filament stability.
Schematic representation of ATPase-independent stabilization of the Rad51-ssDNA filament by Rad54. The exact interaction of Rad54 with the Rad51-ssDNA filament, laterally or terminally, monomer or multimer is unknown.
Multi-faceted evidence from yeast and mammalian cells strongly suggests that Rad54 stabilizes Rad51-presynaptic filaments as well in vivo in an ATP-independent fashion. Cytological data show that Rad51 filament formation at IR-induced DSBs is independent of Rad54 [84]. However, closer examination by chromatin-immunoprecipitation revealed that Rad51 filament assembled at the MAT locus after HO-mediated DSB formation in ScRad54-deficient cells were not as robust as in wild type cells [107, 113]. Stabilization of Rad51 filaments in vivo was independent of ATP-hydrolysis and was also observed in ATPase-deficient ScRad54-K341R mutant cells, congruent with the biochemical observations [108, 111]. Also in mammalian cells it was observed that Rad51 filaments formed in RAD54 deficient cells are less robust and sensitive to fixation conditions [64, 71]. In fact, an elaborate study using the mouse knock-in of the RAD54 ATPase-deficient mutant demonstrated slow accumulation of RAD51 foci at break sites, consistent with a stability defect. Importantly, this RAD54 function was independent of its ability to hydrolyze ATP, as the ATP-deficient mutant cells showed normal RAD51 focus formation [49]. In yeast, Rad51 filaments do form in the absence of ScRad54 and are competent for homology search, possibly with diminished efficiency [107, 109]. Results from yeast with the ScRad54 ATPase-deficient mutants and the recombination-dependent lethality of the srs2 rad54 double mutant discussed earlier suggest that the ATP-independent functions may be important, but not essential for HR, and that the critical ATPase-dependent function of ScRad54 is after assembly of the Rad51-ssDNA filament. In summary, stabilization of the Rad51 presynaptic filament appears to be a conserved, ATPase-independent function of Rad54 proteins, which may be important but is not essential for HR.,
6.3. Homology search and DNA strand invasion
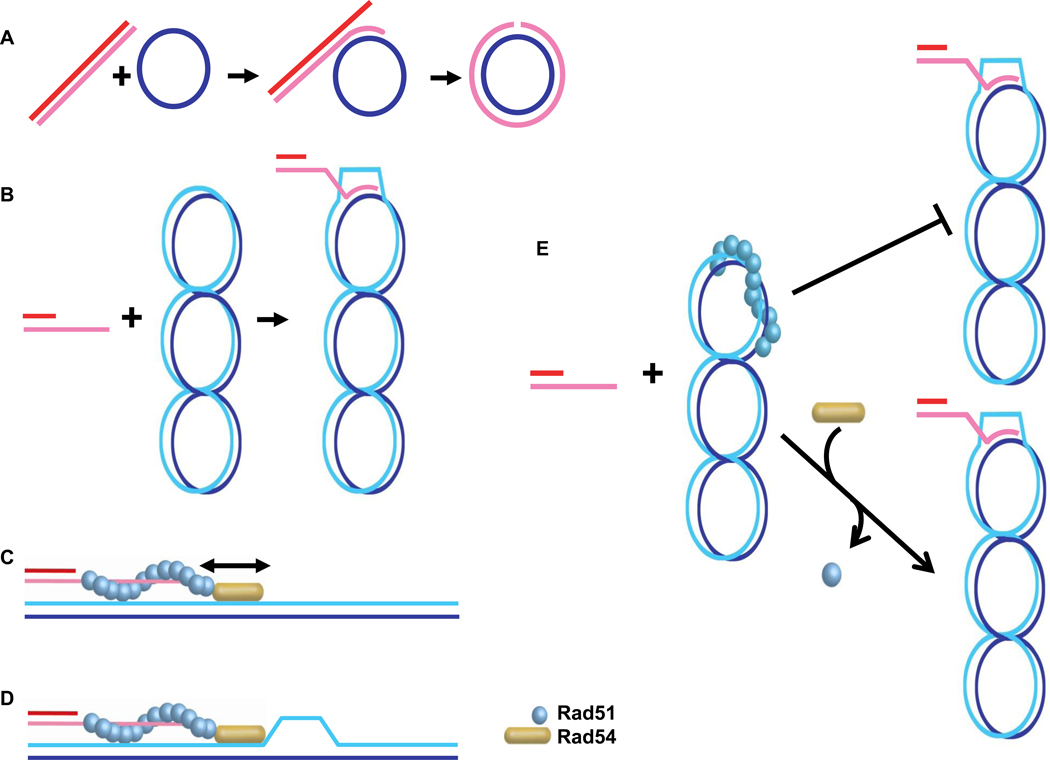
The Rad51-ssDNA filament performs homology search and DNA strand invasion, the key steps in HR. Through its association with the presynaptic filament, Rad54 is targeted to the pairing site, where it may bind duplex DNA, which support the Rad54 ATPase activity and ability to translocate on dsDNA [69, 70]. In fact, the seminal discovery that ScRad54 stimulates the in vitro recombination activity of Rad51 protein opened the field of mechanistic studies with Rad54 protein [114]. Sub-stoichiometric amounts of Rad54 stimulate Rad51 in the three-strand DNA strand exchange reaction (circular ssDNA and linear dsDNA; Fig. 5A) and the D-loop reaction (linear ssDNA and supercoiled duplex DNA; Fig. 5B) in all systems examined [37, 102, 114–116]. The stimulation depends on the Rad54 ATPase activity and requires species-specific protein interactions between the Rad51 and Rad54 proteins, suggesting that the observed in vitro effect is of biological importance [6–8].
Figure 5. Homology search and DNA strand invasion.
A. Scheme of the three-stranded DNA strand exchange reaction between linear duplex DNA and circular single-stranded DNA. B. Scheme of the D-loop reaction between supercoiled, circular duplex DNA and a single-stranded DNA oligonucleotide, which is depicted with a heterologous duplex DNA tail. C. The dsDNA sliding model envisions that the Rad54 motor protein slides duplex DNA along the Rad51 presynaptic filament to enhance homology search. D. The dsDNA strand-opening model suggests that Rad54 translocation on the duplex target DNA results in strand opening that facilitates the conversion to stable joint molecules. In both models, Rad54 protein needs to transition from an ATPase inactive form when associated with the Rad51-ssDNA filament to an active ATPase on duplex DNA, a process which is not depicted in the figure. E. The Rad51-dsDNA complex dissociation model maintains that in the in vitro reaction some Rad51 will bind to the duplex DNA, which inhibits D-loop formation, unless Rad51 is dissociated from duplex DNA by Rad54 and entirely redistributed to bind to the ssDNA.
The mechanism of how Rad54 stimulates Rad51-mediated recombination remains to be determined. Since the Rad54-mediated stimulation is well recognized on non-chromatin substrates, chromatin remodeling by Rad54 cannot be the primary mechanism. Several models can be entertained: first, the dsDNA translocation activity of Rad54 may help by sliding the target duplex DNA along the Rad51 filament along for improved homology search [114] (Fig 5C). In vivo chromatin-immunoprecipitation experiments, however, show that Rad51 filaments formed at the MAT locus can perform homology search and target Rad51 to the HML donor locus in the absence of ScRad54 [107], arguing against such a homology search model. However, a contribution to homology search by Rad54 cannot be entirely excluded by these experiments, because the low kinetic resolution in these experiments (1 hr time points) may have missed a subtle effect. Second, the Rad54 translocation activity may lead to transient strand opening on the duplex DNA to facilitate joint formation [68–70] (Fig. 5D). Consistent with this proposal is the ability of Rad51-ssDNA filaments to perform homology search in vivo and in vitro without Rad54 [105, 107]. Importantly, in the biochemical experiments, the ScRad54 ATPase activity was required to convert protein-stabilized, homology-dependent joints mediated by Rad51 in a D-loop assay into stable DNA joints that could be analyzed by gel electrophoresis after deproteinization. This transition may be a reflection of duplex opening by Rad54 to allow intertwining of the invading strand with the target template DNA. Alternatively, it is possible that Rad54 is required to dissociate Rad51 from the duplex target DNA in the in vitro reaction, as Rad51 bound to the target DNA blocks DNA strand invasion [117] (Fig. 5E). It is interesting to observe that yeast Rad51 is completely dependent on ScRad54 for D-loop formation [114], whereas human RAD51 can form D-loops in the absence of hRAD54 [115, 116]. This suggests that Rad54 function may relate to the DNA binding properties of its cognate Rad51 protein [118–120]. At present, it is difficult to distinguish between the latter two models. The strand-opening model predicts that Rad54 protein is required for D-loop formation in vivo, whereas the Rad51 dissociation model predicts that in vivo D-loops can be formed in the absence of Rad54. There are presently no tools to demonstrate D-loop intermediates in vivo and indirect measures, such as chromatin-immunoprecipitation, cannot distinguish between successful homology search and DNA strand invasion.
6.4. D-loop dissolution and branch migration
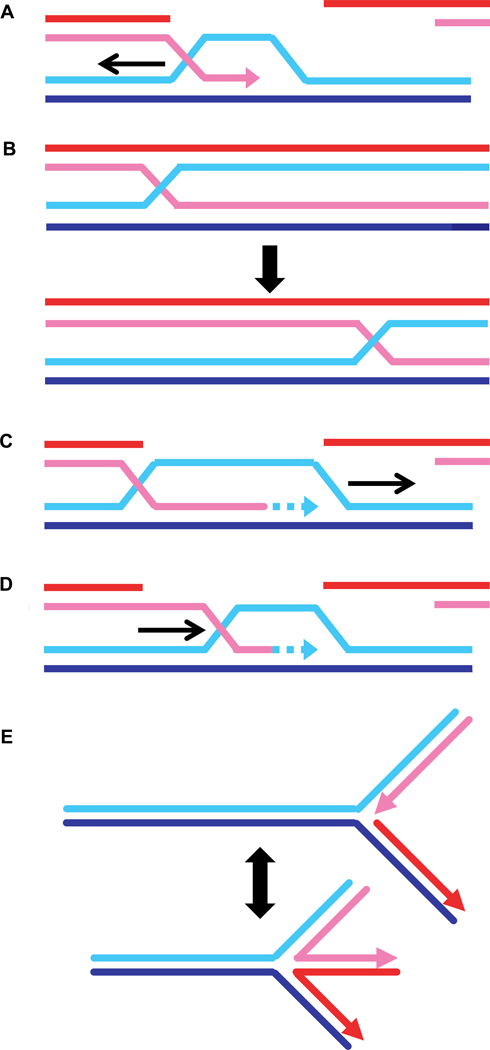
Branch migration describes the movement of recombination-mediated joints, which appears to be isoenergetic, as it involves breaking and forming the same number of Watson-Crick base pairs (Fig. 1). To achieve directionality, however, ATP-dependent branch migration motor proteins are required. The paradigm for such a motor is the RuvB motor, which is targeted to Holliday junctions through its partner protein RuvA [121]. Branch migration can occur in several different contexts: first, branch migration could enlarge the D-loop and generate longer heteroduplex DNA (Fig. 6A) or second, branch migration could move nicked junctions or classical Holliday junctions containing four uninterrupted strands (Fig. 6B). Both processes affect the length of heteroduplex DNA. However, the initial resection of the DSB also determines heteroduplex DNA length [5]. In bacteria, branch migration moves junctions to the preferred cleavage site of the RuvC Holliday junction resolvase [121]. The recombination junction nucleases known in eukaryotes, Mus81-Mms4 (human MUS81-EME1), Slx1-Slx4, Rad1-Rad10 (human XPF-ERCC1), and Yen1 (human GEN1), have not been reported to display any sequence preference. Therefore, there is no compelling need for branch migration. It is currently unclear whether branch migration occurs during HR in eukaryotes [5, 18, 19], but genetic data suggest that heteroduplex length is defined by more than just end resection [122]. Rad54 has also been suggested to be involved in other processes that resemble branch migration, such as D-loop migration during recombination-associated DNA synthesis (Fig. 6C) or D-loop dissolution (Fig. 6D). The interested reader is also referred to a previous detailed review on the role of Rad54 in branch migration [8].
Figure 6. Branch migration, D-loop dissociation, migrating D-loop during DNA synthesis, and fork regression.
Schematic representation of the potential roles of Rad54 in branch migration: A) enlarging D-loops, B) moving nicked (not shown) or closed Holliday junctions, C) migrating D-loops during recombination-associated DNA synthesis, D) dissociating D-loops, and E) regressing replication forks. For details see text.
In reconstituted recombination reactions and using specifically designed synthetic substrates, Rad54 exhibits ATP-dependent branch migration activity in vitro. The first indication came from experiments using the three-strand DNA strand exchange reaction (Fig. 5A) [123]. Rad54 protein enhanced branch migration in this assay by about five-fold with defined directionality and in a species-specific manner, as yeast Rad54 has no effect on the RecA-mediated reaction. These findings were confirmed and extended to the related four-strand reaction, where migration of a four-stranded DNA junction is measured, for the yeast and human Rad54 proteins [124]. hRAD54 (and ScRad54) were found to bind with preference to partial X or X junctions (see Fig. 8) compared to linear duplex DNA and were able to migrate junction substrates reconstituted from oligonucleotides [124, 125]. Binding to its preferred substrate also stimulated the ATPase activity of hRAD54 about three to five-fold over dsDNA [124, 125]. Inclusion of hRAD51 protein also stimulated hRAD54 branch migration activity [126], likely through the stimulation of the Rad54 ATPase activity [75, 116]. hRAD54 was also able to drive branch migration through regions of heterology when using junction substrates reconstituted from oligonucleotides or in reconstituted recombination reactions with hRAD51 [124, 127]. While the biochemical data clearly support a role of Rad54 in branch migration, there are presently no specific in vivo data to demonstrate biological significance.
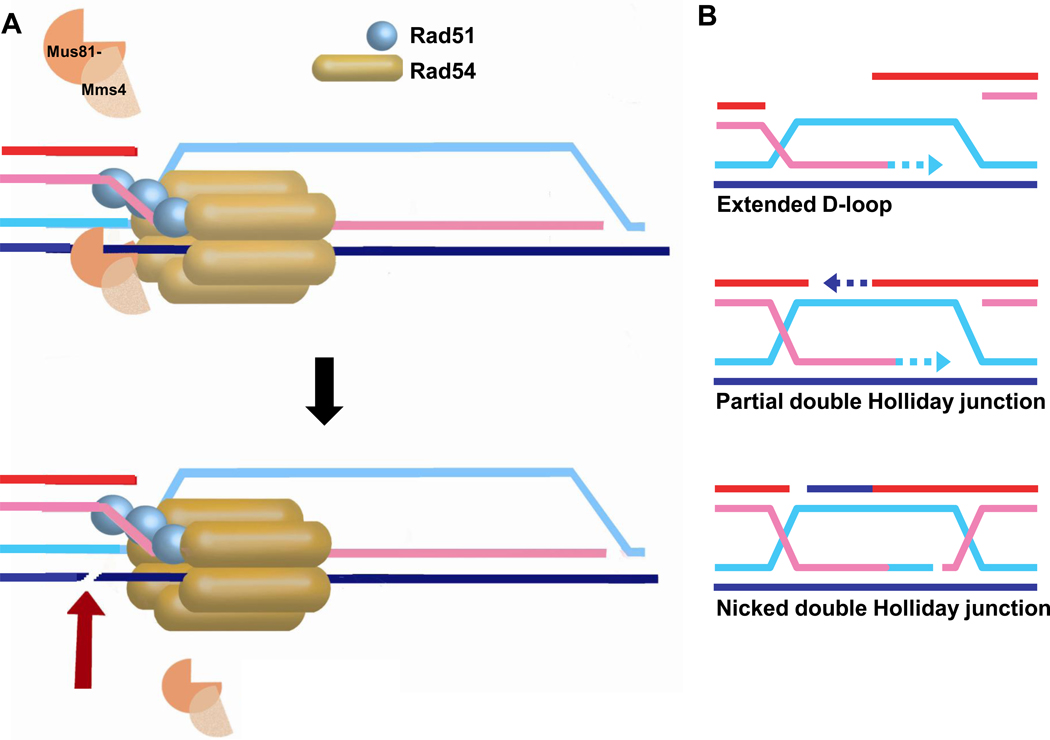
Figure 8. Recruitment of the Mus81-Mms4 structure-selective endonuclease.
A) Recruitment of the Mus81-Mms4 endonuclease by Rad54 to a D-loop and B) other recombination-mediated junctions by Rad54, including a DNA synthesis extended D-loop, a partial double Holliday junction after second end capture, or a nicked double Holliday junction. Note that all three junction intermediates depicted are excellent substrates for Mus81-Mms4 cleavage in vitro, whereas the mature double Holliday junction with four full-length ligated strands has not been reported to be cleaved by Mus81-Mms4 and a single Holliday junction is either not cleaved at all or very inefficiently [32, 152].
D-loop migration during recombination-associated DNA synthesis is mediated by the Dda helicase in the phage T4 recombination/replication system, where the process was termed bubble migration (Fig. 6C) [128]. The role of Rad54 in recombination-associated DNA synthesis, extension of the invading strand in the D-loop, has been directly tested in reconstituted reactions using yeast proteins (RPA, Rad51, ScRad54, DNA polymerase delta, PCNA, and RFC), but the Rad54 motor protein was unable to migrate the D-loop during DNA synthesis [27].
ScRad54 is required for D-loop formation in vitro catalyzed by the S. cerevisiae Rad51 protein [114], and hRAD54 stimulates D-loop formation by hRAD51 [115]. During a D-loop time course in reactions including Rad54, a decrease in initial product formation can be noted, which is due to ATP-dependent disruption of the D-loop by Rad54 [129]. It has been proposed that this D-loop-dissociation activity (Fig 6D) is biologically significant [129], because overexpression of ATPase-deficient ScRad54 (Rad54-K342R/A) leads to an increase in conversion tract length [130]. It was interpreted that the absence of Rad54 motor activity leads to longer heteroduplex length because of a defect in D-loop dissociation [129]. However, this result could also reflect other mechanisms, for example the failure to dissociate Rad51 from heteroduplex DNA after DNA strand invasion (see 6.5.). D-loop dissolution occurs during SDSA (Fig. 1), but there is no direct evidence suggesting that Rad54 protein is required specifically for SDSA, and other motor proteins such as Mph1/Fml1/FANCM, Srs2 and BLM/Sgs1 have been implicated [131–133]. hRAD54 protein has also been implicated in the process of replication fork regression, a mechanism to provide lesion bypass when the replication fork encounters a blocking lesion (Fig. 6E) [134]. In reactions utilizing oligonucleotide-based replication fork substrates, Rad54 was shown to migrate such substrates in an ATP-dependent fashion. This activity is reminiscent of the branch migration mediated by Rad54 on similar substrates, but there is presently no evidence to support the biological significance of this activity.
In conclusion, Rad54 is a potent motor protein that tracks on dsDNA and is capable of moving junctions that are encountered as it translocates on duplex DNA in vitro. Although branch migration is a standard feature of HR models, there is actually no direct evidence that it occurs in eukaryotes. At present, the biological significance of the branch migration, D-loop dissociation, or fork regression activities of the Rad54 protein are unclear.
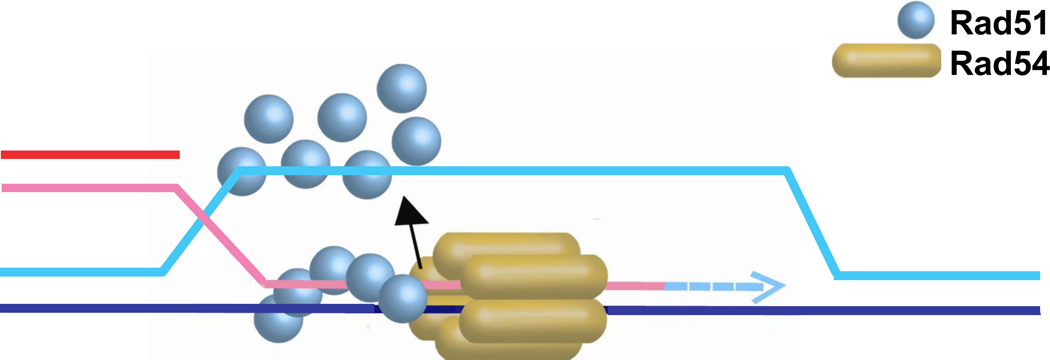
6.5. Dissociation of Rad51 from heteroduplex DNA to allow extension of the invading 3’-OH end by DNA polymerase
After DNA strand exchange Rad51 remains bound to the heteroduplex (double-stranded) DNA [135], much like the bacterial RecA protein, which serves as a paradigm for Rad51 [136] (Fig. 7). RecA requires ATP-dependent turnover and release of the product duplex DNA before DNA polymerase III holoenzyme can access the invading 3’-OH end for recombination-associated DNA synthesis [137]. Rad51, however, displays about 200-fold lower ATPase activity on dsDNA than RecA [138, 139], resulting in very low dynamics of Rad51 cycling off duplex DNA [140]. ScRad54 specifically dissociates Rad51 from dsDNA in a time- and ATP-dependent manner [117, 140]. This reaction displays strong species preference, as human RAD51 is very poorly displaced by ScRad54 [117]. Furthermore, the ScRad54 ATPase activity was enhanced up to 6-fold on dsDNA substrates covered with sub-saturating amounts of Rad51 protein, again displaying significant preference for the cognate Rad51 protein [75, 140]. Hence, the absence of a Rad54-like HR protein in bacteria is consistent with bacterial RecA being capable of autonomous turnover driven by its intrinsic nucleotide cofactor cycle of ATP-binding and hydrolysis, whereas eukaryotic Rad51 may have come to rely on an extrinsic turnover factor, namely Rad54, to release duplex DNA. Electron microscopic (EM) analysis of the interaction between Rad54 and Rad51-dsDNA filaments suggests that Rad54 is either directly recruited to the ends of Rad51-dsDNA filaments or translocates along dsDNA to dock at the terminus of a Rad51-dsDNA filaments before dissociating the filament [74]. Efficient dissociation required the Rad54 ATPase activity, likely to power Rad54 translocation on dsDNA, but also the intrinsic Rad51 ATPase activity. While filaments of the ATPase-deficient Rad51-K191R mutant protein on dsDNA stimulated the Rad54 ATPase activity like wild type Rad51, their dissociation became very inefficient [140]. Furthermore, Rad54 dissociated Rad51-dsDNA filaments containing Rad51-ADP protomers much more efficiently than ATP-bound Rad51 protomers, suggesting that Rad54 may target the Rad51-filament end with a terminal ADP-bound subunit [140]. These conclusions are consistent with extensive single molecule studies of human RAD51 that demonstrate slow release of RAD51 from dsDNA even under conditions allowing ATP hydrolysis [141–143]. In summary, the results from the biochemical and EM analysis demonstrate that the Rad51-dsDNA filament is a specific substrate for the Rad54 motor protein.
Figure 7. Rad51 dissociation from heteroduplex DNA.
Schematic representation of Rad54 dissociating Rad51 from heteroduplex DNA after DNA strand invasion. The stippled blue arrow indicates new DNA synthesis by a DNA polymerase that gains access after Rad51 dissociation.
Using Klenow polymerase as a tool to probe access to the invading 3’-OH end, Rad54 and its ATPase activity were shown to be essential for DNA synthesis from the invading strand in reconstituted recombination reactions using yeast proteins [135]. Although human RAD51 is proficient in D-loop formation, no DNA synthesis was observed in the presence of ScRad54, suggesting that species-specific protein interactions are necessary. These reactions used D-loop formed between linear DNA molecules to circumvent the need for Rad54 in D-loop formation (as is the case when circular dsDNA is used), allowing analysis of Rad54 function in steps that occur after synapsis [135]. Examination of reconstituted recombination-associated DNA synthesis with the cognate DNA polymerase delta and its co-factors PCNA and RFC also revealed a dependence on Rad54 and Rad54 ATPase activity [27]. However, these experiments used the classic D-loop assay with supercoiled dsDNA, so that Rad54 was first required for D-loop formation and, as inferred from the studies with linear D-loops, later for D-loop extension.
What is the evidence in vivo that Rad54 is specifically required to allow extension of the invading 3’-OH end in the D-loop? Chromatin-immunoprecipitation experiments monitoring protein occupancy at a single DSB site in yeast (the MAT locus) and its recombination target site (HML) showed that Rad51 filaments can form in the absence of ScRad54 in budding yeast [107], although these filaments show diminished stability as discussed above under 6.2. Rad51-ssDNA filament stability. Live-cell imaging data with fluorescently tagged proteins also supports the conclusion that IR-induced Rad51 filaments form in the absence of ScRad54 [84]. Importantly and as noted earlier, Rad51 can be immunoprecipitated at the HML target sequence in ScRad54-deficient cells, suggesting that ScRad54 is not essential for homology search [107]. ScRad54-deficient cells are unable to perform recombination-associated DNA synthesis, measured by a PCR-assay that requires less than 100 nt of DNA synthesis [107]. Recent work showed that ScRad54 is required for the mobility of the positioned nucleosome L6 at the HML target region during recombination [109]. However, these results cannot distinguish whether the Rad54-dependent step is DNA strand invasion (D-loop formation) or D-loop extension or both. The question is whether Rad51 can form D-loops in vivo in the absence of ScRad54. However, the tools to detect D-loops in vivo are lacking. Consistent with the notion that Rad51 displacement from heteroduplex DNA is required for DNA synthesis is the observation that recombination-associated DNA synthesis in meiotic recombination, measured as BrdU incorporation, is never associated with Rad51 foci, suggesting that Rad51 dissociation is required before recombination-associated DNA synthesis can take place [144]. This important result supports the notion that Rad51 must be displaced from heteroduplex DNA, likely by Rad54, before recombination-associated DNA synthesis can commence from the D-loop. In summary, the present data are consistent with an ATP-dependent in vivo role of ScRad54 in D-loop formation and/or D-loop extension and further work is needed to clarify the Rad54-dependent step(s).
6.6. Stimulation of Mus81-Mms4/EME1 nuclease activity
A two-hybrid screen using ScRad54 protein as a bait led to the original identification of Mus81, the catalytic subunit of the Mus81-Mms4 structure-selective endonuclease [145]. Binding of Mus81 to ScRad54 potentially recruits the nuclease to junction intermediates during HR, specifically the D-loop during synapsis or later junction intermediates that mature from second-end capture and branch migration (Figs. 1, 8). Consistent with this model, the activity of low and limiting amounts of yeast Mus81-Mms4 (0.25 nM) was stimulated by ScRad54 achieving a maximum at 20 or more fold molar excess of ScRad54 protein [146]. Similar results were found with the human proteins, using 2.5 nM MUS81-EME1 with a maximal nuclease stimulation at 40-fold molar excess of hRAD54 [147]. Whether this effect depends on species-specific protein interaction is unclear, because hRAD54 stimulated the yeast nuclease Mus81-Mms4 [146], whereas ScRad54 did not stimulate, rather inhibited, human MUS81-EME1 [147]. In both studies, ScRad54 or hRAD54 did not change the substrate preference for the cognate endonuclease [146, 147]. In particular, there was no significant cleavage of the intact Holliday junction in the presence of ScRad54/hRAD54 [146, 147], suggesting that even when recruited by the Rad54 motor protein, Mus81-Mms4/hMUS81-EME1 maintains its preference for junctions containing at least one strand interruption [32, 148–152]. The ScRad54 Walker-A box mutant proteins (ScRad54-K341R/A) stimulated Mus81-Mms4 cleavage like wild type ScRad54, suggesting that Mus81-Mms4 stimulation occurred independent of Rad54 ATP binding or hydrolysis [146]. Using non-hydrolysable ATP analogs, it was concluded instead that hRAD54 requires ATP binding but not hydrolysis to stimulate human MUS81-EME1 [147]. It is unclear whether this is a difference between the yeast and human enzymes or whether nucleotide cofactor analogs affect the MUS81-EME1 catalytic activity. In vivo, Mus81 forms foci in about 5% of yeast cells under normal growth conditions, which is increased to 9% after IR (40 Gy) [146]. Importantly, deletion of RAD54, but not the Walker A-box mutants, abolished the IR-induced Mus81 foci. The RAD54 deletion had no effect on spontaneous Mus81 foci, whereas the Walker A-box mutants increased the frequency of spontaneous Mus81 foci to 8–13%. This suggests that ScRad54 recruits Mus81-Mms4 to sites of DNA damage in an ATPase-independent fashion [146]. Recruitment of Mus81-Mms4 is the second ATPase-independent function of Rad54, but it is unclear when this recruitment occurs during the HR process in wild type cells: during Rad51 presynaptic filament formation, homology search, DNA strand invasion, or even subsequent steps?
6.7. Turnover of Rad51-dsDNA dead-end complexes
Unlike bacterial RecA protein, yeast or human Rad51 readily bind dsDNA [118, 153]. This biochemical property poses a significant problem for the cell, as Rad51 may form dead-end complexes on chromosomal DNA, which exists in vast excess over sites of DNA damage, where Rad51 is destined to assemble on ssDNA [117] (Fig. 9). This problem was first experimentally demonstrated with Dmc1, the meiosis-specific Rad51 paralog (see 7. Functions of Rad54 paralogs) [154]. As discussed above, Rad54 protein dissociates Rad51 bound to dsDNA in a time- and ATP-dependent manner [117], but how much Rad54 contributes to the turnover of Rad51-dsDNA dead-end complexes in vivo is uncertain. Genetic and cytological analysis of yeast strains with defects in ScRad54, Rdh54, and Uls1 suggested that ScRad54 primarily turns over Rad51 from DNA damage-induced foci, whereas Rdh54, and to lesser degree Uls1, turns over spontaneous Rad51 foci [155]. This may be interpreted that ScRad54 primarily removes Rad51 from dsDNA in the context of DNA strand invasion, i.e. from the heteroduplex DNA, whereas Rdh54 (and Uls1) dissociate Rad51-dsDNA dead-end complexes. However, the nature of spontaneous Rad51 foci in vegetative (somatic) yeast cells remains unclear - they may represent dead-end complexes or sites of spontaneous DNA damage with Rad51 in the presynaptic filament, D-loop, or on the heteroduplex DNA. An analysis of Rad51 foci in meiotic cells may clarify the role of ScRad54 protein in the turnover of dead-end Rad51 complexes (see [154]). The turnover of Rad51 from duplex DNA by Rad54, either in dead-end complexes or heteroduplex DNA (see 6.5.), is very different from the anti-recombination mechanism mediated by the Srs2 helicase, which translocates on ssDNA to dissociate Rad51 from the presynaptic filament [51, 52].
Figure 9. Turnover of Rad51-dsDNA dead-end complexes.
Schematic representation of Rad54 dissociating Rad51 dead-end complex on chromosomal duplex DNA.
6.8. Regulation of Rad54 function
HR is a key pathway to maintain genomic stability; however, uncontrolled HR can lead to loss-of-heterozygosity, genomic rearrangements, and toxic recombination intermediates that lead to cell death [14]. Hence the cell must achieve a careful balance in regulating HR, and Rad54 appears to be a target of HR regulation (Fig. 10). HR favors the use of the sister chromatid as a template in DNA repair in somatic (vegetative) cells, and in S. cerevisiae HR-mediated DSB repair is down-regulated in the G1 phase of the cell cycle by limiting end resection [156–158]. This problem is exacerbated in haplontic organisms, such as the fission yeast Schizosaccharomyces pombe, where accurate DSB repair by HR is impossible in the G1 phase for lack of an appropriate template. One mechanism to downregulate HR in the G1 phase of fission yeast is targeting Rad54 (termed Rhp54 in S. pombe) for ubiquitin-mediated proteolysis [159]. This mechanism is also operational in meiosis. Rad54 is primarily involved in sister chromatid recombination during meiosis, and failure to downregulate Rad54 (Rhp54) using a non-degradable Rhp54 mutant shifted recombination from interhomolog to intersister events [159].
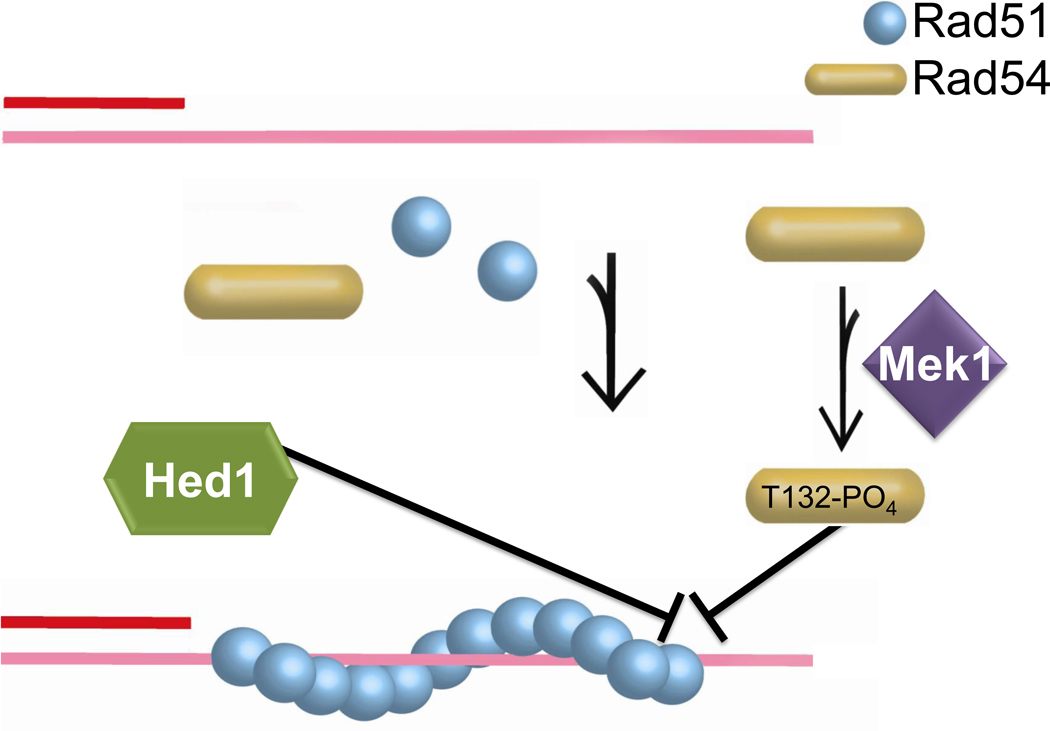
Figure 10. Regulation of the Rad54-Rad51 protein interaction.
Inhibition of the Rad54-Rad51 interaction by Mek1-mediated phosphorylation of Rad54-T132 and Hed1 binding to Rad51.
Rad54 protein is also targeted for regulation during meiosis in budding yeast, involving two distinct mechanisms to inhibit the functionally critical interaction between Rad51 and ScRad54 (Fig. 10). The meiosis-specific kinase Mek1 is required to bias meiotic recombination towards interhomolog instead of intersister interactions [160]. Mek1 kinase phosphorylates ScRad54 in vitro and T132 is found phosphorylated on ScRad54 isolated from cells arrested in meiotic prophase [161]. The meiotic phenotype of the non-phosphorylatable allele rad54-T132A is consistent with being a direct Mek1 target site and mimics the mek1 phenotype in derepressing inter-sister recombination during meiosis. ScRad54-T132D mimics the phosphorylated form of ScRad54 and provided a valuable tool to identify the mechanisms involved. The T132D mutation significantly weakened the ScRad54-Rad51 protein interaction, reducing the stimulation of the ScRad54 ATPase activity by Rad51 and, importantly, the formation of D-loop in reconstituted recombination reactions with Rad51 compared to the wild type ScRad54 protein. These effects were specific to the Rad51 interaction, as the ScRad54 ATPase activity in the absence of ScRad51 was the same between the wild type and ScRad54-T132D mutant proteins. The ScRad54-T132D mutant protein also exhibited a DNA repair defect when expressed in vegetative cells [161]. Interestingly, Rad53, a DNA damage checkpoint kinase that is related to Mek1, was also found to target ScRad54-T132 in vegetative yeast cells [162], suggesting that ScRad54 may also be subject to negative control outside the meiotic context.
The Rad51-ScRad54 interaction is also the target of an independent regulatory mechanism during budding yeast meiosis involving Hed1, a meiosis-specific Rad51 binding protein that prevents binding of ScRad54 to Rad51 [163, 164]. Analogous to ScRad54-T132 phosphorylation, Hed1 inhibits the protein interaction between Rad51 and ScRad54, the Rad51-stimulation of the ScRad54 ATPase activity, and the functional interaction between Rad51 and ScRad54 in D-loop formation in vitro [163]. The interaction defect is also evident in chromatin-immunoprecipitation experiments monitoring the recruitment of ScRad54 to a DSB at the MAT locus. Hed1 also inhibits the interaction of Rad51 with ScRdh54, but to a lesser degree than the Rad51-ScRad54 interaction [163]. When ectopically expressed in vegetative cells, Hed1 inhibits recombinational DNA repair, suggesting that no other meiosis-specific protein is needed for the inhibitory effect [164]. The inhibition of ScRad54 function during meiosis is likely to be only temporary, as ScRad54 is required for successful meiosis. It is possible that ScRad54 is transiently inhibited to bias HR events towards homologs, and that after the establishment of inter-homolog events that lead to the essential meiotic crossovers, repression by Hed1 is relieved and ScRad54 functions in the repair of residual meiotically-induced DSBs by sister chromatid repair [5].
Hence, in budding yeast two mechanisms, Mek1-mediated ScRad54-T132 phosphorylation and Hed1, target the Rad51-ScRad54 interaction during meiosis, and genetic results suggest that they operate independently of each other [161]. Based on the biochemical data, these inhibitory mechanisms will prevent Rad51-mediated recombination at the level of D-loop formation or D-loop extension, and direct chromatin-immunoprecipitation experiments suggest that absence of Hed1 does not affect Rad51 filament formation/stability [163]. Currently, the effect of ScRad54-T132 phosphorylation on Rad51-ssDNA filament formation or stability is unknown.
7. Functions of the Rad54 paralogs, ScRdh54 and hRAD54B
The biochemical properties of ScRad54 and ScRdh54 are, where comparable, highly similar, as discussed above. It appears that both proteins derive a degree of specificity through differential protein interactions. ATP-dependent chromatin remodeling activity has been directly demonstrated for ScRdh54 using mono-nucleosomal substrates, showing nucleosome sliding and enhancement of restriction enzyme accessibility to chromatinized substrates [90]. The specificity of this activity remains to be established, and it is unknown whether ScRdh54 directly interacts with nucleosomal histones or whether its ATPase activity is stimulated by nucleosomal substrates. Similar to ScRad54, ScRdh54 enables yeast Rad51 to form D-loops, both on protein-free and chromatinized substrates [90, 165]. Stimulation of Dmc1-mediated D-loop formation by Rdh54 on protein-free substrates was also observed [166], suggesting that ScRdh54 could be a co-factor for Rad51 and Dmc1. Chromatin-immunoprecipitation experiments show that ScRdh54 is recruited to DSBs during HR-mediated repair in a Rad51- and Rad52-dependent manner [90]. However, the absence of an obvious DNA repair defect in ScRdh54-deficient cells demonstrates that Rad54 is the predominant Rad51-cofactor in vegetative yeast cells. ScRdh54 efficiently dissociates Dmc1 bound to duplex DNA in vitro [166], and cytological and immunoprecipitation experiments in meiotic cells demonstrated that Dmc1 accumulates on chromatin in ScRdh54-deficient cells [154]. Importantly, Dmc1 accumulated on chromatin in a DSB-independent fashion, strongly supporting the model that ScRdh54 turns over dead-end complexes of Dmc1 on undamaged chromosomal DNA [154]. ScRdh54 has a similar function in vegetative cells, turning over spontaneous Rad51 foci [155]. It is not entirely clear whether these Rad51 foci represent sites of spontaneous DNA damage or dead-end complexes, but the absence of colocalizing RPA favors the interpretation that they represent dead-end complexes [155]. Adding a mutation in RAD54 did not enhance this phenotype, but a triple mutant with a deletion in the ULS1 gene, encoding another yeast Snf2/Swi2-like protein, further increased spontaneous Rad51 focus formation [154]. IR-induced Rad51/RPA foci require ScRad54 and Uls1 for their turnover, and to a far lesser degree Rdh54 [154]. These data led to the model that ScRdh54 primarily targets Dmc1 and Rad51 dead-end complexes, whereas ScRad54 targets Rad51 bound to heteroduplex DNA after DNA strand invasion [154]. Although Uls1 is a member of the Snf2/Swi2 family, it shows no similarity to Rad54 outside the motor domain. Uls1 is a SUMO-targeted ubiquitin ligase (StUbL) that is involved in proteolytic control of sumoylated substrates and functions in transcriptional silencing of the silent mating type locus HMR [167, 168]. Uls1 was also identified as Tid4 in a two-hybrid screen with Dmc1, but the significance of this interaction still needs to be tested [88]. The specific function of the Uls1 protein in these processes remains to be established, whether it acts as a motor protein on DNA or via its StUbL activity. It will be interesting to understand how ScRad54 and ScRdh54 can distinguish between the different functional contexts of the Rad51-dsDNA complexes. In summary, the in vivo phenotypes of ScRad54- and ScRdh54-deficient cells demonstrate functional separation with evidence for partial overlap, but the underlying biochemical mechanisms that distinguish both proteins still need to be established.
Much less is known about the biochemical mechanisms of the human Rad54 paralog, hRAD54B. hRAD54B interacts with human RAD51 and DMC1, and enhances the D-loop formation of both proteins [64, 169, 170]. Further work is needed to understand the functional specification between hRAD54 and hRAD54B in vertebrates. While ScRad54 and hRAD54 are clearly functional homologs, this does not extend to ScRdh54 and hRAD54B. Genetic analysis in mouse ES cells and animals of single and double RAD54 and RAD54B mutants argues strongly against this possibility, because single RAD54B-deficient mouse ES cells display DNA repair phenotypes, unlike yeast single RDH54 mutants [64]. It is possible that vertebrates contain yet another RAD54 paralog among their cohort of Snf2/Swi2-like proteins. Such a protein is expected to exert a meiotic function, because the mouse RAD54 RAD54B double knockout displays no obvious meiotic phenotype [64], whereas the RAD54 RDH54 double mutant in budding yeast displays a strong meiotic defect [44].
8. Summary
Biochemical analysis generated a multitude of potential functions for Rad54 protein during HR, including mediator function in Rad51 filament assembly, chromatin remodeling, plectonemic joint formation, branch migration, D-loop dissolution, turnover of Rad51-dsDNA filaments after DNA strand invasion, dissociation of dead-end Rad51-dsDNA complexes, and recruitment of the Mus81-Mms4 endonuclease. Multi-facetted evidence from the yeast and vertebrate systems supports a role of Rad54 in Rad51 filament assembly/stability as a first function of Rad54 in vivo. However, this function is independent of the Rad54 ATPase activity, leaving open the critical question of what is the essential, ATP-dependent function of Rad54 protein in HR in vivo. A key question is whether Rad54 is required in vivo for D-loop formation, D-loop extension or both, but we presently lack the tools to measure D-loop formation in vivo. Recent progress to detect physical HR intermediates during DSB repair in somatic yeast cells provides hope that this limitation will be overcome [171]. Such novel in vivo approaches will be needed to help establish the biological significance of the biochemical results. We are also still lacking a high-resolution structure of the full-length Rad54 protein or its paralogs, in particular in conjunction with its binding partner Rad51. The combination of structural and single-molecule studies may also answer the question of whether the active form of Rad54 is a hexameric or double-hexameric ring, as suggested by the initial single molecule experiments [73]. Lastly, more work is needed to understand the functional overlap and separation between Rad54 and its paralogs ScRdh54 and hRAD54B in yeast and vertebrates. Since at least Rad54 is forming multimeric assemblies, it is possible that Rad54 forms a mixed multimer with its paralog. We look forward to the resolution of these problems and the identification of novel facets of Rad54 function in the future.
Highlights.
This review will focus on Rad54, its paralogs, and their function in recombination.
Homologous recombination is a key pathway to maintain genomic integrity.
Rad54 is a core component of the recombination machinery in all eukaryotes.
The review focuses on mechanistic studies with the yeast and human enzymes.
We relate the biochemical findings to genetic and cytological studies of Rad54.
Acknowledgements
We thank members of the Heyer (Clare Fasching, Damon Meyer, Erin Schwartz, Jie Liu, Kirk Ehmsen, Rinti Mukherjee, Ryan Janke, William Wright, Xiao-Ping Zhang) and the Nicolas labs (Aurèle Piazza, Jean-Baptiste Boulé, Gael Millot) for critical comments on the manuscript and John Ceballos for help on the artwork. This work was supported by grants from the US National Institutes of Health (GM58015, CA92276) and US Department of Defense (W81XWH-09-1-0116). SJC was supported by NIH training grant (T32GM007377).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 3.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Lambert S, Froget B, Carr AM. Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair. 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 5.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Homologous Recombination. Berlin-Heidelberg: Springer-Verlag; 2007. pp. 381–441. [Google Scholar]
- 6.Tan TLR, Kanaar R, Wyman C. Rad54, a Jack of all trades in homologous recombination. DNA Repair. 2003;2:787–794. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 7.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. Rad54, the motor of homologous recombination. DNA Repair. 2010;9:286–302. doi: 10.1016/j.dnarep.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Koonin EV. Helicases - amino acid sequence comparisons and structure function relationships. Curr Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 10.Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 11.Auble DT, Hansen KE, Mueller CGF, Lane WS, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 12.Flaus A, Martin DMA, Barton GJ, Owen-Hughes T. Identification of multiple distinctSnf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippo JS, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu.Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 16.West SC. Molecular views of recombination proteins and their control. Nature Rev. Mol.Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 17.Krogh BO, Symington LS. Recombination proteins in yeast. Annu.Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 18.Heyer WD. Biochemistry of eukaryotic homologous recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Berlin-Heidelberg: Springer-Verlag; 2007. pp. 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehmsen KT, Heyer WD. Biochemistry of meiotic recombination. In: Richard Egel DL, editor. Recombination and Meiosis. Berlin-Heidelberg: Springer-Verlag; 2008. pp. 91–164. [Google Scholar]
- 20.Hopfner KP, Michaelis J. Mechanisms of nucleic acid translocases: lessons fromstructural biology and single-molecule biophysics. Curr OpinStruct Biology. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Current Opinion in Genetics & Development. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimitou EP, Symington LS. DNA end resection-Unraveling the tail. DNA Repair. 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases takeon regulatory functions. Nature Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Stith CM, Burgers PM, Heyer W-D. PCNA is required for initiating recombination-associated DNA synthesis by DNA polymeraseδ. Mol Cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 29.McIlwraith MJ, Vaisman A, Liu YL, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol.Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz EK, Heyer WD. DNA structure-selective endonucleases in homologous recombination. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu LJ, Hickson ID. The Bloom's syndrome helicase suppresses crossing-over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 34.Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukhodolets MV, Jin DJ. Interaction between RNA polymerase and RapA, a bacterial homolog of the SWI/SNF protein family. J. Biol. Chem. 2000;275:22090–22097. doi: 10.1074/jbc.M000056200. [DOI] [PubMed] [Google Scholar]
- 36.Yawn B, Zhang L, Mura C, Sukhodolets MV. RapA, the SWI/SNF subunit of Escherichia coli RNA polymerase, promotes the release of nascent RNA from transcription complexes. Biochemistry. 2009;48:7794–7806. doi: 10.1021/bi9004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haseltine CA, Kowalczykowski SC. An archaeal Rad54 protein remodels DNA and stimulates DNA strand exchange by RadA. Nucleic Acids Res. 2009;37:2757–2770. doi: 10.1093/nar/gkp068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dürr H, Körner C, Müller M, Hickmann V, Hopfner KP. X-Ray Structures of theSulfolobus solfataricus SWI2/SNF2 ATPase Core and Its Complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 39.Game JC, Mortimer RK. A genetic study of X-ray sensitive mutants in yeast. Mutat. Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 40.Snow R. Mutants of yeast sensitive to ultraviolet light. J. Bact. 1967;94:571–575. doi: 10.1128/jb.94.3.571-575.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suslova NG, Zakharov IA. Genetic control of radiosensitivity in yeast. VII. Identification of the genes determining the sensitivity to X-rays, Genetika. 1970;6:158. [Google Scholar]
- 42.Schmuckli-Maurer J, Heyer W-D. The Saccharomyces cerevisiae RAD54 gene is important but not essential for natural homothallic mating-type switching. Mol. Gen. Genet. 1999;260:551–558. doi: 10.1007/s004380050928. [DOI] [PubMed] [Google Scholar]
- 43.Lettier G, Feng Q, de Mayolo AA, Erdeniz N, Reid RJD, Lisby M, Mortensen UH, Rothstein R. The role of DNA double-strand breaks in spontaneous homologous recombinationin S. cerevisiae. Plos Genetics. 2006;2:1773–1786. doi: 10.1371/journal.pgen.0020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmuckli-Maurer J, Rolfsmeier M, Nguyen H, Heyer W-D. Genome instability inRAD54 mutants of Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:1013–1023. doi: 10.1093/nar/gkg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmuckli-Maurer J, Heyer WD. Meiotic recombination in RAD54 mutants ofSaccharomyces cerevisiae. Chromosoma. 2000;109:86–93. doi: 10.1007/s004120050415. [DOI] [PubMed] [Google Scholar]
- 47.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotesRad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 48.Clever B, Schmuckli-Maurer J, Sigrist M, Glassner B, Heyer W-D. Specific negative effects resulting from elevated levels of the recombinational repair protein Rad54p inSaccharomyces cerevisiae. Yeast. 1999;15:721–740. doi: 10.1002/(SICI)1097-0061(19990630)15:9<721::AID-YEA414>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 49.Agarwal S, van Cappellen WA, Guenole A, Eppink B, Linsen SE, Meijering E, Houtsmuller A, Kanaar R, Essers J. ATP-dependent and independent functions of Rad54 in genome maintenance. J. Cell Biol. 2011;192:735–750. doi: 10.1083/jcb.201011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 51.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 52.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 53.Palladino F, Klein HL. Analysis of mitotic and meiotic defects in Saccharomyce scerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heude M, Chanet R, Fabre F. Regulation of the Saccharomyces cerevisiae Srs2 helicaseduring the mitotic cell cycle, meiosis and after irradiation. Mol. Gen. Genet. 1995;248:59–68. doi: 10.1007/BF02456614. [DOI] [PubMed] [Google Scholar]
- 55.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Essers J, Hendriks RW, Swagemakers SMA, Troelstra C, deWit J, Bootsma D, Hoeijmakers JHJ, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 57.Bezzubova O, Silbergleit A, YamaguchiIwai Y, Takeda S, Buerstedde JM. Reduced X-ray resistance and homologous recombination frequencies in a RAD54(−/−) mutant of the chickenDT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 58.Kanaar R, Troelstra C, Swagemakers SMA, Essers J, Smit B, Franssen JH, Pastink A, Bezzubova OY, Buerstedde JM, Clever B, Heyer WD, Hoeijmakers JHJ. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: Evidence for functional conservation. Curr. Biol. 1996;6:828–838. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- 59.Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Essers J, van Steeg H, de Wit J, Swagemakers SMA, Vermeij M, Hoeijmakers JHJ, Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Scie. USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin JA, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, Ghazvini M, Tafel AA, Heath H, Galjart N, Essers J, Grootegoed JA, Arnheim N, Bezzubova O, Buerstedde JM, Sung P, Kanaar R. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emery HS, Schild D, Kellogg DE, Mortimer RK. Sequence of RAD54, a Saccharomyces cerevisiae gene involved in recombination and repair. Gene. 1991;104:103–106. doi: 10.1016/0378-1119(91)90473-o. [DOI] [PubMed] [Google Scholar]
- 66.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 67.Swagemakers SMA, Essers J, deWit J, Hoeijmakers JHJ, Kanaar R. The human Rad54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 68.Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNAcomplex provides evidence for protein translocation along DNA. Proc. Natl. Acad. Sci. USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 70.Mazin AV, Bornarth CJ, Solinger JA, Heyer W-D, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 71.Tan TLR, Essers J, Citterio E, Swagemakers SMA, de Wit J, Benson FE, Hoeijmakers JHJ, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 72.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 73.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with theRad51-dsDNA filament. Proc. Natl. Acad. Sci. USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kiianitsa K, Solinger JA, Heyer WD. Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. J. Biol. Chem. 2002;277:46205–46215. doi: 10.1074/jbc.M207967200. [DOI] [PubMed] [Google Scholar]
- 76.Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer WD. Recombinational repair in yeast: Functional interactions between Rad51 and Rad54 proteins. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Golub EI, Kovalenko OV, Gupta RC, Ward DC, Radding CM. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang H, Xie YQ, Houston P, Stemke-Hale K, Mortensen UH, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 79.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 80.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 81.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis R, Durr H, Hopfner KP, Michaelis J. Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic Acids Res. 2008;36:1881–1890. doi: 10.1093/nar/gkn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raschle M, Van Komen S, Chi P, Ellenbeger T, Sung P. Multiple interactions with the Rad51 recombinase govern the homologous recombination function of Rad54. J. Biol. Chem. 2004;279:51973–51980. doi: 10.1074/jbc.M410101200. [DOI] [PubMed] [Google Scholar]
- 84.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 85.Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JHJ, Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwon YH, Seong C, Chi P, Greene EC, Klein H, Sung P. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J. Biol. Chem. 2008;283:10445–10452. doi: 10.1074/jbc.M800082200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee SE, Pellicioli A, Malkova A, Foiani M, Haber JE. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol. 2001;11:1053–1057. doi: 10.1016/s0960-9822(01)00296-2. [DOI] [PubMed] [Google Scholar]
- 92.Lee SE, Pellicioli A, Vaze MB, Sugawara N, Malkova A, Foiani M, Haber JE. Yeast Rad52 and Rad51 recombination proteins define a second pathway of DNA damage assessment in response to a single double-strand break. Mol. Cell. Biol. 2003;23:8913–8923. doi: 10.1128/MCB.23.23.8913-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. Single-molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J Biol Chem. 2007;282:30776–30784. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- 95.Tanaka K, Hiramoto T, Fukuda T, Miyagawa K. A novel human rad54 homologue, Rad54B, associates with Rad51. J Biol Chem. 2000;275:26316–26321. doi: 10.1074/jbc.M910306199. [DOI] [PubMed] [Google Scholar]
- 96.Miyagawa K, Tsuruga T, Kinomura A, Usui K, Katsura M, Tashiro S, Mishima H, Tanaka K. A role for RAD54B in homologous recombination in human cells. Embo J. 2002;21:175–180. doi: 10.1093/emboj/21.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McManus KJ, Barrett IJ, Nouhi Y, Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc Natl Acad Sci U S A. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, Doye V, Nicolas A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeastGenetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair. 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 99.Tanaka K, Kagawa W, Kinebuchi T, Kurumizaka H, Miyagawa K. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 2002;30:1346–1353. doi: 10.1093/nar/30.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sarai N, Kagawa W, Fujikawa N, Saito K, Hikiba J, Tanaka K, Miyagawa K, Kurumizaka H, Yokoyama S. Biochemical analysis of the N-terminal domain of human RAD54B. Nucleic Acids Res. 2008;36:5441–5450. doi: 10.1093/nar/gkn516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clapier CR, Cairns BR. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 102.Alexiadis V, Kadonaga JT. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2003;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by a Rad51-ssDNA nucleoprotein filament. Nature Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Z, Fan HY, Goldman JA, Kingston RE. Homology-driven chromatin remodeling by human RAD54. Nat Struct Mol Biol. 2007;14:397–405. doi: 10.1038/nsmb1223. [DOI] [PubMed] [Google Scholar]
- 105.Sinha M, Peterson CL. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol Cell. 2008;30:803–810. doi: 10.1016/j.molcel.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kwon Y, Chi P, Roh DH, Klein H, Sung P. Synergistic action of the Saccharomyces cerevisiae homologous recombination factors Rad54 and Rad51 in chromatin remodeling. DNA Repair. 2007;6:1496–1506. doi: 10.1016/j.dnarep.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 108.Wolner B, Peterson CL. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- 109.Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cubonova L, Sandman K, Hallam SJ, Delong EF, Reeve JN. Histones in crenarchaea. Journal of Bacteriology. 2005;187:5482–5485. doi: 10.1128/JB.187.15.5482-5485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein - Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 112.Van Komen S, Petukhova G, Sigurdsson S, Sung P. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J. Biol. Chem. 2002;277:43578–43587. doi: 10.1074/jbc.M205864200. [DOI] [PubMed] [Google Scholar]
- 113.Wolner B, van Komen S, Sung P, Peterson CL. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 114.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 115.Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 116.Mazin OM, Mazin AW. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J. Biol. Chem. 2004;279:52041–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- 117.Solinger JA, Kiianitsa K, Heyer W-D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 118.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 119.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair. 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 120.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu YL, West SC. Timeline - Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 2004;5 doi: 10.1038/nrm1502. 937-U921. [DOI] [PubMed] [Google Scholar]
- 122.Keelagher RE, Cotton VE, Goldman ASH, Borts RH. Separable roles for Exonuclease I in meiotic DNA double-strand break repair. DNA Repair. 2011;10:126–137. doi: 10.1016/j.dnarep.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 123.Solinger JA, Heyer W-D. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl. Acad. Sci. USA. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 125.Mazina OM, Rossi MJ, Thomaa NH, Mazin AV. Interactions of human Rad54 protein with branched DNA molecules. J Biol Chem. 2007;282:21068–21080. doi: 10.1074/jbc.M701992200. [DOI] [PubMed] [Google Scholar]
- 126.Rossi MJ, Mazin AV. Rad51 protein stimulates the branch migration activity of Rad54 protein. J. Biol. Chem. 2008;283:24698–24706. doi: 10.1074/jbc.M800839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Urena DE, Zhang ZQ, Tsai YC, Wang YZ, Chen JH. From Strand Exchange to Branch Migration; Bypassing of Non-homologous Sequences by Human Rad51 and Rad54. J. Mol. Biol. 2011;405:77–91. doi: 10.1016/j.jmb.2010.10.042. [DOI] [PubMed] [Google Scholar]
- 128.Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: Characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 129.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nature Struct Mol Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 130.Kim PM, Paffett KS, Solinger JA, Heyer WD, Nickoloff JA. Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54. Nucleic Acids Res. 2002;30:2727–2735. doi: 10.1093/nar/gkf413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, Ira G. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li X, Heyer WD. RAD54 controls access to the invading 3-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–646. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rosselli W, Stasiak A. Energetics of RecA-mediated recombination reactions. J. Mol. Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 137.Xu L, Marians KJ. A dynamic RecA filament permits DNA polymerase-catalyzed extension of the invading strand in recombination intermediates. J. Biol. Chem. 2002;277:14321–14328. doi: 10.1074/jbc.M112418200. [DOI] [PubMed] [Google Scholar]
- 138.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 139.Kowalczykowski SC. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- 140.Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hilario J, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Nat. Acad. Sci. USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJG, Wuite GJL, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 143.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJG, Wuite GJL. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Terasawa M, Ogawa H, Tsukamoto Y, Shinohara M, Shirahige K, Kleckner N, Ogawa T. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proc. Natl. Acad. Sci. USA. 2007;104:5965–5970. doi: 10.1073/pnas.0611490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Interthal H, Heyer WD. MUS81 encodes a novel Helix-hairpin-Helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 146.Matulova P, Marini V, Burgess RC, Sisakova A, Kwon Y, Rothstein R, Sung P, Krejci L. Cooperativity of Mus81.Mms4 with Rad54 in the Resolution of Recombination and Replication Intermediates. J. Biol. Chem. 2009;284:7730–7742. doi: 10.1074/jbc.M806192200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147.Mazina OM, Mazin AV. Human Rad54 protein stimulates human Mus81-Eme1 endonuclease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18249–18254. doi: 10.1073/pnas.0807016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 149.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ciccia A, Constantinou A, West SC. Identification and characterization of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 151.Taylor ER, McGowan CH. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci U S A. 2008;105:3757–3762. doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 153.Tombline G, Heinen CD, Shim KS, Fishel R. Biochemical characterization of the human RAD51 protein - III. - Modulation of DNA binding by adenosine nucleotides. J. Biol. Chem. 2002;277:14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- 154.Holzen TM, Shah PP, Olivares HA, Bishop DK. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006;20:2593–2604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shah PP, Zheng XZ, Epshtein A, Carey JN, Bishop DK, Klein HL. Swi2/Snf2- Related Translocases Prevent Accumulation of Toxic Rad51 Complexes during Mitotic Growth. Mol. Cell. 2010;39:862–872. doi: 10.1016/j.molcel.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan LH, Hollingsworth NM, Haber JE, Foiani M. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. Embo J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Trickey M, Grimaldi M, Yamano H. The anaphase-promoting complex/cyclosome controls repair and recombination by ubiquitylating Rhp54 in fission yeast. Mol. Cell. Biol. 2008;28:3905–3916. doi: 10.1128/MCB.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, Shokat KM, Gygi SP, Hollingsworth NM. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou HL. A Proteome-wide Analysis of Kinase-Substrate Network in the DNA Damage Response. J. Biol. Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chi P, Kwon Y, Moses DN, Seong C, Sehorn MG, Singh AK, Tsubouchi H, Greene EC, Klein HL, Sung P. Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair. 2009;8:279–284. doi: 10.1016/j.dnarep.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang ZM, Buchman AR. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. Ubiquitindependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 169.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 170.Sarai N, Kagawa W, Kinebuchi T, Kagawa A, Tanaka K, Miyagawa K, Ikawa S, Shibata T, Kurumizaka H, Yokoyama S. Stimulation of Dmc1-mediated DNA strand exchange by the human Rad54B protein. Nucleic Acids Res. 2006;34:4429–4437. doi: 10.1093/nar/gkl562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]