Abstract
T-box (TBX) transcription factors are an ancient gene family with critical roles in embryogenesis. Currently, TBX3, TBX5, and TBX20 are TBX genes defined to have multiple protein isoforms created by alternative splicing and characterized by expression and functional studies. These proteins are important for development as mutations lead to severe developmental disorders in humans and mice. Cumulative studies suggest that alternative splicing of these genes can regulate TBX activities during multiple biological processes including cardiogenesis, limb development, and cancer mechanisms. This mini-review focuses on how alternative splicing adds complexity to transcriptional regulation of target genes controlled by TBX transcription factors.
Keywords: alternative splicing, T-box, TBX3, TBX5, TBX20
1. Introduction
T-box (TBX) transcription factors are an ancient gene family important for organogenesis and embryogenesis. Genome searches for genes sharing the T-box DNA-binding domain revealed numerous highly-related transcription factor genes [1]. TBX transcription factors recognize a 20–24 nucleotide palindromic sequence called the T-site or half of the sequence, the T/2 site [2]. The T-box family is thought to have evolved from a primordial gene that gained diversity through tandem duplication and then cluster dispersion resulting in TBX genes maintaining similar functions and loci [3]. While the TBX transcription factors can be expressed in similar developmental times and tissues, they provide distinct functions through control of expression levels, timing and localization of expression, and interaction with different cofactors. Isoforms of the same TBX gene can have different subcellular localizations, expression levels, and functional characteristics. Cumulative studies have shown that alternative splicing provides a potential critical role in regulating TBX protein activities.
Alternative splicing creates different protein products from a single gene and is estimated to affect approximately 90% of the genes expressed in humans [4,5,6]. Multiple isoforms can increase the function and the complexity of the genome. There are multiple types of alternative splicing, with the most common being exon skipping [7,8]. The control of alternative splicing is defined by the pre-mRNA sequence and the availability of spliceosome proteins [4,9]. Alternative splicing is regulated in a developmental stage-specific or tissue-specific manner depending on the localization, timing, and type of factors expressed.
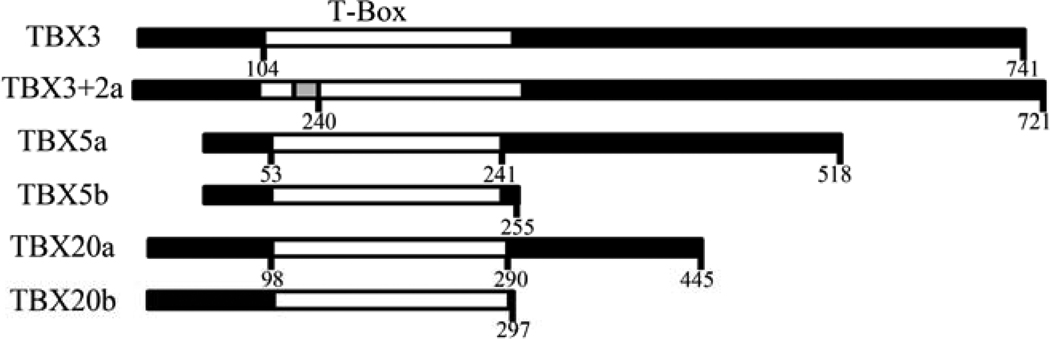
This mini-review highlights the alternatively spliced isoforms found within the TBX transcription factor family. Alternatively spliced isoforms have been described in expression and function analyses in three TBX transcription factor genes: TBX3, TBX5 and TBX20 (Figure 1). All three genes are critical for development as mutations or deletions lead to severe developmental disorders in humans and mice. Determining how these transcription factors are regulated is important to fully understand complex processes such as cardiogenesis, limb development, and cancer mechanisms. From recent studies, expression and functional analyses have revealed a potential regulatory role of protein isoforms on TBX transcription factor functions.
Figure 1. Schematic of TBX protein isoforms.
The protein structures of the murine TBX3, TBX5, and TBX20 protein isoforms are depicted with the T-box DNA-binding domain in white. The 20 aa insertion within the T-box of TBX3+2a is shown in grey.
2. TBX3 and TBX3+2a
TBX3 is a transcriptional repressor important for the development of multiple tissues. Haploinsufficiency of TBX3 causes ulnar-mammary syndrome (UMS, OMIM ID: 181450), an autosomal dominant disorder affecting 1 in 100,000 live births [10,11]. UMS is characterized by defects in upper limb, apocrine gland, mammary gland, dental, and genital development and the defects are highly variable within families. TBX3 is expressed primarily in the tissues affected in UMS. In addition, TBX3 is overexpressed in different cancer cells including malignant primary breast cancer tumors and immortalized cancer cell lines of breast cancer, bladder carcinoma, and melanoma [12,13,14]. In most studies, TBX3 functions as inhibiting senescence and thereby promoting cell proliferation and immortalization. Overexpression of TBX3 in primary cells leads to immortalization [12,15,16]. Ablation of TBX3 expression leads to a decrease in proliferation with a cell type-dependent increase in apoptosis [13,17,18]. TBX3 regulation of proliferation is cell-type dependent as TBX3 inhibits proliferation during cardiogenesis [14,19,20,21]. Other functions of TBX3 include potential roles in bone development, where a decrease in TBX3 expression reduced the differentiation of osteoblasts and the osteogenic-differentiation of human adipose stromal cells (hADSC) [13,17]. These studies place TBX3 function as important for the proliferation and specification of cells and tissues critical for development.
TBX3 isoforms are designated TBX3 and TBX3+2a [22]. Alternative splicing of exon 2a results in a 60 nucleotide (nt) insertion incorporating 20 amino acids (aa) within the T-box DNA-binding domain (Figure 1) [11,12]. The transcripts of TBX3 and TBX3+2a are found in multiple tissues in mice and humans. This insertion is highly conserved within other TBX3 mammalian orthologs and is absent in the avian genome. In other vertebrate and invertebrate genomes, insertions at the exon 2a site are observed, however they do not exhibit similarity to the mammalian transcripts. Currently, there are no other insertions of this nature within the TBX family [14]. The ratio of the transcripts varies between tissues and species indicating tissue-specific and species-specific regulation of expression. Both isoforms are expressed in breast cancer cell lines, and the ratio between isoforms is altered between cell lines, indicating a potential role in cancer mechanisms [12]. In hADSC, TBX3 is the predominant isoform and upon differentiation with osteogenic medium, the isoform ratio is altered with increasing amounts of TBX3+2a expressed, indicating a potential role in differentiation [17]. With the altered isoform levels, future studies should concentrate on determining the importance of isoform amounts. It will be important to identify if isoform-specific target genes exist as a subset of these genes can be alternatively regulated depending on isoform protein levels.
With the 20 aa insertion within the T-box DNA-binding domain, it was important to determine whether both isoforms could bind to DNA. Two separate studies offered conflicting results. In the first study, analysis by electrophoretic mobility shift assay (EMSA) was used to determine if the amount of T-box binding element (TBE) DNA would increase when incubated with nuclear extracts of mouse embryonic fibroblast (MEF) cells overexpressing TBX3 or TBX3+2a. The addition of TBX3 resulted in an increase of bound DNA, indicating DNA-binding ability. However, the addition of TBX3+2a did not result in any discernable increase of bound DNA [12]. While TBX3+2a was not able to bind DNA in this study, another EMSA study was modified to strictly test the DNA-binding capabilities of both isoforms. TBX3 and TBX3+2a proteins were purified from bacterial cultures and incubated with the consensus TBE and the TBE within the Nppa promoter region. This assay resulted in TBX3 and TBX3+2a binding to DNA. Through comparison to TBX3/DNA structure [23], they propose that the 20 aa insert does not contact the DNA and most likely does not affect DNA-binding [14]. The direct DNA-binding experiments and structural analysis more conclusively show that both isoforms can bind to DNA. In future studies, DNA-binding affinities and confirmation of DNA-binding within relevant cell types should be determined for both isoforms.
TBX3 and TBX+2a share common transcriptional targets that function in inhibiting senescence. In one study, overexpression of TBX3 in MEF cells resulted in continuous and increased growth, while TBX3+2a resulted in decreased lifespan. However, it was later noticed that the TBX3+2a isoform was unknowingly used in a similar functional study resulting in increased proliferation [14,24]. In luciferase reporter assays, TBX3 and TBX3+2a inhibited the expression of p21CIP1, a tumor suppressor gene involved in p53-mediated senescence [14]. Future studies should focus on identifying additional target genes with roles in proliferation and senescence. Analyzing target gene expression between primary and immortalized cell lines can establish a connection between TBX3 isoform protein levels and target gene expression.
In vivo analysis of TBX3 isoform functions has been analyzed in the developing hearts of mice. Transgenic mice were created with TBX3 or TBX3+2a overexpressed in the myocardium of the developing heart. Embryos overexpressing TBX3 or TBX3+2a resulted in abnormalities or failure in chamber formation and heart looping. In addition, the ectopic expression resulted in a severe decrease in expression of the chamberspecific markers of Cx40 and Nppa [14]. In luciferase reporter assays, TBX3 and TBX3+2a decreased the NKX2-5/TBX5-driven activation of the Nppa promoter. In addition, both isoforms physically interact with NKX2-5 via the T-box DNA-binding domain [14]. While these studies do not support isoform-specific functions during heart development, more relevant cell types will need to be tested. Since TBX3 is critical for mammary and limb development, future studies should include functional analysis within these developmental systems.
3. TBX5a and TBX5b
TBX5 is critical for forelimb development and cardiogenesis [25,26,27,28,29]. Haploinsufficiency of TBX5 causes Holt-Oram Syndrome (HOS, OMIM ID: 142900), an autosomal dominant disorder characterized by upper limb malformations and cardiac septation defects, which occurs in 1 in 100,000 live births [30,31]. Analysis of TBX5 expression identified an alternatively spliced isoform [32]. The longer TBX5 isoform (518 aa) is designated TBX5a. Alternative splicing of exon 8 inserts 40 nt after the T-box DNA-binding domain creating 4 aa and a stop codon resulting in a 255 aa protein designated TBX5b (Figure 1). Both isoforms are expressed in different tissues and cell lines. During heart development, the ratio is altered with TBX5a being more prominent in embryonic hearts and TBX5b in adult hearts. In transfected cell studies, TBX5a is strictly localized to the nucleus and TBX5b in both the nucleus and cytoplasm. These localizations are seen in various cell types including cardiac, skeletal muscle, and fibroblast cells [32].
Both isoforms can bind to DNA with TBX5a showing a stronger binding affinity. TBX5a can bind to the Nppa promoter and activate transcription. However, due to the lack of the C-terminal transcriptional activation domain on the TBX5b isoform, TBX5b cannot activate the Nppa promoter in luciferase reporter assays in cell lines or cardiomyocytes. It is noteworthy that increasing amounts of TBX5b does not attenuate TBX5a-driven activation of Nppa transcription. Both TBX5 isoforms physically interact with GATA4 and can collaborate to activate transcription of GATA4 target genes. However, TBX5a has a stronger binding affinity for GATA4 and is the only isoform to interact with NKX2-5, resulting in isoform-specific activation of Nppa [32].
Examining TBX5 isoform amounts throughout heart development revealed that TBX5a is more prominent in proliferative developing cells while TBX5b is more prominent in terminally differentiated cells. To identify potential isoform-specific functions, TBX5a was overexpressed in adult hearts using tamoxifen-inducible α-myosin heavy chain-driven Cre transgenic mice [32]. Treatment increased TBX5a expression in the ventricles with a concomitant upregulation of Nppa. Overexpression of TBX5a resulted in cardiac hypertrophy with immunohistochemical analysis showing an increase in myocyte growth without an increase in proliferation. To study TBX5b function, the proliferative myoblast cell line, C2C12, transfected with TBX5b resulted in cell morphology changes and significant cell death due to increased apoptosis. Similar overexpression studies with TBX5a did not result in changes of cell morphology or apoptosis. These studies support the roles of TBX5a regulating cardiomyocyte growth and TBX5b regulating cardiomyocyte growth arrest. Future studies should include conditional isoform-specific knockout mouse model strategies to determine developmental time point and cell type requirements for each isoform.
4. TBX20a and TBX20b
TBX20 is a critical cardiogenic transcription factor important for proliferation, chamber specification, and valvulogenesis. Missense mutations have been identified in human patients with congenital heart defects and adult cardiomyopathies [33,34,35]. TBX20 physically interacts with NKX2-5, GATA4, and GATA5 and synergizes with these cofactors to regulate target genes [36]. TBX20 can activate or repress cardiac target genes important for cardiac chamber specification and extracellular matrix formation within endocardial cushions [37,38,39].
Characterization of TBX20 identified multiple isoforms expressed in several species. One study comprehensively characterized TBX20 isoforms designated TBX20a-d [36]. The two most-studied isoforms are TBX20a and TBX20b (Figure 1). In mice, both transcripts contain exons 1–6 which encode for the entire T-box DNA-binding domain. TBX20a is the full length protein of 445 aa encoded by exons 1–6, 9, and 10. TBX20b is truncated after the T-box DNA-binding domain at 297 aa due to alternative splicing of exon 7 which contains a termination codon [36].
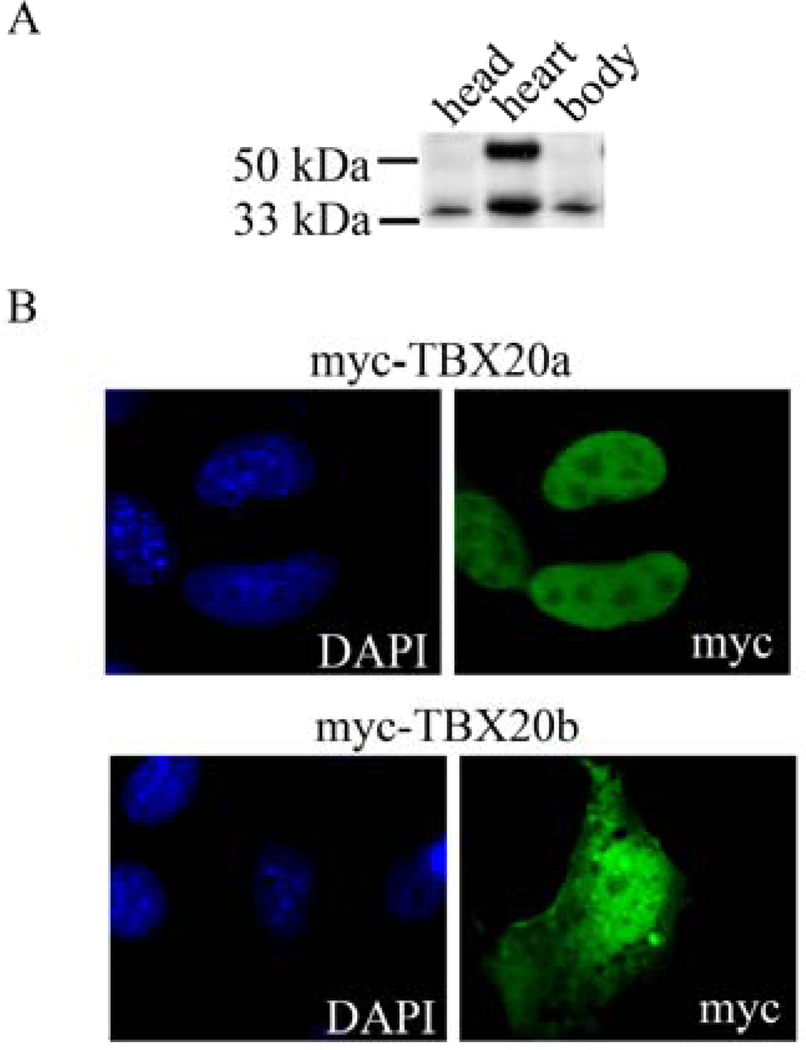
The TBX20 isoforms are coexpressed during heart development with TBX20a expressed at higher levels in both mice and humans [36,40]. One study examined TBX20a-specific expression in E9.0-12.0 mouse hearts. While general TBX20 expression is seen throughout the myocardium, TBX20a is restricted to the developing outflow tract with less myocardial expression [37]. In our laboratory, we determined the presence of TBX20 isoforms by western blot analysis of protein lysates collected from the head, heart, and body of E12.5 mice (Figure 2a). TBX20a was only expressed in the heart while TBX20b was expressed in all three samples. While this was a brief look into TBX20 expression during development, it does reveal the need for careful isoform-specific expression analysis. We also examined the subcellular localization of myc-tagged TBX20 isoforms by immunofluorescent staining of transfected COSM6 cells. Similar to TBX5, TBX20a is localized exclusively in the nucleus while TBX20b has cytoplasmic and nuclear localization (Figure 2b). Future studies of heart development will need to define isoform-specific role within the heart, specifically in determining cell-type specific expression of TBX20 isoforms. Conditional ablation of each TBX20 isoform in knockout mouse models will help define cell type-specific roles. In addition, future studies involving the development of other embryonic structures should concentrate on the functions of TBX20b.
Figure 2. TBX20 isoforms show distinct tissue expression and subcellular localizations.
(A) Tissue from E12.5 ICR mice was dissected into head, heart, and body lysates. Lysates were analyzed by SDS-PAGE and western blot analysis. Antibody used was TBX20 (Sigma, HPA008192). TBX20a is 55 kDa. TBX20b is 33 kDa.
(B) COSM6 cells were plated on glass coverslips in a 24 well plate and transfected with 0.8 ug of DNA using Lipofectamine 2000 according to manufacturer’s protocol (Invitrogen). TBX20a and TBX20b were cloned into the pCMV-Tag3 construct to create a myc-tagged fusion protein (Stratagene). Cells were processed for immunocytochemistry using antibodies for myc (Cell Signaling). Coverslips were mounted onto glass microscope slides with DAPI counterstaining mounting solution (Vectashield).
The TBX20 isoforms have similar and dynamic protein interactions and transcriptional activities. Both isoforms can bind to DNA, interact with cofactors NKX2-5, GATA4, and GATA5, and activate transcription. TBX20b can synergistically increase activation of the NKX2-5-binding sites and the Nppa promoter when coexpressed with cofactors NKX2-5, GATA4, and GATA5. While TBX20a can activate transcription with the same cofactors, the effect is additive instead of synergistic, most likely due to the C-terminal repression domain [36]. A recently published study from our laboratory identified muskelin as a novel interacting partner to only the TBX20b isoform [41]. Muskelin is an intracellular protein involved in protein complexes of nucleocytoplasmic shuttling and signal transduction machinery [42,43,44,45,46,47]. As protein interactions are critical for maintaining a cardiac transcription factor network, future studies should identify more isoform-specific protein interactions and test their role in regulating TBX20 activities.
To determine potential functional differences between the TBX20 isoforms, Xenopus embryos were injected with TBX20a or TBX20b mRNA [36]. Ectopic overexpression of TBX20a resulted in changes in cell migration of the anterior/posterior axis. The formation of a protrusion resembling a secondary anterior/posterior axis or tail was observed and analysis revealed induction of lateral mesoderm and endoderm. Injections of TBX20b resulted in no change. This study suggests that within the Xenopus system, the activities of TBX20a cannot be replaced by TBX20b.
5. Conclusions
The TBX genes are important for proper embryogenesis with mutations or deletions causing developmental disorders in humans and mice. Regulation of TBX transcription factor activity has been characterized through protein interactions and DNA binding affinities. An emerging mechanism of regulation is the production of different protein isoforms by alternative splicing. This mini-review highlights the role of alternative splicing within the TBX genes, TBX3, TBX5, and TBX20, and how alternative splicing adds another level of complexity of TBX functions. Future studies should determine the prevalence of alternative splicing of the other TBX genes. Most expression analyses show general expression without defining isoform specificity. Determining isoform-specific expression throughout development will help define cell-type specific functions. The expression analyses can then be supported by the use of isoform-specific knockout mouse model strategies to further define isoform-specific functional significance. In addition, with tissue-specific expression evident with these TBX genes, it will be important to determine the upstream regulators of alternative splicing, potentially tissue-specific expression of splicing proteins. Finally, these TBX proteins have significant roles in human development and determining whether the pathogenic mutations affect alternative splicing or isoform-specific function could have clinical significance.
T-box genes TBX3, TBX5, and TBX20 have alternatively spliced transcripts.
Alternative splicing of TBX genes provide a potential regulatory role on function.
Future studies of TBX proteins should include isoform-specific functional analysis.
Acknowledgments
We regret that we couldn’t reference all studies in this area due to space limitations. The work in KJ’s lab is supported by a UAB faculty development grant and a NHLBI R01 grant (HL095783-01A1) awarded to KJ.
Abbreviations
- aa
amino acids
- EMSA
electrophoretic mobility shift assay
- hADSC
human adipocyte stromal cells
- HOS
Holt-Oram syndrome
- MEF
mouse embryonic fibroblast
- nt
nucleotide
- TBE
T-box binding element
- TBX
T-box
- UMS
ulnar-mammary syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Minguillon C, Logan M. The comparative genomics of T-box genes. Brief Funct Genomic Proteomic. 2003;2:224–233. doi: 10.1093/bfgp/2.3.224. [DOI] [PubMed] [Google Scholar]
- 2.Müller C, Herrmann B. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 3.Agulnik S, Garvey N, Hancock S, Ruvinsky I, Chapman D, Agulnik I, Bollag R, Papaioannou V, Silver L. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144:249–254. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 9.David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–285. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 11.Bamshad M, Le T, Watkins WS, Dixon ME, Kramer BE, Roeder AD, Carey JC, Root S, Schinzel A, Van Maldergem L, Gardner RJ, Lin RC, Seidman CE, Seidman JG, Wallerstein R, Moran E, Sutphen R, Campbell CE, Jorde LB. The spectrum of mutations in TBX3: Genotype/Phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet. 1999;64:1550–1562. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004;64:5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- 13.Govoni KE, Lee SK, Chadwick RB, Yu H, Kasukawa Y, Baylink DJ, Mohan S. Whole genome microarray analysis of growth hormone-induced gene expression in bone: T-box3, a novel transcription factor, regulates osteoblast proliferation. Am J Physiol Endocrinol Metab. 2006;291:E128–E136. doi: 10.1152/ajpendo.00592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoogaars W, Barnett P, Rodriguez M, Clout D, Moorman A, Goding C, Christoffels V. TBX3 and its splice variant TBX3 + exon 2a are functionally similar. Pigment Cell Melanoma Res. 2008;21:379–387. doi: 10.1111/j.1755-148X.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- 15.Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10:2403–2413. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- 16.Platonova N, Scotti M, Babich P, Bertoli G, Mento E, Meneghini V, Egeo A, Zucchi I, Merlo GR. TBX3, the gene mutated in ulnar-mammary syndrome, promotes growth of mammary epithelial cells via repression of p19ARF, independently of p53. Cell Tissue Res. 2007;328:301–316. doi: 10.1007/s00441-006-0364-4. [DOI] [PubMed] [Google Scholar]
- 17.Lee HS, Cho HH, Kim HK, Bae YC, Baik HS, Jung JS. Tbx3, a transcriptional factor, involves in proliferation and osteogenic differentiation of human adipose stromal cells. Mol Cell Biochem. 2007;296:129–136. doi: 10.1007/s11010-006-9306-4. [DOI] [PubMed] [Google Scholar]
- 18.Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 2005;219:105–112. doi: 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 19.Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 20.Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 21.Mesbah K, Harrelson Z, Théveniau-Ruissy M, Papaioannou VE, Kelly RG. Tbx3 is required for outflow tract development. Circ Res. 2008;103:743–750. doi: 10.1161/CIRCRESAHA.108.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bamshad M, Le T, Watkins W, Dixon M, Kramer B, Roeder A, Carey J, Root S, Schinzel A, Van Maldergem L, Gardner R, Lin R, Seidman C, Seidman J, Wallerstein R, Moran E, Sutphen R, Campbell C, Jorde L. The spectrum of mutations in TBX3: Genotype/Phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet. 1999;64:1550–1562. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coll M, Seidman JG, Müller CW. Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure. 2002;10:343–356. doi: 10.1016/s0969-2126(02)00722-0. [DOI] [PubMed] [Google Scholar]
- 24.Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R. TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem. 2002;277:6567–6572. doi: 10.1074/jbc.M110492200. [DOI] [PubMed] [Google Scholar]
- 25.Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125:2499–2509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- 26.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130:5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- 28.Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol. 2000;223:169–180. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi JK, Koshiba-Takeuchi K, Matsumoto K, Vogel-Höpker A, Naitoh-Matsuo M, Ogura K, Takahashi N, Yasuda K, Ogura T. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 1999;398:810–814. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- 30.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 31.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 32.Georges R, Nemer G, Morin M, Lefebvre C, Nemer M. Distinct expression and function of alternatively spliced Tbx5 isoforms in cell growth and differentiation. Mol Cell Biol. 2008;28:4052–4067. doi: 10.1128/MCB.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirk E, Sunde M, Costa M, Rankin S, Wolstein O, Castro M, Butler T, Hyun C, Guo G, Otway R, Mackay J, Waddell L, Cole A, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler G, Zorn A, Feneley M, Winlaw D, Harvey R. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Shen A, Li X, Jiao W, Zhang X, Li Z. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51:580–587. doi: 10.1016/j.ejmg.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Posch M, Gramlich M, Sunde M, Schmitt K, Lee S, Richter S, Kersten A, Perrot A, Panek A, Al Khatib I, Nemer G, Mégarbané A, Dietz R, Stiller B, Berger F, Harvey R, Ozcelik C. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet. 2010;47:230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stennard F, Costa M, Elliott D, Rankin S, Haast S, Lai D, McDonald L, Niederreither K, Dolle P, Bruneau B, Zorn A, Harvey R. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi J, Mileikovskaia M, Koshiba-Takeuchi K, Heidt A, Mori A, Arruda E, Gertsenstein M, Georges R, Davidson L, Mo R, Hui C, Henkelman R, Nemer M, Black B, Nagy A, Bruneau B. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 38.Shelton E, Yutzey K. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev Biol. 2007;302:376–388. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai C, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld M, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammer S, Toenjes M, Lange M, Fischer J, Dunkel I, Mebus S, Grimm C, Hetzer R, Berger F, Sperling S. Characterization of TBX20 in human hearts and its regulation by TFAP2. J Cell Biochem. 2008;104:1022–1033. doi: 10.1002/jcb.21686. [DOI] [PubMed] [Google Scholar]
- 41.Debenedittis P, Harmelink C, Chen Y, Wang Q, Jiao K. Characterization of the novel interaction between muskelin and TBX20, a critical cardiogenic transcription factor. Biochem Biophys Res Commun. 2011;409:338–343. doi: 10.1016/j.bbrc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams J, Seed B, Lawler J. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 1998;17:4964–4974. doi: 10.1093/emboj/17.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasegawa H, Katoh H, Fujita H, Mori K, Negishi M. Receptor isoform-specific interaction of prostaglandin EP3 receptor with muskelin. Biochem Biophys Res Commun. 2000;276:350–354. doi: 10.1006/bbrc.2000.3467. [DOI] [PubMed] [Google Scholar]
- 44.Umeda M, Nishitani H, Nishimoto T. A novel nuclear protein, Twa1, and Muskelin comprise a complex with RanBPM. Gene. 2003;303:47–54. doi: 10.1016/s0378-1119(02)01153-8. [DOI] [PubMed] [Google Scholar]
- 45.Ledee D, Gao C, Seth R, Fariss R, Tripathi B, Zelenka P. A specific interaction between muskelin and the cyclin-dependent kinase 5 activator p39 promotes peripheral localization of muskelin. J Biol Chem. 2005;280:21376–21383. doi: 10.1074/jbc.M501215200. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi N, Yang J, Ueda A, Suzuki T, Tomaru K, Takeno M, Okuda K, Ishigatsubo Y. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 47.Valiyaveettil M, Bentley A, Gursahaney P, Hussien R, Chakravarti R, Kureishy N, Prag S, Adams J. Novel role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator of cell morphology regulation. J Cell Biol. 2008;182:727–739. doi: 10.1083/jcb.200801133. [DOI] [PMC free article] [PubMed] [Google Scholar]